Introduction
Glioma is the most common malignant tumor among
primary tumors of the central nervous system in adults and is
divided into 4 pathological grades (I–IV) based on the
classification criteria published by the World Health Organization
(WHO). The most malignant type of glioma is glioblastoma (1). Although glioma can be treated by
comprehensive regimens, including postoperative radiotherapy in
combination with temozolomide chemotherapy, excessive glioma cell
proliferation, apoptotic resistance, advanced invasion and
infiltration, and chemotherapeutic drug resistance can result in
unsatisfactory treatment outcomes. Patients with diffused
astrocytoma have the best prognosis, with a median postoperative
survival of 6–8 years, while glioblastoma patients have the poorest
prognosis (median survival of 12–15 months) (2,3).
Hence, identification of molecular targets that can effectively
suppress the malignant activity of glioma cells or be used to
determine glioma development and prognosis may have important
clinical value for treating glioma.
MicroRNAs (miRNAs) are endogenous small (~22
nucleotide) non-coding RNAs that regulate post-transcriptional gene
expression primarily through full or partial integration with the
3′-untranslated region (3-UTR) of target mRNAs (4). Recent studies have revealed a number
of miRNAs that are involved in the incidence, development,
metastasis and drug resistance of glioma. High or low expression of
miRNAs plays similar roles to oncogenes or tumor suppressor genes,
which are often associated with glioma staging, thus demonstrating
the potential for miRNAs to act as therapeutic targets (5–8). The
miR-130 family includes miR-130a and miR-130b, which are located in
the 22q11 locus. Recently, miR-130b was demonstrated to have tumor
suppressive and cancer-promoting properties depending on the tumor
type. miR-130b is highly expressed in melanoma (9), gastric (10), bladder (11) and colorectal cancer (12), esophageal squamous cell carcinoma
(13) and glioma (14), but significantly and poorly
expressed in papillary thyroid carcinomas (15), endometrial cancer (16), pituitary adenomas (17) and pancreatic cancer (18). Previously, miR-130b was observed to
be overexpressed in glioma tissues, while the suppression of the
expression of miR-130b via peroxisome proliferator-activated
receptor-γ (PPAR-γ) inhibited the proliferation and invasive
activity of glioma (14).
The present study utilized quantitative PCR to
detect miR-130b expression in 129 glioma specimens, assessing the
correlation between miR-130b expression and the clinicopathological
characteristics of glioma patients. In addition, both the
Kaplan-Meier method and Cox's proportional hazard model were used
to analyze the prognostic potential of miR-130b expression in
glioma. Target gene prediction software was used to predict a
potential target gene of miR-130b, cylindromatosis (CYLD)
gene, this was followed by the construction of a wild-type (WT)
CYLD gene and a dual-luciferase reporter plasmid with a
mutant 3′-UTR sequence. A Dual-Luciferase Reporter Assay System was
used to determine the effect of miR-130b on CYLD expression.
The role of CYLD in cell proliferation, the cell cycle, cell
migration and cell invasion were investigated using a miR-130b
mimic and inhibitor in U87 and U251 cells, and by upregulating and
downregulating the expression of miR-130b, as well as by utilizing
a CYLD-targeted siRNA.
Materials and methods
Clinical samples
Glioma specimens from 129 glioma patients and normal
brain tissues from 20 patients with severe traumatic brain injury
and craniotomy were collected from the Department of Neurosurgery
at Fuzhou General Hospital (Fuzhou, China) from January 2006 to
December 2010. The research protocol was approved by the Ethics
Committee of Fuzhou General Hospital, and all patients or their
families signed informed consent forms before participating in the
study. Pathological and diagnostic results of the patients were
assessed and confirmed by two experienced neuropathologists. The
glioma malignancy of the patients was categorized according to the
classification criteria published by the WHO in 2007, including 45
cases of grades I/II and 84 cases of grades III/IV gliomas. None of
the patients had undergone preoperative chemotherapy or
radiotherapy. All tissue specimens were rapidly frozen in liquid
nitrogen after resection for subsequent analysis. Follow-up with
the patients was terminated in December 2015.
RNA extraction and quantitative
real-time PCR (RT-PCR)
TRIzol reagent was used to extract total RNA from
glioma tissues and cells, as described in the manufacturer's
instructions. miScript Reverse Transcription kit (Qiagen) was used
for cDNA synthesis, followed by quantitative RT-PCR using the PCR
reaction mixture (20 µl/reaction) recommended by the miScript
SYBR-Green PCR kit instructions. After reverse transcription,
primers specific to miR-130b, or an internal reference (U6) were
used in the PCR system. A Prism 7500 RT-PCR system was used to
perform qRT-PCR using the following amplification conditions: 95°C
denaturation for 10 sec; 40 cycles of 95°C denaturation for 5 sec
and 60°C annealing for 34 sec. The experiment was repeated 3 times.
The 2−ΔΔCt method was used to calculate the relative
expression of the target gene (miR-130b) based on the internal
reference gene expression.
Cell culture and transfection
Glioma cell lines were purchased from the Shanghai
Institute of Biochemistry and Cell Biology (Shanghai, China).
Glioma cells were cultured in Dulbecco's modified Eagle's medium
(DMEM) (containing 10% fetal bovine serum), and incubated at 37°C
in a humid atmosphere with 5% CO2 humidity. Conventional
replacement of culture medium was performed when cells reached
logarithmic growth and a 90% fusion rate, to continue culturing
cells. Twenty-four hours before transfection, 1×105
glioma U87 and U251 cells under optimized growth conditions were
independently seeded into the wells of a 6-well culture plate. Upon
reaching 80% confluency, transfection was performed using
Lipofectamine™ 2000 as described in the manufacturer's instructions
to transfect NC, miR-130b mimic, miR-130b inhibitor, and
CYLD-targeted siRNA at a 100 nM final concentration (Shanghai
GenePharma Co., Ltd., Shanghai, China) in U87 and U251 cells.
Standard culture medium was replaced to continue culturing the
cells 6 h after transfection. The transfection was repeated in
triplicate.
Luciferase reporter assay
The TargetScan database was used to predict
miRNA/target interaction sites between miR-130b and
CYLD. GenBank was used to determine the 3′-UTR sequence of
the CYLD gene. The CYLD-WT 3′-UTR and
CYLD-mutant 3′-UTR genes were synthesized at Sangon Biotech
Co., Ltd. (Shanghai, China) with NotI and XhoI
restriction sites at the two ends. The genes were subsequently
inserted into the multiple cloning sites of psi-CHECK2, a
dual-luciferase reporter plasmid, to prepare WT and mutant clones,
followed by sequencing to confirm gene identity. HEK293A cells were
seeded into the wells of a 24-well plate. Each well contained
8×104 cells which were ready for transfection once the
cell fusion rate reached 80%. Based on the instructions for
Lipofectamine™ 2000, 0.2 µg of psi-CHECK2 and 100 nM miR-130b
inhibitor or negative control were co-transfected with HEK293A
cells. Five hours after transfection, complete DMEM was used to
replace the cell culture media. After culturing for 48 h, the
HEK293A cells were lysed according to the manufacturers (Promega,
Madison, WI, USA) instructions and labeled with the luciferase
substrate to detect the relative activity of luciferase in the
cells.
Cell Counting Kit-8 (CCK-8)
After trypsinization, ~5×103 cells were
seeded into the wells of a 96-well plate. Each sample was seeded
into 5-wells, followed by the addition of cell culture medium and
CCK-8 working solution. Blank controls contained only culture
medium and CCK-8 working solution. The optical densities (ODs) of
each group of cells were assessed at 450 nm using a microplate
reader at 24, 48, 72 and 96 h after transfection. The following
formula was used to calculate growth inhibition and to prepare a
growth inhibition curve: Cell growth inhibition = (1 - OD of the
experimental group/OD of the control group) × 100%. This experiment
was repeated 3 times.
Cell cycle assay
Freshly-prepared 70% pre-chilled ethanol was used
for fixing cells and permeabilization occurred at 4°C overnight.
Each group of cells was digested and mixed to form a single cell
suspension. The collected cells were washed once in
phosphate-buffered saline (PBS) and fixed with 70% pre-chilled
ethanol at 4°C overnight. After being washed once with PBS, the
cell suspension was filtered through a 400-mesh filter. RNase A was
incubated with the cell suspension for 30 min, followed by the
staining of the cells with propidium iodide (PI) for 30 min in the
dark. The cell cycle distribution was assessed by flow cytometry
and the experiment was repeated in triplicate.
Invasion and migration assays
Invasion assay
Matrigel matrix was dissolved at 4°C overnight,
then, diluted at a 1:3 ratio with pre-chilled serum-free medium by
adding 40 µl of the diluent into a pre-chilled Transwell culture
plate, followed by the covering of the plate with a polycarbonate
resin film and incubation at 37°C for 2 h to solidify the Matrigel
matrix. A 100 µl cell suspension (5×104 cells) was added
into the Transwell culture plate, followed by 200 µl of serum-free
medium. Chemokine (500 µl) was added to the lower chamber of the
Transwell culture plate, and the cells were incubated at 37°C for
24 h in the incubator containing 5% CO2. The upper
Transwell chamber was removed in such a way so that the culture
medium could be aspirated from inside the chamber. The residual
cells and Matrigel matrix were wiped onto the upper Transwell
chamber. The culture medium of the lower Transwell chamber was
aspirated, followed by immerse washing of the chamber twice with
PBS. Paraformaldehyde (4%) was used to fix the chamber membrane for
15 min, and then the chamber was washed twice with PBS and stained
with 0.1% crystal violet solution at room temperature for 15 min.
After being washed 3 times with PBS, the membrane of each small
chamber was collected and mounted onto a glass slide for imaging
under microscopy.
Migration assay
For migration assays, the Transwell upper chamber
was not coated with Matrigel matrix. The remaining procedures were
similar to the invasion assay. Both the invasion and migration
assays were repeated 3 times.
Tumor inoculation in nude mice
Pre-transfected cells in each group were prepared as
previously described. After transfection of the cells for 48 h, the
cell suspension in each group was prepared to adjust the single
cell suspension to 4×106 cells/200 µl. The left armpit
of each nude mouse was used as a culturing site for the
disinfection with ethanol and the subcutaneous injection of the 200
µl single cell suspension, followed by acupressure to avoid
overflow of the cell suspension from the culturing site. A Vernier
caliper was used to assess and record the tumor diameter (a) and
width (b) every 7 days for 42 days. Each nude mouse was euthanized
via decapitation to assess the tumor size, after which the tumor
was completely resected and the tumor was preserved at −80°C for
later use in the immunoblot analysis. The tumor volume (V) was
calculated according to the following equation: V =
(ab2)/2. The animal studies were approved by the
Institutional Animal Care and Use Committee of the Fuzhou General
Hospital, Fuzhou, China.
Immunoblot analysis
U87 and U251 glioma cells were collected 48 h after
transfection and added to RIPA protein lysate for total protein
extraction, which was quantified via BCA assay. Total protein (40
µg) extracted from each culture were separated using SDS-PAGE and
transferred onto a polyvinylidene fluoride (PVDF) membrane. Each
blot was blocked with 5% skim milk at room temperature for 1 h,
followed by incubation with anti-CYLD monoclonal antibody (1:1,000)
at 4°C overnight. After washing with Tris-buffered saline with
Tween-20 (TBST) the blot was incubated with horseradish
peroxidase-labeled secondary antibody (1:2,000) at room temperature
for 1 h. Finally, the blot was washed with TBST washing, developed
with enhanced chemiluminescence (ECL) reagent, and photographed to
visualize CYLD on the blot. The relative protein content of CYLD
was assessed based on the gray scale value of the protein band.
Statistical analysis
The SPSS 19.0 software (SPSS, Inc., Chicago, IL,
USA) was used for statistical analysis. All data are presented
using the mean ± standard deviation. ANOVA was used for the
comparison between groups. The Kaplan-Meier method and log-rank
test were used to calculate and compare patient survival,
respectively. Multivariate Cox regression analysis was used to
assess predictors related to survival. P<0.05 was considered to
be statistically significant. All experiments were independently
repeated in triplicate.
Results
miR-130b upregulation is associated
with unfavorable clinicopathological parameters in human
glioma
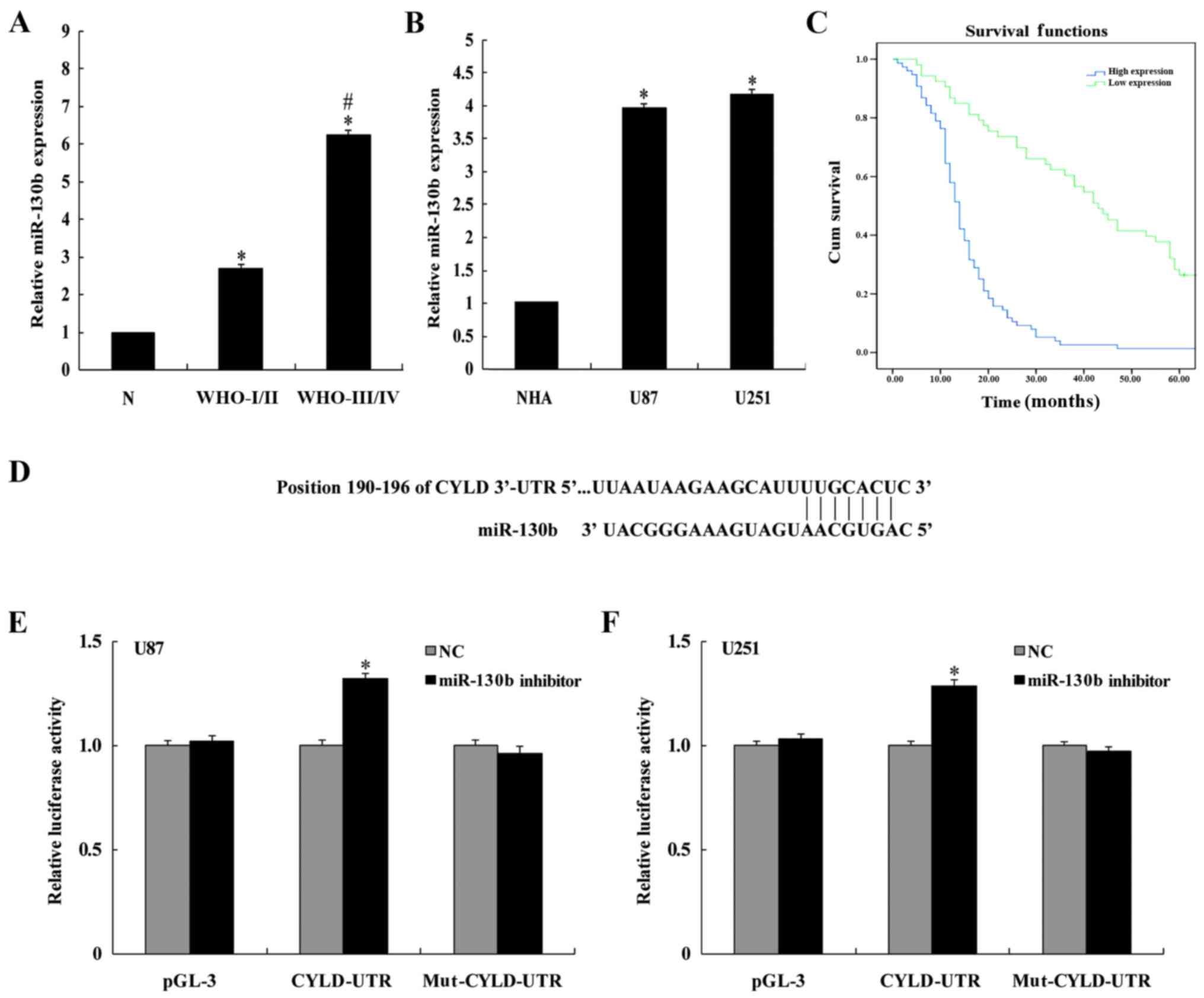
The relative miR-130b expression was assessed by
RT-PCR using miRNAs isolated from 129 glioma specimens, 26 normal
brain tissues, and the U87 and U251 glioma cell lines. The relative
miR-130b expression in glioma tissue and cell lines was
significantly higher than in normal brain tissues and primary
normal human astrocytes (NHA; P<0.05; Fig. 1A and B), suggesting that miR-130b
may play a role as an oncogene in glioma. A correlation analysis
between miR-130 expression and clinicopathological features/glioma
prognosis indicated that miR-130 expression was not associated with
the sex and age of glioma patients (P>0.05), but it was
significantly associated with the WHO glioma classification grading
scale and the Karnofsky performance status (KPS) scale (P<0.05;
Table I). The median survival of
patients with upregulated miR-130b expression was significantly
lower than patients with low miR-130b expression (14±0.717 and
43±4.094, respectively; P<0.05). The 5-year survival of patients
with upregulated miR-130b expression was significantly lower than
patients with low miR-130b expression (P<0.001; Fig. 1C). Multivariate Cox's model analysis
indicated a high pathological grade for glioma (P=0.002) and
miR-130b overexpression (P=0.000), which are two independent
factors used to predict the survival of glioma patients (Table II).
 | Table I.Relationship between miR-130b
expression and clinicopathological features. |
Table I.
Relationship between miR-130b
expression and clinicopathological features.
|
|
| miR-130b expression
median value |
|
|---|
|
|
|
|
|
|---|
| Clinicopathological
features | Pts. | Low (n) | High (n) | P-value |
|---|
| Age (years) |
|
|
| 0.193 |
|
<50 | 50 | 17 | 33 |
|
|
≥50 | 79 | 36 | 43 |
|
| Sex |
|
|
| 0.307 |
|
Male | 75 | 25 | 29 |
|
|
Female | 54 | 28 | 47 |
|
| WHO grade |
|
|
| 0.000a |
|
Low | 45 | 36 | 9 |
|
|
High | 84 | 17 | 67 |
|
| KPS score |
|
|
| 0.010a |
|
<90 | 61 | 43 | 38 |
|
|
≥90 | 68 | 10 | 28 |
|
 | Table II.Multivariate analyses of prognostic
parameters in the glioma patients using Cox regression
analysis. |
Table II.
Multivariate analyses of prognostic
parameters in the glioma patients using Cox regression
analysis.
|
| Multivariable
analysis |
|---|
|
|
|
|---|
| Variable | HR | 95% CI | P-value |
|---|
| Age (years) | 1.547 | 0.982–2.276 | 0.097 |
| Sex | 1.074 | 0.738–1.564 | 0.71 |
| WHO grade | 2.992 | 1.509–5.931 | 0.002a |
| KPS score | 1.602 | 0.903–2.841 | 0.107 |
| miR-130b | 2.910 | 1.736–4.875 | 0.000a |
CYLD is a potential downstream target
of miR-130b
Using a prediction algorithm through the TargetScan
bioinformatics database, the 3′-UTR of CYLD contained several
miR-130b interaction sites, including a seed sequence. This seed
sequence is located at the 190–196th nucleotides of the phosphatase
and tensin homolog 3′-UTR (Fig.
1D). A reporter construct was designed by ligating the 3′-UTR
of the CYLD fragment (containing the predicted miR-130b
interaction sites) downstream of a luciferase reporter gene.
Transfection of the construct demonstrated a significant increase
(P<0.05) in luciferase activity of the WT reporter plasmid after
co-transfection with the miR-130b inhibitor. In contrast, the
luciferase activity using the mutant plasmid was not significantly
different from the control group (P>0.05; Fig. 1E and F).
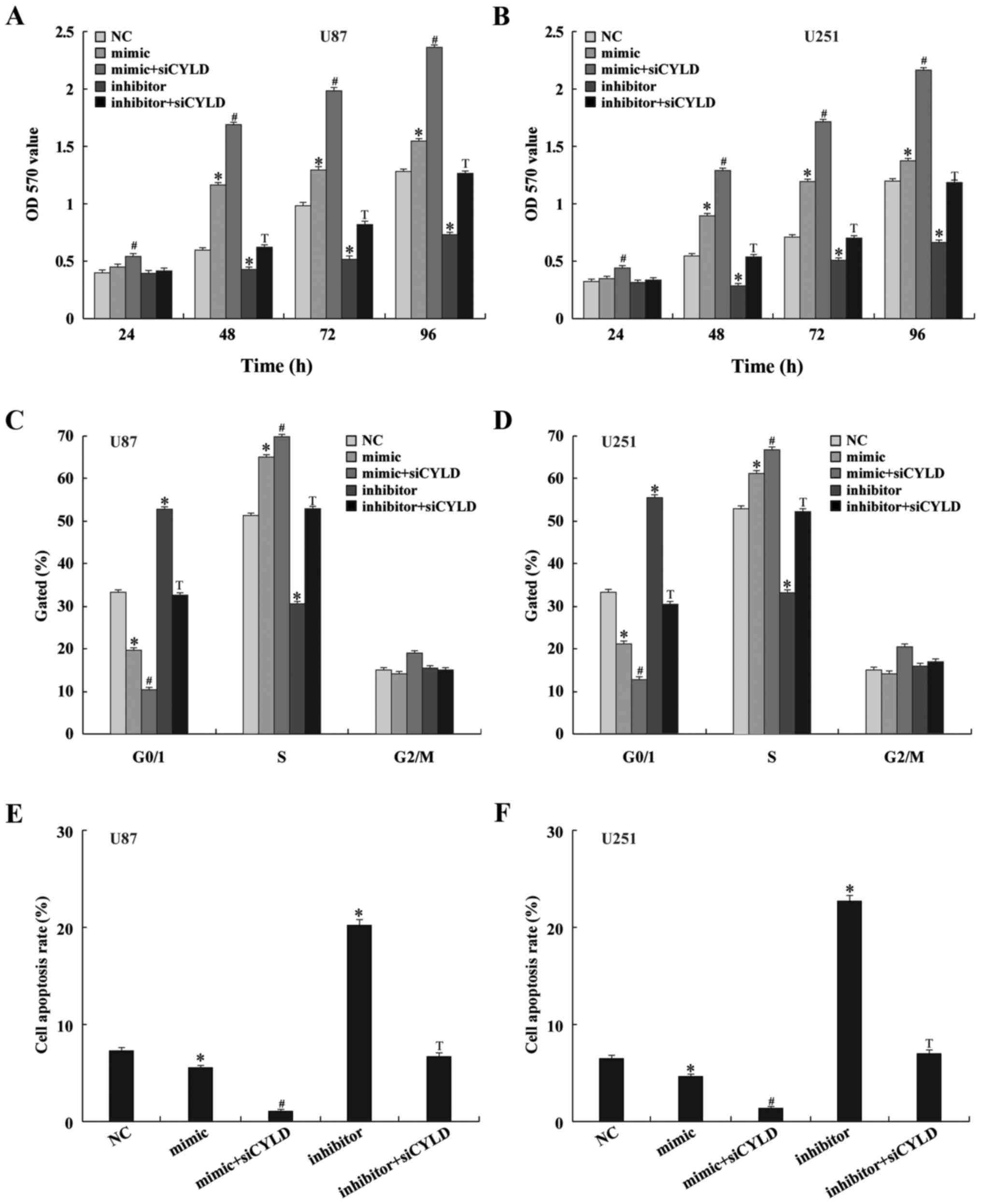
miR-130b regulates the proliferation
of U87 and U251 cells
Both CCK-8 and colony formation assays were utilized
to validate the impact of miR-130b on cell proliferation in U87 and
U251 cell lines. Twenty-four hours post-transfection, the glioma
cells were collected and the cell proliferation was assessed at 24,
48, 72 and 96 h after the cells were replaced. Glioma cell
proliferation was significantly enhanced at 48, 72 and 96 h after
transfection with the miR-130b mimic compared to the negative
control (P<0.05; Fig. 2A and B).
Additionally, 24 h after co-transfection with CYLD siRNA, the
miR-130b mimic-induced cell proliferation was enhanced. In
contrast, cell proliferation of glioma cells was significantly
suppressed at 48, 72 and 96 h after transfection with the miR-130b
inhibitor; while co-transfection with CYLD siRNA suppressed the
activity of the miR-130b inhibitor at these time-points. Flow
cytometric analysis indicated that glioma cell counts were
decreased during the G0/G1 phase, but were increased in number in
the S phase after transient transfection with the miR-130b mimic.
Co-transfection with CYLD siRNA significantly decreased the number
of cells in the G0/G1 phase, while also increasing the number of
cells in the S phase. In contrast, the number of cells at the G0/G1
phase was arrested and the cells in the S phase were decreased
after transfection with the miR-130b inhibitor (Fig. 2C and D); co-transfection with the
miR-130b inhibitor and CYLD siRNA counteracted the effect. These
in vitro experiments revealed that miR-130b affected cell
proliferation by regulating the G1/S transition in the cell
cycle.
miR-130b regulates apoptosis in U87
and U251 cells
In order to ascertain the effect of miR-130b on
apoptosis in glioma cells, an Annexin V-FITC/PI double staining
assay was performed on cells transfected with the miR-130b mimic.
Compared to the negative control group, the number of apoptotic
cells in the U87 and U251 cell lines was significantly decreased
(P<0.05). In contrast, the number of apoptotic cells in the U87
and U251 cell lines was significantly enhanced after transfecting
the cells with the miR-130b inhibitor (P<0.05). Co-transfection
with CYLD siRNA enhanced the anti-apoptotic ability of the miR-130b
mimic, while also decreasing the enhanced apoptosis of glioma cells
that was induced by the miR-130b inhibitor (Fig. 2E and F).
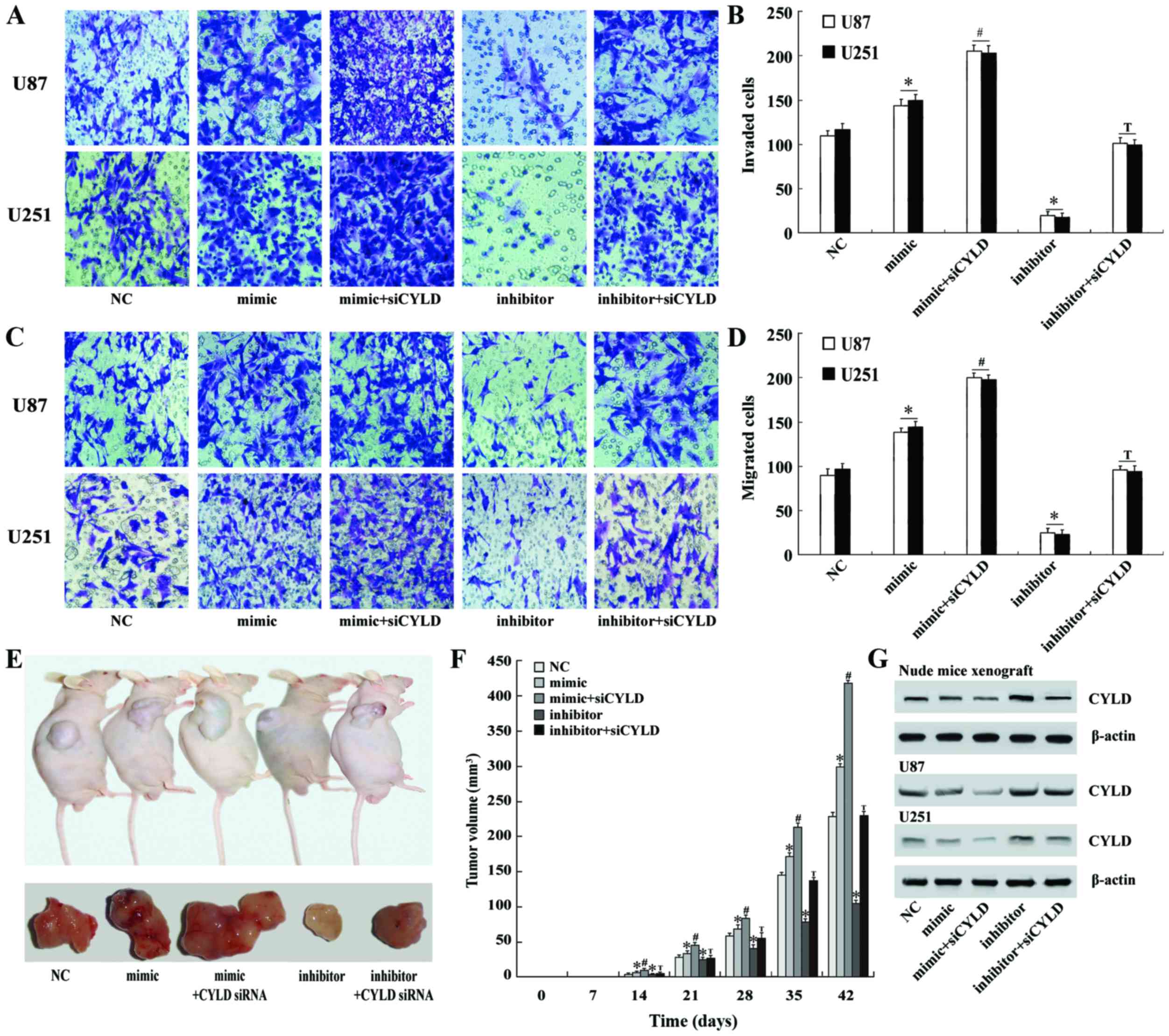
miR-130b regulates the invasiveness
and metastasis of U87 and U251 cells
Using invasion and migration assays, the impact of
miR-130b on cell invasion and migration in U87 and U251 cell lines
was assessed. Twenty-four hours after miR-130b mimic transfection,
the amount of U87 and U251 cells that passed through the Transwell
membrane was significantly higher than the negative control group
(P<0.05; Fig. 3A and B);
co-transfection with siRNA further increased these invasion
results. In contrast, U87 and U251 cells that were transfected with
the miR-130b inhibitor had decreased invasion in comparison to the
negative control, while co-transfection with CYLD siRNA
counteracted the suppressive effect of the miR-130b inhibitor on
the invasiveness of glioma cells (Fig.
3A and B). The experimental results obtained with the migration
assay were similar to the invasive assay results (Fig. 3C and D).
miR-130b affects glioma growth in nude
mice
Nude mice were subcutaneously inoculated with in
vitro-cultured U251 cells once the transfection rate reached
>90%. Small tumor nodules were found at the inoculation sites of
the nude mice 1–2 weeks later, with a tumor formation rate of 100%,
and in vivo glioma growth was monitored weekly. Tumor
nodules which appeared earlier, with rapid glioma growth and large
tumor volumes were found in the nude mice inoculated with glioma
cells transfected with the miR-130b mimic. In contrast, tumor
nodules which appeared later, with slow glioma growth and smaller
tumor volumes were found in the nude mice inoculated with glioma
cells transfected with the miR-130b inhibitor. Significant
differences in tumor volume were observed (P<0.05; Fig. 3E and F). Immunoblot analysis on the
glioma specimens demonstrated that CYLD expression in the glioma
tissues of the miR-130b mimic group was significantly decreased,
while CYLD expression in the glioma tissues of the miR-130b
inhibitor group was significantly enhanced (Fig. 3G).
miR-130b regulates CYLD protein
expression in U87 and U251 cells
Twenty-four hours after transient transfection,
changes in the expression of CYLD were assessed from total
extracted protein isolated from U87 and U251 glioma cells. In
comparison to the negative control group, the protein expression of
CYLD was significantly decreased after the expression of miR-130b
was upregulated, while the protein expression of CYLD was
significantly increased after downregulation of the expression of
miR-130b. The protein expression of CYLD was further decreased in
glioma cells after co-transfection with CYLD siRNA and the miR-130b
mimic. However, co-transfection with CYLD siRNA counteracted the
effects of the miR-130b inhibitor in enhancing CYLD expression in
glioma cells (Fig. 3G).
Discussion
miRNAs, a class of short non-coding small molecular
RNAs which are comprised of 19–25 nucleotides, are extremely
important biomolecules in the regulation of gene expression,
affecting ~30% of human genome transcription through full or
partial integration with the 3′-UTR of target mRNAs. This results
in mRNA degradation or inhibition of post-transcriptional
translation, thereby altering processes related to the cell cycle,
apoptosis, cell proliferation and cell migration (19). miRNA-130b is located on chromosome
22 and the mature sequence is 22 nucleotides in length. A variety
of genes such as PPAR-γ (20), STAT3 (18), PTEN (13) and Fmr1 (21), that are associated with the
incidence and development of cancer are also target genes for
miR-130b. However, recent studies have demonstrated that miR-130b
expression is either enhanced or decreased in different types of
tumors, demonstrating that it serves a role as either a tumor
promoter or suppressor, and may serve as a potential biomarker for
tumor development and prognosis. Wu et al (22) observed upregulated miR-130b
expression in clear-cell renal cell carcinoma and
clinicopathological analysis indicated that miR-130b was associated
with tumor metastasis. A long-term follow-up study of patients that
had enhanced miR-130 expression suggested an association with poor
prognoses. Zhao et al (18)
observed that miR-130b expression was significantly decreased in
pancreatic cancer, and a long-term follow-up study suggested that
pancreatic cancer patients with low miR-130b expression had poor
prognoses compared to pancreatic cancer patients with high miR-130b
expression, suggesting that miR-130b shares features with
tumor-suppressor genes.
In the present study, miR-130b was expressed at
significantly higher levels in glioma tissue and cells in
comparison to normal brain tissues and NHA. The pathological
grading of glioma that had upregulated miR-130b expression was
significantly higher than glioma with low miR-130 expression,
suggesting that miR-130b played a similar role to cancer-promoting
genes during the incidence of gliomas. A correlation analysis
between clinicopathological features and prognosis of gliomas
indicated that miR-130b overexpression was closely associated with
a high WHO glioma grade, low KPS and short cumulative survival of
glioma patients. Analysis by Cox's proportional hazard model
demonstrated that the hazard ratio (HR) of glioma patients with
miR-130b overexpression was 2.910-fold higher than glioma patients
with low miR-130b expression. In addition, the HR of glioma
patients with high pathological grading was 2.922-fold higher than
glioma patients with low pathological grading, suggesting that
miR-130b overexpression and high pathological grading were two
independent parameters in predicting poor prognosis in glioma
patients. Thus, this clinical study suggested that miR-130b could
act as a marker for predicting the prognosis of glioma
patients.
CYLD, a tumor-suppressor gene, is extensively
expressed in humans and consists of 956 amino acid residues,
including a CAP-Gly and a C-terminal domain (23). Numerous studies have demonstrated
that CYLD can act as a de-ubiquitination enzyme and that this
activity inhibits the activation of the NF-κB, JNK and
WNT/β-catenin signaling pathways. Thus, CYLD plays an important
regulatory role in various processes, including cell cycle
regulation, mediating apoptosis and inhibiting tumor incidence
(24–26). Multiple studies have shown that
deficiency or downregulation of CYLD is involved in the
development of melanoma, T cell leukemia, incidence and development
of colon cancer and hepatocellular carcinoma (27–29).
Song et al (30) observed
downregulated CYLD expression in 14 glioma tissues and 15 glioma
cell lines; CYLD expression was negatively associated with the
pathological WHO grading of glioma. In addition, glioma patients
with low CYLD expression had shorter postoperative survival than
glioma patients with higher CYLD expression. Moreover, CYLD
expression was negatively associated with CD31, Ki67 and MMP-9
expression. These findings confirm that decreased CYLD expression
is related to the incidence and development of glioma. Recently, an
in-depth study of miRNAs in tumors revealed that CYLD is a target
gene among all miRNA targets that regulate cell proliferation,
apoptosis and invasion of cancers. For example, miR-454 promotes
human colon cancer cell proliferation via CYLD gene
expression (31); miR-182 regulates
glioma cell proliferation and invasiveness by targeting the
CYLD gene (30); miR-130b
inhibits the proliferation and mediated apoptosis of stomach cancer
cells by targeting CYLD (10).
Using the TargetScan bioinformatics database,
CYLD was a predicted target gene for miR-130b. The putative
mRNA 3′-UTR ‘seed sequence’ region of CYLD was ligated into
the 3′ end of a pGL-3 luciferase reporter gene, allowing the
authenticity of the targets to be confirmed by co-transfection of
cells with the miRNA and a modified pGL-3 luciferase reporter gene
vector, simulating the miRNA regulating gene expression. This
experiment confirmed that CYLD was a downstream target of
miR-130b and additional in vitro experiments demonstrated
that glioma cells transfected with the miR-130b mimic and inhibitor
were able to suppress or promote the protein expression of
CYLD. Increased miR-130b expression promoted cell
proliferation and cells in the G0/G1 phase and entering the S
phase, arresting apoptosis while promoting tumor invasion and
metastasis. Conversely, decreased miR-130b expression suppressed
cell proliferation and cells in the G0/G1 phase and entering the S
phase, induced apoptosis, and inhibited tumor invasion and
metastasis. Co-transfection of the miR-130b inhibitor and
CYLD siRNA reversed the inhibitory effect of the miR-130b
inhibitor on tumor cell proliferation and invasiveness, while
co-transfection of the miR-130 mimic and CYLD siRNA enhanced
the ability of the miR-130 mimic to promote tumor cell
proliferation and invasiveness. The present study demonstrated that
miR-130b was associated with the activation of the CYLD signaling
pathway during the incidence and development of glioma.
In conclusion, the present study confirmed that the
upregulation of miR-130b expression in glioma was associated with a
positive disease prognosis. miR-130b negatively regulated the
expression of CYLD, a tumor-suppressor gene, in glioma and played
an important role in the proliferation, apoptosis, and invasiveness
of glioma cells. Therefore, miR-130b has the potential to serve as
prognostic marker and therapeutic target for glioma.
References
|
1
|
Ricard D, Idbaih A, Ducray F, Lahutte M,
Hoang-Xuan K and Delattre JY: Primary brain tumours in adults.
Lancet. 379:1984–1996. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lee JH, Jung TY, Jung S, Kim IY, Jang WY,
Moon KS and Jeong EH: Performance status during and after
radiotherapy plus concomitant and adjuvant temozolomide in elderly
patients with glioblastoma multiforme. J Clin Neurosci. 20:503–508.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wen PY and Kesari S: Malignant gliomas in
adults. N Engl J Med. 359:492–507. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Calin GA, Liu CG, Sevignani C, Ferracin M,
Felli N, Dumitru CD, Shimizu M, Cimmino A, Zupo S, Dono M, et al:
MicroRNA profiling reveals distinct signatures in B cell chronic
lymphocytic leukemias. Proc Natl Acad Sci USA. 101:11755–11760.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Weber F, Teresi RE, Broelsch CE, Frilling
A and Eng C: A limited set of human MicroRNA is deregulated in
follicular thyroid carcinoma. J Clin Endocrinol Metab.
91:3584–3591. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Jun GJ, Zhong GG and Ming ZS: miR-218
inhibits the proliferation of glioma U87 cells through the
inactivation of the CDK6/cyclin D1/p21Cip1/Waf1 pathway. Oncol
Lett. 9:2743–2749. 2015.PubMed/NCBI
|
|
7
|
Jiang L, Wang C, Lei F, Zhang L, Zhang X,
Liu A, Wu G, Zhu J and Song L: miR-93 promotes cell proliferation
in gliomas through activation of PI3K/Akt signaling pathway.
Oncotarget. 6:8286–8299. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hou SX, Ding BJ, Li HZ, Wang L, Xia F, Du
F, Liu LJ, Liu YH, Liu XD, Jia JF, et al: Identification of
microRNA-205 as a potential prognostic indicator for human glioma.
J Clin Neurosci. 20:933–937. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sand M, Skrygan M, Sand D, Georgas D,
Gambichler T, Hahn SA, Altmeyer P and Bechara FG: Comparative
microarray analysis of microRNA expression profiles in primary
cutaneous malignant melanoma, cutaneous malignant melanoma
metastases, and benign melanocytic nevi. Cell Tissue Res.
351:85–98. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sun B, Li L, Ma W, Wang S and Huang C:
MiR-130b inhibits proliferation and induces apoptosis of gastric
cancer cells via CYLD. Tumour Biol. 37:7981–7987. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Egawa H, Jingushi K, Hirono T, Ueda Y,
Kitae K, Nakata W, Fujita K, Uemura M, Nonomura N and Tsujikawa K:
The miR-130 family promotes cell migration and invasion in bladder
cancer through FAK and Akt phosphorylation by regulating PTEN. Sci
Rep. 6:205742016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Colangelo T, Fucci A, Votino C, Sabatino
L, Pancione M, Laudanna C, Binaschi M, Bigioni M, Maggi CA, Parente
D, et al: MicroRNA-130b promotes tumor development and is
associated with poor prognosis in colorectal cancer. Neoplasia.
15:1086–1099. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yu T, Cao R, Li S, Fu M, Ren L, Chen W,
Zhu H, Zhan Q and Shi R: MiR-130b plays an oncogenic role by
repressing PTEN expression in esophageal squamous cell carcinoma
cells. BMC Cancer. 15:292015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gu JJ, Zhang JH, Chen HJ and Wang SS:
MicroRNA-130b promotes cell proliferation and invasion by
inhibiting peroxisome proliferator-activated receptor-γ in human
glioma cells. Int J Mol Med. 37:1587–1593. 2016.PubMed/NCBI
|
|
15
|
Dettmer MS, Perren A, Moch H, Komminoth P,
Nikiforov YE and Nikiforova MN: MicroRNA profile of poorly
differentiated thyroid carcinomas: New diagnostic and prognostic
insights. J Mol Endocrinol. 52:181–189. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li BL, Lu C, Lu W, Yang TT, Qu J, Hong X
and Wan XP: miR-130b is an EMT-related microRNA that targets DICER1
for aggression in endometrial cancer. Med Oncol. 30:4842013.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Leone V, Langella C, D'Angelo D, Mussnich
P, Wierinckx A, Terracciano L, Raverot G, Lachuer J, Rotondi S,
Jaffrain-Rea ML, et al: Mir-23b and miR-130b expression is
downregulated in pituitary adenomas. Mol Cell Endocrinol. 390:1–7.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhao G, Zhang JG, Shi Y, Qin Q, Liu Y,
Wang B, Tian K, Deng SC, Li X, Zhu S, et al: MiR-130b is a
prognostic marker and inhibits cell proliferation and invasion in
pancreatic cancer through targeting STAT3. PLoS One. 8:e738032013.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Mei Q, Li F, Quan H, Liu Y and Xu H:
Busulfan inhibits growth of human osteosarcoma through miR-200
family microRNAs in vitro and in vivo. Cancer Sci. 105:755–762.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lee EK, Lee MJ, Abdelmohsen K, Kim W, Kim
MM, Srikantan S, Martindale JL, Hutchison ER, Kim HH, Marasa BS, et
al: miR-130 suppresses adipogenesis by inhibiting peroxisome
proliferator-activated receptor gamma expression. Mol Cell Biol.
31:626–638. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gong X, Zhang K, Wang Y, Wang J, Cui Y, Li
S and Luo Y: MicroRNA-130b targets Fmr1 and regulates embryonic
neural progenitor cell proliferation and differentiation. Biochem
Biophys Res Commun. 439:493–500. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wu X, Weng L, Li X, Guo C, Pal SK, Jin JM,
Li Y, Nelson RA, Mu B, Onami SH, et al: Identification of a
4-microRNA signature for clear cell renal cell carcinoma metastasis
and prognosis. PLoS One. 7:e356612012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Trompouki E, Hatzivassiliou E, Tsichritzis
T, Farmer H, Ashworth A and Mosialos G: CYLD is a deubiquitinating
enzyme that negatively regulates NF-kappaB activation by TNFR
family members. Nature. 424:793–796. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sun SC: CYLD: A tumor suppressor
deubiquitinase regulating NF-kappaB activation and diverse
biological processes. Cell Death Differ. 17:25–34. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Xue L, Igaki T, Kuranaga E, Kanda H, Miura
M and Xu T: Tumor suppressor CYLD regulates JNK-induced cell death
in Drosophila. Dev Cell. 13:446–454. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tauriello DV, Haegebarth A, Kuper I,
Edelmann MJ, Henraat M, Canninga-van Dijk MR, Kessler BM, Clevers H
and Maurice MM: Loss of the tumor suppressor CYLD enhances
Wnt/beta-catenin signaling through K63-linked ubiquitination of
Dvl. Mol Cell. 37:607–619. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Massoumi R, Kuphal S, Hellerbrand C, Haas
B, Wild P, Spruss T, Pfeifer A, Fässler R and Bosserhoff AK:
Down-regulation of CYLD expression by Snail promotes tumor
progression in malignant melanoma. J Exp Med. 206:221–232. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Espinosa L, Cathelin S, D'Altri T,
Trimarchi T, Statnikov A, Guiu J, Rodilla V, Inglés-Esteve J,
Nomdedeu J, Bellosillo B, et al: The Notch/Hes1 pathway sustains
NF-κB activation through CYLD repression in T cell leukemia. Cancer
Cell. 18:268–281. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hellerbrand C, Bumes E, Bataille F,
Dietmaier W, Massoumi R and Bosserhoff AK: Reduced expression of
CYLD in human colon and hepatocellular carcinomas. Carcinogenesis.
28:21–27. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Song L, Liu L, Wu Z, Li Y, Ying Z, Lin C,
Wu J, Hu B, Cheng SY, Li M, et al: TGF-β induces miR-182 to sustain
NF-κB activation in glioma subsets. J Clin Invest. 122:3563–3578.
2012. View
Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liang HL, Hu AP, Li SL, Xie JP, Ma QZ and
Liu JY: MiR-454 prompts cell proliferation of human colorectal
cancer cells by repressing CYLD expression. Asian Pac J Cancer
Prev. 16:2397–2402. 2015. View Article : Google Scholar : PubMed/NCBI
|