Introduction
Osteosarcoma is the most common bone malignancy
among patients in the pediatric age group (1). Although therapeutic interventions for
osteosarcoma have improved over the past decade, the 5-year overall
survival rate is still low (2,3). The
molecular pathogenesis of osteosarcoma remains unclear, which has
hampered efforts to improve treatment. Therefore, a better
understanding of the molecular pathogenesis of osteosarcoma is
crucial for the development of effective therapies for this
disease.
MicroRNAs (miRNAs) are a class of endogenous small
RNAs of ~22 nucleotides in length, and they negatively modulate
gene expression by targeting the 3′-untranslated region (3-UTR) of
mRNAs, leading to translational inhibition (4,5). By
post-transcriptionally regulating target genes, miRNAs are involved
in various cellular processes, including cell proliferation,
apoptosis, migration and invasion (6). In recent years, a growing body of
evidence suggests that miRNAs with deregulated expression patterns
are critical regulators in tumorigenesis (7–9).
Various miRNAs have been found to participate in the pathogenesis
of osteosarcoma, regulating numerous target genes and critical
pathways (10–12). These deregulated miRNAs have the
potential to serve as biomarkers for diagnosis and prognosis
(10–12). Targeting miRNAs has also shown
promise for the treatment of osteosarcoma. A better understanding
of the role of miRNAs in osteosarcoma may aid in the development of
miRNA-targeted therapies.
High-mobility group nucleosome-binding domain 5
(HMGN5), also known as nucleosome-binding protein 1, has been
suggested as an potential oncogene in several types of cancers
(13,14). HMGN5 can regulate histone
modification, DNA replication, DNA repair, and gene transcription
through binding to chromatin regulators (15). HMGN5 is found in a variety of
tissues (16,17), and regulates the expression of
numerous genes (18). However,
dysregulation of HMGN5 is associated with cancer progression and
development (13). To date, high
expression of HMGN5 has been linked to many types of cancers
including gliomas (19), prostate
cancer (20), renal cell carcinoma
(21), breast (22) and bladder cancer (23). Furthermore, overexpression of HMGN5
is also found in cases of osteosarcoma that are associated with
cell proliferation, metastasis, and drug resistance (24). HMGN5 has the potential to serve as a
molecular target for the prevention and treatment of
osteosarcoma.
MicroRNA-495 (miR-495) is associated with
tumorigenesis in various types of cancers, through the regulation
of numerous target genes (25–28).
It may be also involved in osteosarcoma. However, no data have yet
been reported. The aim of the present study was to determine
whether miR-495 is involved in osteosarcoma, and to investigate the
potential molecular mechanism of its involvement. We found that
miR-495 was significantly downregulated in osteosarcoma tissues and
cell lines. Overexpression of miR-495 inhibited proliferation,
induced apoptosis and repressed the invasion of osteosarcoma cells.
Notably, we identified HMGN5 as a potential target gene of miR-495
in osteosarcoma cells. miR-495 also regulated the expression of
cyclin B1, Bcl-2 and matrix metalloproteinase 9 (MMP9) which are
the downstream targets of HMGN5 involved in regulating cancer cell
proliferation, apoptosis and invasion (20,21,23).
The restoration of HMGN5 abrogated the miR-495-induced antitumor
effects. Taken together, our findings indicate that miR-495 exerts
antitumor effects in osteosarcoma by targeting HMGN5, providing
novel insight into the molecular pathogenesis of osteosarcoma and
suggesting a potential molecular target for the development of an
miRNA-targeted therapeutic strategy for osteosarcoma.
Materials and methods
Clinical samples
Fifteen paired human osteosarcoma and adjacent
normal tissues (located >3 cm from the tumor) were obtained from
the Department of Orthopaedics at The Second Hospital of Jilin
University. The resected tissues were immediately frozen in liquid
nitrogen and stored at −80°C for use. Use of the clinical tissue
samples was approved by the Institutional Human Experiment and
Ethics Committee of The Second Hospital of Jilin University, with
written informed consent from all of the participants.
Cell lines
Human osteosarcoma cell lines (143B, SaOS-2, U2OS
and MG63), normal osteoblastic cell line hFOB and 293T cells were
purchased from the American Type Culture Collection (ATCC;
Manassas, VA, USA). Cells were cultured in Dulbecco's modified
Eagles medium (DMEM; Invitrogen, Carlsbad, CA, USA), supplemented
with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin
solution (both from Gibco, Rockville, MD, USA), and maintained in a
humidified incubator containing 5% CO2 at 37°C.
RNA extraction and real-time
quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted using TRIzol reagent
(Invitrogen), according to the recommended protocols. The RNA
samples were pretreated with DNase I (Takara, Dalian, China) before
cDNA synthesis. To detect HMGN5 mRNA expression, cDNA was generated
by Moloney murine leukemia virus (M-MLV) reverse transcriptase
(BioTeke, Corporation, Beijing, China). To detect miR-495
expression, cDNA was generated with the TaqMan MicroRNA reverse
transcription kit (Applied Biosystems, Foster City, CA, USA).
RT-qPCR was conducted with SYBR-Green PCR Master Mix with a 7900HT
Fast Real-Time PCR System (both from Applied Biosystems). U6 and
glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were used for
normalization. Relative gene expression was calculated by the
2−ΔΔCt method. The fold-change of gene expression was
obtained by normalization against the control group. The primer
sequences were as follows: miR-495, 5′-AAACAAACAUGGUGCACUUCUU-3′
and 5′-GAAGUGCACCAUGUUUGUUUUU-3′; U6,
5′-GCTTCGGCAGCACATATACTAAAAT-3′ (forward) and
5′-CGCTTCACGAATTTGCGTGTCAT-3′ (reverse); HMGN5,
5′-GCAGTCAGGCAGTGACTGCCTTCG-3′ (forward) and
5′-CCCTTTTCTGTGGCATCTTC-3′ (reverse); and GAPDH,
5′-TGTGTCCGTCGTGGATCTGA-3′ (forward) and
5′-TTGCTGTTGAAGTCGCAGGAG-3′ (reverse).
Cell transfection
The miR-495 mimics and negative control (miR-NC)
were purchased from RiboBio (Guangzhou, China) and transfected into
cells using Lipofectamine 2000 (Invitrogen) following the
manufacturer's instructions. The HMGN5 cDNA without 3′-UTR was
cloned into a pcDNA3.1 vector (RiboBio) to generate a
pcDNA3.1/HMGN5 overexpressing vector. The vector was transfected
into cells by Lipofectamine 2000. The transfection efficacy was
assessed using RT-qPCR or western blot analysis after a 48-h
transfection period.
MTT assay
Cells were seeded into 96-well plates at a density
of 5×103 cells/well and cultured overnight. Cells were
transfected with either miR-495 mimics or miR-NC for 48 h. Then,
the medium was refreshed and 20 µl of
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT;
5 mg/ml; Sigma-Aldrich, St. Louis, MO, USA) was added to each well.
The cells were cultured for 4 h and 200 µl of dimethyl sulfoxide
(DMSO; Sigma-Aldrich) was added to dissolve the formazan crystals.
After 15 min, the absorbance was detected with a wavelength of 490
nm by an ELISA reader (Bio-Rad, Hercules, CA, USA).
Colony formation assay
The transfected cells were plated into 6-well plates
at a density of 100 cells/well and cultured in growth medium
containing 0.3% noble agar (Sigma-Aldrich) for 10 days. Afterwards,
the colonies were stained with 1% crystal violet (Sigma-Aldrich).
The colonies were counted under a microscope (Olympus, Tokyo,
Japan) and analyzed.
Cell cycle assay
After treatment, cells were fixed with 70% ethanol
and washed with phosphate-buffered saline (PBS). Cells were then
treated with 100 µg/ml of propidium iodide (PI; Sigma-Aldrich) and
10 µg/ml of RNase A (Roche Applied Science, Indianapolis, IN, USA)
for 30 min at room temperature in a dark place. Cells were examined
by flow cytometer BD FACSCalibur (BD Biosciences, San Jose, CA,
USA). The data were collected by CellQuest software (BD
Biosciences).
Annexin V/PI apoptosis assay
Cell apoptosis was detected using an Annexin V/PI
double staining kit (Beyotime Biotechnology, Haimen, China). Cells
were harvested and digested with 2.5 g/l of trypsin. After washing
with PBS, cells were re-suspended in 200 µl of binding buffer, and
treated with 10 µl of Annexin V for 30 min followed by incubation
with 5 µl of PI solution for 5 min in a dark place. Then, cells
were examined by flow cytometry (BD Biosciences).
Caspase-3 activity assay
Caspase-3 activity was detected using a commercial
kit (Roche Applied Science), following the manufacturer's
recommended protocols. Cells were lysed and incubated with DEVD-pNA
substrate at 37°C for 2 h. The absorbance at 405 nm was determined
using an ELISA reader (Bio-Rad).
Invasion assay
Cell invasion was detected using Transwell assay.
The upper chamber of the Transwell inserts (Costar, Corning, NY,
USA) was precoated with Matrigel (BD Biosciences). After
transfection, 1×105 cells were re-suspended in 500 µl of
serum-free medium, and added to the upper chamber. Meanwhile, the
lower chamber was filled with 500 µl of growth medium containing
10% FBS. The cells were cultured at 37°C for 24 h. Afterwards, the
filters were removed, fixed with 20% methanol, and stained with 1%
crystal violet (Sigma-Aldrich). The invaded cells were counted
under a microscope (Olympus).
Western blot analysis
Proteins were extracted using lysis buffer (Beyotime
Biotechnology), and equivalent amounts of proteins were loaded on
12.5% sodium dodecyl sulfate polyacrylamide gels for separation,
and then, transferred to a nitrocellulose membrane (Bio-Rad). After
being blocked with 3% non-fat milk, the membrane was blotted with
primary antibodies at 4°C overnight, and then incubated with
horseradish peroxidase-conjugated secondary antibodies (1:2,000;
Bioss, Beijing, China). The protein bands were developed using
enhanced chemiluminescence (Millipore, Boston, MA, USA). The
intensity of the protein bands was detected using Image-Pro Plus
6.0 software (Media Cybernetics, Inc., Rockville, MD, USA). The
fold-change of protein expression was obtained by normalization
with GAPDH, and then compared with the control group. The primary
antibodies, including anti-HMGN5, anti-cyclin B1, anti-Bcl-2,
anti-MMP9 and anti-GAPDH, were purchased from Santa Cruz
Biotechnology (Santa Cruz, CA, USA).
Luciferase reporter assays
HMGN5-3′-UTR containing either the predicted seed
sequence of miR-495 or a mutated binding site of the 3′-UTR were
inserted into a pmirGLO Dual-Luciferase plasmid (Promega, Madison,
WI, USA). The recombinant vector was then cotransfected with either
miR-495 mimics or miR-NC, into 293T cells using Lipofectamine 2000.
After 48 h, the relative luciferase activity was analyzed using the
Dual-Luciferase Assay kit (Promega).
Data analysis
Data are expressed as means ± standard deviation.
Statistical analyses were performed using the SPSS package (version
11.5; SPSS, Inc., Chicago, IL, USA). Differences were analyzed
using Student's t-test or one-way analysis of variance followed by
Bonferronis post hoc test. Values of p<0.05 were regarded as
statistically significant.
Results
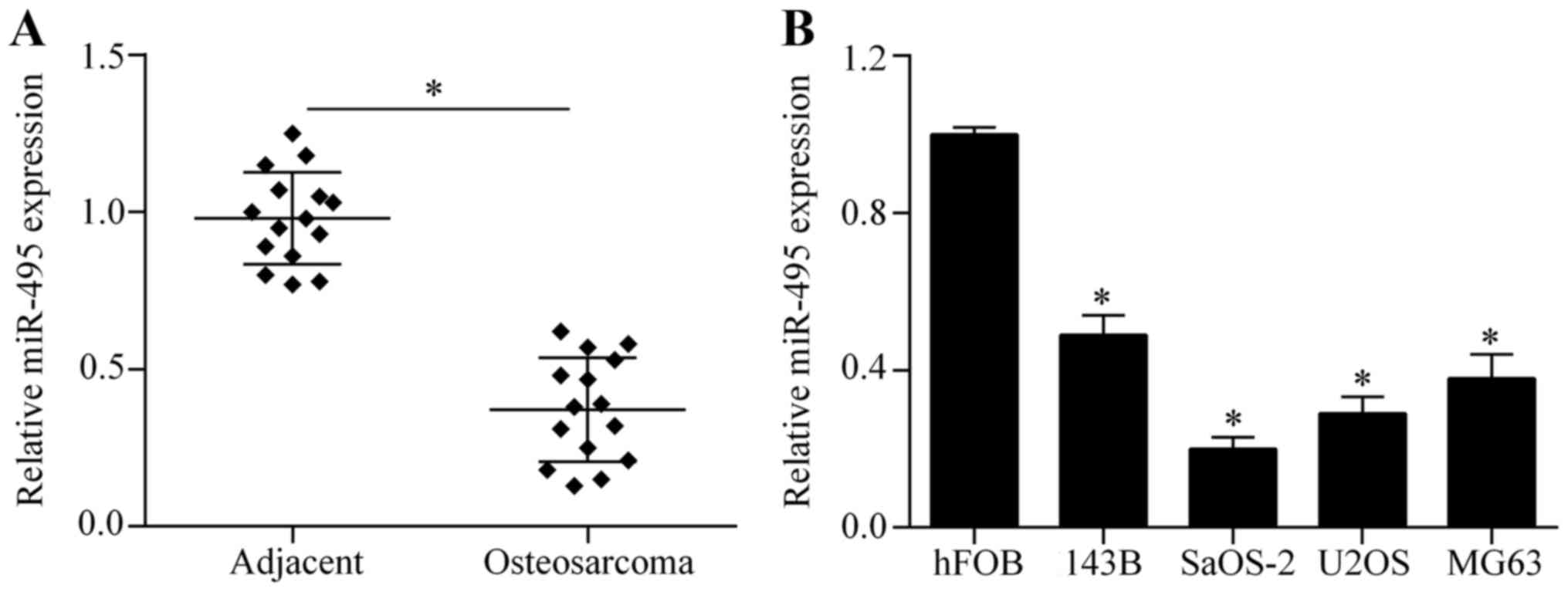
The expression of miR-495 is
downregulated in osteosarcoma tissues and cell lines
To investigate the possible role of miR-495 in
osteosarcoma, we first investigated the expression of miR-495 in
osteosarcoma tissues by RT-qPCR. The results showed that miR-495
was significantly downregulated in osteosarcoma tissues compared to
corresponding adjacent normal tissues (Fig. 1A). Next, we further examined the
expression pattern of miR-495 in four osteosarcoma cell lines:
143B, SaOS-2, U2OS and MG63. We found that miR-495 was markedly
decreased in the osteosarcoma cell lines compared with the normal
osteoblastic cell line, hFOB (Fig.
1B). Taken together, these findings indicate that miR-495 acts
in a tumor-suppressing role in cases of osteosarcoma.
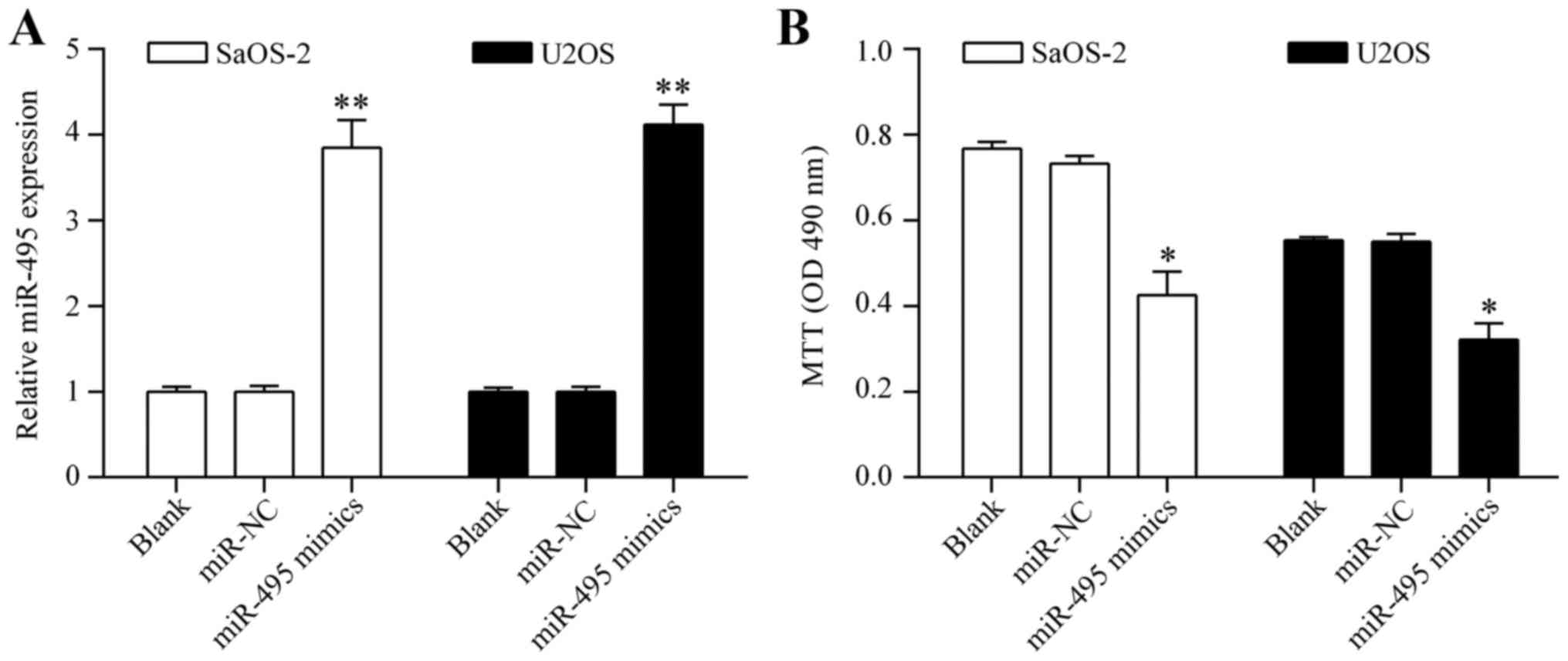
Overexpression of miR-495 inhibits
osteosarcoma cell proliferation
To investigate whether miR-495 shows an antitumor
effect on osteosarcoma cells, we overexpressed miR-495 in
osteosarcoma by transiently transfecting miR-495 mimics into SaOS-2
and U2OS cells (Fig. 2A). The
effect of miR-495 overexpression on cell proliferation was detected
by MTT assay. The results showed that overexpression of miR-495
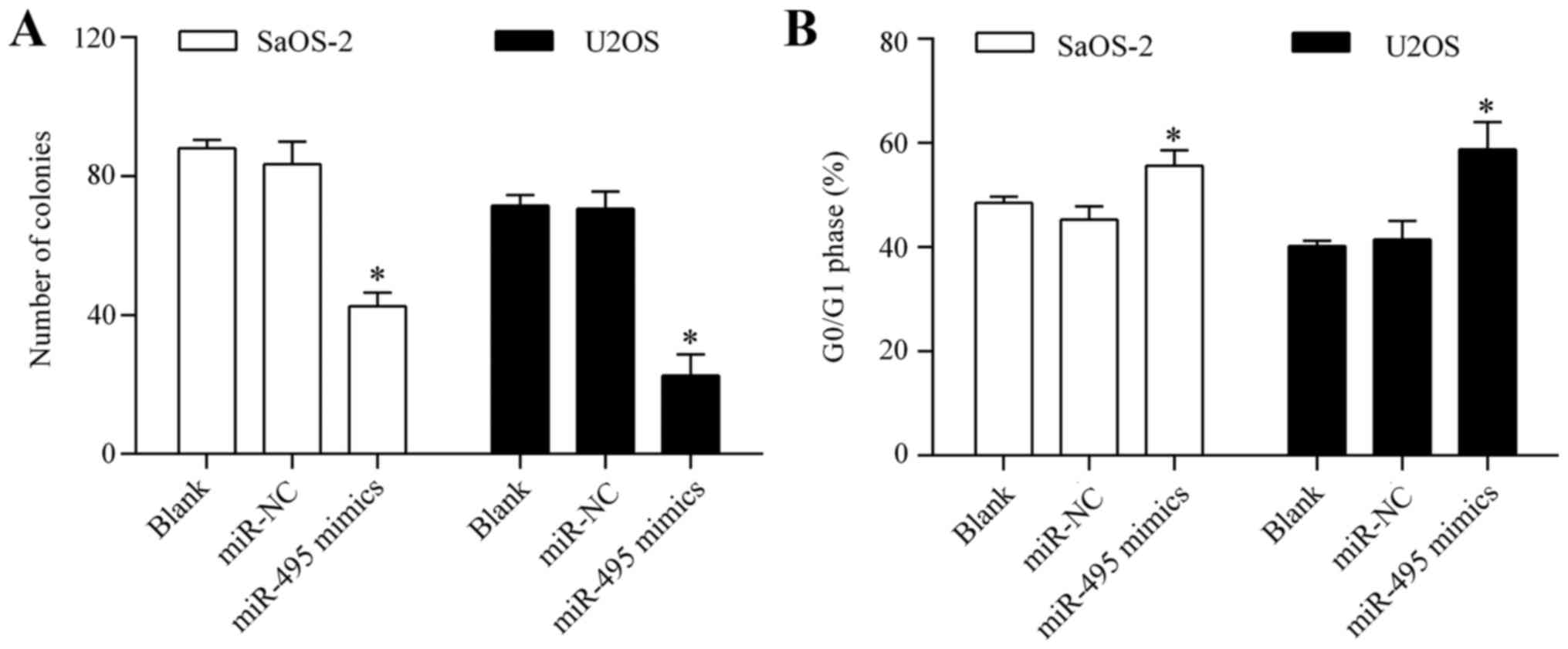
significantly inhibited osteosarcoma cell proliferation (Fig. 2B). Colony formation assay showed
that overexpression of miR-495 significantly suppressed the
colony-forming capacity of the osteosarcoma cells (Fig. 3A). Analysis of the cell cycle
distribution showed that miR-495 overexpression markedly induced
cell cycle arrest in the G0/G1 phase (Fig. 3B). Overall, these results suggest
that miR-495 inhibits osteosarcoma cell proliferation.
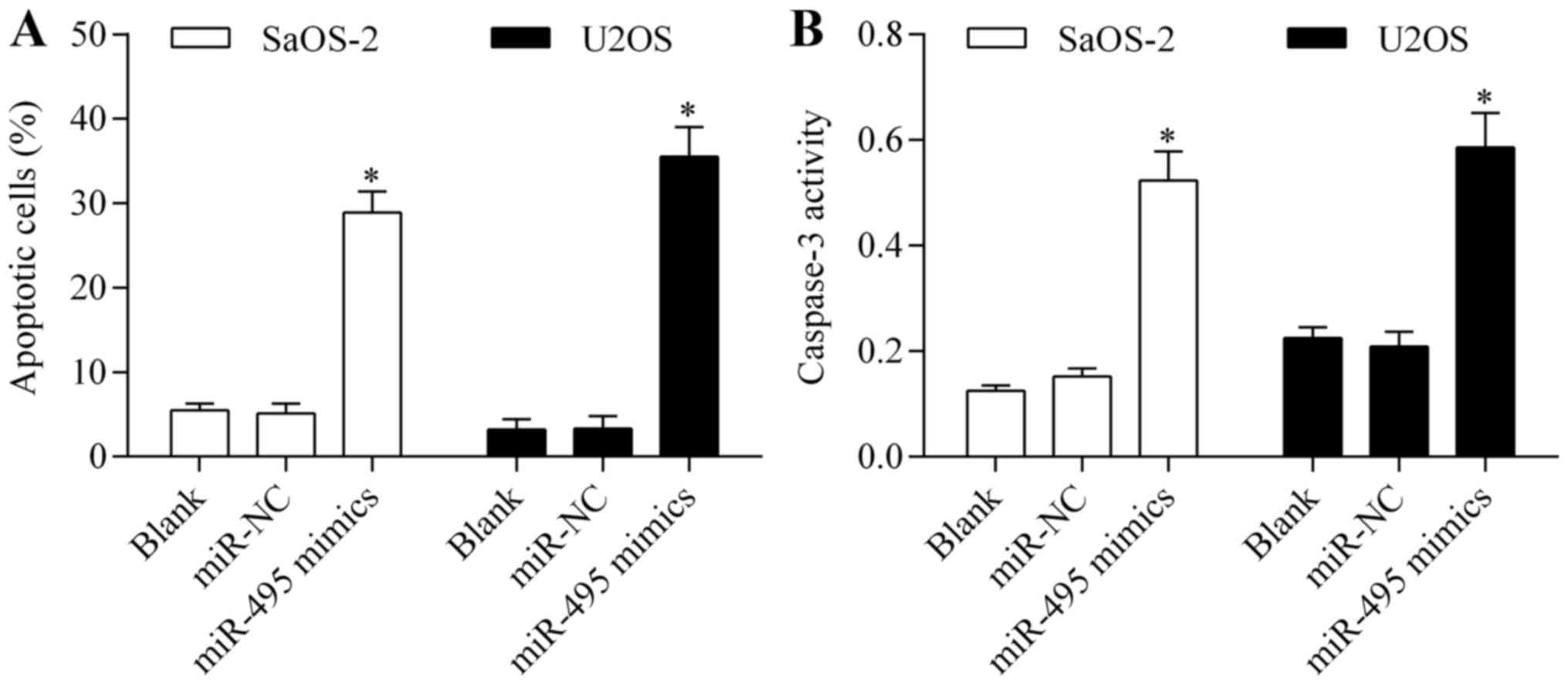
Overexpression of miR-495 induces
osteosarcoma cell apoptosis
To confirm the antitumor effect of miR-495 on
osteosarcoma cells, we examined the effect of miR-495
overexpression on cell apoptosis. Annexin V/PI apoptosis assay
showed that overexpression of miR-495 significantly promoted
osteosarcoma cell apoptosis (Fig.
4A). Moreover, miR-495 overexpression markedly increased the
activity of caspase-3 (Fig. 4B).
These data imply that miR-495 induces osteosarcoma cell
apoptosis.
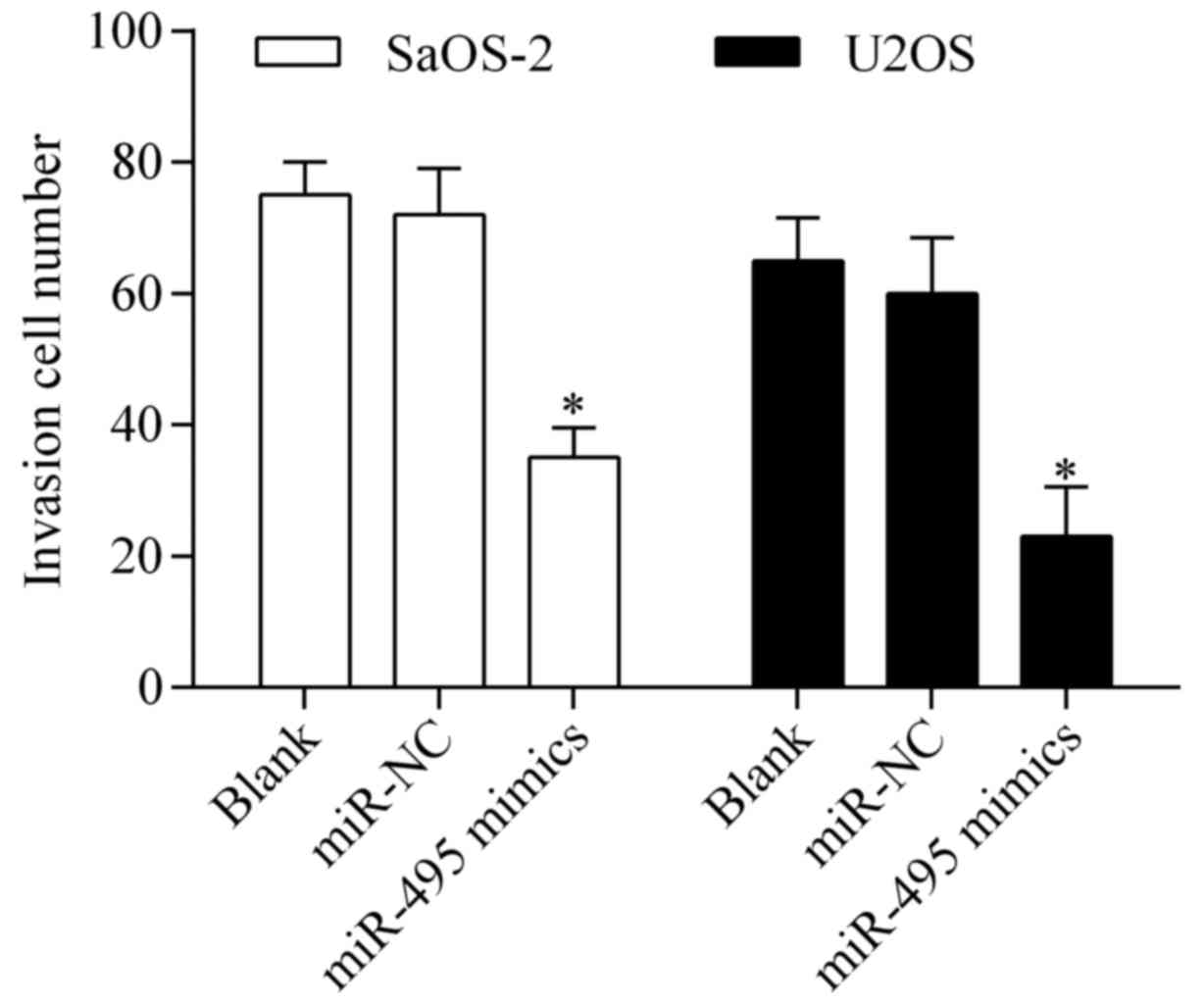
Overexpression of miR-495 suppresses
osteosarcoma cell invasion
To further investigate the antitumor effect of
miR-495, we detected the effect of miR-495 overexpression on
osteosarcoma cell invasion by Transwell invasion assay. The results
showed that overexpression of miR-495 significantly repressed the
invasive potential of SaOS-2 and U2OS cells (Fig. 5), indicating that miR-495 inhibits
osteosarcoma cell invasion.
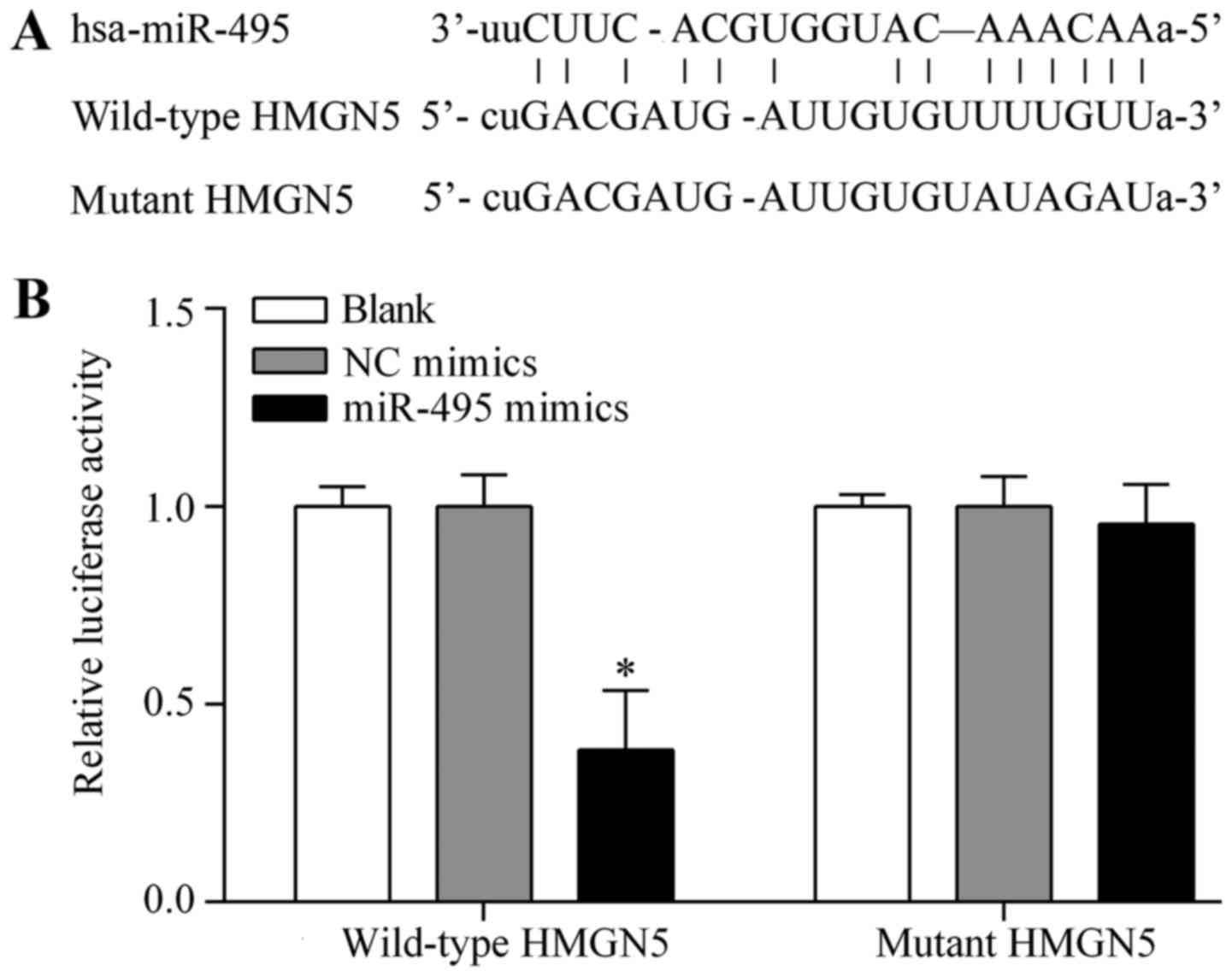
HMGN5 is the potential target gene of
miR-495 in osteosarcoma cells
To investigate the underlying mechanism by which
miR-495 exerts an antitumor effect, we performed bioinformatics
analysis to identify the target gene of miR-495. HMGN5, a
well-known oncogene associated with osteosarcoma tumorigenesis
(13), was predicted as the target
gene of miR-495. The putative binding sites of miR-495 within the
wild-type 3′-UTR of HMGN5 are described in Fig. 6A. The complementary seed sequences
were mutated to generate the mutant 3′-UTR of HMGN5 (Fig. 6A). To confirm the interaction
between miR-495 and HMGN5 3′-UTR, dual-luciferase assay was
performed using pmirGLO dual-luciferase containing either the
wild-type 3′-UTR of HMGN5 or the mutant 3′-UTR of HMGN5. The
results showed that the luciferase activity of the reporter vector
with the wild-type 3′-UTR of HMGN5 was significantly suppressed by
miR-495 overexpression (Fig. 6B).
However, miR-495 overexpression showed no significant effect on
luciferase activity of the reporter vector with the mutant 3′-UTR
of HMGN5 (Fig. 6B), indicating that
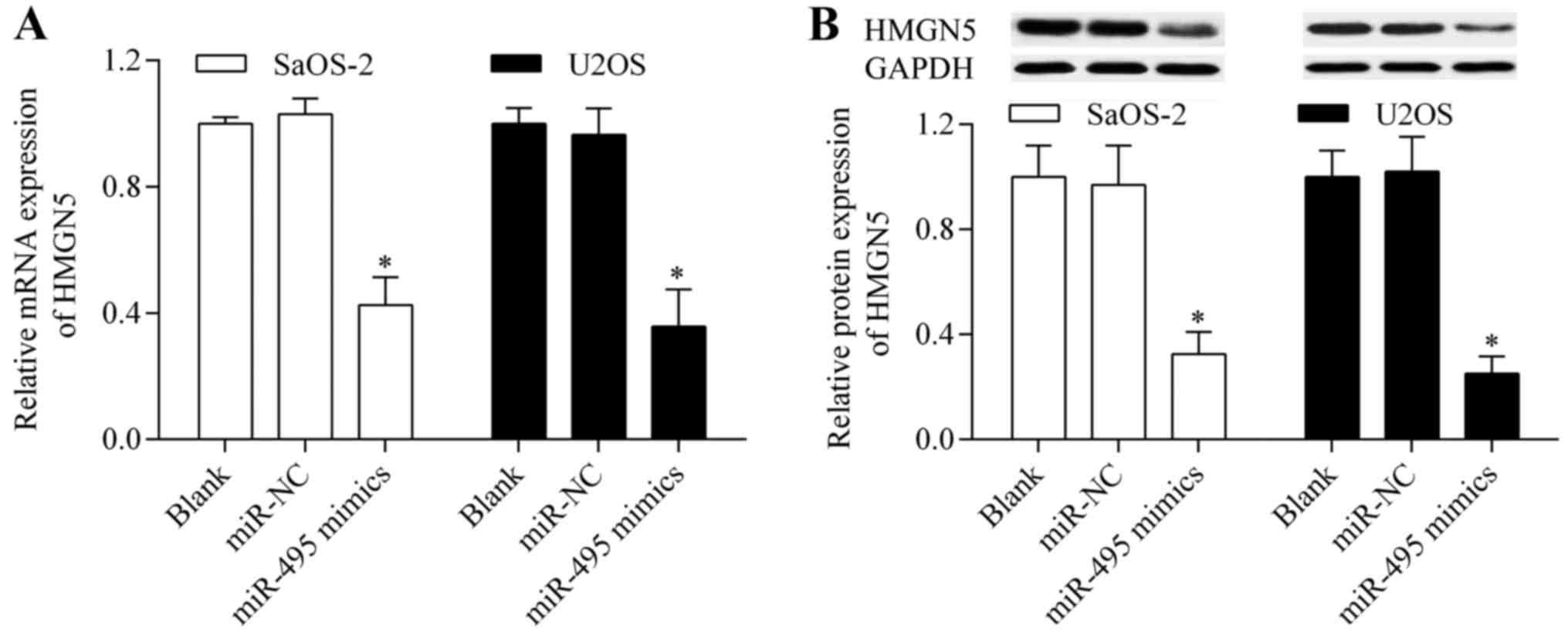
miR-495 directly targeted the 3′-UTR of HMGN5. To further verify
that HMGN5 is the target gene of miR-495, we examined the effect of
miR-495 on HMGN5 expression. The results showed that HMGN5 mRNA
(Fig. 7A) and protein (Fig. 7B) expression levels were
significantly reduced by miR-495 overexpression. Taken together,
these results suggest that HMGN5 is the potential target gene of
miR-495.
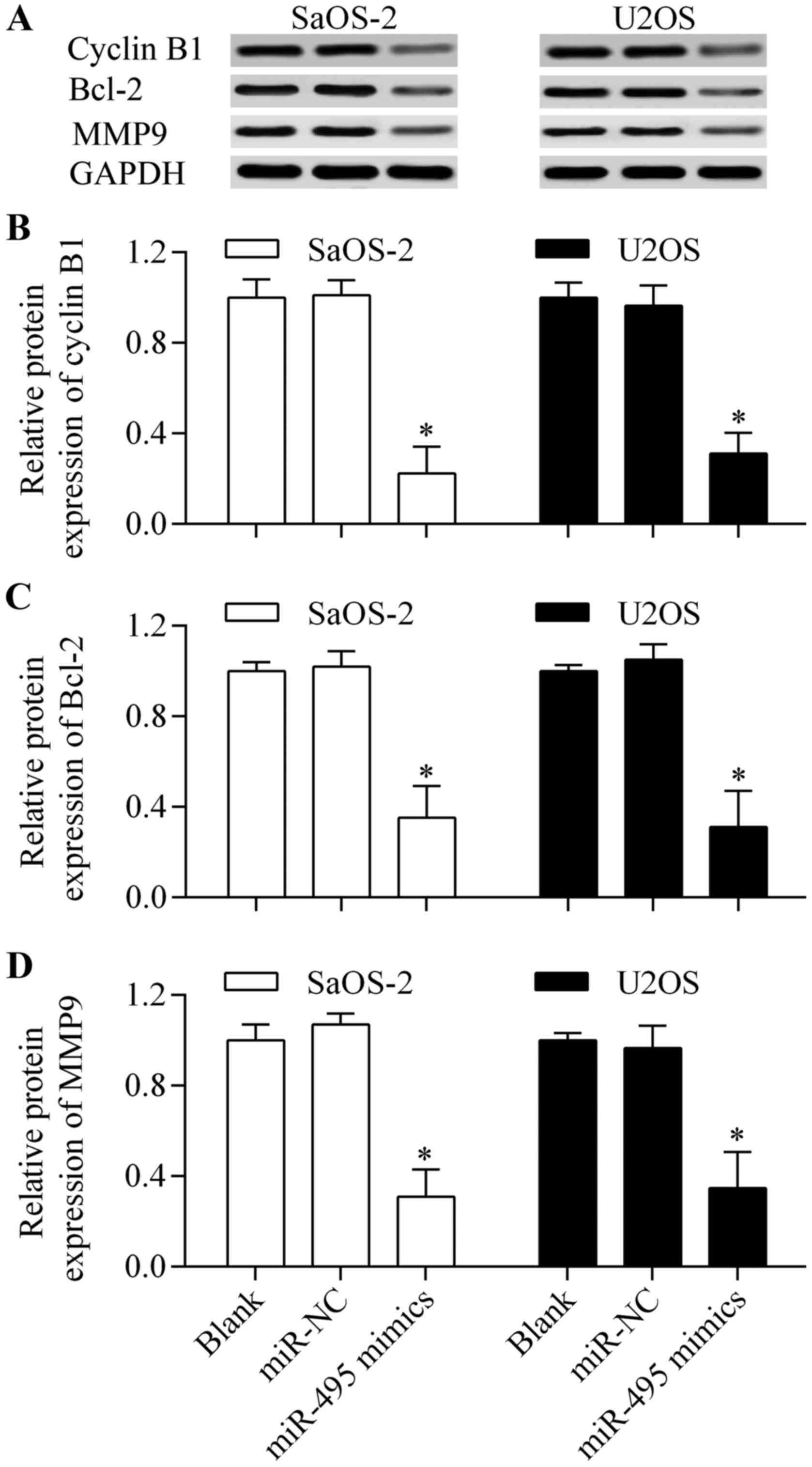
Overexpression of miR-495 affects the
downstream genes of HMGN5
To further investigate the molecular mechanism of
miR-495-induced antitumor effects, we detected the effect of
miR-495 on cyclin B1, Bcl-2 and MMP9 expression which are the
characterized downstream genes of HMGN5 (20,21,23).
The results showed that the protein expression levels of cyclin B1,
Bcl-2 and MMP9 (Fig. 8A-D) were
significantly decreased by miR-495 overexpression. These results
indicate that miR-495-induced antitumor effects are associated with
the inhibition of cyclin B1, Bcl-2 and MMP9.
HMGN5 is involved in miR-495-induced
antitumor effects
To validate whether miR-495 exerts antitumor effects
through HMGN5, we performed rescue experiments using pcDNA3.1/HMGN5
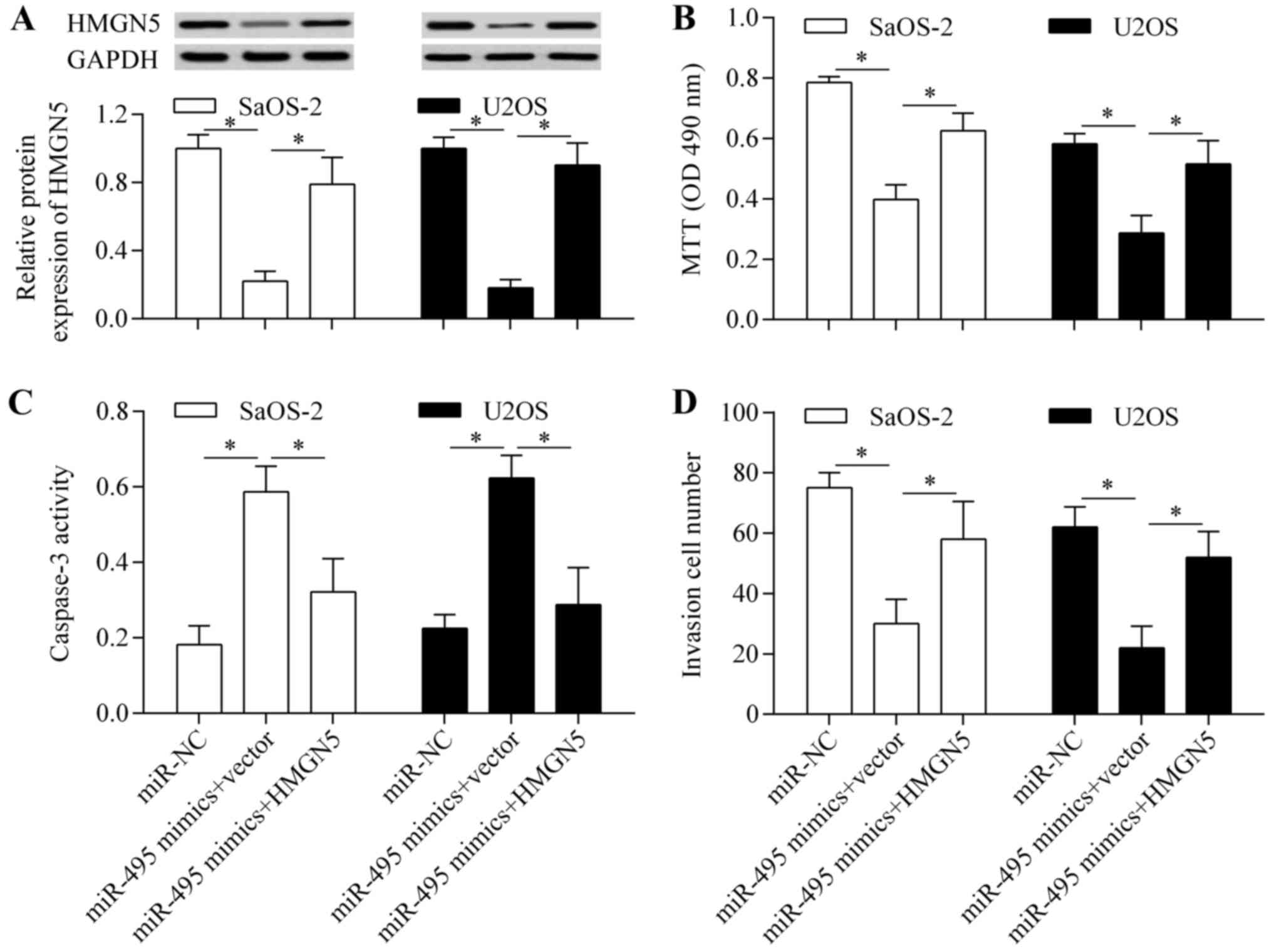
as an overexpressing vector. The results showed that the decreased
HMGN5 expression induced by miR-495 overexpression was markedly
restored by transfection of the pcDNA3.1/HMGN5 vector (Fig. 9A). We then evaluated the effect of
HMGN5 restoration on miR-495-induced antitumor effects. The results
showed that HMGN5 restoration significantly reversed the effects of
miR-495 overexpression on cell proliferation (Fig. 9B), apoptosis (Fig. 9C) and invasion (Fig. 9D). Overall, these findings indicate
that miR-495 exerts antitumor effects by targeting and inhibiting
HMGN5.
Discussion
A growing body of evidence suggests that miRNAs are
critical regulators in osteosarcoma (12). Deregulated miRNAs can be used as
biomarkers for the diagnosis and prognosis of cancer (10–12).
Targeting miRNAs has shown promise for the treatment of
osteosarcoma. Therefore, it is of great importance to gain a better
understanding of how miRNAs act in osteosarcoma. In the present
study, we revealed that miR-495 is a novel miRNA involved in
osteosarcoma. We found that miR-495 was downregulated in
osteosarcoma and that overexpression of miR-495 inhibited
osteosarcoma cell proliferation and invasion and promoted cell
apoptosis. These findings suggest that miR-495 may be associated
with the development and progression of osteosarcoma, and could be
a potential molecular target for the development of miRNA-targeted
therapies.
The deregulated expression of miR-495 has been found
in numerous types of cancers (29–32).
Li et al reported that miR-495 inhibited gastric cancer cell
migration and invasion through targeting phosphatase of
regenerating liver-3 (33).
Similarly, gastric cancer cell migration and invasion can be
suppressed though demethylation treatment, which upregulates
miR-495 (34). Numerous studies
have reported that miR-495 is decreased in brain tumors and can be
used to inhibit tumor progression by targeting cyclin-dependent
kinase 6 (35), Glut1 (36), Gfi1 (37) and v-myb avian myeloblastosis viral
oncogene homolog (38).
Downregulation of miR-495 was found in non-small cell lung cancer
tissues and cells, and overexpression of miR-495 inhibited cell
proliferation and migration by targeting metastasis-associated
protein 3 (25). Moreover,
overexpression of miR-495 promoted the sensitivity of non-small
cell lung cancer cells to platinum by inhibiting
copper-transporting P-type adenosine triphosphatase A (39). Xu et al reported that miR-495
inhibited cell growth and migration in endometrial cancer by
targeting Forkhead box C1 (40).
Overexpression of miR-495 also inhibited prostate cancer cell
migration and invasion by targeting Akt and the mammalian target of
rapamycin signaling (28).
Additionally, miR-495 induced cell cycle arrest in breast cancer
cells by inhibiting B cell-specific Moloney murine leukemia virus
integration site 1 (27). Overall,
these findings suggest that miR-495 has a tumor suppressive role.
However, an oncogenic role of miR-495 has also been reported. In
breast cancer stem cells, miR-495 was found to be overexpressed,
and promoted cell proliferation and invasion in hypoxia (41). In addition, overexpression of
miR-495 induced breast cancer cell migration by targeting
junctional adhesion molecule A (26). Under hypoxic conditions, miR-495 was
found to be overexpressed and promote proliferation and tumor
angiogenesis of gastric cancer cells by inhibiting Runt-related
transcription factor 3 (42). In
the present study, we found that miR-495 was significantly
decreased in osteosarcoma tissues and cell lines. Overexpression of
miR-495 suppressed osteosarcoma cell proliferation and invasion and
induced cell apoptosis of osteosarcoma cells, supporting a
tumor-suppressive role of miR-495 in osteosarcoma. The apparent
discrepancy in the role of miR-495 in tumorigenesis implies that
the precise biological role of miR-495 may be dependent on cell
type and condition. Thus, the role of miR-495 in tumorigenesis
requires further investigation.
To investigate the underlying mechanism responsible
for miR-495-mediated antitumor effects, we characterized the
functional target gene of miR-495 in osteosarcoma cells. We found
that HMGN5, a well-known oncogene (14), is a potential target of miR-495. A
reduction in HMGN5 was found to inhibit proliferation and induce
apoptosis of prostate cancer cells (20,43),
and suppression of HMGN5 induce cell cycle arrest in glioma cells
(19). High expression of HMGN5 is
also found in many other cancer types including renal cell
carcinoma (21), breast (22), bladder (23,44)
and lung cancer (45), and it is
associated with tumor progression. HMGN5 promotes tumorigenesis
through regulating cyclin B1, Bcl-2 and MMP9 (20,21,23).
Cyclin B1 regulates the G2/M transition (46), Bcl-2 is a strong anti-apoptotic
protein (47), and MMP9 promotes
cancer cell metastasis (48). By
promoting the expression of these genes, HMGN5 contributes to
cancer development and progression. HMGN5 has been found to be
highly expressed in osteosarcoma tissues associated with pathologic
staging (24). Suppression of HMGN5
inhibited the invasion, induced cell cycle arrest, and promoted the
sensitivity to doxorubicin-induced cell apoptosis in osteosarcoma
cells (24). Suppression of HMGN5
also increased apoptosis gene expression and decreased Akt, cyclin
B1 and MMP9 gene expression (24).
In line with these findings, Yang et al reported that HMGN5
overexpression promoted drug resistance by upregulating autophagy
in osteosarcoma cells (49). These
findings indicate that HMGN5 plays an important role in
osteosarcoma cells. In the present study, we demonstrated that
inhibition of HMGN5 by miR-495 overexpression inhibited
osteosarcoma cell proliferation and invasion, and induced cell
apoptosis of osteosarcoma cells, indicating the potential for the
development of a novel strategy for the treatment of osteosarcoma
by targeting HMGN5.
The epigenetic regulation of HMGN5 by miRNAs has
been reported in several studies (50–52).
Yao et al reported that miR-186 inhibited the growth and
metastasis of bladder cancer by targeting HMGN5 (50). By targeting HMGN5, miR-340 repressed
the tumorigenic potential of prostate cancer cells (51). In addition, inhibition of HMGN5 by
miR-326 impeded non-small cell lung cancer cell proliferation and
invasion (52). In line with our
findings, these studies indicate that HMGN5 underwent epigenetic
regulation during tumor progression. Targeting HMGN5 with specific
miRNAs shows promise for the development of cancer therapies.
The present study found, for the first time, that
miR-495 plays a critical role in osteosarcoma. Decreased levels of
miR-495 were observed in osteosarcoma and miR-495 overexpression
showed obvious antitumor effects. Investigation of the underlying
mechanism showed that miR-495 targeted HMGN5 and inhibited HMGN5
expression. The present study provides novel insights into the
molecular pathogenesis of osteosarcoma and suggests a potential
molecular target for the development of miRNA-based therapy for
osteosarcoma.
Glossary
Abbreviations
Abbreviations:
|
miRNAs
|
microRNAs
|
|
RT-qPCR
|
real-time quantitative polymerase
chain reaction
|
|
MTT
|
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
|
|
UTR
|
untranslated region
|
|
HMGN5
|
high-mobility group nucleosome-binding
domain 5
|
|
MMP9
|
matrix metalloproteinase 9
|
References
|
1
|
Botter SM, Neri D and Fuchs B: Recent
advances in osteosarcoma. Curr Opin Pharmacol. 16:15–23. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Meyers PA, Schwartz CL, Krailo MD, Healey
JH, Bernstein ML, Betcher D, Ferguson WS, Gebhardt MC, Goorin AM,
Harris M, et al: Children's Oncology Group: Osteosarcoma: The
addition of muramyl tripeptide to chemotherapy improves overall
survival - a report from the Children's Oncology Group. J Clin
Oncol. 26:633–638. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hughes DP: Strategies for the targeted
delivery of therapeutics for osteosarcoma. Expert Opin Drug Deliv.
6:1311–1321. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Winter J, Jung S, Keller S, Gregory RI and
Diederichs S: Many roads to maturity: MicroRNA biogenesis pathways
and their regulation. Nat Cell Biol. 11:228–234. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Thomson DW, Bracken CP and Goodall GJ:
Experimental strategies for microRNA target identification. Nucleic
Acids Res. 39:6845–6853. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Manikandan J, Aarthi JJ, Kumar SD and
Pushparaj PN: Oncomirs: The potential role of non-coding microRNAs
in understanding cancer. Bioinformation. 2:330–334. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Pang JC, Kwok WK, Chen Z and Ng HK:
Oncogenic role of microRNAs in brain tumors. Acta Neuropathol.
117:599–611. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Silber J, James CD and Hodgson JG:
microRNAs in gliomas: Small regulators of a big problem.
Neuromolecular Med. 11:208–222. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kushlinskii NE, Fridman MV and Braga EA:
Molecular mechanisms and microRNAs in osteosarcoma pathogenesis.
Biochemistry. 81:315–328. 2016.PubMed/NCBI
|
|
11
|
Zhang J, Yan YG, Wang C, Zhang SJ, Yu XH
and Wang WJ: MicroRNAs in osteosarcoma. Clin Chim Acta. 444:9–17.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sampson VB, Yoo S, Kumar A, Vetter NS and
Kolb EA: MicroRNAs and potential targets in osteosarcoma: Review.
Front Pediatr. 3:692015.Review. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Rochman M, Malicet C and Bustin M:
HMGN5/NSBP1: A new member of the HMGN protein family that affects
chromatin structure and function. Biochim Biophys Acta. 1799:86–92.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Shi Z, Tang R, Wu D and Sun X: Research
advances in HMGN5 and cancer. Tumour Biol. 37:1531–1539. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hock R, Furusawa T, Ueda T and Bustin M:
HMG chromosomal proteins in development and disease. Trends Cell
Biol. 17:72–79. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Shirakawa H, Landsman D, Postnikov YV and
Bustin M: NBP-45, a novel nucleosomal binding protein with a
tissue-specific and developmentally regulated expression. J Biol
Chem. 275:6368–6374. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
King LM and Francomano CA:
Characterization of a human gene encoding nucleosomal binding
protein NSBP1. Genomics. 71:163–173. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Postnikov Y and Bustin M: Regulation of
chromatin structure and function by HMGN proteins. Biochim Biophys
Acta. 1799:62–68. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Qu J, Yan R, Chen J, Xu T, Zhou J, Wang M,
Chen C, Yan Y and Lu Y: HMGN5: A potential oncogene in
gliomas. J Neurooncol. 104:729–736. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jiang N, Zhou LQ and Zhang XY:
Downregulation of the nucleosome-binding protein 1 (NSBP1) gene can
inhibit the in vitro and in vivo proliferation of prostate cancer
cells. Asian J Androl. 12:709–717. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ji SQ, Yao L, Zhang XY, Li XS and Zhou LQ:
Knockdown of the nucleosome binding protein 1 inhibits the growth
and invasion of clear cell renal cell carcinoma cells in vitro and
in vivo. J Exp Clin Cancer Res. 31:222012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Weng M, Song F, Chen J, Wu J, Qin J, Jin T
and Xu J: The high-mobility group nucleosome-binding domain 5 is
highly expressed in breast cancer and promotes the proliferation
and invasion of breast cancer cells. Tumour Biol. 36:959–966. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wahafu W, He ZS, Zhang XY, Zhang CJ, Yao
K, Hao H, Song G, He Q, Li XS and Zhou LQ: The nucleosome binding
protein NSBP1 is highly expressed in human bladder cancer and
promotes the proliferation and invasion of bladder cancer cells.
Tumour Biol. 32:931–939. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhou X, Yuan B, Yuan W, Wang C, Gao R and
Wang J: The expression and clinical significance of high mobility
group nucleosome binding domain 5 in human osteosarcoma. Tumour
Biol. 35:6539–6547. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chu H, Chen X, Wang H, Du Y, Wang Y, Zang
W, Li P, Li J, Chang J, Zhao G, et al: MiR-495 regulates
proliferation and migration in NSCLC by targeting MTA3. Tumour
Biol. 35:3487–3494. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Cao M, Nie W, Li J, Zhang Y, Yan X, Guan
X, Chen X, Zen K, Zhang CY, Jiang X, et al: MicroRNA-495 induces
breast cancer cell migration by targeting JAM-A. Protein Cell.
5:862–872. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang L, Liu JL, Yu L, Liu XX, Wu HM, Lei
FY, Wu S and Wang X: Downregulated miR-495 [corrected] inhibits the
G1-S phase transition by targeting Bmi-1 in breast cancer.
Medicine. 94:e7182015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li JZ, Wang ZL, Xu WH, Li Q, Gao L and
Wang ZM: MicroRNA-495 regulates migration and invasion in prostate
cancer cells via targeting Akt and mTOR signaling. Cancer Invest.
34:181–188. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Widodo Djati MS and Rifa'i M: Role of
MicroRNAs in carcinogenesis that potential for biomarker of
endometrial cancer. Ann Med Surg. 7:9–13. 2016. View Article : Google Scholar
|
|
30
|
Mishra S, Srivastava AK, Suman S, Kumar V
and Shukla Y: Circulating miRNAs revealed as surrogate molecular
signatures for the early detection of breast cancer. Cancer Lett.
369:67–75. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Formosa A, Markert EK, Lena AM, Italiano
D, Finazzi-Agro' E, Levine AJ, Bernardini S, Garabadgiu AV, Melino
G and Candi E: MicroRNAs, miR-154, miR-299-5p, miR-376a, miR-376c,
miR-377, miR-381, miR-487b, miR-485-3p, miR-495 and miR-654-3p,
mapped to the 14q32.31 locus, regulate proliferation, apoptosis,
migration and invasion in metastatic prostate cancer cells.
Oncogene. 33:5173–5182. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Jiang X, Huang H, Li Z, He C, Li Y, Chen
P, Gurbuxani S, Arnovitz S, Hong GM, Price C, et al: miR-495 is a
tumor-suppressor microRNA down-regulated in MLL-rearranged
leukemia. Proc Natl Acad Sci USA. 109:19397–19402. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Li Z, Cao Y, Jie Z, Liu Y, Li Y, Li J, Zhu
G, Liu Z, Tu Y, Peng G, et al: miR-495 and miR-551a inhibit the
migration and invasion of human gastric cancer cells by directly
interacting with PRL-3. Cancer Lett. 323:41–47. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Li Z, Zhang G, Li D, Jie Z, Chen H, Xiong
J, Liu Y, Cao Y, Jiang M, Le Z, et al: Methylation-associated
silencing of miR-495 inhibit the migration and invasion of human
gastric cancer cells by directly targeting PRL-3. Biochem Biophys
Res Commun. 456:344–350. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chen SM, Chen HC, Chen SJ, Huang CY, Chen
PY, Wu TW, Feng LY, Tsai HC, Lui TN, Hsueh C, et al: MicroRNA-495
inhibits proliferation of glioblastoma multiforme cells by
downregulating cyclin-dependent kinase 6. World J Surg Oncol.
11:872013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Nie S, Li K, Huang Y, Hu Q, Gao X and Jie
S: miR-495 mediates metabolic shift in glioma cells via targeting
Glut1. J Craniofac Surg. 26:e155–e158. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wang C, Yun Z, Zhao T, Liu X and Ma X:
MiR-495 is a predictive biomarker that downregulates GFI1
expression in medulloblastoma. Cell Physiol Biochem. 36:1430–1439.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhang B, Yuan F, Liu J, Li Y, Zhou F, Liu
X, Hao Z, Li Q, Zheng Y and Wang W: Hsa-miR-495 acts as a tumor
suppressor gene in glioma via the negative regulation of MYB. Mol
Med Rep. 14:977–982. 2016.PubMed/NCBI
|
|
39
|
Song L, Li Y, Li W, Wu S and Li Z: miR-495
enhances the sensitivity of non-small cell lung cancer cells to
platinum by modulation of copper-transporting P-type adenosine
triphosphatase A (ATP7A). J Cell Biochem. 115:1234–1242. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Xu YY, Tian J, Hao Q and Yin LR:
MicroRNA-495 downregulates FOXC1 expression to suppress cell growth
and migration in endometrial cancer. Tumour Biol. 37:239–251. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Hwang-Verslues WW, Chang PH, Wei PC, Yang
CY, Huang CK, Kuo WH, Shew JY, Chang KJ, Lee EY and Lee WH: miR-495
is upregulated by E12/E47 in breast cancer stem cells, and promotes
oncogenesis and hypoxia resistance via downregulation of E-cadherin
and REDD1. Oncogene. 30:2463–2474. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Lee SH, Jung YD, Choi YS and Lee YM:
Targeting of RUNX3 by miR-130a and miR-495 cooperatively increases
cell proliferation and tumor angiogenesis in gastric cancer cells.
Oncotarget. 6:33269–33278. 2015.PubMed/NCBI
|
|
43
|
Zhang XY, Guo ZQ, Ji SQ, Zhang M, Jiang N,
Li XS and Zhou LQ: Small interfering RNA targeting HMGN5 induces
apoptosis via modulation of a mitochondrial pathway and Bcl-2
family proteins in prostate cancer cells. Asian J Androl.
14:487–492. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Gan Y, Tan J, Yang J, Zhou Y, Dai Y, He L,
Yao K and Tang Y: Knockdown of HMGN5 suppresses the viability and
invasion of human urothelial bladder cancer 5637 cells in vitro and
in vivo. Med Oncol. 32:1362015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Chen P, Wang XL, Ma ZS, Xu Z, Jia B, Ren
J, Hu YX, Zhang QH, Ma TG, Yan BD, et al: Knockdown of HMGN5
expression by RNA interference induces cell cycle arrest in human
lung cancer cells. Asian Pac J Cancer Prev. 13:3223–3228. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Norbury C and Nurse P: Animal cell cycles
and their control. Annu Rev Biochem. 61:441–470. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Aizawa K, Ueki K, Suzuki S, Yabusaki H,
Kanda T, Nishimaki T, Suzuki T and Hatakeyama K: Apoptosis and
Bbcl-2 expression in gastric carcinomas: Correlation
withclinicopathological variables, p53 expression, cell
proliferation and prognosis. Int J Oncol. 14:85–91. 1999.PubMed/NCBI
|
|
48
|
Belotti D, Paganoni P, Manenti L, Garofalo
A, Marchini S, Taraboletti G and Giavazzi R: Matrix
metalloproteinases (MMP9 and MMP2) induce the release of vascular
endothelial growth factor (VEGF) by ovarian carcinoma cells:
Implications for ascites formation. Cancer Res. 63:5224–5229.
2003.PubMed/NCBI
|
|
49
|
Yang C, Gao R, Wang J, Yuan W, Wang C and
Zhou X: High-mobility group nucleosome-binding domain 5 increases
drug resistance in osteosarcoma through upregulating autophagy.
Tumour Biol. 35:6357–6363. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Yao K, He L, Gan Y, Zeng Q, Dai Y and Tan
J: MiR-186 suppresses the growth and metastasis of bladder cancer
by targeting NSBP1. Diagn Pathol. 10:1462015. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Wei P, Qiao B, Li Q, Han X, Zhang H, Huo Q
and Sun J: microRNA-340 suppresses tumorigenic potential of
prostate cancer cells by targeting high-mobility group
nucleosome-binding domain 5. DNA Cell Biol. 35:33–43. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Li D, Du X, Liu A and Li P: Suppression of
nucleosome-binding protein 1 by miR-326 impedes cell proliferation
and invasion in non-small cell lung cancer cells. Oncol Rep.
35:1117–1124. 2016.PubMed/NCBI
|