Introduction
Melanoma, a cancer derived from melanocytes, is the
most aggressive skin cancer and responsible for 80% of skin
cancer-related deaths (1).
According to a report from American Cancer Society, there are an
estimated 76,380 new cases of melanoma and 10,130 melanoma-related
deaths in the United States in 2016 (2). In China, it is estimated that
approximately 3,200 Chinese individuals died from melanoma in 2015
(3). The identification of
activating mutations in BRAF has led to the development of targeted
therapies to treat melanoma patients bearing these mutations.
However, although patients exhibit an initial response to targeted
therapies, a majority of them finally develop recurrences due to
onset of acquired drug resistance (4). Therefore, there is an urgent need to
elucidate the molecular basis of melanoma development and
progression to facilitate the development of an effective strategy
to treat this disease.
INPP4B was initially characterized as an inositol
polyphosphate phosphatase hydrolyzing PtdIns(3,4)P2 to
PtdIns(3)P. As the intracellular
PI(3,4)P2 is required for the full activation of
Akt, a critical oncogene in various cancers, INPP4B can restrain
the PI3K/Akt signaling (5).
Recently, INPP4B was shown to play a tumor suppressor role in
variety of cancers, including lung, prostate, bladder and
melanocytic cancers (6–9). The tumor suppressive mechanism of
INPP4B has been attributed to its negative regulation role in
PI3K/Akt signaling. Nevertheless, increasing evidence is
accumulating that INPP4B has an oncogene role. INPP4B was reported
to be overexpressed in acute myeloid leukemia (AML) and high levels
of INPP4B are predictive of poor clinical outcome and
chemoresistance (10). In a subset
of breast cancers with low Akt, INPP4B mediated SGK3 activation
drives tumorigenesis (11,12). These studies demonstrate that the
role in carcinogenesis is cell type- and context-dependent. In
melanoma, one recent study showed that INPP4B is overexpressed in
melanoma tissue and functions as an oncogenic driver through
activating SGK3 kinase (13). SGK3,
a member of the AGC family of kinases, possesses certain shared
substrates with Akt and is also activated by PI3K involving PDK1
and mTORC2 (14). Similar to Akt,
abnormal activation of SGK3 is implicated in the induction and
progression of multiple cancers, such as breast cancer, prostate
cancer, glioblastoma and hepatocellular carcinoma (15–18).
More recently, SGK3 was reported to contribute to the development
of melanoma as a key mediator of PDK1 activity (19).
MicroRNAs (miRNAs), a class of small non-coding
RNAs, can modulate gene expression at the post-transcription level
by enhancing mRNA degradation or repressing mRNA translation
through binding to the 3′-untranslated region (3′-UTR) of its
target mRNA. Accumulating evidence demonstrates that miRNAs
participate in various cellular processes, including cell
proliferation, migration, apoptosis and differentiation (20). Due to the crucial role of miRNAs in
these biological processes, their aberrant expression is involved
in the initiation and progression of numerous cancers. miRNAs can
function as either oncogenes or tumor suppressors, depending on
their target genes. In human melanoma, oncogenic miRNAs, such as
miR-148, miR-182 and miR-221, were reported to be upregulated in
melanoma and increase tumor progression (21–23).
In contrast, miRNAs, including miR-101, miR-137 and miR-200c, are
downregulated in melanoma and exhibit tumor-suppressive roles
(24–26).
miR-605 was first identified as a positive regulator
of p53 activity through repressing the expression of mdm2, which
targets p53 for degradation (27).
Growing evidence demonstrate that the genetic variants or
deregulation of miR-605 participates in carcinogenesis. The genetic
variants of miR-605 has been reported to be associated with the
susceptibility of various cancers, including gastric cancer,
prostate cancer and lung cancer (28–30).
miR-605 was shown to be downregulated in intrahepatic
cholangiocarcinoma specimens and repress tumor progression by
directly targeting PSMD10 (31).
However, the role of miR-605 in melanoma is still unknown. In this
study, we provide evidence that miR-605 is downregulated in
melanoma cells and tissues and suppresses melanoma cell growth and
tumorigenesis. Moreover, we show that the tumor suppressor role of
miR-605 is mediated by its direct repression of INPP4B expression,
which leads to the inactivation of SGK3 kinase.
Materials and methods
Antibodies and reagents
Antibodies against NDRG1, pNDRG1 (T346) and pSGK3
(Thr320) were purchased from Cell Signaling Technology (Beverly,
MA, USA). Antibodies from Santa Cruz Biotechnology (Santa Cruz, CA,
USA) included INPP4B, SGK3 and tubulin. Chemically synthesized
miR-605 mimics and antagomiR-605 were obtained from RiboBio
(Guangzhou, China).
Tissue samples
This study was approved by the ethics committee of
the Affiliated Hospital of Guiyang Medical University. Tissue
samples used for this study were obtained with informed consent
from the participants at the affiliated hospital of Guiyang Medical
University. Pathology of the tissue samples was confirmed, and then
frozen in liquid nitrogen.
Cells and cell culture
HEMn-MP cells were cultured in Medium 254 (Cascade
Biologics, Portland, OR, USA) containing human melanocyte growth
supplement (HMGS; Life Technologies, Carlsbad, CA, USA). All the
melanoma cell lines were maintained in Dulbecco's modified Eagle's
medium (DMEM; Invitrogen, Carlsbad, CA, USA) containing 5% fetal
calf serum (FCS; Gibco-Life Technologies). Cells were grown in a
37°C incubator at 5% CO2.
To generate Mel-RM cells stably overexpressing
miR-605, cells were infected with Lentivirus-harboring hsa-miR-605
(GeneChem Co., Shanghai, China) and selected by treatment with 1
μg/ml puromycin (Sigma-Aldrich, St. Louis, MO, USA) for 10
days.
Quantitative real-time PCR
miRNA was isolated using a mirVana miRNA isolation
kit (Ambion, Carlsbad, CA, USA) according to the manufacturer's
suggestions. Expression of miR-605 was measured with the
PrimeScript miRNA RT-PCR kit (Takara, Dalian, China) and the level
of U6 was used as an endogenous control. The forward primer for
miR-605 is: 5′-TGCGGTAAATCCCATGGTGCCTTC-3′; the reverse primer for
miRNAs is the UnimiRqPCR Primer (Takara). The forward primer for U6
is: 5′-GCGCGTCGTGAAGCGTTC-3′; the reverse primer for U6 is
5′-GTGCAGGGTCCGAGGT-3′.
Total RNA was extracted using TRIzol (Invitrogen,
Grand Island, NY, USA) and reverse-transcribed into cDNA with M-MLV
Reverse Transcriptase (Promega, Madison, WI, USA). The qRT-PCR was
performed in an ABI 7500 system using a Takara SYBR RT-PCR kit
(Takara) and the level of β-actin mRNA was used as an endogenous
control. The primer sequences were as follows: INPP4B (forward:
5′-CCCCGGGTACTGAGGCTTCG-3′; reverse:
5′-CTTTGTATTCTCTCCCGGAGGCG-3′); β-actin (forward:
5′-GCACAGAGCCTCGCCTT-3′; reverse: 5′-GTTGTCGACGACGAGCG-3′).
Transient transfection of miR-605
mimics or inhibitors
miR-605 mimics or antagomiR-605 transfections were
performed with Lipofectamine RNAiMAX Transfection Reagent
(Invitrogen) according to the manufacturer's protocols and used for
subsequent measurements 24 or 48 h after transfection.
Cell proliferation assays
Transfected melanoma cells were plated in 96-well
plates (2,000 cells/well). Cell proliferation was determined using
Cell Counting Kit-8 (CCK-8) (Dojindo Laboratories, Tokyo, Japan)
according to the manufacturer's instructions. Assays were performed
in triplicate, and the results are presented as means ± standard
deviation (SD).
Colony formation assay
Transfected melanoma cells were seeded in a 6-well
plate (2,000 cells/well) containing DMEM/5% FBS medium. Cells were
allowed to grow for two weeks and then fixed with methanol and
stained with 0.5% crystal violet solution. The colony numbers were
measured using ImageJ (MD, USA). Assays were performed in
triplicate, and the results are presented as means ± standard
deviation (SD).
Soft agar assay
The cells were seed in a layer of 0.35% agarose
containing RPMI-1640/10% FBS medium at 5,000 cells per well, which
was on top of a base layer of 0.5% agarose in 6-well culture
plates. Two weeks after plating, colonies were photographed and
counted under a light microscope.
Cloning and luciferase reporter
assay
The wild-type or mutant INPP4B 3′-UTR was cloned
into pGL3 luciferase reporter plasmid (Promega) following the
manufacturer's instructions. All constructs were validated by
sequencing.
The cells were plated into 24-well
plates (3×104 cell/per well)
The following day, pGL3 luciferase reporter plasmid
with the wild-type or mutant INPP4B 3′-UTR was co-transfected into
melanoma cells along with miR-605 mimics or antagomiR-605 using
Lipofectamine 2000 (Invitrogen). The cells were harvested 48 h
post-transfection and luciferase activities were measured with the
Dual-Luciferase Reporter Assay system (Promega) according to the
manufacturer's suggestions. The pRL-TK vector was used as an
internal control. Assays were performed in triplicate and results
are presented as means ± standard deviation (SD).
Western blotting
Western blotting was performed as previously
described (32).
In vivo tumorigenesis assay
Five-week-old male BALB/c nude mice were obtained
from the Animal Center for Vitalriver (Beijing, China). To measure
the role of miR-605 overexpression on the tumor growth, Mel-RM
cells stably overexpressing miR-605 or control cells in 0.1 ml
OptiMEM were subcutaneously injected into the right flank of nude
mice, respectively. To detect the effect of miR-605 inhibition on
the tumorigenic ability of melanoma cells, SK-MEL-28 cells were
subcutaneously injected into the right flank of nude mice. When the
tumor reached an average volume of 100 mm3, the nude
mice bearing tumor were randomly divided to 2 groups (n=6/group)
according to tumor volumes and body weights and received
intratumoral injection of antagomir-605 (10 nM of antagomir-605
diluted in 50 µl PBS) or antagomir-NC for 3 weeks (three times per
week). Tumor growth were monitored at indicated times.
Statistical analysis
Statistical analysis was performed with the unpaired
Student's t-test using the SPSS 17.0 software (SPSS, Chicago, IL,
USA). The data are presented as mean ± standard deviation (SD).
P-values of <0.05 were considered statistically significant.
Results
miR-605 is significantly downregulated
in melanoma cells and tissues
To explore the significance of miR-605 in the
initiation and progression of melanoma, we first determined the
expression of miR-605 in a panel of BRAF/NRAS wild-type (SK-MEL-31,
ME4405, WM1321 and Me1007), NRAS mutant (Mel-RM, SK-MEL-2,
SK-MEL-103 and WM1366) and BRAF mutant (Mel-RMU, WM278, A375, MM200
and SK-Mel-28) melanoma cell lines. The results show that miR-605
exhibits various expression levels in three types of melanoma cell
lines. Although miR-605 expression was significantly lower in
melanoma cell lines compared to melanocyte cell line, there was no
significant difference in the three types of melanoma cell lines
(Fig. 1A).
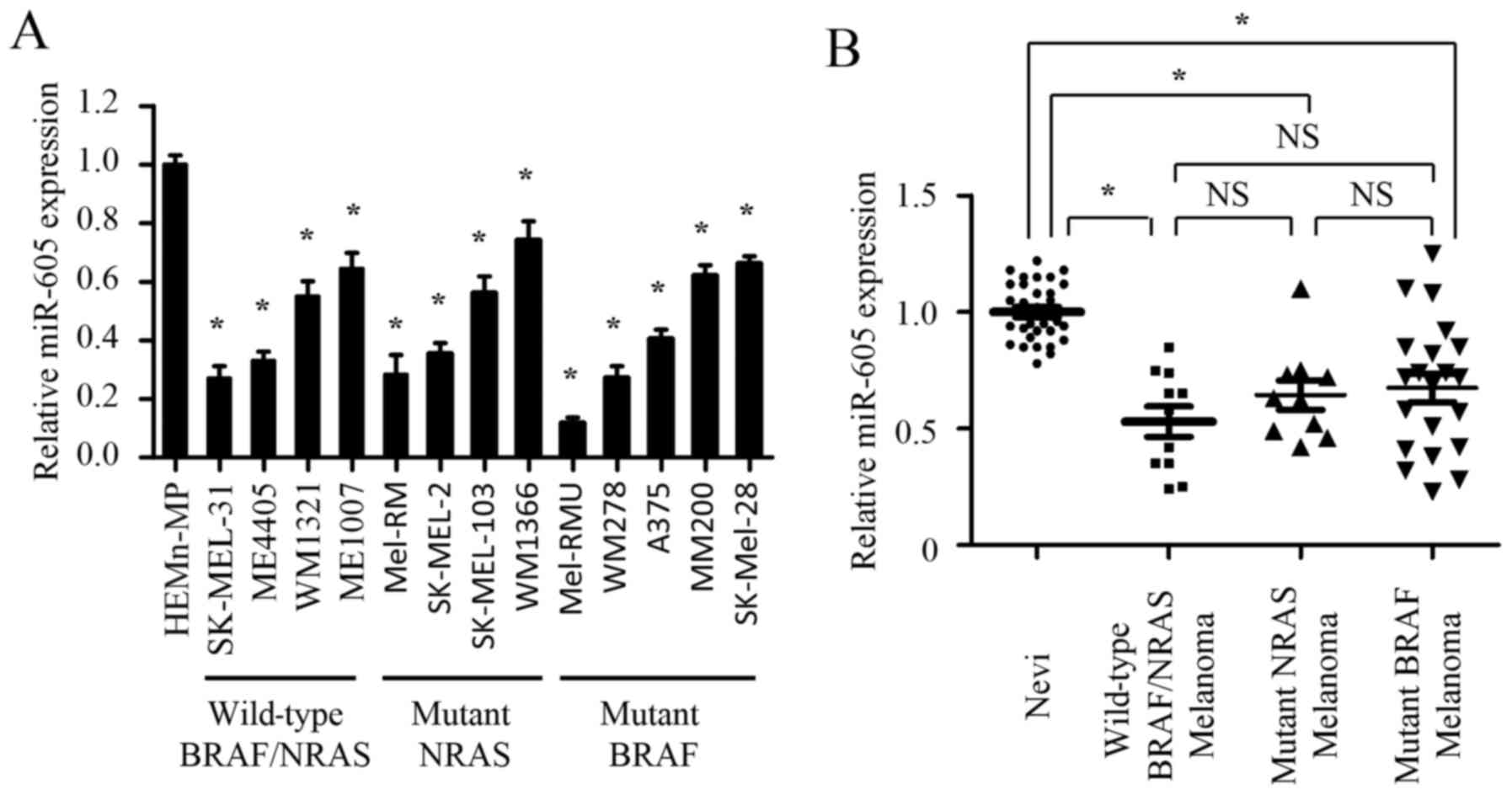 | Figure 1.miR-605 is significantly
downregulated in melanoma cells and tissues. (A) Relative
expression of miR-605 in melanocyte cell line (HEMn-MP), BRAF/NRAS
wild-type (SK-MEL-31, ME4405, WM1321 and Me1007), NRAS mutant
(Mel-RM, SK-MEL-2, SK-MEL-103 and WM1366) and BRAF mutant (Mel-RMU,
WM278, A375, MM200 and SK-Mel-28) melanoma cell lines. The
expression of miR-605 was normalized to U6 snRNA (*P<0.05). (B)
Comparison of miR-605 expression among nevi (n=31), BRAF/NRAS
wild-type melanomas (n=11), NRAS mutant melanomas (n=10) and BRAF
mutant melanomas (n=21) determined by qRT–PCR. The expression of
miR-605 was normalized to U6 snRNA (*P<0.05). |
Next, we investigated whether miR-605 is
downregulated in clinical melanoma samples. The abundance of
miR-605 was examined in 73 clinical samples, including nevi (n=31),
nevi (n=31), BRAF/NRAS wild-type melanomas (n=11), NRAS mutant
melanomas (n=10) and BRAF mutant melanomas (n=21). In agree with
the results in cell lines, the expression of miR-605 in three types
of melanomas was markedly decreased compared with that in nevi.
However, miR-605 expression exhibited no statistical difference in
three types of melanomas clinical samples (Fig. 1B). Together, our data demonstrate
that miR-605 is significantly downregulated in melanoma cells and
tissues but shows no correlation with the mutational status of
melanoma cell lines or the clinical specimens, suggesting the
miR-605 may play a potential role in the initiation and progression
of melanomas. Furthermore, we examined the expression levels of
miR-605 in 73 clinical samples.
Upregulation of miR-605 suppresses the
growth of melanoma cells
The reduced expression of miR-605 in melanomas
indicates that it may function as tumor suppressor. To access the
role of miR-605, the miR-605 mimics were transfected into Mel-RM
and ME4405 cells, which expressed relatively low levels of miR-605
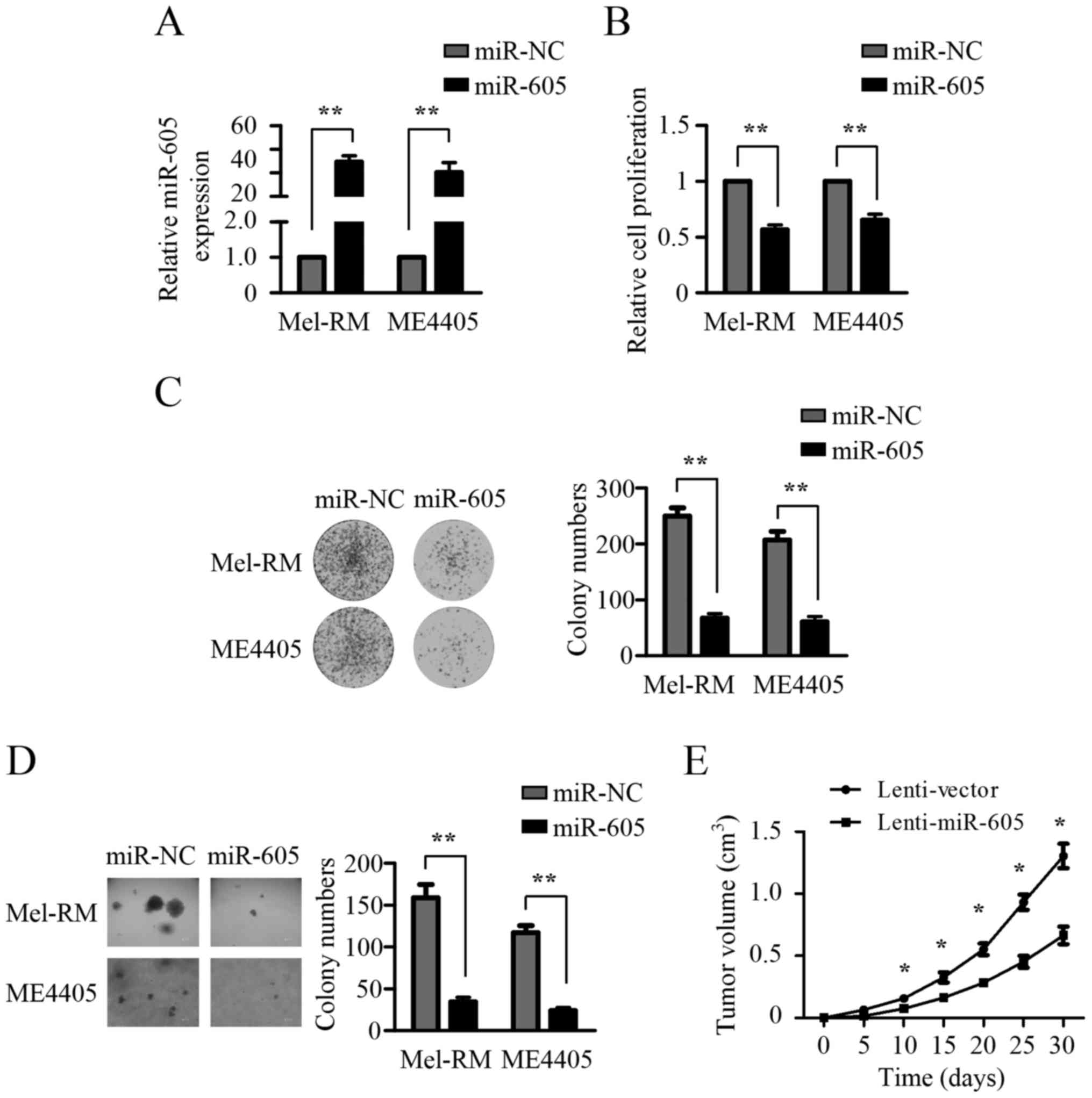
(Fig. 1A). As shown in Fig. 2A, the expression of miR-605 in
Mel-RM and ME4405 cells was significantly increased by miR-605
mimics transfection. CCK8 assays showed that ectopic expression of
miR-605 resulted in a significant inhibition in the growth of both
Mel-RM and ME4405 cells (Fig. 2B).
Furthermore, the clonogenic potential of Mel-RM and ME4405 cells
was markedly suppressed upon miR-605 overexpression (Fig. 2C). Soft agar assays demonstrated
that Mel-RM and ME4405 cells expressing miR-605 both exhibited
substantially decreased anchorage-independent growth ability
(Fig. 2D). These observations
indicated that enforced expression of miR-605 inhibited the growth
of melanomas cells in vitro, and we next detected the effect
of miR-605 overexpression on the growth of melanoma cells in
vivo. Mel-RM cells stably overexpressing miR-605 were
established and injected into the flanks of nude mice. As shown in
Fig. 2E, miR-605 overexpression
profoundly suppressed the growth of subcutaneous xenograft tumors.
Taken together, these findings suggest that miR-605 has a
suppressor role in the growth of melanomas cells.
Inhibition of miR-605 promotes the
growth of melanoma cells
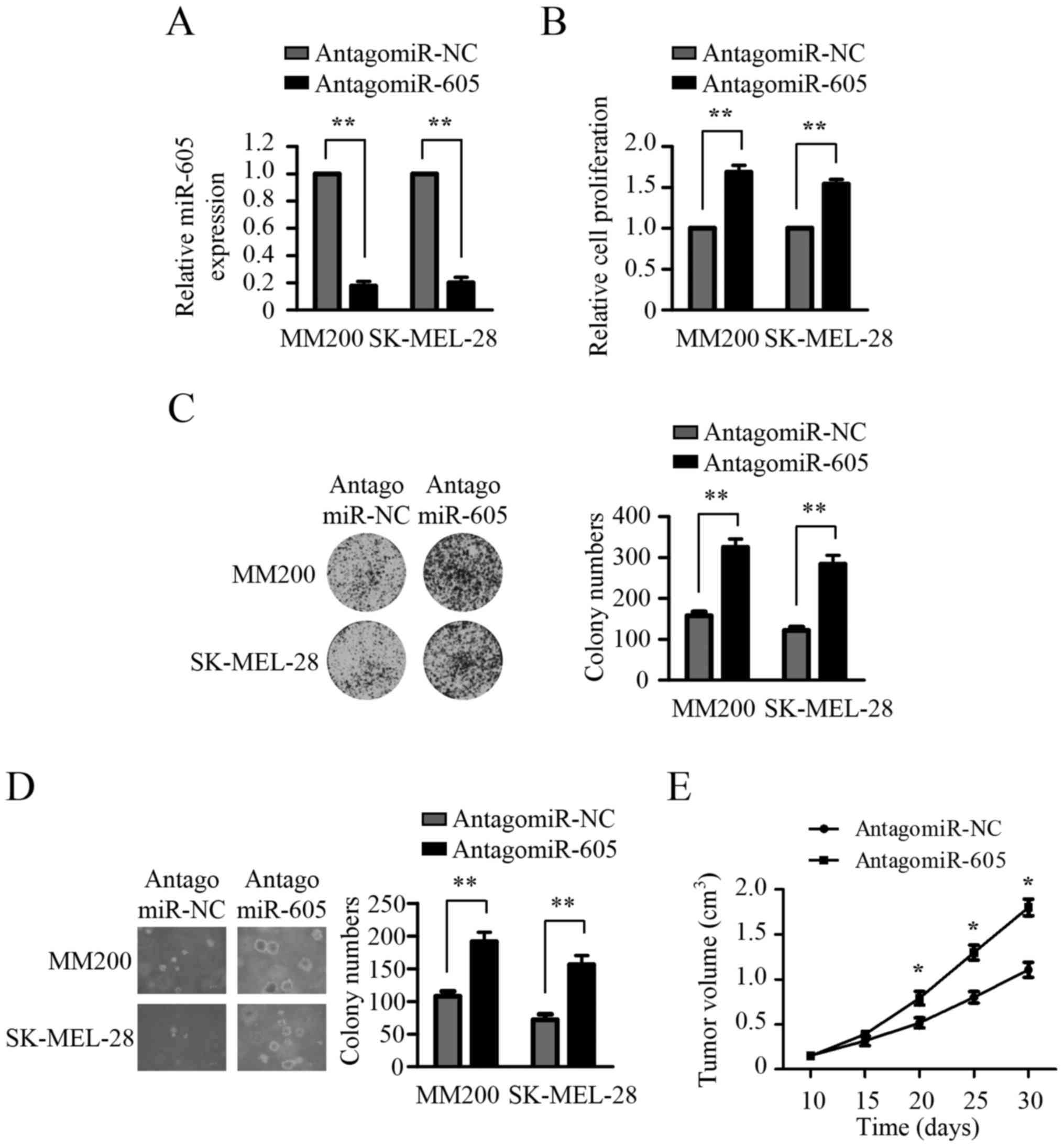
To confirm the inhibitory role of miR-605 in the
growth of melanoma cells, we next evaluated the impact of miR-605
inhibition on the growth of melanoma cells. To silence the
expression of miR-605, Antagomir-605 was transfected into MM200 and
SK-MEL 28 cells, which showed relatively higher expression levels
of miR-605 compared with Mel-RM and ME4405 cells (Fig. 1A). As shown in Fig. 3A, antago-miR-605 transfection
profoundly reduced endogenous miR-605 expression in both MM200 and
SK-MEL 28 cells. CCK8 assays revealed that inhibition of miR-605
significantly promoted the growth of both MM200 and SK-MEL 28 cells
(Fig. 3B). Colony formation assays
demonstrated that MM200 and SK-MEL 28 cells transfected with
antagomir-605 showed a marked increase in both the size and number
of colonies compared to the cells transfected with antagomir-NC
(Fig. 3C). Soft agar assays showed
that miR-605 depletion strikingly promoted anchorage-independent
growth ability of both MM200 and SK-MEL 28 cells (Fig. 3D). To validate the in vitro
finding that silencing the expression of miR-605 enhanced the
growth of melanoma cells, we subsequently determined the effect of
miR-605 inhibition on the tumorigenic ability of melanoma cells.
SK-MEL-28 cells were injected subcutaneously into nude mice. After
two weeks, nude mice bearing tumors were randomly divided to 2
groups and injected with antagomir-605 or negative control for
three weeks. As shown in Fig. 3E,
antagomir-605 treatment profoundly increased tumorigenic ability of
SK-MEL 28 cells in vivo. These results suggest that
inhibition of miR-605 promotes the growth of melanoma cells in
vitro and in vivo.
INPP4B is a direct target of
miR-605
To investigate the molecular mechanisms by which
miR-605 inhibits the growth of melanomas cells, we searched for its
target genes using two bioinformatics tools, miRanda and
TargetScan. INPP4B, one of putative target genes of miR-605,
attracted our attention due to its oncogenic activity in melanomas
reported recently and one possible binding site of miR-605 in its
3′-untranslated region (3′-UTR) (Fig.
4A). To determine whether miR-605 targets INPP4B, we cloned the
3′-UTR of wild-type and mutant INPP4B (mutations in miR-605 binding
sites) into a luciferase reporter plasmid and performed the
luciferase activity assay. As shown in Fig. 4B, miR-605 transfection significantly
suppressed luciferase activity in both Mel-RM and ME4405 cells,
whereas this inhibitory effects was abolished by the mutation of
the potential miR-605 binding sequence in the 3′-UTRs of INPP4B,
suggesting that INPP4B is a direct target of miR-605. The opposite
result was obtained in MM200 and SK-MEL-28 cells transfected with
antagomir-605 (Fig. 4C), revealing
that the 3′-UTRs of INPP4B was inhibited by endogenous miR-605.
In addition, western blot showed that ectopic
expression of miR-605 suppressed its protein expression in Mel-RM
and ME4405 cells, whereas inhibition of endogenous miR-605
increased the expression of INPP4B in MM200 and SK-MEL-28 cells
(Fig. 4F and G). However, miR-605
expression levels did not alter the mRNA expression of INPP4B
(Fig. 4D and E), suggesting that
miR-605 negatively regulates INPP4B expression by repressing its
mRNA translation, but not enhancing its mRNA degradation. Taken
together, these results suggest that miR-605 negatively regulates
the expression of INPP4B through directly targeting its 3′-UTR. To
explore the correlation between miR-605 and INPP4B levels in
clinical specimens, we collected 10 pairs of nevi and melanomas
tissues from the same patients and examined miR-605 and INPP4B
protein expression. As shown in Fig.
4H, when the relative expression levels of INPP4B
(melanomas/nevi) were plotted against that of miR-605
(melanomas/nevi) in each patient, a significant inverse correlation
was found (P<0.0322; r= −0.675). These data indicate that
miR-605 downregulation is associated with the increase of INPP4B
protein levels in melanomas.
miR-605 suppresses the growth of
melanoma cells by inhibiting INPP4B
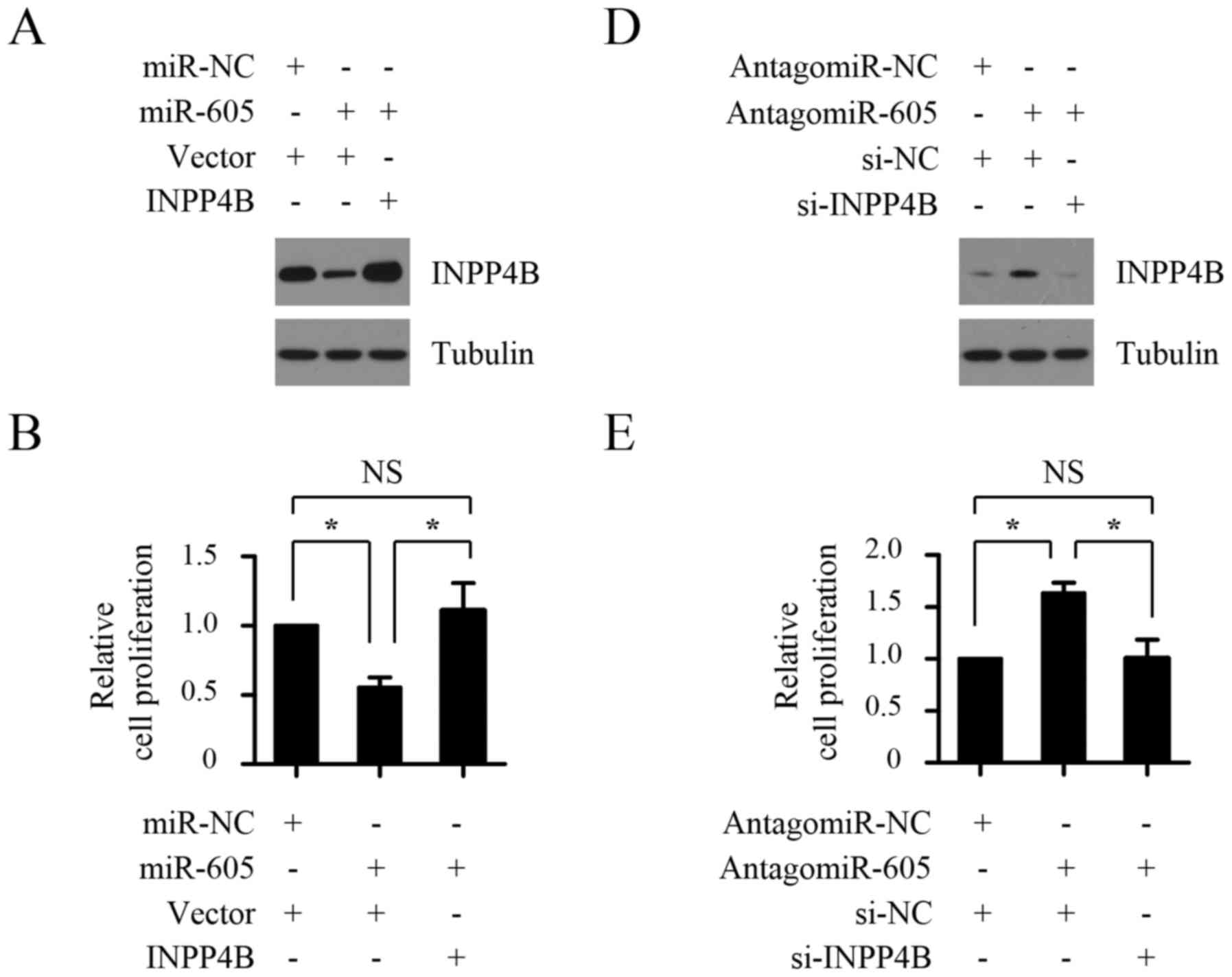
To investigate the functional significance of INPP4B
in the growth of melanomas cells suppressed by miR-605, 3′
UTR-deleted INPP4B plasmid was introduced into Mel-RM cells
transfected with miR-605 mimics, and then cell proliferation and
anchorage-independent growth ability of melanomas cells were
measured by CCK8 assays and soft agar growth assay, respectively.
As shown in Fig. 5A-C, miR-605
mimics transfection suppressed proliferation of Mel-RM cells and
resulted in decreased anchorage-independent growth ability, whereas
introduction with INPP4B plasmid rescued the phenotypic alteration
caused by miR-605 overexpression. To confirm that INPP4B is a
functional target of miR-605, we next detected the impact of INPP4B
silencing on antagomir-605-mediated promotion of proliferation and
anchorage-independent growth ability of melanomas cells. As shown
in Fig. 5D-F, MM200 cells
transfected with antagomir-605 exhibited markedly increased cell
proliferation and anchorage-independent growth ability, whereas
INPP4B knockdown abrogated the increase. Taken together, these
results suggest that miR-605 suppresses the growth of melanomas
cells by inhibiting INPP4B.
The inhibition of SGK3 activity is
crucial for the suppressive role of miR-605 on melanoma cell
growth
Given that one recent study demonstrated that SGK3
activation was critical for INPP4B-mediated melanoma cell
proliferation (13) and the above
results revealed that INPP4B is a functional target of miR-605, we
presumed that the inhibition of SGK3 activity may mediate the
suppressive role of miR-605 on melanomas cell growth. To test this
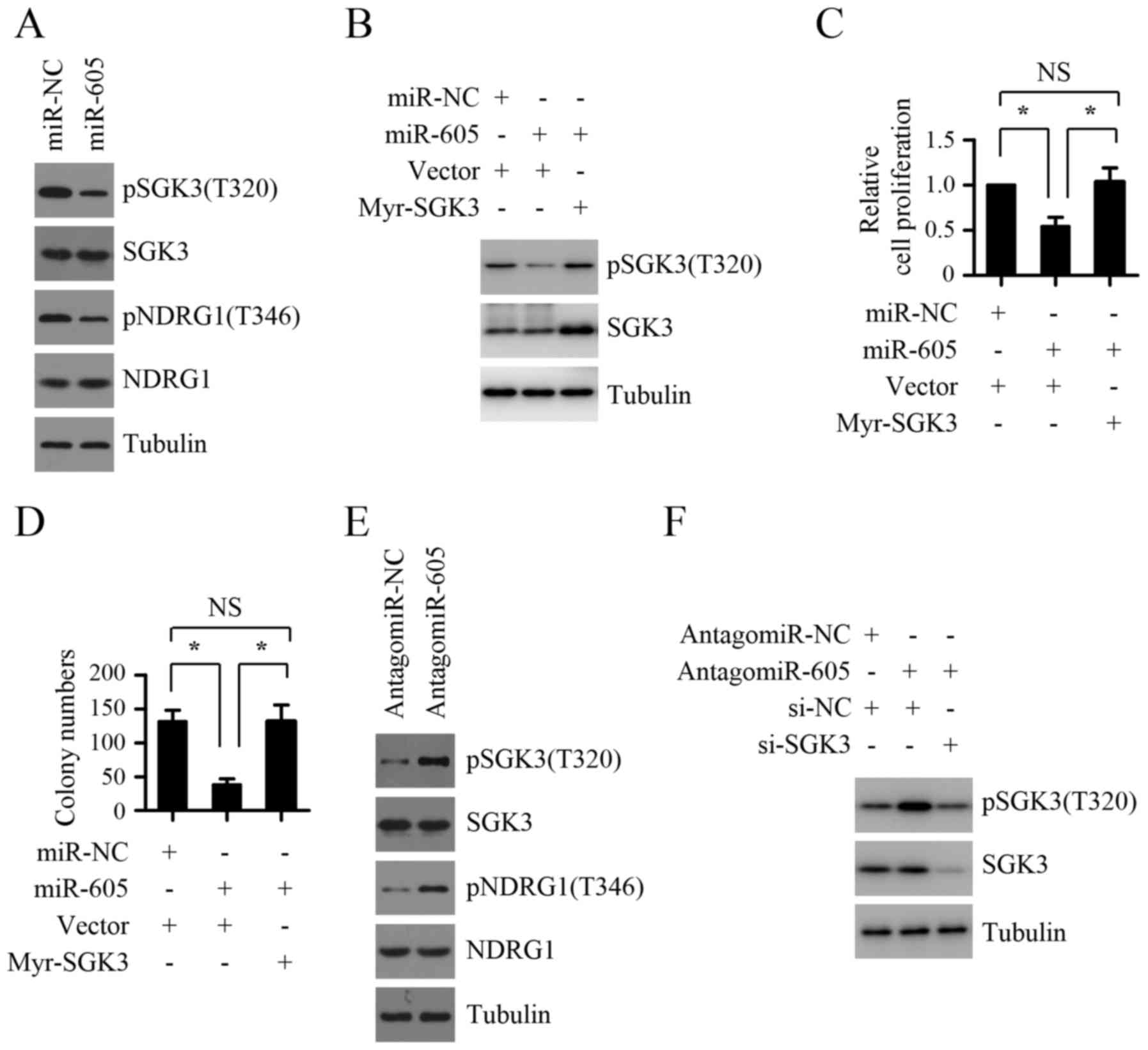
hypothesis, we first detected the effect of miR-605 expression
levels on the activity of SGK3. As shown in Fig. 6A, miR-605 overexpression in Mel-RM
cells inhibited the phosphorylation levels of SGK3 as well as
NDRG1, one downstream substrate of SGK3. In contrast, silencing the
expression endogenous miR-605 using antagomir-605 significantly
enhanced the phosphorylation levels of SGK3 and NDRG1 in MM200
cells (Fig. 6E), indicating that
miR-605 inhibits the activity of SGK3 in melanoma cells. We next
explored whether the inhibition of SGK3 activity contributes to the
suppressive role of miR-605 on melanoma cell growth. As shown in
Fig. 6B-D, introduction of miR-605
mimics led to inhibition of proliferation and anchorage-independent
growth ability in Mel-RM cells, which was abolished by
co-introduction of exogenous myr-SGK3. Furthermore, the increase of
proliferation and anchorage-independent growth ability in Mel-RM
cells caused by miR-605 inhibition could be markedly attenuated by
SGK3 depletion (Fig. 6F and G).
Taken together, these results suggest that the inhibition of SGK3
activity is crucial for the suppressive role of miR-605 on melanoma
cell growth.
Discussion
Increasing evidence demonstrates that miRNAs display
altered expression levels in a variety of cancer types and play a
key role in the initiation and progression of cancer. Therefore,
miRNAs have been extensively investigated to identify novel
biomarkers for cancer diagnosis and prognosis and develop effective
therapeutic strategy to treat cancer patients. In melanoma, many
miRNAs exhibit abnormal expression and are involved in tumor
progression. miR-182 was reported to be upregulated in melanoma
cell lines and tissues and its expression levels were associated
with melanoma progression and malignancy. Moreover, miR-182
contributed to melanoma development through directly targeting
FOXO3 (22). miR-137 was the first
identified tumor suppressor in melanoma. miR-137 was downregulated
in melanoma and its reduced expression correlated with reduced
overall survival in stage IV melanoma patients. miR-137 exerts
tumor suppressor role by targeting multiple oncogenes, including
c-Met, EZH2, PAK2 and AURKA (25,33,34).
miR-605 was originally identified as a positive
regulator of P53 through post-transcriptionally repressing the
expression of Mdm2, which facilitates rapid accumulation of p53 in
response to cellular stress (27).
The studies of miR-605 in cancer mainly focus on the association of
the genetic variants in miR-605 with cancer susceptibility. Chen
et al reported a decreased risk of breast cancer in miR-605
rs2043556*A allele carriers in Asia (35). It was shown that AG and GG genotype
carriers of miR-605 rs2043556 who has exposure to cooking oil fumes
displayed an increased risk of lung cancer compared with AA
genotype carriers without exposure to cooking oil fumes (30). Zhang et al reported that
miR-605 AG/GG genotype carriers with the habit of smoke inhalation
predicted elevated risk of gastric cancer (28). Recent studies also revealed the
abnormal expression of miR-605 in cancers. miR-605 showed a
significantly decreased expression in very high-risk (VHR) prostate
cancer patient serum samples when compared with low-risk (LR)
prostate cancer patient serum samples (36). It was reported that miR-605 showed a
decreased expression in intrahepatic cholangiocarcinoma (ICC)
specimens and suppressed ICC cell proliferation and invasion by
directly targeting PSMD10 (31).
However, the expression status of the miR-605 in melanoma tissues
and its role in melanoma progression are unclear. In this study, we
found that miR-605 showed decreased expression level in melanomas
when compared with nevi, suggesting that miR-605 is associated with
melanomagenesis and it may act as a tumor suppressor. Further
studies revealed that miR-605 inhibited anchorage-dependent and
-independent growth of melanoma cells. In addition, in vivo
studies demonstrated that miR-605 overexpression caused retardation
in melanoma growth in a xenograft model. All these studies indicate
that miR-605 exhibits tumor suppressor role in melanomagenesis.
Through in silico algorithms analyses, we
identified INPP4B as a putative target. Reporter assays
demonstrated that miR-605 inhibited the expression of INPP4B by
directly binding to its 3′-UTR. Further analysis showed that
miR-605 suppressed the protein expression level of INPP4B, but had
little effect on its mRNA expression level, suggesting that miR-605
negatively regulates INPP4B expression by repressing its mRNA
translation. Although numerous studies demonstrate that INPP4B acts
as a tumor suppressor through inhibition of PI3K/Akt signaling in
many types of cancers, recent reports show its oncogenic role in
some cancers. INPP4B showed markedly elevated expression in colon
cancer tissues when compared with paired adjacent noncancerous
colon tissues and its overexpression significantly promoted colon
cancer cell proliferation and colon cancer xenograft growth
(37). High levels of INPP4B were
observed in acute myeloid leukemia patient samples and predicted
poor clinical outcome (10). A
recent study pointed to INPP4B as an essential effector of
oncogenic PIK3CA activated breast cancer (12). In particular, INPP4B was recently
reported to be highly expressed in melanoma and to promote
proliferation of melanoma cells (13), suggesting an oncogenic role of
INPP4B in melanoma. Our results showed that enforced expression of
INPP4B effectively reversed reduced cell proliferation and
anchorage-independent growth caused by miR-605 overexpression,
indicating the functional significance of INPP4B in mediating the
tumor suppresser role of miR-605 and further confirming the
oncogenic role of INPP4B in melanoma. Whether miR-605 and INPP4B
show the reciprocal expression in clinical melanoma tissues remains
to be clarified.
The activation of SGK3 is controlled by cellular
PtdIns(3)P, which binds to the
N-terminal PX domain of SGK3 and mediates its translocation to
early endosomes for phosphorylation by its upstream PDK1 kinase
(14). Therefore, INPP4B mediated
PtdIns(3)P generation may
contribute to the activation of SGK3. SGK3 has been reported to
mediate the oncogenic role of INPP4B in many cancers, including
lung cancer, breast cancer and melanoma (6,12,13).
In either melanoma cell lines or fresh melanoma isolates with
various levels of INPP4B, the phosphorylation levels of SGK3 are
positively correlated with the expressing levels of INPP4B. INPP4B
depletion significantly inhibited the phosphorylation levels of
SGK3 and cell proliferation in melanoma cells and introduction of
an active form of SGK3 (myr-SGK3) can abolished the suppressive
effect of INPP4B knockdown on melanoma cell proliferation,
revealing SGK3 activation is essential for INPP4B-induced melanoma
cell proliferation. As we have validated that miR-605 suppresses
the growth of melanoma cells by inhibiting INPP4B, we speculated
that miR-605 might affect the activation of SGK3. Our results
showed that miR-605 overexpression profoundly inhibited the
phosphorylation levels of SGK3 and its target NDRG1, which verified
our hypothesis. Further studies demonstrate that co-introduction of
exogenous myr-SGK3 markedly attenuated suppression of proliferation
resulting from miR-605 overexpression, suggesting that the
inhibition of SGK3 activity is required for miR-605-mediated
melanoma cell growth suppression. A recent report showed that SGK3
could activating mTORC1 signaling through phosphorylating TSC2
(38). As mTORC1 signaling plays a
critical role in the initiation and progression of melanoma and
contributes to the development of resistance to BRAF inhibitors
(39), it would be of interest to
investigate the association of miR-605 repression with mTORC1
signaling activation and the resistance to anti-BRAF therapies in
melanoma.
In summary, in the present study we present evidence
that miR-605 functions as a tumor suppressor by repressing INPP4B
expression and SGK3 activity in melanoma progression. miR-605 was
significantly downregulated in melanoma cells and tissues and
suppressed the growth of melanoma cells in vitro and in
vivo. Although additional studies are required to address the
mechanism involved in the reduced expression of miR-605, our
results indicate that miR-605 provide novel insight into molecular
basis regulating melanoma malignancy and are helpful to develop
novel therapeutic approach to treat melanoma patients.
Acknowledgements
This study was supported by Guizhou Province Chinese
Native Medicine Modernization Special Project (20125018 to Y.C.)
and Guiyang Science and Technology Bureau Science and Technology
Innovation Platform Project (2012303 to Y.C.).
Glossary
Abbreviations
Abbreviations:
|
miRNAs
|
microRNAs
|
|
miR-605
|
microRNA-605
|
|
3′-UTR
|
the 3′-untranslated region
|
References
|
1
|
Arrangoiz R, Dorantes J, Cordera F, Juarez
MM and Paquentin EM: Melanoma review: Epidemiology, risk factors,
diagnosis and staging. J Cancer Treat Res. 4:1–15. 2016.doi:
10.11648/j.jctr.20160401.11. View Article : Google Scholar
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen W: Cancer statistics: updated cancer
burden in China. Chin J Cancer Res. 27:12015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Holderfield M, Deuker MM, McCormick F and
McMahon M: Targeting RAF kinases for cancer therapy: BRAF-mutated
melanoma and beyond. Nat Rev Cancer. 14:455–467. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Agoulnik IU, Hodgson MC, Bowden WA and
Ittmann MM: INPP4B: The new kid on the PI3K block. Oncotarget.
2:321–328. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhang L, Zeng D, Chen Y, Li N, Lv Y, Li Y,
Xu X and Xu G: miR-937 contributes to the lung cancer cell
proliferation by targeting INPP4B. Life Sci. 155:110–115. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Rynkiewicz NK, Fedele CG, Chiam K, Gupta
R, Kench JG, Ooms LM, McLean CA, Giles GG, Horvath LG and Mitchell
CA: INPP4B is highly expressed in prostate intermediate cells and
its loss of expression in prostate carcinoma predicts for
recurrence and poor long term survival. Prostate. 75:92–102. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hsu I, Yeh CR, Slavin S, Miyamoto H, Netto
GJ, Tsai YC, Muyan M, Wu XR, Messing EM, Guancial EA, et al:
Estrogen receptor alpha prevents bladder cancer via INPP4B
inhibited akt pathway in vitro and in vivo. Oncotarget.
5:7917–7935. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Perez-Lorenzo R, Gill KZ, Shen CH, Zhao
FX, Zheng B, Schulze HJ, Silvers DN, Brunner G and Horst BA: A
tumor suppressor function for the lipid phosphatase INPP4B in
melanocytic neoplasms. J Invest Dermatol. 134:1359–1368. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Dzneladze I, He R, Woolley JF, Son MH,
Sharobim MH, Greenberg SA, Gabra M, Langlois C, Rashid A, Hakem A,
et al: INPP4B overexpression is associated with poor clinical
outcome and therapy resistance in acute myeloid leukemia. Leukemia.
29:1485–1495. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Fedele CG, Ooms LM, Ho M, Vieusseux J,
O'Toole SA, Millar EK, Lopez-Knowles E, Sriratana A, Gurung R,
Baglietto L, et al: Inositol polyphosphate 4-phosphatase II
regulates PI3K/Akt signaling and is lost in human basal-like breast
cancers. Proc Natl Acad Sci USA. 107:22231–22236. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gasser JA, Inuzuka H, Lau AW, Wei W,
Beroukhim R and Toker A: SGK3 mediates INPP4B-dependent PI3K
signaling in breast cancer. Mol Cell. 56:595–607. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chi MN, Guo ST, Wilmott JS, Guo XY, Yan
XG, Wang CY, Liu XY, Jin L, Tseng HY, Liu T, et al: INPP4B is
upregulated and functions as an oncogenic driver through SGK3 in a
subset of melanomas. Oncotarget. 6:39891–39907. 2015.PubMed/NCBI
|
|
14
|
Bruhn MA, Pearson RB, Hannan RD and
Sheppard KE: AKT-independent PI3-K signaling in cancer - emerging
role for SGK3. Cancer Manag Res. 5:281–292. 2013.PubMed/NCBI
|
|
15
|
Wang Y, Zhou D, Phung S, Masri S, Smith D
and Chen S: SGK3 is an estrogen-inducible kinase promoting
estrogen-mediated survival of breast cancer cells. Mol Endocrinol.
25:72–82. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang Y, Zhou D and Chen S: SGK3 is an
androgen-inducible kinase promoting prostate cancer cell
proliferation through activation of p70 S6 kinase and up-regulation
of cyclin D1. Mol Endocrinol. 28:935–948. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu H, Li C, Shen C, Yin F, Wang K, Liu Y,
Zheng B, Zhang W, Hou X, Chen X, et al: MiR-212-3p inhibits
glioblastoma cell proliferation by targeting SGK3. J Neurooncol.
122:431–439. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Liu M, Chen L, Chan TH, Wang J, Li Y, Li
Y, Zeng TT, Yuan YF and Guan XY: Serum and glucocorticoid kinase 3
at 8q13.1 promotes cell proliferation and survival in
hepatocellular carcinoma. Hepatology. 55:1754–1765. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Scortegagna M, Lau E, Zhang T, Feng Y,
Sereduk C, Yin H, De SK, Meeth K, Platt JT, Langdon CG, et al: PDK1
and SGK3 contribute to the growth of BRAF-mutant melanomas and are
potential therapeutic targets. Cancer Res. 75:1399–1412. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Di Leva G, Garofalo M and Croce CM:
MicroRNAs in cancer. Annu Rev Pathol. 9:287–314. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Haflidadóttir BS, Bergsteinsdóttir K,
Praetorius C and Steingrímsson E: miR-148 regulates Mitf in
melanoma cells. PLoS One. 5:e115742010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Segura MF, Hanniford D, Menendez S, Reavie
L, Zou X, Alvarez-Diaz S, Zakrzewski J, Blochin E, Rose A,
Bogunovic D, et al: Aberrant miR-182 expression promotes melanoma
metastasis by repressing FOXO3 and microphthalmia-associated
transcription factor. Proc Natl Acad Sci USA. 106:1814–1819. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Felicetti F, Errico MC, Bottero L,
Segnalini P, Stoppacciaro A, Biffoni M, Felli N, Mattia G, Petrini
M, Colombo MP, et al: The promyelocytic leukemia zinc
finger-microRNA-221/−222 pathway controls melanoma progression
through multiple oncogenic mechanisms. Cancer Res. 68:2745–2754.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Luo C, Merz PR, Chen Y, Dickes E, Pscherer
A, Schadendorf D and Eichmüller SB: MiR-101 inhibits melanoma cell
invasion and proliferation by targeting MITF and EZH2. Cancer Lett.
341:240–247. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Luo C, Tetteh PW, Merz PR, Dickes E,
Abukiwan A, Hotz-Wagenblatt A, Holland-Cunz S, Sinnberg T, Schittek
B, Schadendorf D, et al: miR-137 inhibits the invasion of melanoma
cells through downregulation of multiple oncogenic target genes. J
Invest Dermatol. 133:768–775. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liu S, Tetzlaff MT, Cui R and Xu X:
miR-200c inhibits melanoma progression and drug resistance through
down-regulation of BMI-1. Am J Pathol. 181:1823–1835. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Xiao J, Lin H, Luo X, Luo X and Wang Z:
miR-605 joins p53 network to form a p53:miR-605:Mdm2 positive
feedback loop in response to stress. EMBO J. 30:524–532. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhang MW, Jin MJ, Yu YX, Zhang SC, Liu B,
Jiang X, Pan YF, Li QI, Ma SY and Chen K: Associations of
lifestyle-related factors, hsa-miR-149 and hsa-miR-605 gene
polymorphisms with gastrointestinal cancer risk. Mol Carcinog.
51:(Suppl 1). E21–E31. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
Huang SP, Levesque E, Guillemette C, Yu
CC, Huang CY, Lin VC, Chung IC, Chen LC, Laverdière I, Lacombe L,
et al: Genetic variants in microRNAs and microRNA target sites
predict biochemical recurrence after radical prostatectomy in
localized prostate cancer. Int J Cancer. 135:2661–2667. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yin Z, Li H, Cui Z, Ren Y, Li X, Wu W,
Guan P, Qian B, Rothman N, Lan Q, et al: Polymorphisms in pre-miRNA
genes and cooking oil fume exposure as well as their interaction on
the risk of lung cancer in a Chinese nonsmoking female population.
Onco Targets Ther. 9:395–401. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li J, Tian F, Li D, Chen J, Jiang P, Zheng
S, Li X and Wang S: MiR-605 represses PSMD10/Gankyrin and inhibits
intrahepatic cholangiocarcinoma cell progression. FEBS Lett.
588:3491–3500. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chen L, Liu T, Tu Y, Rong D and Cao Y:
Cul1 promotes melanoma cell proliferation by promoting DEPTOR
degradation and enhancing cap-dependent translation. Oncol Rep.
35:1049–1056. 2016.PubMed/NCBI
|
|
33
|
Hao S, Luo C, Abukiwan A, Wang G, He J,
Huang L, Weber CE, Lv N, Xiao X, Eichmüller SB, et al: miR-137
inhibits proliferation of melanoma cells by targeting PAK2. Exp
Dermatol. 24:947–952. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Chang X, Zhang H, Lian S and Zhu W:
miR-137 suppresses tumor growth of malignant melanoma by targeting
aurora kinase A. Biochem Biophys Res Commun. 475:251–256. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chen QH, Wang QB and Zhang B: Ethnicity
modifies the association between functional microRNA polymorphisms
and breast cancer risk: a HuGE meta-analysis. Tumour Biol.
35:529–543. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Alhasan AH, Scott AW, Wu JJ, Feng G, Meeks
JJ, Thaxton CS and Mirkin CA: Circulating microRNA signature for
the diagnosis of very high-risk prostate cancer. Proc Natl Acad Sci
USA. 113:10655–10660. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Guo ST, Chi MN, Yang RH, Guo XY, Zan LK,
Wang CY, Xi YF, Jin L, Croft A, Tseng HY, et al: INPP4B is an
oncogenic regulator in human colon cancer. Oncogene. 35:3049–3061.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Bago R, Sommer E, Castel P, Crafter C,
Bailey FP, Shpiro N, Baselga J, Cross D, Eyers PA and Alessi DR:
The hVps34-SGK3 pathway alleviates sustained PI3K/Akt inhibition by
stimulating mTORC1 and tumour growth. EMBO J. 35:1902–1922. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Flaherty KT, Robert C, Hersey P, Nathan P,
Garbe C, Milhem M, Demidov LV, Hassel JC, Rutkowski P, Mohr P, et
al: METRIC Study Group: Improved survival with MEK inhibition in
BRAF-mutated melanoma. N Engl J Med. 367:107–114. 2012. View Article : Google Scholar : PubMed/NCBI
|