Introduction
Breast cancer (BC) has become a modern global
epidemic among women as its morbidity has markedly increased over
the past few decades (1). According
to the Cancer Statistics 2016, BC is the most frequently diagnosed
malignancy and the second leading cause of tumor-related death in
females in the US, with 246,660 new diagnoses and 40,450 deaths
projected to have occurred in 2016 (2). Recently, accumulating evidence
indicates that early diagnosis and treatment result in favorable
prognoses; however, current clinical tumor biomarkers lack
sensitivity and specificity for early detection of BC (3). Consequently, identification of novel
diagnostic biomarkers with high sensitivity and specificity for
differentiating breast cancer from benign breast disease and normal
samples is urgently needed. Previous studies have reported that
microRNAs (miRNAs) play vital roles in the initiation and
progression of BC and that certain miRNAs can serve as potential
diagnostic biomarkers (4,5).
miRNAs are short non-coding RNA molecules of 18–25
nucleotides that play essential roles in regulating gene expression
by binding to target mRNAs and directing RNA-induced silencing
complexes (RISCs) to degrade target mRNAs or repress mRNA
translation (6,7). Notably, miRNAs are reportedly linked
to the occurrence and development of a variety of malignant tumors.
Numerous studies have indicated that miRNAs could serve as
oncogenes or tumor suppressors by regulating cellular biological
processes, such as cell differentiation, proliferation and
apoptosis (4,8,9).
Moreover, miRNAs are also involved in multiple cancer-related cell
signaling pathways in various malignancies, including breast cancer
(10,11). Previous studies have reported that
many miRNAs are aberrantly expressed in BC and thus potentially act
as biomarkers for cancer diagnosis. Fu et al (12) showed that miR-141, miR-145, miR-183,
miR-21 and miR-638, which are differentially expressed in BC and
normal tissues, function as potential diagnostic indicators to
distinguish tumor cases from normal cases. However, most of the
previous studies focused on the diagnostic value of a single miRNA.
Additionally, the small sample size of current studies and the lack
of sensitivity and specificity of candidate miRNAs limit the
clinical application of these biomarkers. In recent years, the
development of high-throughput technologies and bioinformatics has
promoted the development of such diagnostic markers.
The Cancer Genome Atlas (TCGA) provides open access
to many comprehensive miRNA-sequencing datasets. In the present
study, we analyzed miRNA expression data published by TCGA to
identify miRNAs differentially expressed between breast tumor and
normal tissues. Ultimately, we determined a nine-miRNA signature
for diagnosing breast cancer. Furthermore, we predicted the targets
of these miRNAs through in silico algorithms and conducted
gene oncology annotation and pathway enrichment analyses to
determine the potential biological functions of the nine miRNAs in
the signature.
Materials and methods
Data collection from TCGA
The level three miRNASeq datasets of 1,110 BC and
104 normal samples were collected from the TCGA (http://cancergenome.nih.gov/). The miRNA expression
profiles for both breast tumor and normal tissues were analyzed
using Illumina HiSeq systems (781 tumor specimens and 87 normal
specimens) or Illumina Genome Analyzer systems (324 tumor samples
and 17 normal samples). There were no ethical issues since the data
were retrieved from TCGA. The present study, was conducted in
accordance with the publication guidelines proposed by TCGA
(http://cancergenome.nih.gov/publications/publicationguidelines).
Processing of miRNA expression
data
After downloading miRNASeq datasets, we first
excluded miRNAs with no expression. Then, the expression data of
the remaining miRNAs were log2-transformed for downstream analyses.
To screen differentially expressed miRNAs, we conducted two-sample
t-tests to compare miRNA expression levels in BC and normal breast
tissues. In addition, we calculated log2 fold-change
(log2FC) ratios. To reduce the false-positive rates
(FPR) of our diagnostic tests, we set p<0.001 and
|log2FC| >1 (13) as
standards for identifying aberrantly expressed miRNAs. Statistical
analyses were conducted through SPSS 20 (IBM, Armonk, NY, USA), and
graphs were produced by GraphPad Prism 5 (GraphPad Software, Inc.,
La Jolla, CA, USA).
Selection of candidate diagnostic
biomarkers
We selected differentially expressed miRNAs that
were analyzed using both Illumina HiSeq and Illumina Genome
Analyzer systems. Next, we generated receiver operating
characteristic (ROC) curves, calculated the area under the curve
(AUC) with a 95% confidence interval (95% CI) and calculated the
diagnostic sensitivity and specificity using MedCalc software
(14). miRNAs with AUC >0.9 were
identified as potential diagnostic biomarkers.
Target prediction and enrichment
analysis
The potential targets of candidate miRNAs were
predicted by 10 online databases: TarBase, miRTarBase, TargetScan,
TargetMiner, microRNA.org, RNA22, PicTar-vert, miRDB,
PITA and PolymiRTS. Only target genes predicted by at least four
algorithms were chosen, and genes targeted by more than one miRNA
were selected for further analysis. Gene Ontology (GO) functional
annotation and KEGG pathway analyses were conducted through the
DAVID online tool (https://david.ncifcrf.gov/) and visualized by
Cytoscape 3.3.0 software and the R package ‘ggplot2’ (http://www.inside-r.org/packages/cran/ggplot2).
Results
Selection of differentially expressed
miRNAs and candidate diagnostic biomarkers
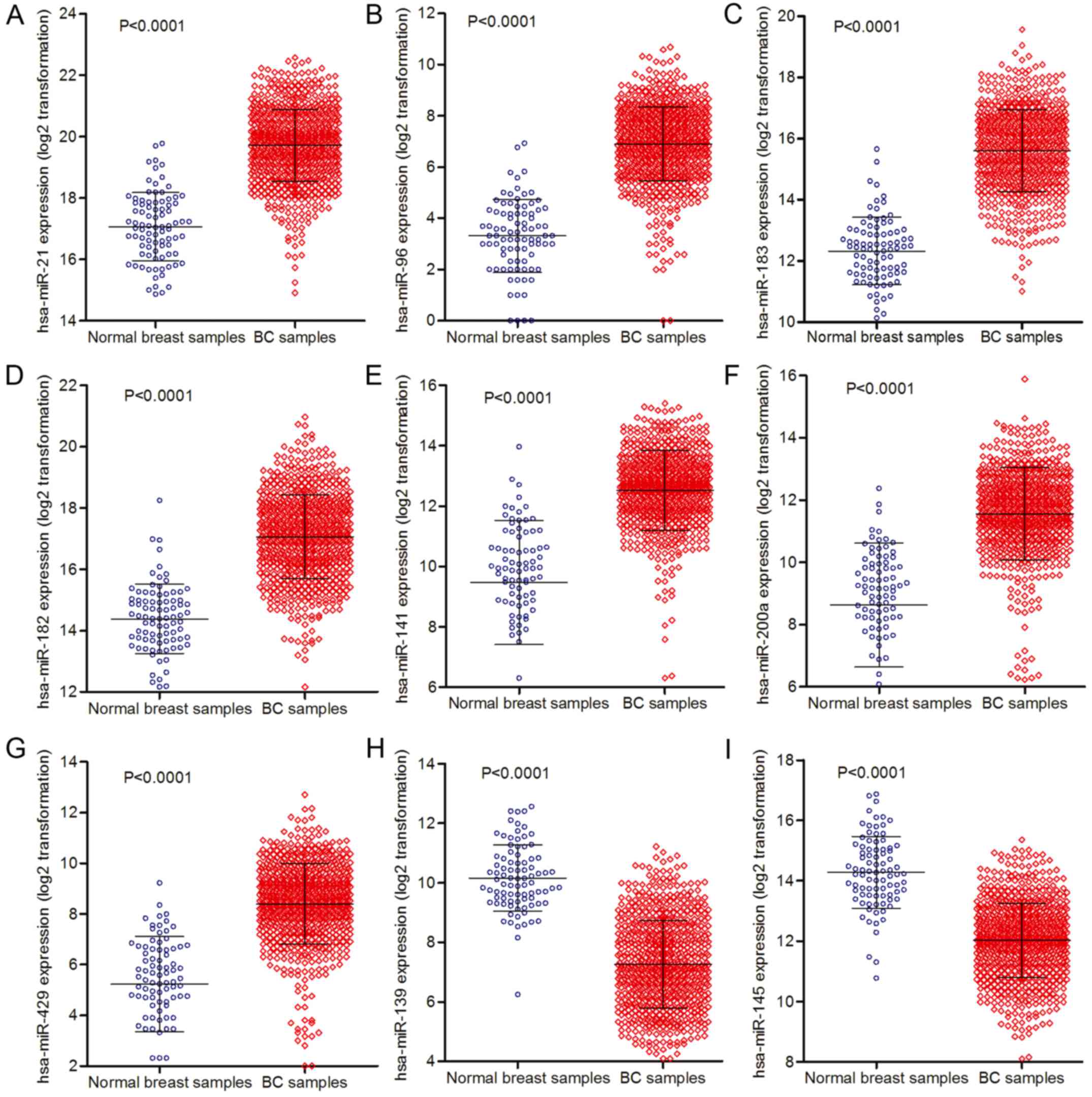
A total of 66 differentially expressed miRNAs were
identified (p<0.001, |log2FC| >1) after miRNA
expression profiles were analyzed in BC and normal breast samples.
Of these, 13 miRNAs were downregulated and 53 miRNAs were
upregulated in BC specimens (data not shown). We performed ROC
analysis of the 66 miRNAs and selected nine dysregulated miRNAs as
candidate diagnostic markers since they showed high diagnostic
accuracy (AUC >0.9). As shown in Table I, these nine dysregulated miRNAs
included seven upregulated miRNAs (hsa-miR-21, hsa-miR-96,
hsa-miR-183, hsa-miR-182, hsa-miR-141, hsa-miR-200a and
hsa-miR-429) and two downregulated miRNAs (hsa-miR-139 and
hsa-miR-145). Differential analysis of the expression data of the
selected nine miRNAs indicated that these miRNAs exhibited
significant statistical differences (p<0.0001,
|log2FC| >1) (Table
I, Fig. 1). Among these nine
miRNAs, both hsa-miR-429 and hsa-miR-200a are located on chromosome
1p36.33, while hsa-miR-183, hsa-miR-96 and hsa-miR-182 are located
on chromosome 7q32.2.
 | Table I.Characteristics of the nine miRNA
candidates. |
Table I.
Characteristics of the nine miRNA
candidates.
|
|
|
| Two-sample
t-test |
|
|---|
|
|
|
|
|
|
|---|
| Gene name | Chromosome | Expression | t-test | P-value |
|log2FC| |
|---|
| hsa-miR-21 | 17q23.1 | Upregulation | 25.27 | <0.0001 | 2.652 |
| hsa-miR-96 | 7q32.2 | Upregulation | 23.92 | <0.0001 | 3.542 |
| hsa-miR-183 | 7q32.2 | Upregulation | 21.34 | <0.0001 | 3.392 |
| hsa-miR-182 | 7q32.2 | Upregulation | 20.14 | <0.0001 | 2.784 |
| hsa-miR-141 | 12p13.31 | Upregulation | 20.12 | <0.0001 | 2.426 |
| hsa-miR-200a | 1p36.33 | Upregulation |
20.9 | <0.0001 | 2.568 |
| hsa-miR-429 | 1p36.33 | Upregulation | 20.33 | <0.0001 | 2.908 |
| hsa-miR-139 | 11q13.4 | Downregulation | 9.211 | <0.0001 | 2.578 |
| hsa-miR-145 | 5q32 | Downregulation | 8.422 | <0.0001 | 2.216 |
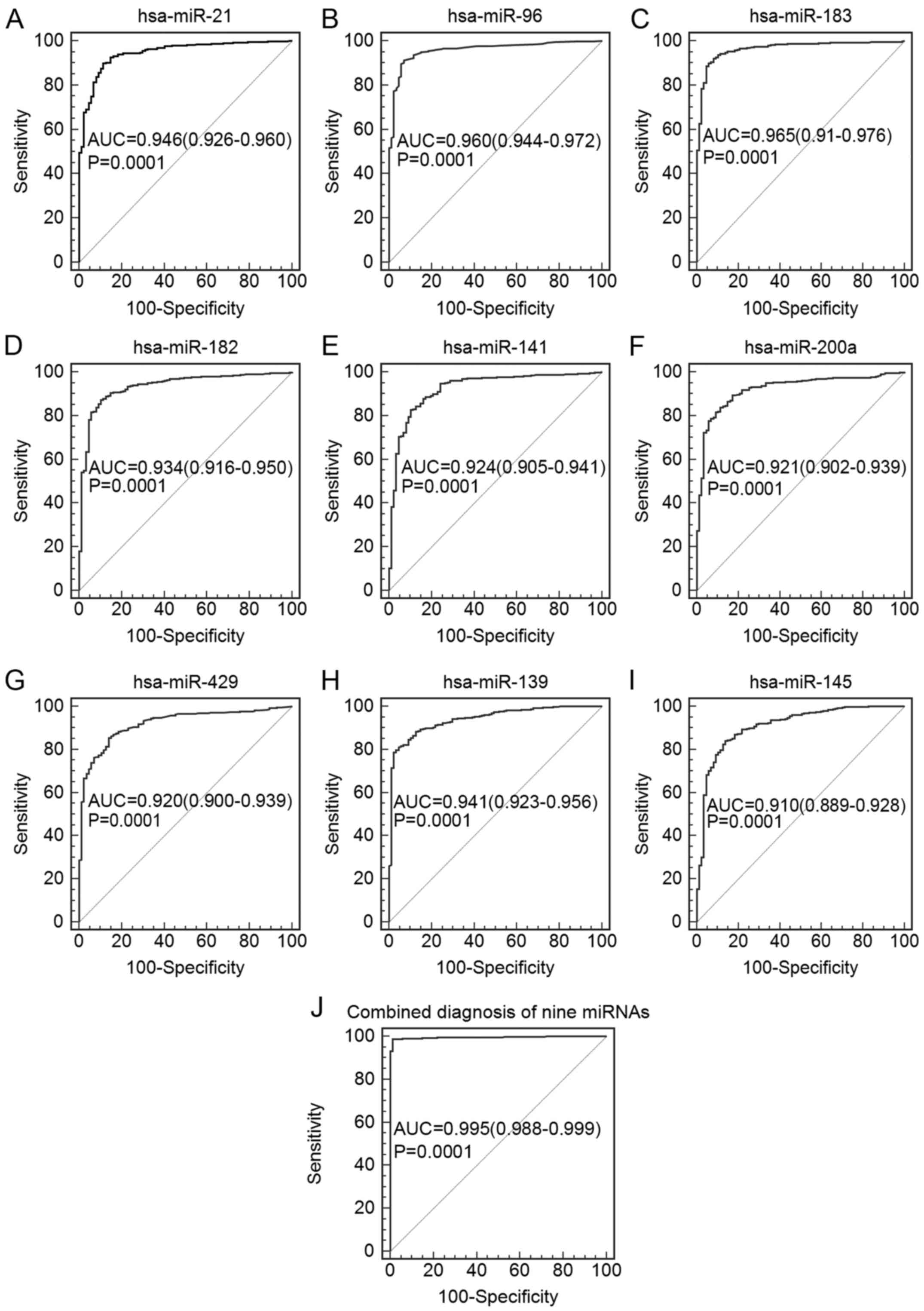
Diagnostic performance of selected
miRNAs
Among the seven upregulated miRNAs, the AUC values
with 95% CI of hsa-miR-21, hsa-miR-183, hsa-miR-96, hsa-miR-182,
hsa-miR-141, hsa-miR-200a and hsa-miR-429 were 0.946 (0.926–0.960),
0.965 (0.951–0.976), 0.960 (0.944–0.972), 0.934 (0.916–0.950),
0.924 (0.905–0.941), 0.921 (0.902–0.939) and 0.920 (0.900–0.937),
respectively. For the two downregulated miRNAs, the AUC values with
95% CI of hsa-miR-139 and hsa-miR-145 were 0.941 (0.923–0.956) and
0.910 (0.889–0.928), respectively (Table II, Fig.
2). Furthermore, we constructed a binary logistic regression
model to evaluate the combination of these nine miRNAs. The
diagnostic performance of the combination was improved
significantly. The results showed that the AUC reached 0.995 (95%
CI=0.988–0.999), and the diagnostic sensitivity and specificity
reached 98.7 and 98.9%, respectively (Table II, Fig.
2).
 | Table II.Diagnostic performance of the nine
selected miRNAs. |
Table II.
Diagnostic performance of the nine
selected miRNAs.
|
| ROC |
|---|
|
|
|
|---|
| Gene name | AUC | 95% CI | P-value | Sensitivity
(%) | Specificity
(%) |
|---|
| hsa-miR-21 | 0.946 | 0.926–0.960 | 0.0001 | 90.0 | 88.5 |
| hsa-miR-96 | 0.960 | 0.944–0.972 | 0.0001 | 91.4 | 93.0 |
| hsa-miR-183 | 0.965 | 0.951–0.976 | 0.0001 | 90.0 | 94.3 |
| hsa-miR-182 | 0.934 | 0.916–0.950 | 0.0001 | 87.2 | 89.7 |
| hsa-miR-141 | 0.924 | 0.905–0.941 | 0.0001 | 82.8 | 89.7 |
| hsa-miR-200a | 0.921 | 0.902–0.939 | 0.0001 | 83.9 | 88.5 |
| hsa-miR-429 | 0.920 | 0.900–0.937 | 0.0001 | 85.4 | 86.0 |
| hsa-miR-139 | 0.941 | 0.923–0.956 | 0.0001 | 81.2 | 95.4 |
| hsa-miR-145 | 0.910 | 0.889–0.928 | 0.0001 | 84.1 | 86.2 |
|
Combineda | 0.995 | 0.988–0.999 | 0.0001 | 98.7 | 98.9 |
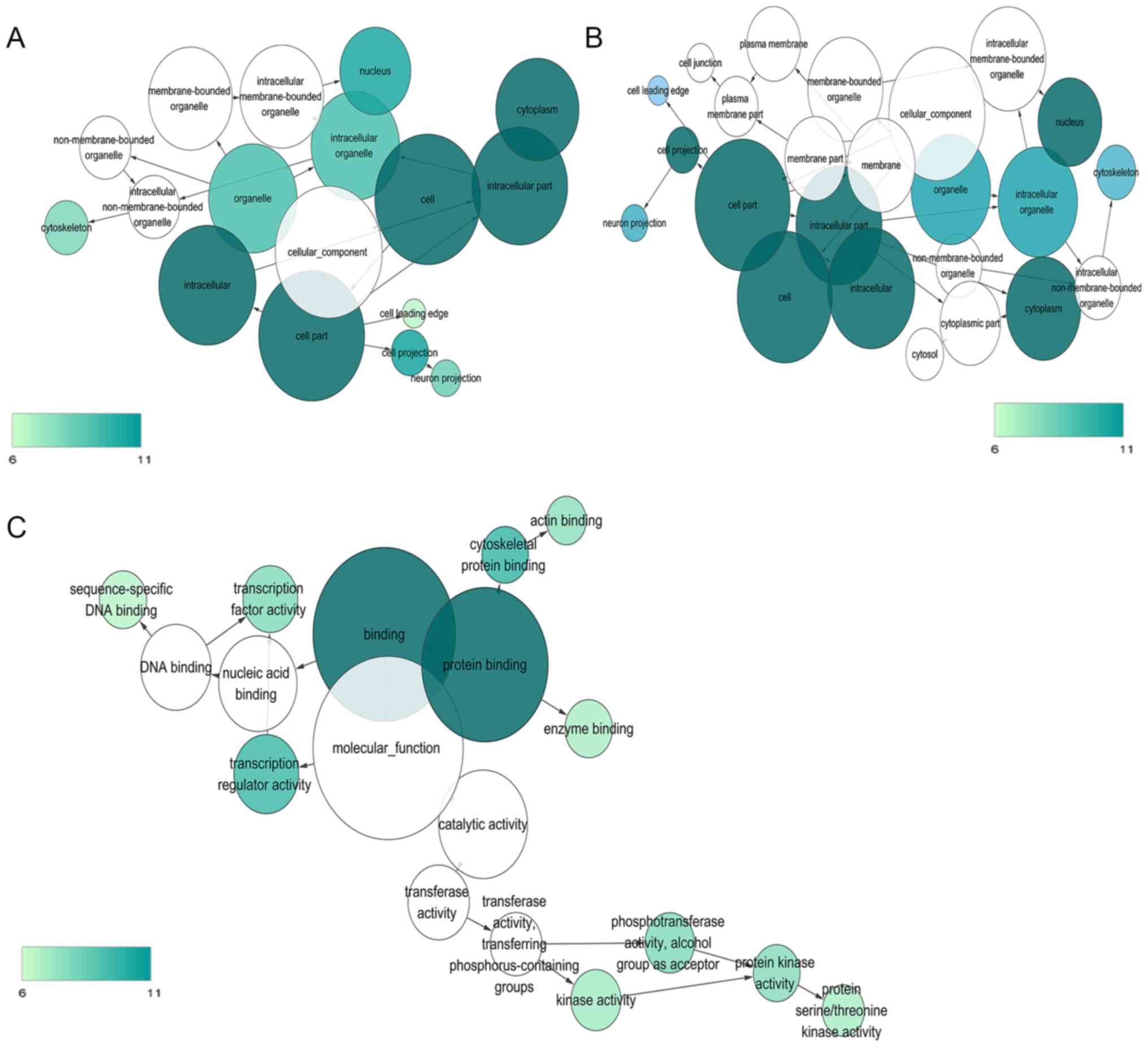
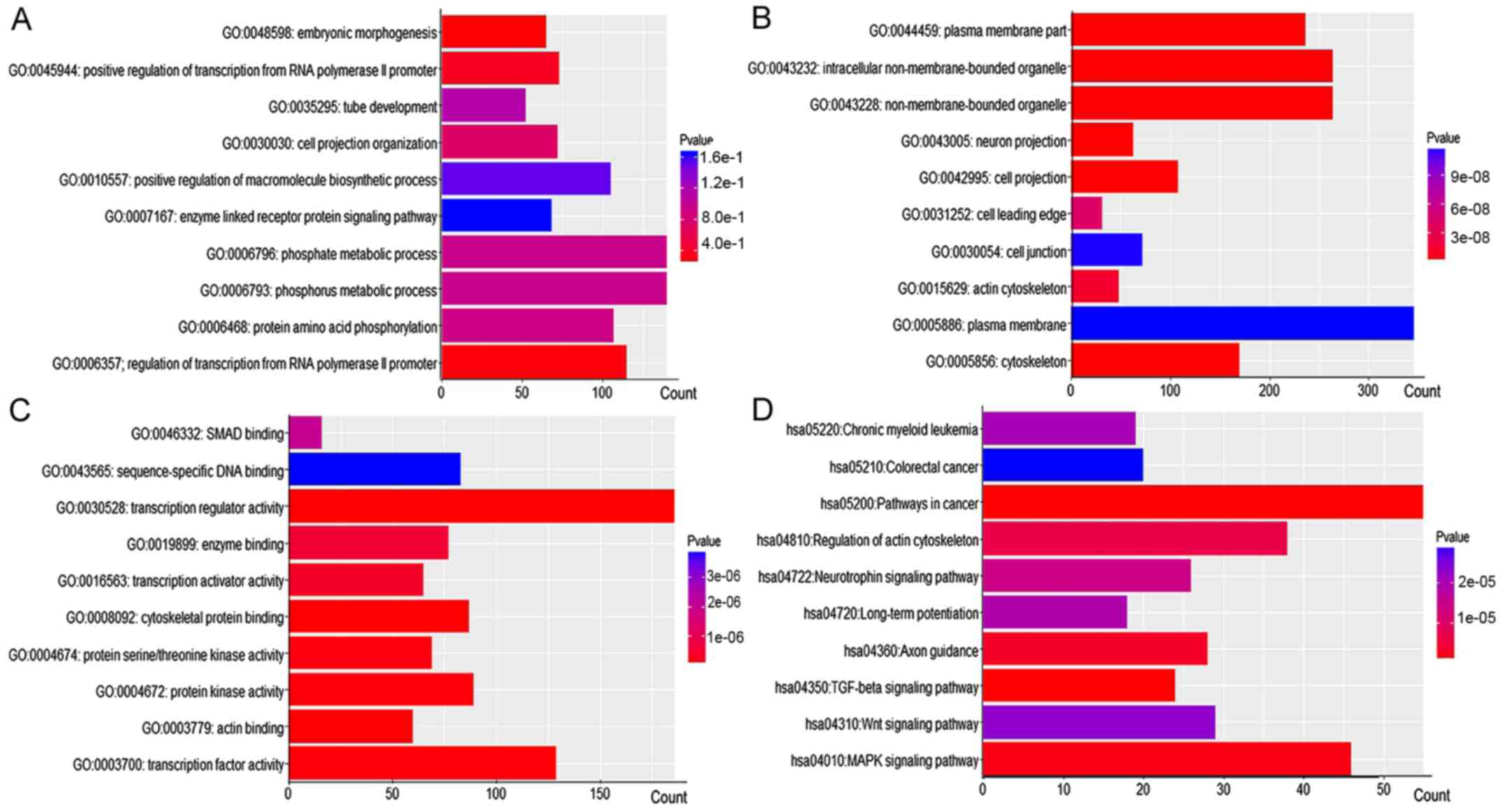
Target prediction and enrichment
analysis
We gathered target genes predicted by at least four
algorithms, and then chose those that were targets of multiple
miRNAs for GO and KEGG analyses. The results of target predictions
for the nine miRNAs are presented as supporting information (data
not shown). Eventually, we selected 1,339 target genes for GO
annotation and KEGG pathway analyses. The most strongly enriched GO
processes and KEGG pathways are listed in Fig. 3. The top 10 GO functional
annotations and KEGG pathways are shown in Fig. 4. GO and KEGG enrichment analyses
revealed that the targets of candidate miRNAs were involved in
several critical cancer-related pathways, including pathways in
cancer, colorectal cancer, the TGF-β, the MAPK and the Wnt
signaling pathways.
Discussion
Increasing evidence indicates that dysregulation of
miRNAs may be closely related to many malignances, including breast
carcinoma (BC) (15–17). In the present study, we
comprehensively analyzed high-throughput miRNA expression profiles
of breast cancer specimens from TCGA and selected nine miRNAs with
high diagnostic accuracy, including seven upregulated miRNAs
(hsa-miR-21, hsa-miR-96, hsa-miR-183, hsa-miR-182, hsa-miR-141,
hsa-miR-200a and hsa-miR-429) and two downregulated miRNAs
(hsa-miR-139 and hsa-miR-145).
Among the seven upregulated miRNAs, miR-21 showed
the most obvious overexpression in BC specimens. As one of the
first mammalian onco-miRNAs identified, miR-21 has been
experimentally confirmed to participate in the initiation and
progression of various carcinomas. In BC, miR-21 also plays a vital
role in carcinogenesis. miR-21 is located on chromosome 17q, a
region containing several oncogenes, including HER2, TOP2A and
PPM1D (18–20). miR-21 has been reported to function
as an onco-miRNA by directly regulating several tumor suppressors,
such as tropomyosin 1 (TPM1), programmed cell death 4 (PDCD4),
maspin and phosphatase and tensin homolog (PTEN) (21,22).
Additionally, it has been reported that miR-21 is involved in
certain cancer-related signaling pathways, such as the
PTEN/PI3K/AKT, the ERK1/2/MAPK and the miR-21/smad7/ERK signaling
pathways, thus, promoting tumor growth and metastasis in
vivo and in vitro (21,23–26).
Previous studies have shown that the
miR-183/miR-96/miR-182 cluster is overexpressed in various
malignant tumors, including BC; thus, this cluster may be an
onco-miRNA cluster and could potentially act as a multi-marker for
diagnosing malignancies (27). The
upregulated miR-183/miR-96/miR-182 cluster inhibits transcriptional
factors, such as FOXO1 and FOXO3, and thus, promotes BC cell
proliferation, invasion, and migration and reduces cell apoptosis
in vivo and in vitro (28,29).
As members of the miR-200 family, miR-141, miR-200a
and miR-429 exert multiple complex effects on tumor occurrence and
development in different carcinomas. Among the three miRNAs,
miR-429 and miR-200a are located on chromosome 1p36.33, while
miR-141 is located on chromosome 12p13.31. Several studies suggest
that miR-200a, miR-429 and miR-141 may be tumor suppressors since
they are downregulated in cancer samples (30–32).
However, other studies showed that these three miRNAs may function
as oncogenes since they are upregulated in tumor tissues (33–36).
In the present study, the expression levels of the three miRNAs
were significantly elevated in BC specimens compared with normal
breast tissues. These discrepancies may be partially explained by
the various sources of the cases studied, different detection
techniques and different platforms used. It is reported that
members of the miR-200 family can reverse epithelial-mesenchymal
transition (EMT) by increasing E-cadherin expression and decreasing
ZEB1, ZEB2 and β-catenin expression, further inhibiting cancer cell
invasion and migration (31,37–40).
Notably, EMT mechanisms are distinct in various malignancies, and
the expression of EMT markers (E-cadherin and ZEB) is inconsistent
in diverse types of cancers and different histological types of the
same cancer (41). This could
partially explain the opposing actions of miR-200a, miR-429 and
miR-141 on tumorigenesis.
Real-time PCR and miRNA microarray confirm that
miR-139 is downregulated in breast cancer (42–44),
which is consistent with our results. In vitro, forced
expression of miR-139 in the BC cell line MDA-MD-231 was found to
suppress cell migration and invasion by targeting genes involved in
metastasis-related signaling pathways (42). Functional experiments conducted by
Hua et al (45) revealed
that increased expression of miR-139 reduced cell proliferation in
luminal type BC cells by directly targeting topoisomerase IIα
(TOP2α), an oncogene that can promote cell proliferation in
vitro.
Both previous studies, and our present study,
revealed that miR-145 is downregulated in BC samples (46–50).
Acting as an oncogene, miR-145 promoted cell apoptosis (46), and suppressed cell growth,
proliferation, migration and invasion (47,51)
in vitro. More importantly, the decreased expression of
miR-145 was found to be negatively correlated with several
clinicopathological parameters, such as ER and HER2 status, tumor
size and lymph node metastasis (52), which indicated its potential
diagnostic or prognostic value in the clinic.
Current studies have focused on the diagnostic value
of dysregulated miRNAs in BC (12,53).
However, this is the first identification of a nine-miRNA signature
as a potential multi-marker for diagnosing BC found by analyzing
high-throughput miRNA expression profiles from TCGA data. In the
present study, the ROC curve of the combined nine differentially
expressed miRNAs showed an extremely high diagnostic accuracy with
an AUC of 0.995 (95% CI=0.988–0.999), and diagnostic sensitivity
and specificity of 98.7 and 98.9%, respectively. The diagnostic
value of these nine miRNAs combined was higher than that of the
individual miRNAs, indicating that the nine-miRNA signature could
serve as a potential multi-marker for diagnosing BC.
Previous studies have demonstrated that miRNAs play
vital roles in carcinogenesis through participation in
cancer-related pathways. Therefore, we performed target prediction
of the nine selected miRNAs and subsequently carried out GO and
KEGG pathway enrichment analyses to explore the potential effects
of these nine miRNAs on carcinogenesis. GO and KEGG pathways
revealed that the targets of these dysregulated miRNAs were
involved in many critical cancer-related biological processes and
pathways, such as positive regulation of macromolecule biosynthetic
process, regulation of transcription, regulation of gene
expression, the enzyme-linked receptor protein signaling pathway,
pathways in cancer, the TGF-β signaling pathway, and the MAPK
signaling pathway. The results of enrichment analyses suggested
that these nine miRNAs could regulate important pathways correlated
with breast cancer by targeting oncogenes or tumor-suppressor
genes.
To the best of our knowledge, this is the first
study identifying a nine-miRNA signature as a multi-biomarker for
diagnosing BC based on TCGA datasets. Nevertheless, the present
study has several limitations. Firstly, our data were retrieved
from TCGA and had not been verified in experiments. Therefore,
further multicenter clinical trials are needed in the future to
confirm our findings. Secondly, the expression profiling of these
miRNAs was only based on tissue. It may be of more clinical value
to investigate noninvasive diagnostic markers.
In conclusion, in the present study, we identified a
tumor-specific miRNA signature in BC containing seven upregulated
miRNAs (hsa-miR-183, hsa-miR-96, hsa-miR-21, hsa-miR-182,
hsa-miR-141, hsa-miR-200a and hsa-miR-429) and two downregulated
miRNAs (hsa-miR-139 and hsa-miR-145) by analyzing high-throughput
data from a TCGA dataset. We discovered that these nine aberrantly
expressed miRNAs may play important roles in the initiation and
progression of BC. More importantly, we assessed the combination of
these nine miRNAs, and found that an integrated analysis of these
nine miRNAs improved the accuracy of breast cancer diagnosis.
However, multicenter studies with large sample sizes are needed to
validate our findings before the clinical application of this
nine-miRNA signature.
Acknowledgements
The present study was supported by grants from the
Natural Science Foundation of Guangxi, China (no.
2015GXNSFAA139187) and the Guangxi Provincial Health Bureau
Scientific Research Project (no. Z2014057).
References
|
1
|
Schneider AP II, Zainer CM, Kubat CK,
Mullen NK and Windisch AK: The breast cancer epidemic: 10 facts.
Linacre Q. 81:244–277. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Donepudi MS, Kondapalli K, Amos SJ and
Venkanteshan P: Breast cancer statistics and markers. J Cancer Res
Ther. 10:506–511. 2014.PubMed/NCBI
|
|
4
|
Zhou J, Tian Y, Li J, Lu B, Sun M, Zou Y,
Kong R, Luo Y, Shi Y, Wang K, et al: miR-206 is down-regulated in
breast cancer and inhibits cell proliferation through the
up-regulation of cyclinD2. Biochem Biophys Res Commun. 433:207–212.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gao Y, Cai Q, Huang Y, Li S, Yang H, Sun
L, Chen K and Wang Y: MicroRNA-21 as a potential diagnostic
biomarker for breast cancer patients: A pooled analysis of
individual studies. Oncotarget. 7:34498–34506. 2016.PubMed/NCBI
|
|
6
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Carthew RW and Sontheimer EJ: Origins and
mechanisms of miRNAs and siRNAs. Cell. 136:642–655. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang P, Chen L, Zhang J, Chen H, Fan J,
Wang K, Luo J, Chen Z, Meng Z and Liu L: Methylation-mediated
silencing of the miR-124 genes facilitates pancreatic cancer
progression and metastasis by targeting Rac1. Oncogene. 33:514–524.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Marquardt JU and Galle PR: Epigenetic
regulation of methionine adenosyltransferase 1A: A role for
MicroRNA-based treatment in liver cancer? Hepatology. 57:2081–2084.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xia H, Ooi LL and Hui KM:
MicroRNA-216a/217-induced epithelial-mesenchymal transition targets
PTEN and SMAD7 to promote drug resistance and recurrence of liver
cancer. Hepatology. 58:629–641. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang F, Li L, Chen Z, Zhu M and Gu Y:
MicroRNA-214 acts as a potential oncogene in breast cancer by
targeting the PTEN-PI3K/Akt signaling pathway. Int J Mol Med.
37:1421–1428. 2016.PubMed/NCBI
|
|
12
|
Fu SW, Lee W, Coffey C, Lean A, Wu X, Tan
X, Man YG and Brem RF: miRNAs as potential biomarkers in early
breast cancer detection following mammography. Cell Biosci.
6:62016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Shi H, Chen J, Li Y, Li G, Zhong R, Du D,
Meng R, Kong W and Lu M: Identification of a six microRNA signature
as a novel potential prognostic biomarker in patients with head and
neck squamous cell carcinoma. Oncotarget. 7:21579–21590.
2016.PubMed/NCBI
|
|
14
|
Bland JM and Altman DG: Statistical
methods for assessing agreement between two methods of clinical
measurement. Lancet. 1:307–310. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Riecke L, Sack AT and Schroeder CE:
Endogenous delta/theta sound-brain phase entrainment accelerates
the buildup of auditory streaming. Curr Biol. 25:3196–3201. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cheng Q, Yi B, Wang A and Jiang X:
Exploring and exploiting the fundamental role of microRNAs in tumor
pathogenesis. Onco Targets Ther. 6:1675–1684. 2013.PubMed/NCBI
|
|
17
|
Tan YY, Xu XY, Wang JF, Zhang CW and Zhang
SC: MiR-654-5p attenuates breast cancer progression by targeting
EPSTI1. Am J Cancer Res. 6:522–532. 2016.PubMed/NCBI
|
|
18
|
Jacot W, Fiche M, Zaman K, Wolfer A and
Lamy PJ: The HER2 amplicon in breast cancer: Topoisomerase IIA and
beyond. Biochim Biophys Acta. 1836:146–157. 2013.PubMed/NCBI
|
|
19
|
Zaczek A, Markiewicz A, Supernat A,
Bednarz-Knoll N, Brandt B, Seroczyńska B, Skokowski J, Szade J,
Czapiewski P, Biernat W, et al: Prognostic value of TOP2A gene
amplification and chromosome 17 polysomy in early breast cancer.
Pathol Oncol Res. 18:885–894. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lambros MB, Natrajan R, Geyer FC,
Lopez-Garcia MA, Dedes KJ, Savage K, Lacroix-Triki M, Jones RL,
Lord CJ, Linardopoulos S, et al: PPM1D gene amplification and
overexpression in breast cancer: A qRT-PCR and chromogenic in situ
hybridization study. Mod Pathol. 23:1334–1345. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhu S, Wu H, Wu F, Nie D, Sheng S and Mo
YY: MicroRNA-21 targets tumor suppressor genes in invasion and
metastasis. Cell Res. 18:350–359. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mandal CC, Ghosh-Choudhury T, Dey N,
Choudhury GG and Ghosh-Choudhury N: miR-21 is targeted by omega-3
polyunsaturated fatty acid to regulate breast tumor CSF-1
expression. Carcinogenesis. 33:1897–1908. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yang F, Wang Y, Xue J, Ma Q, Zhang J, Chen
YF, Shang ZZ, Li QQ, Zhang SL and Zhao L: Effect of Corilagin on
the miR-21/smad7/ERK signaling pathway in a schistosomiasis-induced
hepatic fibrosis mouse model. Parasitol Int. 65:308–315. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Liu F, Zheng S, Liu T, Liu Q, Liang M, Li
X, Sheyhidin I, Lu X and Liu W: MicroRNA-21 promotes the
proliferation and inhibits apoptosis in Eca109 via activating
ERK1/2/MAPK pathway. Mol Cell Biochem. 381:115–125. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Han M, Liu M, Wang Y, Mo Z, Bi X, Liu Z,
Fan Y, Chen X and Wu C: Re-expression of miR-21 contributes to
migration and invasion by inducing epithelial-mesenchymal
transition consistent with cancer stem cell characteristics in
MCF-7 cells. Mol Cell Biochem. 363:427–436. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Han M, Wang F, Gu Y, Pei X, Guo G, Yu C,
Li L, Zhu M, Xiong Y and Wang Y: MicroRNA-21 induces breast cancer
cell invasion and migration by suppressing smad7 via EGF and TGF-β
pathways. Oncol Rep. 35:73–80. 2016.PubMed/NCBI
|
|
27
|
Marino AL, Evangelista AF, Vieira RA,
Macedo T, Kerr LM, Abrahão-Machado LF, Longatto-Filho A, Silveira
HC and Marques MM: MicroRNA expression as risk biomarker of breast
cancer metastasis: A pilot retrospective case-cohort study. BMC
Cancer. 14:7392014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Gebremedhn S, Salilew-Wondim D, Hoelker M,
Rings F, Neuhoff C, Tholen E, Schellander K and Tesfaye D:
MicroRNA-183-96-182 cluster regulates bovine granulosa cell
proliferation and cell cycle transition by coordinately targeting
FOXO1. Biol Reprod. 94:1272016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lin H, Dai T, Xiong H, Zhao X, Chen X, Yu
C, Li J, Wang X and Song L: Unregulated miR-96 induces cell
proliferation in human breast cancer by downregulating
transcriptional factor FOXO3a. PLoS One. 5:e157972010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chang L, Guo F, Huo B, Lv Y, Wang Y and
Liu W: Expression and clinical significance of the microRNA-200
family in gastric cancer. Oncol Lett. 9:2317–2324. 2015.PubMed/NCBI
|
|
31
|
Korpal M, Lee ES, Hu G and Kang Y: The
miR-200 family inhibits epithelial-mesenchymal transition and
cancer cell migration by direct targeting of E-cadherin
transcriptional repressors ZEB1 and ZEB2. J Biol Chem.
283:14910–14914. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tan H, Zhu Y, Zhang J, Peng L and Ji T:
miR141 expression is downregulated and negatively correlated with
STAT5 expression in esophageal squamous cell carcinoma. Exp Ther
Med. 11:1803–1808. 2016.PubMed/NCBI
|
|
33
|
Yu SJ, Hu JY, Kuang XY, Luo JM, Hou YF, Di
GH, Wu J, Shen ZZ, Song HY and Shao ZM: MicroRNA-200a promotes
anoikis resistance and metastasis by targeting YAP1 in human breast
cancer. Clin Cancer Res. 19:1389–1399. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Li J, Du L, Yang Y, Wang C, Liu H, Wang L,
Zhang X, Li W, Zheng G and Dong Z: MiR-429 is an independent
prognostic factor in colorectal cancer and exerts its
anti-apoptotic function by targeting SOX2. Cancer Lett. 329:84–90.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yoneyama K, Ishibashi O, Kawase R, Kurose
K and Takeshita T: miR-200a, miR-200b and miR-429 are onco-miRs
that target the PTEN gene in endometrioid endometrial carcinoma.
Anticancer Res. 35:1401–1410. 2015.PubMed/NCBI
|
|
36
|
Zhang X, Li P, Rong M, He R, Hou X, Xie Y
and Chen G: MicroRNA-141 is a biomarker for progression of squamous
cell carcinoma and adenocarcinoma of the lung: Clinical analysis of
125 patients. Tohoku J Exp Med. 235:161–169. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wu CL, Ho JY, Chou SC and Yu DS: MiR-429
reverses epithelial-mesenchymal transition by restoring E-cadherin
expression in bladder cancer. Oncotarget. 7:26593–26603.
2016.PubMed/NCBI
|
|
38
|
Nishijima N, Seike M, Soeno C, Chiba M,
Miyanaga A, Noro R, Sugano T, Matsumoto M, Kubota K and Gemma A:
miR-200/ZEB axis regulates sensitivity to nintedanib in non-small
cell lung cancer cells. Int J Oncol. 48:937–944. 2016.PubMed/NCBI
|
|
39
|
Asakura T, Yamaguchi N, Ohkawa K and
Yoshida K: Proteasome inhibitor-resistant cells cause EMT-induction
via suppression of E-cadherin by miR-200 and ZEB1. Int J Oncol.
46:2251–2260. 2015.PubMed/NCBI
|
|
40
|
Korpal M and Kang Y: The emerging role of
miR-200 family of microRNAs in epithelial-mesenchymal transition
and cancer metastasis. RNA Biol. 5:115–119. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Lee JW, Park YA, Choi JJ, Lee YY, Kim CJ,
Choi C, Kim TJ, Lee NW, Kim BG and Bae DS: The expression of the
miRNA-200 family in endometrial endometrioid carcinoma. Gynecol
Oncol. 120:56–62. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Krishnan K, Steptoe AL, Martin HC,
Pattabiraman DR, Nones K, Waddell N, Mariasegaram M, Simpson PT,
Lakhani SR, Vlassov A, et al: miR-139-5p is a regulator of
metastatic pathways in breast cancer. RNA. 19:1767–1780. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zhang HD, Sun DW, Mao L, Zhang J, Jiang
LH, Li J, Wu Y, Ji H, Chen W, Wang J, et al: MiR-139-5p inhibits
the biological function of breast cancer cells by targeting Notch1
and mediates chemosensitivity to docetaxel. Biochem Biophys Res
Commun. 465:702–713. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Romero-Cordoba S, Rodriguez-Cuevas S,
Rebollar-Vega R, Quintanar-Jurado V, Maffuz-Aziz A, Jimenez-Sanchez
G, Bautista-Piña V, Arellano-Llamas R and Hidalgo-Miranda A:
Identification and pathway analysis of microRNAs with no previous
involvement in breast cancer. PLoS One. 7:e319042012. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Hua W, Sa KD, Zhang X, Jia LT, Zhao J,
Yang AG, Zhang R, Fan J and Bian K: MicroRNA-139 suppresses
proliferation in luminal type breast cancer cells by targeting
Topoisomerase II alpha. Biochem Biophys Res Commun. 463:1077–1083.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
MiR-145 promotes TNF-α-induced apoptosis
by facilitating the formation of RIP1-FADDcaspase-8 complex in
triple-negative breast cancer. Tumour Biol. 37:8599–8607. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Zheng M, Sun X, Li Y and Zuo W:
MicroRNA-145 inhibits growth and migration of breast cancer cells
through targeting oncoprotein ROCK1. Tumour Biol. 37:8189–8196.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Lv J, Xia K, Xu P, Sun E, Ma J, Gao S,
Zhou Q, Zhang M, Wang F, Chen F, et al: miRNA expression patterns
in chemoresistant breast cancer tissues. Biomed Pharmacother.
68:935–942. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Eades G, Wolfson B, Zhang Y, Li Q, Yao Y
and Zhou Q: lincRNA-RoR and miR-145 regulate invasion in
triple-negative breast cancer via targeting ARF6. Mol Cancer Res.
13:330–338. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Sengupta S, Das JK and Gangopadhyay A:
Naevoid psoriasis and ILVEN: Same coin, two faces? Indian J
Dermatol. 57:489–491. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Yan X, Chen X, Liang H, Deng T, Chen W,
Zhang S, Liu M, Gao X, Liu Y, Zhao C, et al: miR-143 and miR-145
synergistically regulate ERBB3 to suppress cell proliferation and
invasion in breast cancer. Mol Cancer. 13:2202014. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Min W, Wang B, Li J, Han J, Zhao Y, Su W,
Dai Z, Wang X and Ma Q: The expression and significance of five
types of miRNAs in breast cancer. Med Sci Monit Basic Res.
20:97–104. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Hou Y, Wang J, Wang X, Shi S, Wang W and
Chen Z: Appraising microRNA-155 as a noninvasive diagnostic
biomarker for cancer detection: A meta-analysis. Medicine.
95:e24502016. View Article : Google Scholar : PubMed/NCBI
|