Introduction
Colorectal cancer is one of the most common
malignant tumors worldwide. In the past few years, the incidence
and mortality of colorectal cancer increased rapidly and the onset
age is much younger (1). It is
promising that therapeutic options for patients have increased
substantially, including earlier diagnosis and treatments such as
surgery, radiotherapy, and chemotherapy (2). However, many colorectal cancers still
remain incurable due to late stages. Therefore, prevention of
progression and early metastasis become critical for colorectal
cancer treatment.
Evidence indicates that several transcription
factors can suppress colorectal cancer cell proliferation or
migration successfully. High-mobility group AT-hook 2 (HMGA2) could
induce the expression of Slug and promote EMT, migration, invasion,
and proliferation of colorectal cancer cells (3). Inhibition of transcription factor Sp1
could suppress the growth of colorectal cancer stem cell and induce
apoptosis (4). Noticeably, recent
studies found that Krüppel-like factor 4 (KLF4) had important roles
in suppressing colorectal cancer proliferation through upregulating
p21WAF1/Cip1 and downregulating cyclin D1 (5). Overexpression of KLF4 in colorectal
cancer cell line RKO could reduce the tumorigenesis ability. Evans
showed that KLF4 was acetylated by p300/CBP to bind with
β-catenin/TCF complex, and inhibited the proliferation effect
induced by β-catenin (6). We are
very curious whether there are other mechanisms of KLF4 in the
suppression effect during colorectal cancer progression.
N-Myc downstream-regulated gene 2 (NDRG2) was
first cloned in our laboratory (7).
We confirmed that NDRG2 was a novel tumor suppressor, with
decreased expression in colorectal tumors and other types of tumor
tissues (6,8). It has been indicated that NDRG2 was
able to promote cell differentiation and suppress tumor cell
proliferation. Our previous work found that NDRG2 can be
transcriptionally regulated by p53, HIF-1α and c-Myc (9–12). To
better understand the function and regulation mechanism of NDRG2,
in this study, we analyzed whether KLF4 could regulated NDRG2
expression in colorectal cancer model. There was three potential
KLF4 binding sites in NDRG2 promoter predicted by
MatInspector software analysis. It had been reported that KLF4
activated NDRG2 expression via binding with NDRG2 promoter.
In our assay, we confirmed a novel binding site of KLF4 within
NDRG2 promoter that KLF4 could transcriptionally activate
NDRG2 using luciferase reporter analysis. With in
vitro and in vivo analysis, we confirmed that KLF4 could
suppress colorectal cancer cell proliferation depending on NDRG2
signaling. In colorectal cancer tissue array, expression level of
KLF4 and NDRG2 was significantly correlated with the overall
survival rate. Our data demonstrated that KLF4 inhibited colorectal
cancer proliferation through transcriptional activation of
NDRG2.
Materials and methods
Cell culture
Two colorectal cancer cell lines, HT-29 and HCT-116
were grown and maintained in McCoy's 5a medium with 10% fetal
bovine serum, respectively. HeLa and HEK-293T cells were also grown
in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine
serum. Cells were maintained at 37°C humidified incubator with 5%
CO2/95% air. All cell lines were sub-cultured at 3-day
intervals. We purchased the HT-29 and HCT-116 cell lines from the
American Type Culture Collection (ATCC). The cell lines were
sub-cultured and stored by our research team and we have confirmed
the genetic background through STR analysis.
Plasmid constructs
The human NDRG2 promoter was amplified from
BAC clone RP11-998D10 (The Children's Hospital of Philadelphia,
Philadelphia, PA, USA). The amplicon was cloned into the pGL3-basic
vector to generate the pGL3-NDRG2-luc plasmid. Various
truncations of the NDRG2 promoter were generated with PCR by
using pGL3-NDRG2-luc plasmid as template. The KLF4 was
amplified from HT-29 cDNA. The resulting amplicon was cloned into
the pcDNA3.1(+) and pFLAG-CMV vector to generate the pcDNA3.1-KLF4
and pFLAG-KLF4 vector. All the constructed plasmids were sequenced
correctly.
Real-time PCR
Total RNA was isolated from parental cells or stable
clones using TRIzol reagent (Takara, Dalian, China) according to
the protocol. After reverse transcription, the resulting cDNA was
used as the template for real-time PCR analysis. Real-time PCR was
performed on an ABI 7500 system (Applied Biosystems). GAPDH
was used as an internal control. Real-time PCR primers were
designed using Primer Express v3.0 Software, and the sequences
were: NDRG2 forward primer: 5′-GAGATATGCTCTTAACCACCCG-3′,
NDRG2 reverse primer: 5′-GCTGCCCAATCCATCCAA-3′; GAPDH
forward primer: 5′-TTCGACAGTCAGCCGCATCTTCTT-3′, GAPDH
reverse primer: 5′-CAGGCGCCAATACGACCAAATC-3′. The PCR reaction
consisted of 12.5 µl of SYBR Green PCR Master Mix, 300 nM each for
forward and reverse primers, and 1.5 µg template cDNA in a total
volume of 25 µl. Thermal cycling conditions were: 95°C for 5 min,
followed by 40 cycles of 95°C for 30 sec and 60°C for 30 sec.
Western blot analysis
Cells were collected from 6-well plates, and lysed
in lysis buffer (0.05 M Tris-HCl pH 7.4, 0.15 M NaCl, 0.25%
deoxycholic acid, 1% Nonidet P-40 (NP-40), 1 mM EDTA, 1 mM
phenylmethylsulfonyl fluoride, 1 mg/ml aprotinin and 1 mg/ml
leupeptin). Protein concentrations were measured using the
Bicinchoninic acid (BCA) protein assay (Pierce, Rockford, IL, USA).
Western blot analysis was carried out with standard protocol using
nitrocellulose (NC) membranes (Amersham Biosciences). For the
immunoblotting, the NC membranes were incubated with following
primary antibodies: anti-NDRG2 (HPA002896; Sigma, St. Louis, MO,
USA), anti-KLF4 (Cell Signaling Technology, #4308), anti-p21 (Cell
Signaling Technology, #2947), anti-Cyclin D1 (Cell Signaling
Technology, #2926), anti-NDRG2 (Cell Signaling Technology, #5667),
and anti-β-actin antibodies (Cell Signaling Technology, #4970).
Then, blots were incubated with horseradish peroxidase-conjugated
secondary antibodies (Promega), and detected using the
chemiluminescence method.
Luciferase reporter gene assays
HeLa cells were cultured in DMEM (with 10% FBS) in
96-well plates with density of 1×104 cells/well
overnight. NDRG2 reporter vectors including WT, and truncated
mutants co-transfected with pcDNA3.1-KLF4 using Lipofectamine-2000
(Invitrogen) for 48 h. pRL-CMV plasmid was transfected to each well
to monitor the transfection efficiency. The luciferase activities
of reporter vectors were determined using the Dual-Luciferase
reporter assay system (Promega).
Methyl thiazolyl tetrazolium (MTT)
assay
All the parental cells and the stable clones were
seeded separately with 1×104 cells/well in 96-well
plates containing 200 µl McCoy's 5a medium (with 10% FBS) and
cultured for 5 days. Five wells from each group were selected for
the MTT (Sigma) assay each day. After incubated with MTT for 4 h,
150 µl of DMSO (Sigma) was added to each well. The percentage of
viable cells was detected by measuring the absorbance at 490 nm on
multiscanner reader (TECAN-spectra mini Grodig).
EdU assay
EdU staining was performed according to the
instruction. Cells were grown in 24-well plate containing McCoy's
5a medium with 10% FBS. After 6 h incubation with EdU (Rui Bo Co.,
Guangzhou, China), cells were fixed with 4% paraformaldehyde for 15
min at room temperature, and permeabilized with 0.5% Triton X-100
for 10 min, then stained with 1X Apollo® for 30 min at
room temperature. Finally, DAPI was used for nuclear staining.
Positively stained cells were counted in five randomly selected
visual fields.
Plate colony formation assay
For colony formation assays, 500 cells were seeded
into 60-mm dishes with McCoy's 5a medium (with 10% FBS). After 2
weeks, the resulting colonies containing at least 50 cells were
fixed with methanol and stained with Giemsa (Sigma). Only clear
colonies were counted. Assays were conducted in duplicate in three
independent experiments.
Tumorigenicity in nude mice
The male nude mice weighing 15–20 g and 4–6 weeks of
age were purchased from laboratory animal research center of the
Fourth Military Medical University. Mice were separated into four
groups of five mice per group. The cells (5×106) were
inoculated subcutaneously into the right flank of the nude mice to
establish xenografts. Tumor sizes were measured every 4 days with a
slide caliper and calculated using the formula: length ×
width2/2. Animals were sacrificed 20 days after
inoculation. All animal studies were performed in accordance with
the international guidelines for the care and treatment of
laboratory animals.
Immunohistochemistry
The study of human samples was approved by
Institutional Ethics Committee (IEC) of the First Affiliated
Hospital of Fourth Military Medical University, and an informed
consent was signed by the patients prior to the study project. All
procedures for study of human samples were performed according to
the relevant guidelines and regulations of the First Affiliated
Hospital of Fourth Military Medical University. Human colon cancer
tissues were collected between year 2008 and 2013 in First
Affiliated Hospital of Fourth Military Medical University. Tumor
tissues were fixed with formalin and embedded in paraffin. The
samples were incubated with polyclonal antibodies of NDRG2 and
monoclonal antibody of KLF4, respectively. Then the sections were
incubated with secondary antibody for 1 h at room temperature.
After washing, the sections were incubated with DAB (ZSGB-
Biotechnology, Beijing, China), and lightly counterstained with
hematoxylin, then observed under a photomicroscope.
Evaluation of IHC staining
Staining was evaluated by scanning the entire tissue
specimen under appropriate magnification. Score of IHC staining was
described previously. The criteria for a sample to be scored was
set to the presence of at least one core containing 50 intact tumor
cells. The internal background was discarded. Based on previous
study, the expression of NDRG2 was mainly localized in the
cytoplasm, so we calculated the cytoplasm expression of NDRG2 as
positive. The median was used as cutoff to define the positive
cases, and samples with below 5% positively stained cells were
considered negative. The staining grade was stratified as absent (0
score), weak (1–4 score), moderate (5–8 score) or strong (9–12
score).
Statistical analysis
Data were generally expressed as mean ± standard
error values. Groups of data were compared by analysis of variance
(ANOVA) and post hoc analysis using Student-Keuls method. The
statistics were performed with SPSS 16.0 software. A value of
P<0.05 was considered to indicate a statistically significant
difference.
Results
Prediction of transcription factors
regulating NDRG2 expression
To further explore the function and transcription
regulation mechanism of NDRG2, we adopted two independent
resources to predict transcription factors regulating NDRG2
expression, including transcription factor (TF) binding site and
gene expression correlation. Combination of these two independent
resources has been shown as an effective way to predict the TFs
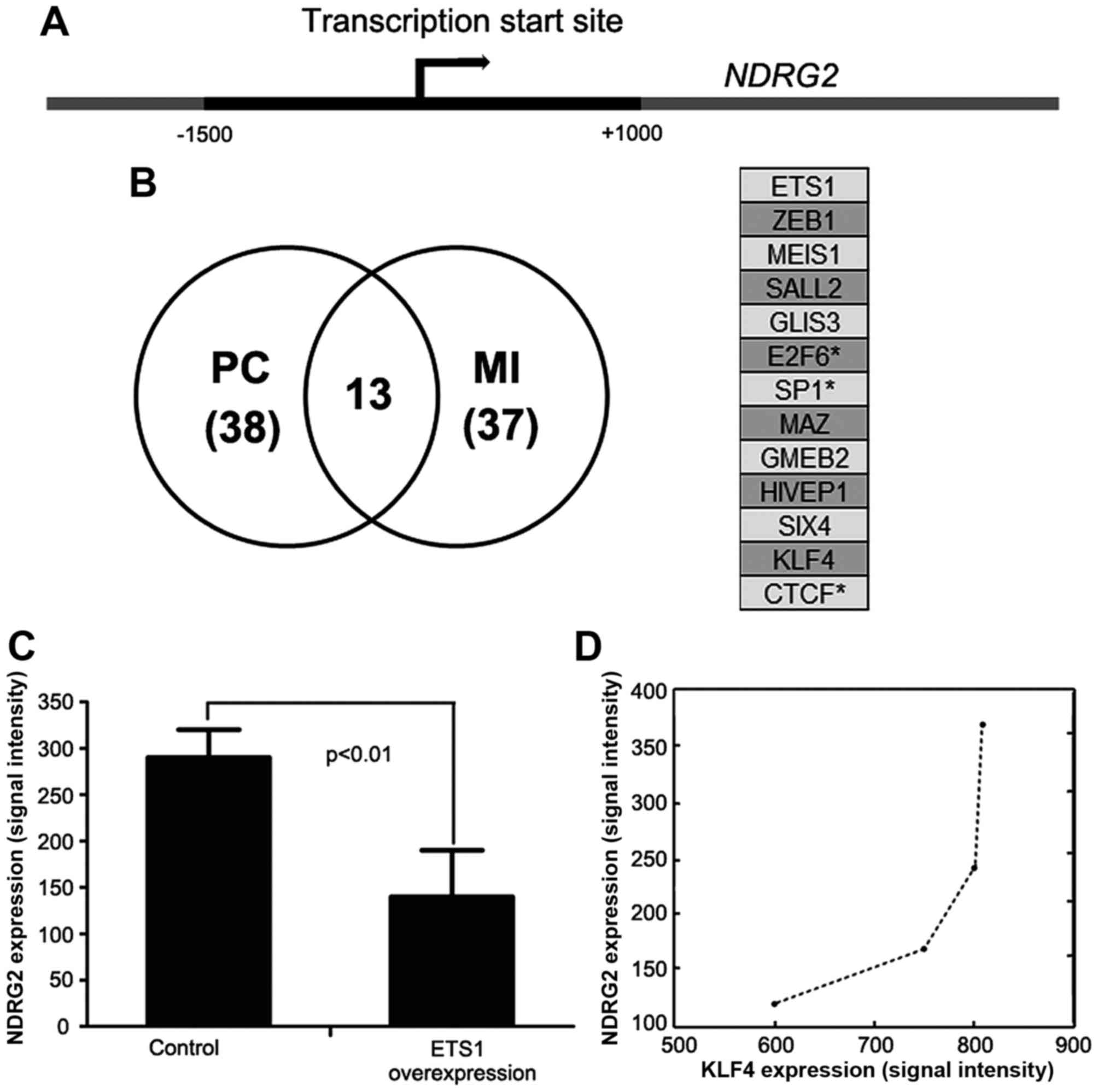
regulating the transcription of a particular gene (13). First, the active transcription
region bound by acetylated H3K27 at the transcription start site of
NDRG2 was extracted from UCSC genome browser (Fig. 1A). Then, the active 2500-bp region
(−1500 bp to +1000 bp) was scanned for transcription factor binding
sites by MatInspector with defaulting parameters. There were 298
transcriptional factors predicted to regulate NDRG2
expression. Generally, not all those predicted transcriptional
factors were involved in NDRG2 expression regulation. Some
factors were false positively predicted. To improve prediction
precision, gene expression correlation was integrated. If a
transcription factor is involved in regulation of NDRG2
expression, its expression variation might lead to NDRG2
expression change. Therefore a significant expression correlation
can be observed between the TFs and NDRG2. The microarray
datasets GSE2350 were employed to find the transcription factors
whose expression levels were significantly correlated with
NDRG2 expression. After removing the TFs not shown in those
microarray datasets, the remaining 139 TFs were accessed for their
expression correlation with NDRG2 expression. We accessed
linear correlation using Pearson correlation and non-linear
correlation using mutual information.
As shown in Fig. 1B,
the numbers of TFs having significant linear and non-linear
correlation with NDRG2 expression are 38 and 37,
respectively. Only 13 TFs show significant correlation with
NDRG2 expression regardless of expression correlation
accessed with Pearson correlation or mutual information. As few of
the TFs were reported to regulate NDRG2 expression, it is
hard to access the performance of our prediction. However, by
combination of TF binding sites and expression correlation, we
indeed predicted several TFs truly regulating NDRG2
expression. For example, SP1 has been shown to be activated by
TGF-β signaling pathway (6),
subsequently promoting NDRG2 expression (Fig. 1B, right panel). CTCF and E2F6
annotated to bind with the active region were also identified by
our method (14). However, the
transcriptional factor WT1 was excluded from candidates for the
reason that its expression was not significantly correlated with
NDRG2 expression (15).
Moreover, c-Myc was also excluded because its binding site was not
identified at the active region by MatInspector. In these
candidates, ETS1 expression has the most significant correlation
with NDRG2 expression. Whether ETS1 can regulate
NDRG2 expression was unclear.
NDRG2 plays as a tumor suppressor gene in various
types of malignant cancers. It has been reported that NDRG2 also
inhibited the proliferation and metastasis of ovarian cancer cells
(9). Herein, we used the microarray
dataset GSE21129 from ovarian cancer, and found that ectopic
expression of ETS1 in HeLa cells was able to reduce NDRG2
expression, implying that ETS1 can directly or indirectly regulate
NDRG2 expression (Fig. 1C). These
results from different datasets suggested that our prediction was
more reliable.
Among these 13 candidates, KLF4 was a crucial TF for
intestinal epithelium differentiation (16), while NDRG2 was upregulated
during the process of intestinal epithelium differentiation
(12), suggesting KLF4 might
participate in upregulating NDRG2 expression. Moreover, by
analysis of the published microarray data GSE4410 (17), we observed a positively correlation
between NDRG2 and KLF4 expression levels when colorectal epithelial
cells were induced to differentiate by sodium butyrate (Spearman's
correlation, r=1, P=0.083) (Fig.
1D). Therefore, we chose KLF4 for further experiment
validation.
KLF4 transcriptionally activates NDRG2
expression
To further explore the molecular mechanism of KLF4
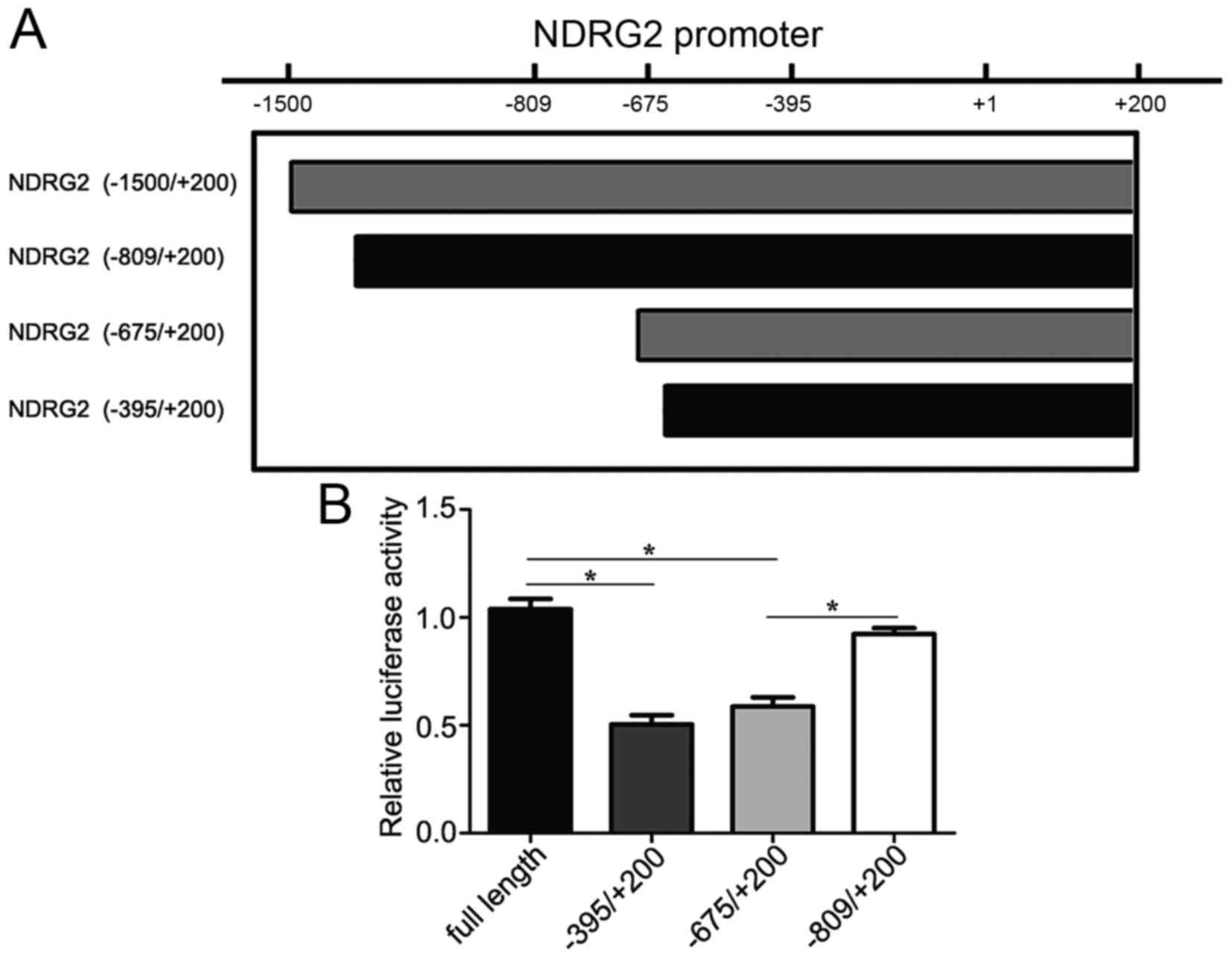
regulating NDRG2 expression, we constructed a series of
different length of NDRG2 promoter (−1500/+200 bp),
including the truncations and mutants. HeLa cells were transfected
with the NDRG2 promoter luciferase reporter gene vector and
the plasmid of pcDNA3.1-KLF4. We detected the higher levels of
NDRG2 promoter activity in the cells transfected with KLF4,
but not in the control (luciferase vector only, data not shown)
(Fig. 2A). Different
transcriptional activities were detected in the truncations and
mutants. Obviously, full length of NDRG2 promoter
(−1500/+200 bp) exhibited the highest activity, and NDRG2
(−809/+200 bp) promoter showed almost the same transcriptional
activity compared with the full length. Otherwise, NDRG2
(−675/+200 bp) exhibited suppressive promoter activity. Moreover,
NDRG2 (−395/+200) showed the inhibitory transcriptional
activity and was almost the same compared with NDRG2
(−675/+200 bp) (Fig. 2B). This
result revealed that KLF4 transcriptionally regulated NDRG2
expression through the promoter located between the region
−809/−675 bp. It was reported that KLF4 transcriptionally regulated
NDRG2 expression via binding to the promoter located between
−133/+55 (18). Our current study
demonstrated a novel binding site for KLF4 within NDRG2
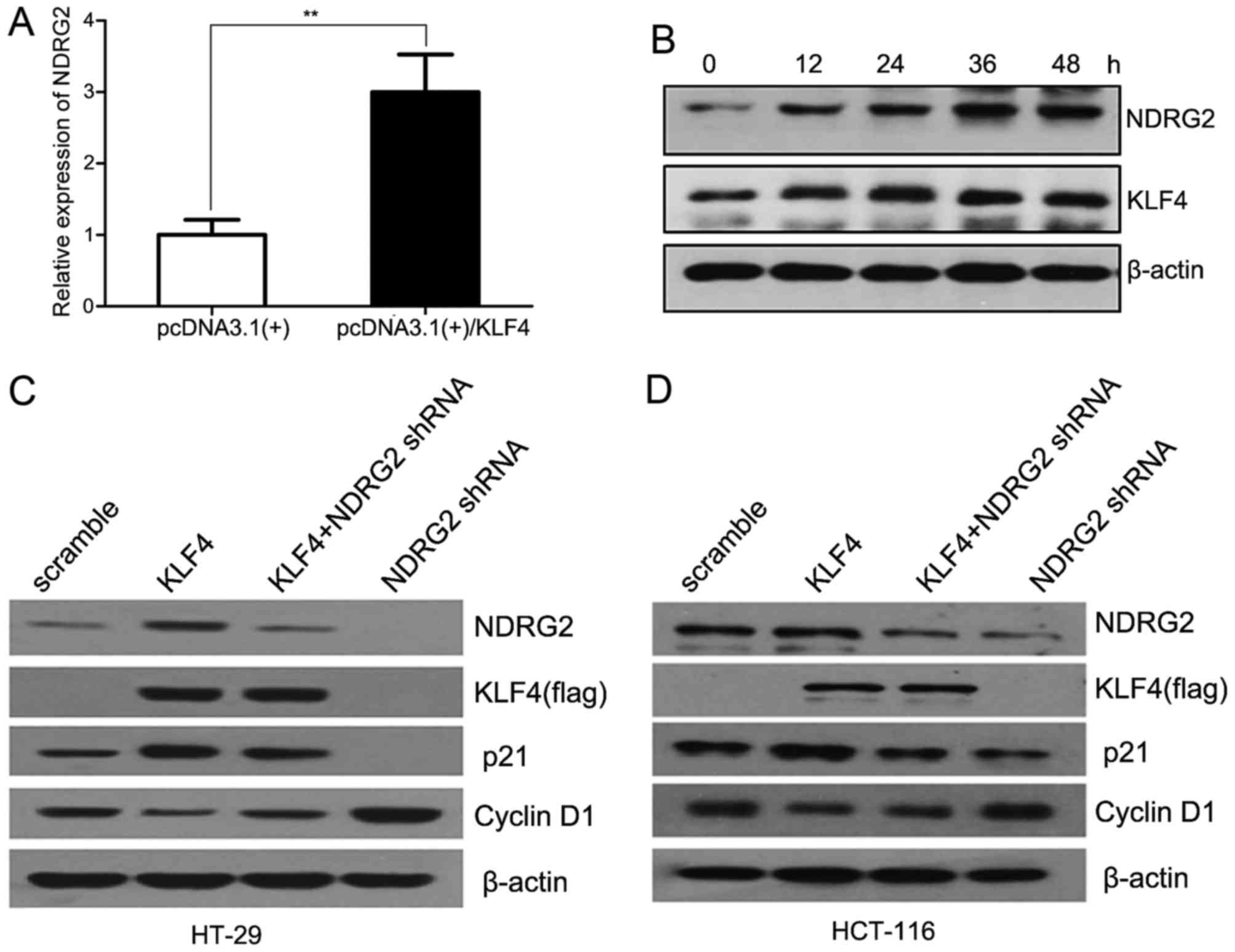
promoter. Simultaneously, we also determined that KLF4 induced the
expression of NDRG2 both at mRNA and protein levels in a
time-dependent manner in HT-29 cells (Fig. 3A and B). Our findings demonstrated
the novel evidence that KLF4 transcriptionally activated
NDRG2 via binding to its promoter.
KLF4 inhibits colorectal cancer cell
proliferation through upregulation of NDRG2
To further elucidate the function of KLF4-NDRG2
signaling, we subsequently analyzed whether KLF4 inhibited the
proliferation of colorectal cancer cells through upregulation of
NDRG2. It has been reported that KLF4 could inhibit cancer cell
proliferation via upregulating p21 expression and suppressing
cyclin D1. In our study, as predicted, we found that KLF4 induced
p21 expression and suppressed cyclin D1 in HT-29 cells. While
shRNA-mediated downregulation of NDRG2 decreased p21 expression and
enhanced cyclin D1, and attenuation of NDRG2 suppressed the
modulation of p21 and cyclin D1 induced by KLF4 (Fig. 3C and D). To further confirm the
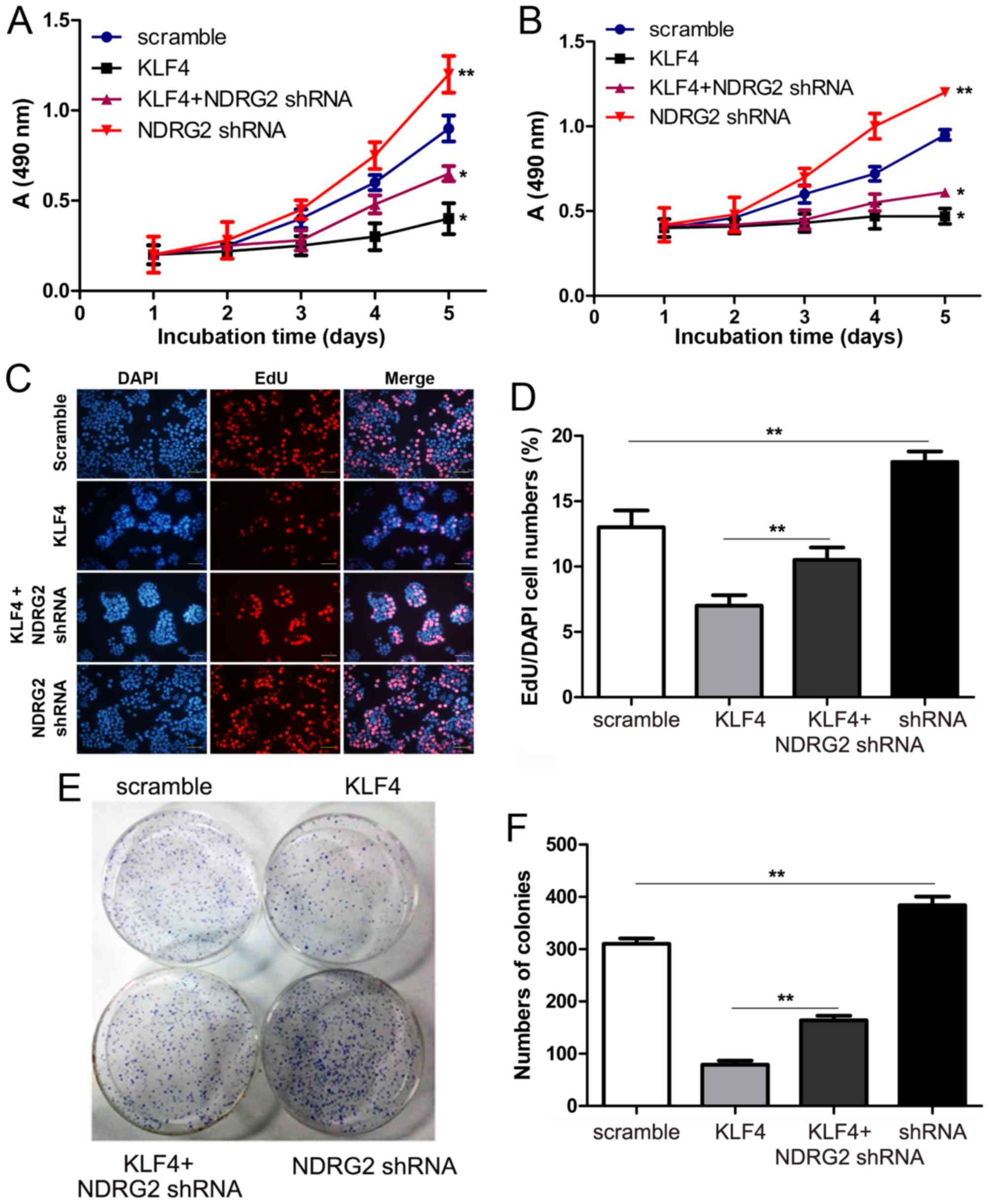
function of KLF4-NDRG2 signaling pathway, we also found that
overexpression of KLF4 could inhibit the proliferation of HT-29 and
HCT-116 cells, while shRNA-mediated attenuation of NDRG2 could
rescue cancer cell proliferation inhibited by KL4. Our data showed
that downregulation of NDRG2 could reverse the inhibitory effect of
KLF4 in the two cell lines. Cell proliferation difference was much
significant between KLF4 and KLF4 + NDRG2 shRNA groups for
the time periods of 4 and 5 days (Fig.
4A and B), suggesting that KLF4 inhibited the proliferation of
colorectal cancer cells dependent on NDRG2. Furthermore, EDU
staining and colony formation assay also confirmed that KLF4
inhibited proliferation of colorectal cancer cell lines via
upregulation of NDRG2 (Fig.
4C-F).
NDRG2 reverses the role of KLF4
inhibiting tumorigenesis in vivo
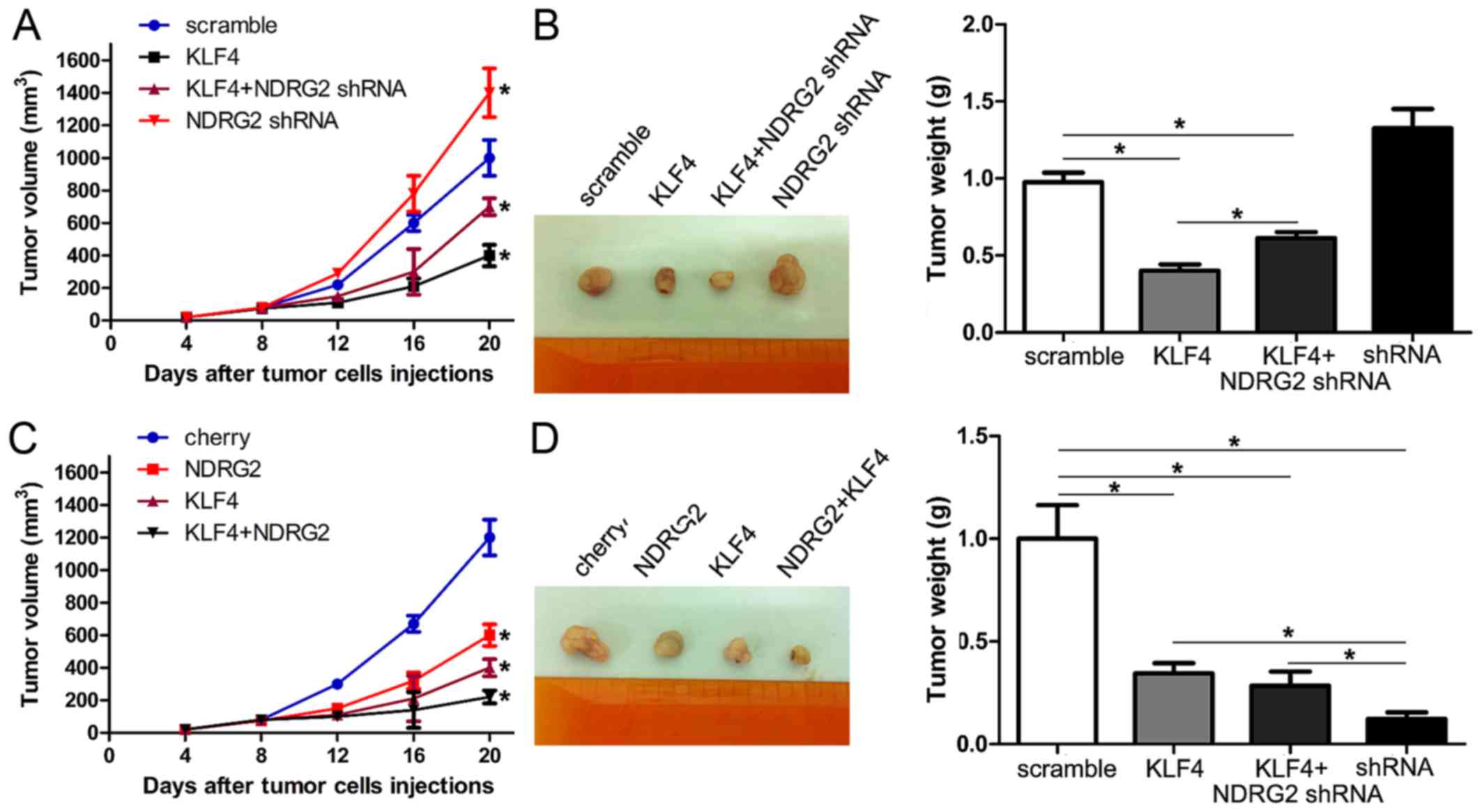
Next, we further evaluated the effect of KLF4-NDRG2
signaling inhibiting tumorigenesis in vivo. Colorectal
cancer cells HT-29 with different expression levels of NDRG2 and
KLF4, including HT-29-Scramble, HT-29-KLF4, HT-29-NDRG2 shRNA and
HT-29-KLF4/NDRG2 shRNA, HT-29-Control, HT-29-NDRG2 and
HT-29-NDRG2/KLF4 cells, were injected into nude mice respectively.
Tumor size was evaluated every 4 days, and on day 20, tumor mass
was weighed.
In our study, the mice injected with HT-29/KLF4
showed a statistically significant decrease in tumor size and tumor
mass compared with the control group. The mice injected with
HT-29-KLF4/NDRG2 shRNA showed a slight decrease in tumor size and
tumor mass (Fig. 5A and B).
Moreover, the mice injected with HT-29-NDRG2 and HT-29-KLF4/NDRG2
decreased the tumor size and tumor mass significantly (Fig. 5C and D). These data demonstrated
that KLF4 inhibited the tumorigenesis of colorectal cancer via
upregulation of NDRG2 expression in vivo.
Decreased expression of KLF4 and NDRG2
correlates with poor overall survival of colorectal cancer
patients
To investigate the clinical significance of KLF4 and
NDRG2 expression in colorectal cancer patients, we used colorectal
cancer tissue array with 101 colorectal cancer samples to analyze
the survival correlation. The characteristics of the 101 colorectal
cancer patients involved in the study cohort are shown in Table I. In the 101 colorectal cancer
patients, there were 62 male (61.4%) and 39 (38.6%) female
patients. The mean age was 64 years, with a range of 16–85. Tumor
with well/moderately/poorly differentiated was 41 (58.4%), 33
(35.6%) and 6 (6%), respectively. According to the International
TNM (Tumor Node Metastasis) Classification, 38 (37.6%), 51 (50.5%),
and 12 (11.9%) of the 101 colorectal cancer patients were
classified as TNM stages I, II, and III, respectively. In all
samples, KLF4 and NDRG2 expression was correlated with TNM grades
and differentiation levels of colorectal cancer (Table I).
 | Table I.Statistical results of the
immunohistology. |
Table I.
Statistical results of the
immunohistology.
|
|
| KLF4 |
| NDRG2 |
|
|---|
|
|
|
|
|
|
|
|---|
| Total | n | − | ± to ++ | P-value | − | ± to ++ | P-value |
|---|
| Sex |
|
Male | 62 | 33 | 29 | 0.231a | 27 | 35 | 0.520a |
|
Female | 39 | 25 | 14 |
| 22 | 17 |
|
| Age |
|
<60 | 53 | 22 | 31 | 0.428a | 20 | 33 | 0.315a |
|
≥60 | 48 | 36 | 12 |
| 29 | 19 |
|
| WHO grade |
| I | 38 | 19 | 19 | 0.012b | 18 | 20 | 0.021b |
| II | 51 | 29 | 22 |
| 23 | 28 |
|
|
III | 12 | 10 | 2 |
| 8 | 4 |
|
| Differentiation
status |
|
Well | 59 | 33 | 26 | 0.035b | 29 | 30 | 0.014b |
|
Moderately | 36 | 20 | 16 |
| 16 | 20 |
|
|
Poor | 6 | 5 | 1 |
| 4 | 2 |
|
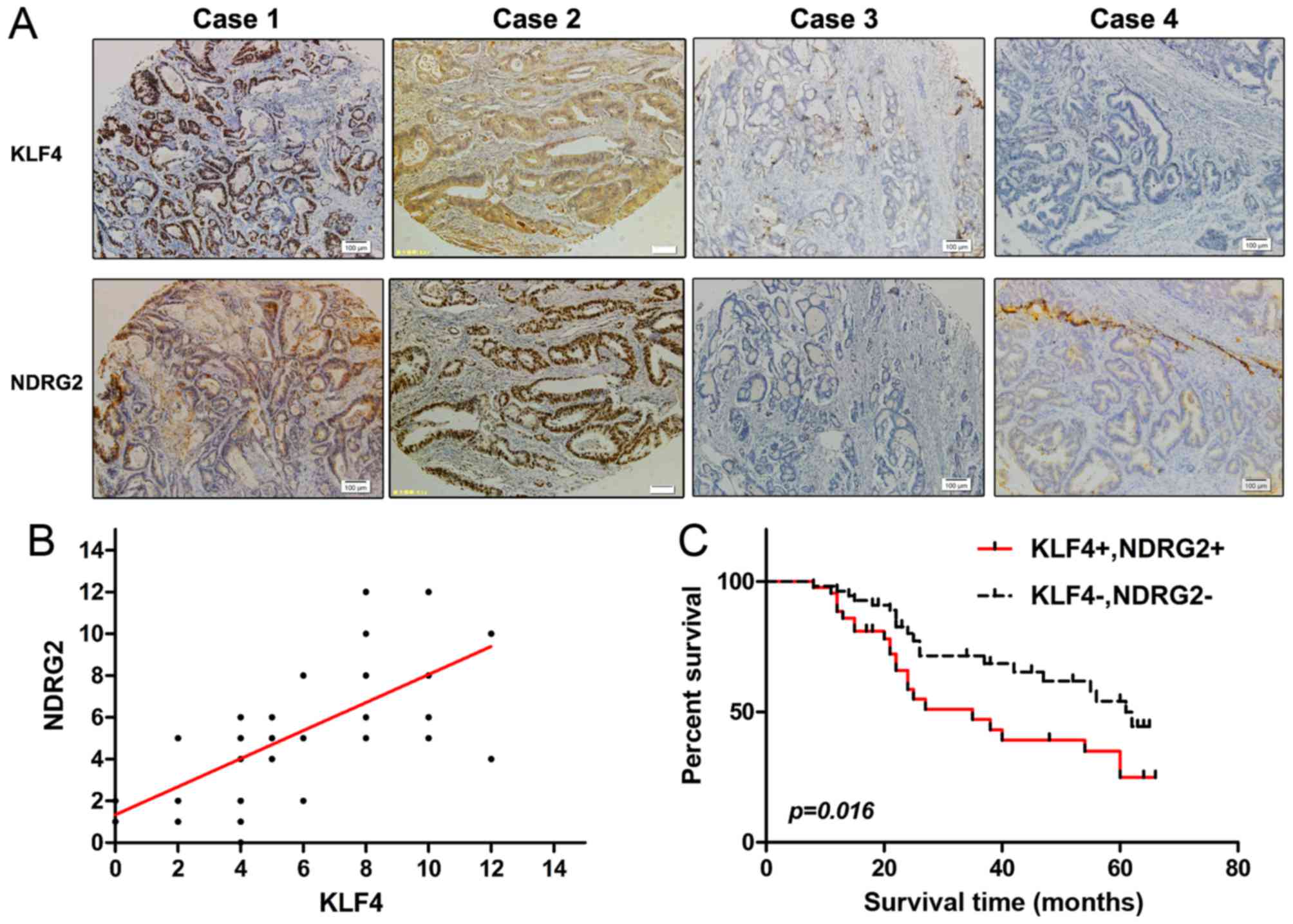
With immunohistochemistry assay, expression of KLF4
was positively correlated with NDRG2 (Fig. 6A and B). We also examined the
correlation of co-expression of KLF4 and NDRG2 with the overall
survival rate. Kaplan-Meier survival curves were applied, and we
found that reduced expression of KLF4 and NDRG2 had a significantly
shorter survival time compared with those with a higher KLF4 and
NDRG2 expression (p<0.05; Fig.
6C). Taken together, NDRG2 expression was positively correlated
with KLF4, and higher NDRG2 expression was associated with better
overall survival rate in colorectal cancer patients.
Discussion
The Krüppel-like factor (KLF) family members are
transcription factors functioned in several biological processes.
The members of KLF family have highly conserved zinc-finger domain,
which can bind to similar DNA binding domain such as CACCC and
GC-rich region (19–22). The SP/KLF transcription factor
family member KLF4 is located in chromatin 9q31 (23), and is highly expressed in the
epithelia of the skin, lungs and intestinal tract and other organs
(24). KLF4 plays important roles
in regulating multiple cellular processes including cell
proliferation, differentiation, apoptosis, inflammation and also
tumor formation. However, it is puzzling that KLF4 can play both
oncogenic and tumor suppressive functions in different tissue types
depending on regulation of various target genes. Over 70% breast
cancers showed high expression level of KLF4, and upregulated KLF4
can enhance tumorigenesis, cell migration and cell invasion
(25). On the contrary, decreased
expression of KLF4 was found in colorectal cancer, gastric cancer,
intestinal adenomas, and pancreatic ductal carcinoma, which
suggested that KLF4 could function as a tumor suppressor gene, and
correlate with inhibitory abilities of cell proliferation, invasion
and tumorigenesis (26–29). KLF4 was identified as an independent
predictor of survival and recurrence of colorectal cancer.
Previously our laboratory identified NDRG2
from normal human brain cDNA library with subtractive hybridization
(7). It belonged to NDRG
gene family together with NDRG1, NDRG3, and
NDRG4, and was involved in cell stress, differentiation and
proliferation (30). We and other
laboratories confirmed that NDRG2 was a novel tumor suppressor gene
with decreased expression in several tumor tissues and cancer cells
such as breast cancer, glioma, and colorectal cancers (31–34).
In a previous study, we found that lower expression of NDRG2 had
strong proliferation and invasion abilities of colorectal cancer
cells, also NDRG2 was a potential independent prognosis biomarker
of human colorectal cancer (35).
In this study, we used bio-information analysis and
found there were three potential KLF4 binding sites locating on
NDRG2 promoter (Fig. 1).
Then we used reporter gene assay to explore whether KLF4
transcriptionally activated NDRG2. We constructed different
truncations based on NDRG2 promoter, and found KLF4 could
upregulate NDRG2 promoter activity especially on −809/−675
and −395/+200 bp sites (Fig. 2),
while −133/+55 bp site had been reported previously, we confirmed a
novel binding site of KLF4 on NDRG2 promoter. In NDRG2
promoter, there might be two different transcription start sites
which were predicted with bio-information system, and this is a
possible reason why our binding site of KLF4 on NDRG2
promoter is not the same.
To further explore the role of NDRG2 in KLF4
suppressing proliferation in colorectal cancer cells, we
downregulated NDRG2 expression in HT29 cells with KLF4
overexpression, and performed in vitro biology experiments
including MTT, EdU staining and colony formation assay. Results
demonstrated that NDRG2 could abrogate the function of KLF4 by
inhibiting colorectal cancer cell proliferation through the
regulation of p21 and cyclin D1 in vitro (Fig. 4). As our previous report, NDRG2
overexpression could induce cell cycle arrest, which might be due
to its regulation of p21 and cyclin D1 expression. Herein, we found
that NDRG2 knockdown caused downregulation of p21 and upregulation
of cyclin D1, which was consistent with our previous finding in
cell cycle analysis (data not shown). Furthermore, in a nude mouse
xenograft model, the tumor sizes and weight of KLF4 and shNDRG2
group were smaller compared with the control group (Fig. 5). All these results revealed that
NDRG2 played an important role in KLF4 signaling of colorectal
cancer proliferation inhibition.
Based on previous studies, KLF4 acts as a tumor
suppressor gene and inhibites the proliferation of various types of
tumor cells via different signaling pathway (27,29).
Moreover, NDRG2 inhibited cell proliferation through upregulation
of p21, p27, and p53 (6,8,9). As
the signaling pathway of NDRG2 and KLF4 was only partially crossed,
it is reasonable to understand the co-overexpression of KLF4 and
NDRG2 could inhibit the proliferation of colorectal cancer cells
more obviously than KLF4 or NDRG2, respectively.
Previous studies have demonstrated that KLF4 and
NDRG2 are both predictors of survival and recurrence for colorectal
cancer. It is not clear whether the association of KLF4/NDRG2
combined expression could benefit us in prediction of better
prognosis for patients with colorectal cancer. In the present
study, we used a colorectal cancer tissue array with 101 colorectal
cancer samples to analyze their expression with tumor prognosis.
There was no significant association between KLF4/NDRG2 expression
and sex or age at diagnosis (Table
I). We observed that lower expression of KLF4 and NDRG2 was
evident in human colorectal tissues compared with normal tissues,
and it was greatly positively related with the TNM grades and
differentiation level of colorectal cancer. Kaplan-Meier analysis
revealed significant difference in prognosis depending on the
status of KLF4/NDRG2 co-expression.
KLF4+/NDRG2+ had better overall survival than
KLF4−/NDRG2−. However, further investigation
in many more cases is still needed to evaluate the potential
application value of KLF4/NDRG2 co-expression in clinical
setting.
In conclusion, our data showed that KLF4 could
transcriptionally upregulate NDRG2 expression by binding
with its promoter. NDRG2 downregulation could interrupt the
function of KLF4 in suppressing colorectal cancer cell
proliferation and tumorigenesis both in vitro and in
vivo. In colorectal cancer tissue array, we found that a
combined detection of KLF4/NDRG2 was positively related with TNM
grades and differentiation levels. The co-expression of KLF4/NDRG2
may be beneficial in predicting the prognosis of colorectal cancer
patients.
Acknowledgements
This work was supported by the National High-tech
R&D Program of China for Young Scientist (2014AA020517), and
National Natural Science Foundation of China (nos. 81172292,
31571437, 81230043, 81372390 and 81421003).
References
|
1
|
Feagins LA, Souza RF and Spechler SJ:
Carcinogenesis in IBD: Potential targets for the prevention of
colorectal cancer. Nat Rev Gastroenterol Hepatol. 6:297–305. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lamprecht SA and Lipkin M: Chemoprevention
of colon cancer by calcium, vitamin D and folate: Molecular
mechanisms. Nat Rev Cancer. 3:601–614. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Li Y, Zhao Z, Xu C, Zhou Z, Zhu Z and You
T: HMGA2 induces transcription factor Slug expression to promote
epithelial-to-mesenchymal transition and contributes to colon
cancer progression. Cancer Lett. 355:130–140. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhao Y, Zhang W, Guo Z, Ma F, Wu Y, Bai Y,
Gong W, Chen Y, Cheng T, Zhi F, et al: Inhibition of the
transcription factor Sp1 suppresses colon cancer stem cell growth
and induces apoptosis in vitro and in nude mouse xenografts.
Oncol Rep. 30:1782–1792. 2013.PubMed/NCBI
|
|
5
|
Shie JL, Chen ZY, Fu M, Pestell RG and
Tseng CC: Gut-enriched Krüppel-like factor represses cyclin D1
promoter activity through Sp1 motif. Nucleic Acids Res.
28:2969–2976. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Shen L, Qu X, Ma Y, Zheng J, Chu D, Liu B,
Li X, Wang M, Xu C, Liu N, et al: Tumor suppressor NDRG2 tips the
balance of oncogenic TGF-β via EMT inhibition in colorectal cancer.
Oncogenesis. 3:e862014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Deng Y, Yao L, Chau L, Ng SS, Peng Y, Liu
X, Au WS, Wang J, Li F, Ji S, et al: N-Myc downstream-regulated
gene 2 (NDRG2) inhibits glioblastoma cell proliferation. Int J
Cancer. 106:342–347. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Xu X, Li J, Sun X, Guo Y, Chu D, Wei L, Li
X, Yang G, Liu X, Yao L, et al: Tumor suppressor NDRG2 inhibits
glycolysis and glutaminolysis in colorectal cancer cells by
repressing c-Myc expression. Oncotarget. 6:26161–26176. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Liu N, Wang L, Li X, Yang Q, Liu X, Zhang
J, Zhang J, Wu Y, Ji S, Zhang Y, et al: N-Myc downstream-regulated
gene 2 is involved in p53-mediated apoptosis. Nucleic Acids Res.
36:5335–5349. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liu N, Wang L, Liu X, Yang Q, Zhang J,
Zhang W, Wu Y, Shen L, Zhang Y, Yang A, et al: Promoter
methylation, mutation, and genomic deletion are involved in the
decreased NDRG2 expression levels in several cancer cell lines.
Biochem Biophys Res Commun. 358:164–169. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang L, Liu N, Yao L, Li F, Zhang J, Deng
Y, Liu J, Ji S, Yang A, Han H, et al: NDRG2 is a new HIF-1 target
gene necessary for hypoxia-induced apoptosis in A549 cells. Cell
Physiol Biochem. 21:239–250. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang J, Li F, Liu X, Shen L, Liu J, Su J,
Zhang W, Deng Y, Wang L, Liu N, et al: The repression of human
differentiation-related gene NDRG2 expression by Myc via
Miz-1-dependent interaction with the NDRG2 core promoter. J Biol
Chem. 281:39159–39168. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Majors BS, Chiang GG and Betenbaugh MJ:
Protein and genome evolution in Mammalian cells for biotechnology
applications. Mol Biotechnol. 42:216–223. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Consortium EP: ENCODE Project Consortium:
An integrated encyclopedia of DNA elements in the human genome.
Nature. 489:57–74. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Svensson E, Vidovic K, Olofsson T,
Vallon-Christersson J, Borg A and Gullberg U: The Wilms' tumor gene
1 (WT1) induces expression of the N-myc downstream regulated gene 2
(NDRG2). DNA Cell Biol. 26:589–597. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yu T, Chen X, Zhang W, Li J, Xu R, Wang
TC, Ai W and Liu C: Krüppel-like factor 4 regulates intestinal
epithelial cell morphology and polarity. PLoS One. 7:e324922012.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tabuchi Y, Takasaki I, Doi T, Ishii Y,
Sakai H and Kondo T: Genetic networks responsive to sodium butyrate
in colonic epithelial cells. FEBS Lett. 580:3035–3041. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li D, Mei H, Pu J, Xiang X, Zhao X, Qu H,
Huang K, Zheng L and Tong Q: Intelectin 1 suppresses the growth,
invasion and metastasis of neuroblastoma cells through
up-regulation of N-myc downstream regulated gene 2. Mol Cancer.
14:472015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Adam PJ, Regan CP, Hautmann MB and Owens
GK: Positive- and negative-acting Kruppel-like transcription
factors bind a transforming growth factor beta control element
required for expression of the smooth muscle cell differentiation
marker SM22alpha in vivo. J Biol Chem. 275:37798–37806. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bourillot PY and Savatier P: Krüppel-like
transcription factors and control of pluripotency. BMC Biol.
8:1252010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Pearson R, Fleetwood J, Eaton S, Crossley
M and Bao S: Krüppel-like transcription factors: A functional
family. Int J Biochem Cell Biol. 40:1996–2001. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yamada T, Park CS, Mamonkin M and
Lacorazza HD: Transcription factor ELF4 controls the proliferation
and homing of CD8+ T cells via the Krüppel-like factors
KLF4 and KLF2. Nat Immunol. 10:618–626. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yet SF, McA'Nulty MM, Folta SC, Yen HW,
Yoshizumi M, Hsieh CM, Layne MD, Chin MT, Wang H, Perrella MA, et
al: Human EZF, a Krüppel-like zinc finger protein, is expressed in
vascular endothelial cells and contains transcriptional activation
and repression domains. J Biol Chem. 273:1026–1031. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Katz JP, Perreault N, Goldstein BG, Lee
CS, Labosky PA, Yang VW and Kaestner KH: The zinc-finger
transcription factor Klf4 is required for terminal differentiation
of goblet cells in the colon. Development. 129:2619–2628.
2002.PubMed/NCBI
|
|
25
|
Yu F, Li J, Chen H, Fu J, Ray S, Huang S,
Zheng H and Ai W: Kruppel-like factor 4 (KLF4) is required for
maintenance of breast cancer stem cells and for cell migration and
invasion. Oncogene. 30:2161–2172. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li T, Lu H, Shen C, Lahiri SK, Wason MS,
Mukherjee D, Yu L and Zhao J: Identification of epithelial stromal
interaction 1 as a novel effector downstream of Krüppel-like factor
8 in breast cancer invasion and metastasis. Oncogene. 33:4746–4755.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Shum CK, Lau ST, Tsoi LL, Chan LK, Yam JW,
Ohira M, Nakagawara A, Tam PK and Ngan ES: Krüppel-like factor 4
(KLF4) suppresses neuroblastoma cell growth and determines
non-tumorigenic lineage differentiation. Oncogene. 32:4086–4099.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhang L, Wali A, Ramana CV and Rishi AK:
Cell growth inhibition by okadaic acid involves gut-enriched
Kruppel-like factor mediated enhanced expression of c-Myc. Cancer
Res. 67:10198–10206. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ghaleb AM, McConnell BB, Nandan MO, Katz
JP, Kaestner KH and Yang VW: Haploinsufficiency of Krüppel-like
factor 4 promotes adenomatous polyposis coli dependent intestinal
tumorigenesis. Cancer Res. 67:7147–7154. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lorentzen A, Vogel LK, Lewinsky RH, Saebø
M, Skjelbred CF, Godiksen S, Hoff G, Tveit KM, Lothe IM, Ikdahl T,
et al: Expression of NDRG2 is down-regulated in high-risk adenomas
and colorectal carcinoma. BMC Cancer. 7:1922007. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Takahashi K and Yamada M, Ohata H, Honda K
and Yamada M: Ndrg2 promotes neurite outgrowth of
NGF-differentiated PC12 cells. Neurosci Lett. 388:157–162. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lee DC, Kang YK, Kim WH, Jang YJ, Kim DJ,
Park IY, Sohn BH, Sohn HA, Lee HG, Lim JS, et al: Functional and
clinical evidence for NDRG2 as a candidate suppressor of liver
cancer metastasis. Cancer Res. 68:4210–4220. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhao H, Zhang J, Lu J, He X, Chen C, Li X,
Gong L, Bao G, Fu Q, Chen S, et al: Reduced expression of N-Myc
downstream-regulated gene 2 in human thyroid cancer. BMC Cancer.
8:3032008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Shi H, Li N, Li S, Chen C, Wang W, Xu C,
Zhang J, Jin H, Zhang H, Zhao H, et al: Expression of NDRG2 in
esophageal squamous cell carcinoma. Cancer Sci. 101:1292–1299.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chu D, Zhang Z, Li Y, Wu L and Zhang J,
Wang W and Zhang J: Prediction of colorectal cancer relapse and
prognosis by tissue mRNA levels of NDRG2. Mol Cancer Ther.
10:47–56. 2011. View Article : Google Scholar : PubMed/NCBI
|