Introduction
Glioma is one of the most common malignant tumor in
adult central nervous system (CNS) (1). Despite recent advances in combination
therapy, which consists mainly of surgical resection, chemotherapy
and radiotherapy, the median survival of glioblastoma remains at
merely 15–17 months (2,3). Therefore, it is urgent to find novel
biomarkers that can identify the biological characteristics of
tumors and predict the prognosis of patients with glioma (4). miRNAs are single-stranded non-coding
RNAs and usually 18–25 nucleotides in length. Mostly miRNAs are
combined with the 3′ end of the untranslated regions (3′UTR) of
target mRNAs, resulting in their degradation or inhibition of
translation (5). Importantly,
miRNAs have acquired extensive attention recently as important
regulators in a wide range of biological processes, such as immune
response (6), angiogenesis
(7), differentiation (8), cell proliferation and metastasis
(9,10). Emerging studies have shown that
dysreglated miRNAs function as either carcinogenic factors or tumor
suppressors in a variety of cancers, such as miR-575 promoting
growth and invasion of non-small cell lung cancer (11), miR-582-5p inhibiting proliferation
of hepatocellular carcinoma by targeting CDK1 and AKT3 (12), miR-22 acting as an antitumor
gatekeeper in de novo acute myeloid leukemia (AML) (13).
miRNA-320b has been identified to play a suppressive
role in various tumors, including nasopharyngeal carcinoma
(14) and colorectal cancer
(15). However, the implication of
miR-320b in glioma is still unclear. In our study, we evaluated the
level of miR-320b in glioma tissues as well as in normal brain
tissues, and explored the correlation between miR-320b with
clinical characteristics and prognosis of glioma patients.
Furthermore, using miRNA-320b mimics, we investigated the potential
molecular mechanisms of miR-320b involved in proliferation,
apoptosis, migration and invasion in glioma U87 and U251 cells.
Materials and methods
Tissue samples
Our study was approved by the Ethics Committee of
The First Affiliated Hospital of Nanchang University (ethics
approval no. 2010-015; date, 12 March 2010), with written informed
consent obtained from all the patients. The glioma tissues in this
study were obtained from 65 patients who had undergone tumor
resection without radiotherapy or chemotherapy from 2010 to 2013 at
The First Affiliated Hospital of Nanchang University (Nanchang,
Jiangxi, China). Sixteen normal brain tissues from cerebral trauma
patients or epilepsy patients were used as controls. All samples
were immediately frozen in liquid nitrogen and stored at −80°C
until RNA extraction. Each patient has experienced a follow-up
period of up to 48 months since surgical resection.
Cell lines and culture conditions
The human glioma cells (U87, U251 and T98G) were
purchased from the Cell Bank of the Shanghai Branch of Chinese
Academy of Sciences and cultured in Dulbecco's modified Eagle's
medium (DMEM) with 10% fetal bovine serum (FBS, Gibco, Carlsbad,
CA, USA). All the cells were incubated at 37°C in a humidified
atmosphere of 5.0% CO2.
RNA extraction and qRT-PCR
Total RNA was isolated from tissue samples and cell
lines with TRIzol reagent (Invitrogen, Carlsbad, CA, USA) following
the manufacturer's instructions. The isolated RNA was reverse
transcribed into cDNA using Thermo Scientifc RevertAid First Strand
cDNA Sythesis kit (Thermo Fisher Scientific). The expression was
qualified by qRT-PCR using SYBR Green real-time PCR kit (Takara,
Dalian, China) on a LightCycler 480 (Roche, USA), with U6 as a
normalizing control and 2−∆Ct as the calculation method.
The forward primers and reverse primers for miR-320b and U6 were
purchased from Ribobio (Guangzhou, China).
Western blot analysis
Glioma tissues and cell lines were lysed using RIPA
buffer (Beyotime Institute of Biotechnology, Shanghai, China) and
protein concentration was quantified with BCA Protein assay kit
(Thermo, USA). The proteins were separated by 10% SDS denaturing
polyacrylamide gel (SDS-PAGE) and then transferred onto a
polyvinylidene fluoride (PVDF) membrane. Then, the primary antibody
to MMP2, MMP9, Cyclin D1, Bax, Bcl-2 or β-actin was incubated with
the membranes overnight at 4°C. After being washed three times with
TBST, the membranes were incubated with horseradish
peroxidase-conjugated secondary antibody for 2 h. The protein bands
were visualized with enhanced chemiluminescence (ECL, GE
Healthcare, USA), with their images captured by Quantity One
software (Bio-Rad, USA). The levels of proteins were represented by
the relative densities of their bands normalized with those of
β-actin. Antibodies against MMP2, MMP9, Cyclin D1, Bax and Bcl-2
were obtained from CST Biotech (USA). Antibody to β-actin from
Sigma (USA) was adopted as a loading control.
Cell proliferation assay
The effect of miR-320b on the proliferation ability
of U87 and U251 cells was determined by MTS (CellTiter 96 aqueous
one solution reagent; Promega, Madison, WI, USA) according to the
manufacturer's instructions. U87 and U251 cells lines were cultured
in 96-well plates (3×103/well) and their proliferation
was assessed at 490 nm on a microplate reader (Bio-Rad
Laboratories, Inc.) every 24 h to 72 h after transfection. Each
experiment was repeated at least three times independently.
Colony formation assay
Briefly, cells (1,000/well) were seeded into 6-well
plates and cultured for 15 days. Then colonies were fixed for 10
min with 4% paraformaldehyde and stained with 0.1% crystal violet
(Beyotime Institute of Biotechnology) for 30 min. The number of
colonies was counted under a microscope and stained colonies were
photographed using a high-resolution camera. The experiment was
performed at least in triplicate.
Flow cytometry analysis of cell cycle
and apoptosis
For cell cycle analysis, all cells were harvested by
trypsinization, washed with PBS and then fixed with ice-cold 70%
ethanol at −20°C overnight. Fixed cells were then suspended in 300
µl staining buffer, 10 µl propidium iodide (PI) and 5 µl RNase
(Beyotime Institute of Biotechnology) for 20 min at room
temperature in the dark, followed by the examination on a FC500
flow cytometer (Beckman Coulter).
Cell apoptosis was detected using Annexin
V-FITC/propidium iodide (PI) Apoptosis Detection kit (BD
Biosciences, San Jose, CA, USA) according to the manufacturer's
instructions. Cells were transfected with miR-320b mimics for 48 h
and harvested by trypsinization before being washed with ice-cold
PBS and suspended with 195 µl binding buffer. Annexin V-FITC (5 µl)
and 5 µl of PI were subsequently added to the suspension and the
cells were mixed for additional 15 min at room temperature in the
dark. The percentage of apoptotic cells was measured by FC500 flow
cytometer, and data analysis was performed with FlowJo software.
The experiments were performed independently three times for each
cell line.
Wound healing assay
Wound healing assay was used to detect cell
migration. In brief, U87 and U251 cells were cultured in 6-well
plates until they reached 90% confluence with cell layers scratched
using a 200 µl plastic pipette tip to form wounded gaps. Then the
detached cells were washed with PBS for three times and cultured in
a medium containing 2% FBS at 37°C. Representative scrape lines
were imaged, along which different stages of wound healing were
observed. At least three separate experiments were carried out.
Transwell invasion assay
Transwell invasion assay was used to determine cell
invasion. U87 and U251 cells transfected with mimic of miR-320b or
non-specific mimic (NC) were resuspended (1×105
cells/ml) in 200 µl serum of 1% FBS medium and were added on a
filter coated with basement membrane Matrigel (BD Bioscience,
Bedford, MA, USA) according to the manufacturer's protocol. After
24 h of incubation, the cells and Matrigel on the upper membrane
surface were removed gently with cotton swabs, and invading cells
on the lower surface were fixed with 4% paraformaldehyde for 10 min
and stained with crystal violet for 30 min. The invading cells were
photographed in randomly selected fields and counted using a light
microscope. The experiments for each group were performed in
triplicate and the results were averaged.
Statistical analysis
Statistical analysis was performed with SPSS 20.0
software system (IBM, SPSS, Chicago, IL, USA). Data on the relative
expression of miRNA were reported as means ± SD and Student's
t-test was used to analyze differences between groups. A Chi-square
test was applied to determine the association of miR-320b levels
with clinicopathologic features. Survival analysis was carried out
with the Graphpad 6. The hazard ratio (HR) was estimated via a Cox
regression model and overall survival curves were plotted from
Kaplan-Meier estimates, while log-rank test was conducted to
compare the survival distributions between two groups. Differences
were considered statistically significant at P<0.05.
Results
miR-320b is significantly
downregulated in glioma tissues
To explore the implication of miRNA-320b in the
development and progression of glioma, the expression levels of
miR-320b in 65 glioma patients without radiotherapy or chemotherapy
were compared against those of 16 normal brain tissues by qRT-PCR.
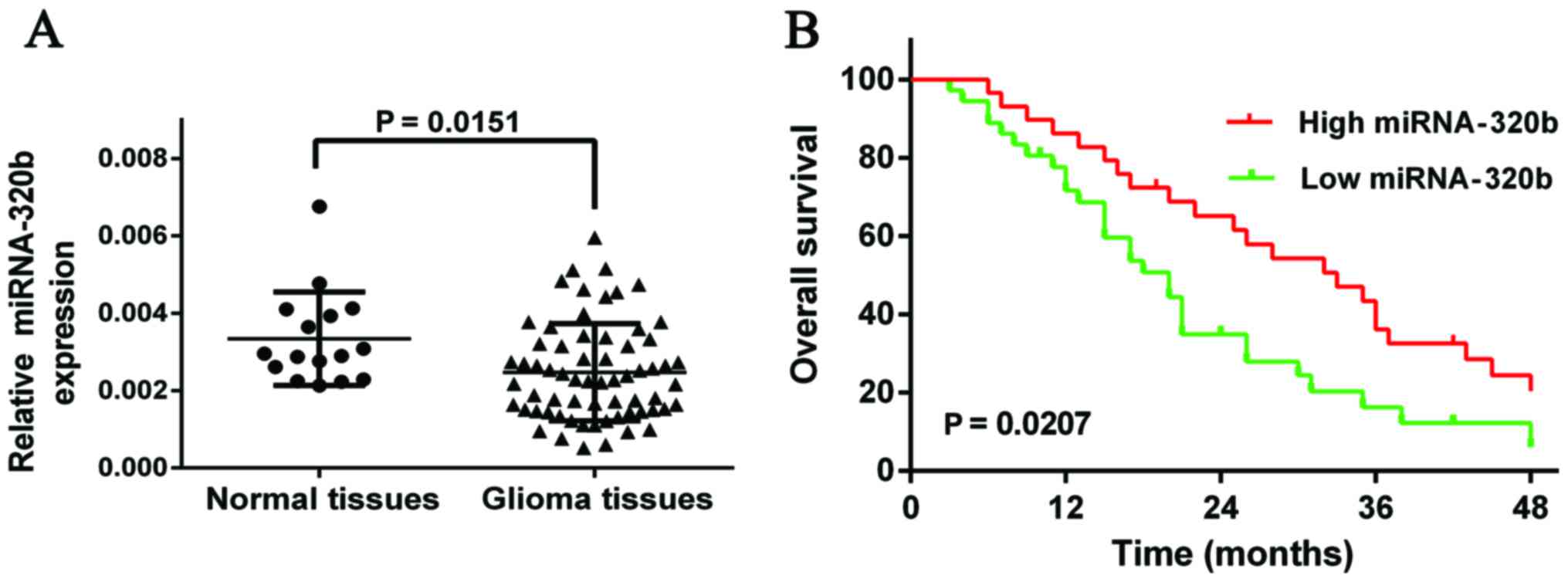
The results demonstrated that miR-320b was significantly
downregulated in glioma tissues compared with normal tissues
(Fig. 1A, P=0.0151).
Low expression of miR-320b was
associated with poor prognosis of glioma patients
In order to further investigate the association of
miRNA-320b expression with prognosis of glioma patients,
Kaplan-Meier analysis and log-rank test were used to evaluate the
effects of miR-320b expression on overall survival, discovering
that patients with low expression of miR-320b might have a worse
overall survival compared with those with highly expressed miR-320b
(Fig. 1B, HR, 1.853; 95% CI,
1.137–3.464; P=0.0207).
Correlations of miR-320b expression
with clinicopathological features of glioma patients
To further elucidate the relationship between
miR-320b with clinical characteristics of glioma patients, 65
glioma patients were divided into two groups, low miR-320b
expression (n=36) and high miR-320b expression (n=29). Our data
revealed that the expression levels of miR-320b was greatly related
to tumor grade of glioma (P=0.0070), while not in association with
age, gender and tumor location (Table
I, P=0.6781, 0.3437 and 0.7849, respectively). Thus, we
speculated that miR-320b might play important roles in the
progression of glioma.
 | Table I.Correlation between miR-320b
expression and clinicopathological features of glioma patients. |
Table I.
Correlation between miR-320b
expression and clinicopathological features of glioma patients.
|
|
| No. of patients |
|
|---|
|
|
|
|
|
|---|
| Clinical
characteristic | No. of patients | High expression
(29) | Low expression
(36) | P-value |
|---|
| Age (years) |
|
<45 | 31 | 13 | 18 | 0.6781 |
|
≥45 | 34 | 16 | 18 |
|
| Gender |
|
Male | 40 | 16 | 24 | 0.3437 |
|
Female | 25 | 13 | 12 |
|
| Clinical stage |
| Low
grades I–II | 35 | 21 | 14 | 0.0070 |
| High
grades III - IV | 30 | 8 | 22 |
|
| Tumor location |
|
Frontal | 19 | 7 | 12 | 0.7849 |
|
Parietal | 6 | 2 | 4 |
|
|
Occipital | 11 | 6 | 5 |
|
|
Temporal | 12 | 5 | 7 |
|
|
Others | 17 | 9 | 8 |
|
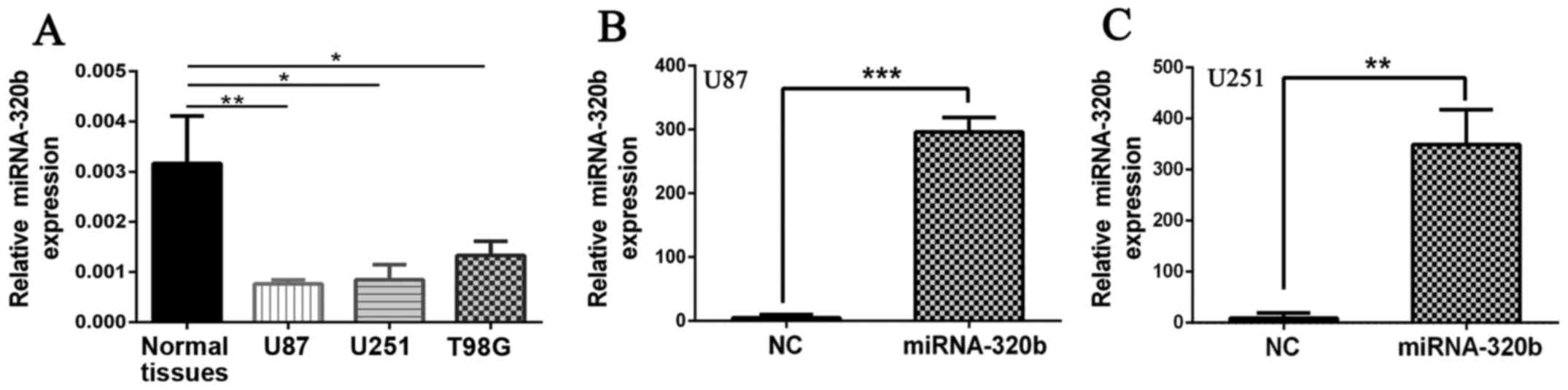
Significant overexpression of miR-320b
with mimics in U87 and U251
To identify whether miR-320b was associated with the
progression of glioma, function analyses were conducted in
vitro. First, we performed qRT-PCR to calculated the expression
of miR-320b in three glioma cell lines (U87, U251, T98G), finding
that similar to the situation for the glioma tissues, miR-320b were
markedly lower in glioma cell lines (U87, U251 and T98G) compared
with normal brain tissues (Fig.
2A). Then, mimic of miR-320b and miR-NC were transfected into
U87 and U251 cells which had the lowest levels of miRNA-320b. As
shown in Fig. 2B and C, miR-320b
was effectively overexpressed in U87 and U251 cell lines compared
with NC groups (Fig. 2B, P<0.001
for U87; 2C, P<0.001 for U251).
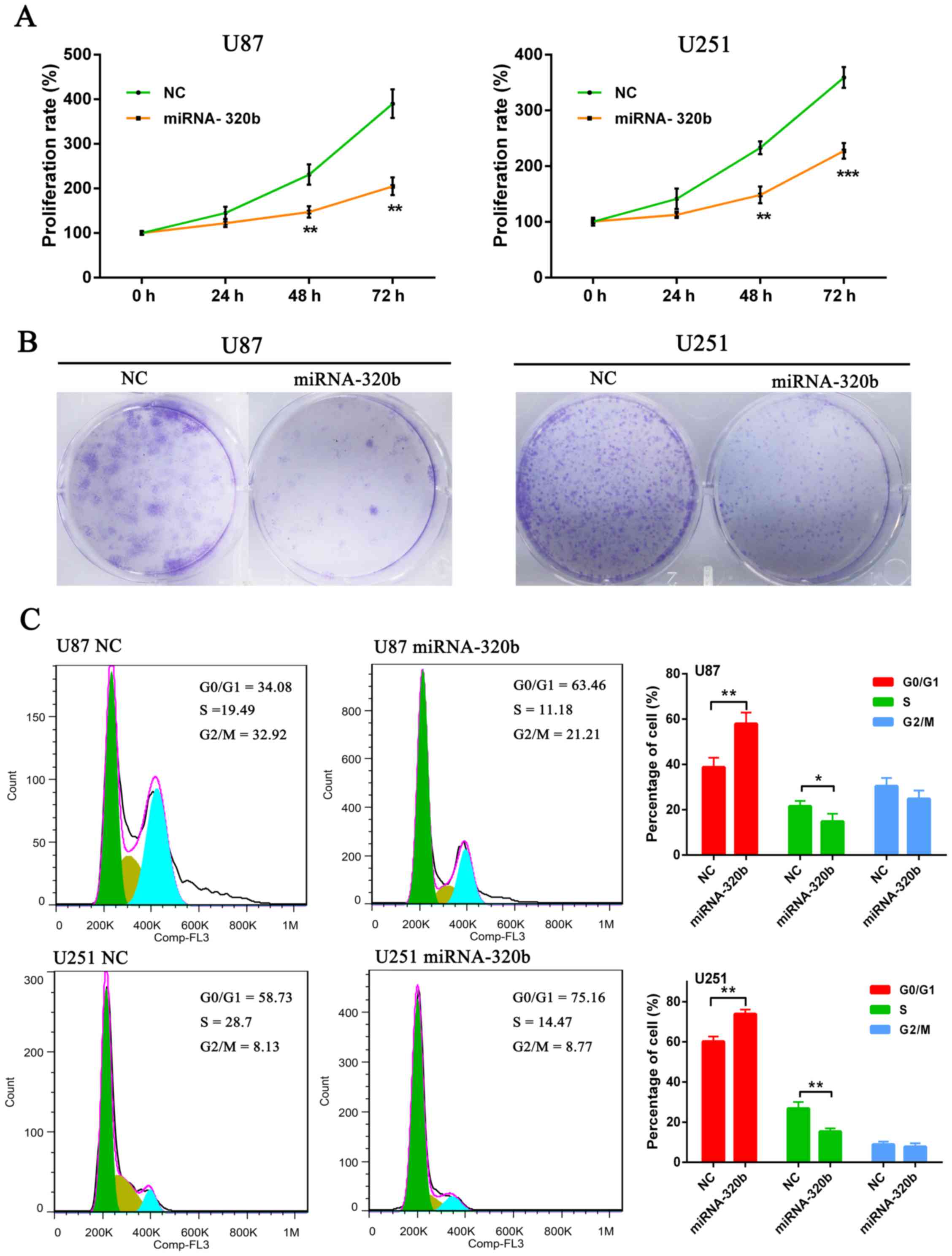
miR-320b mimic inhibits U87 and U251
cell proliferation
In order to further determine the role of miR-320b
in glioma cells, MTS and colony formation assays were carried out
in cultured U87 and U251 cells. MTS results suggested that
overexpression of miR-320b could significantly inhibit cell
viability in contrast with NC groups at 72 h both in U87 and U251
cell lines (Fig. 3A). Moreover,
colony formation assay results demonstrated that the number of U87
and U251 cell colonies was strikingly decreased by the miR-320b
mimic compared with the NC groups (Fig.
3B).
Subsequently, to explore the mechanisms underlying
proliferation suppression after miR-320b mimic transfection, we
examined the effects of miR-320b on cell cycles of U87 and U251
cells by PI staining and flow cytometry. Flow cytometric analysis
indicated that miR-320b mimic could significantly induce
accumulation of cells in G0/G1 phase and correspondingly decrease
the percentage of S phase cells (Fig.
3C). These data suggested that overexpression of miR-320b could
inhibit glioma growth and development by suppressing cell cycle
progression.
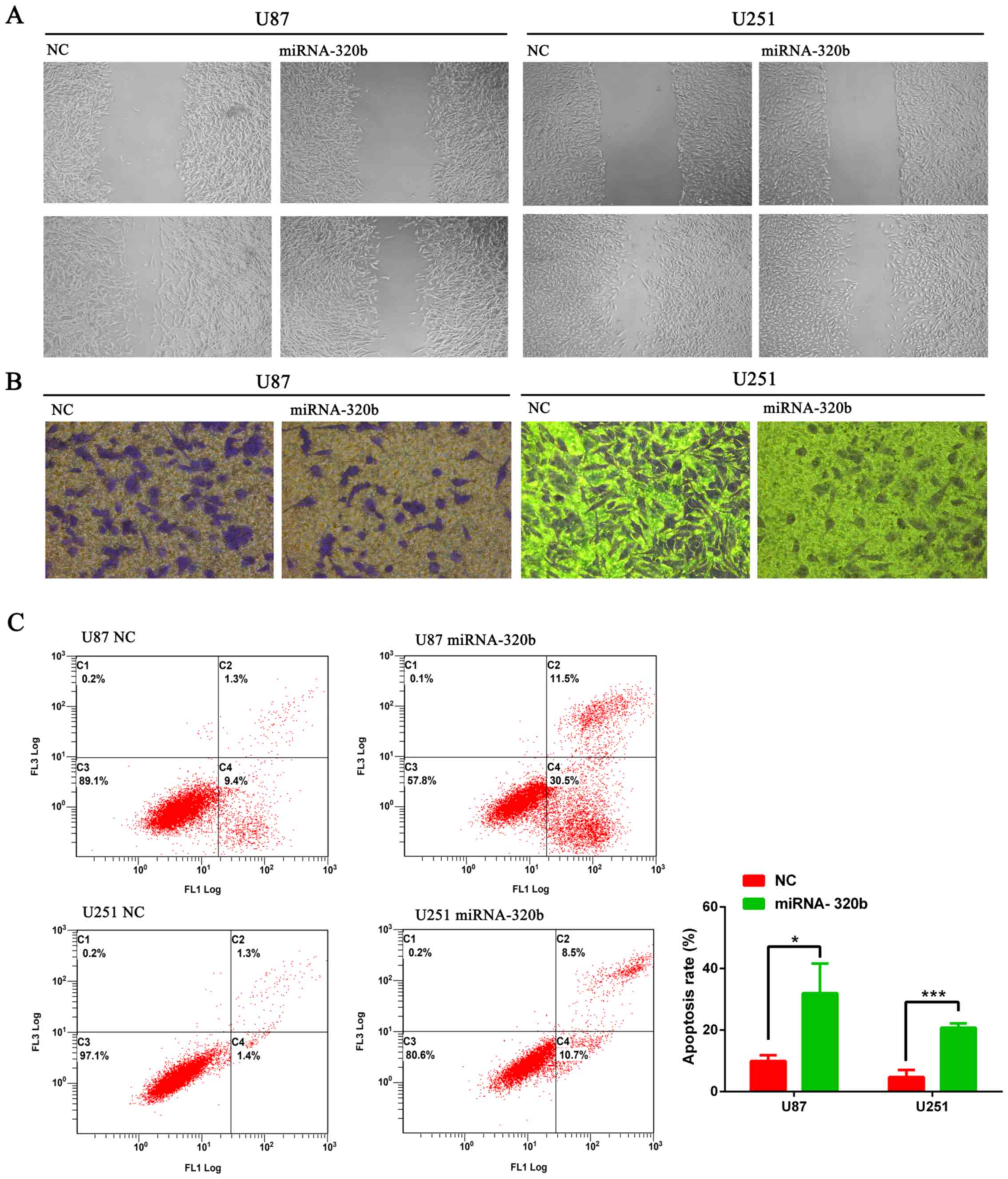
miR-320b mimics inhibit migration and
invasion of U87 and U251 cell lines
Furthermore, to investigate the correlation of
miR-320b expression with the migration and invasion capabilities of
glioma cells, wound healing assay and Transwell invasion assay were
conducted. In the wound healing assay, cell migration was inhibited
after the transfection with miR-320b mimics and incubation for 24 h
(Fig. 4A). Besides, Transwell assay
demonstrated that miR-320b mimics could inhibit invasion abilities
of U87 and U251 cells in comparison with NC groups (Fig. 4B).
miR-320b mimic promotes U87 and U251
apoptosis
As defects in apoptosis could induce carcinogenesis
and cancer progression, we subsequently investigated whether
overexpression of miR-320b could affect apoptosis of glioma cell
lines. Flow cytometry analysis was performed and results suggested
the significant increase of apoptotic rates of U87 and U251 cells
transfected with miR-320b mimics compared with NC groups (Fig. 4C), indicating that miR-320b might
play a crucial role in the apoptosis regulation of human glioma
cells.
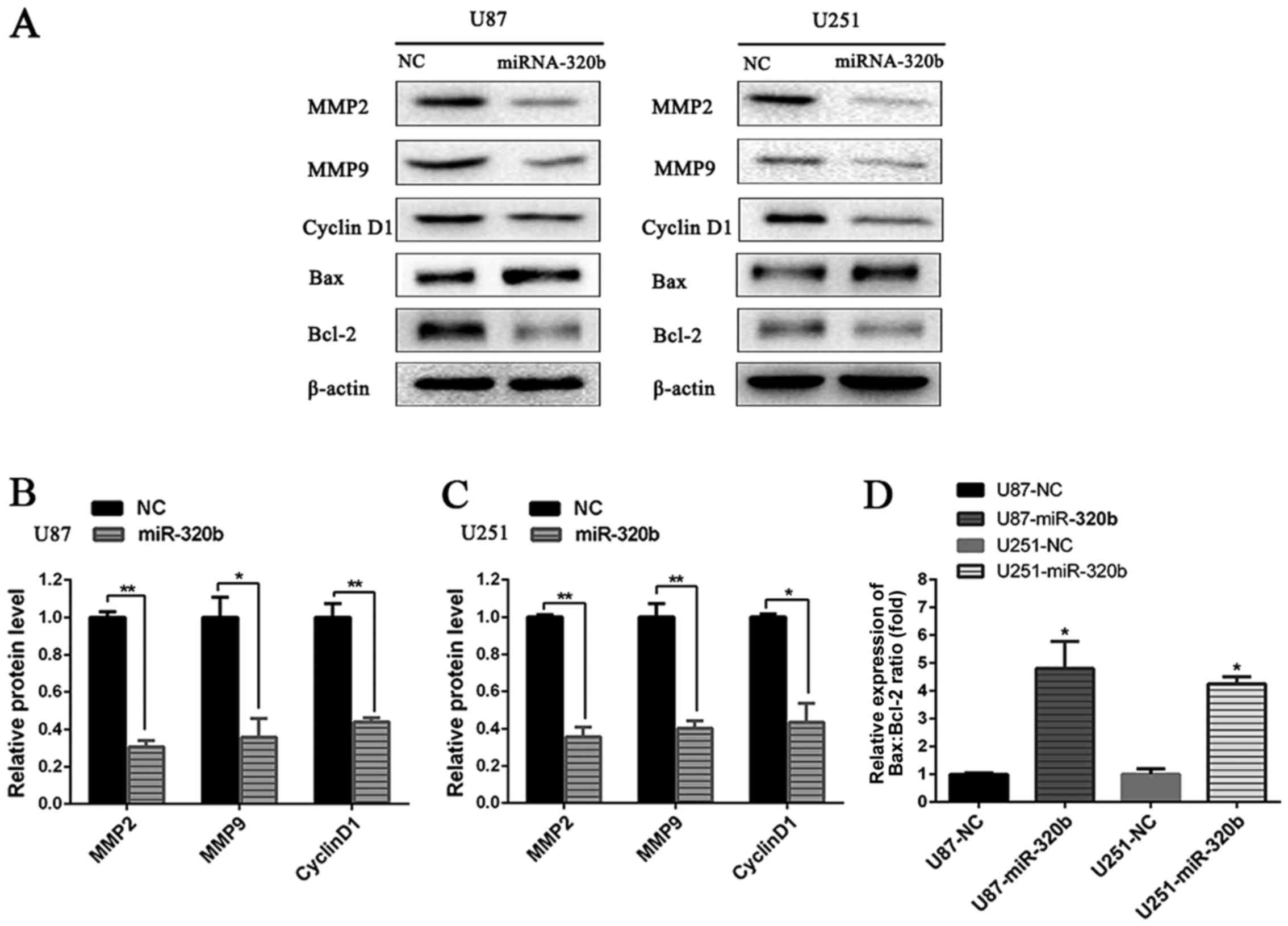
Signaling pathways relative to
proliferation, apoptosis, migration and invasion of U87 and U251
cells
To further explore the molecular mechanisms by which
miRNA-320b promoted glioma progression, western blot analysis was
conducted to detect the target proteins involved in cell cycle,
apoptosis, invasion and metastasis of tumor. As shown in Fig. 5, compared with NC groups, the
expression of MMP2, MMP9, Cyclin D1 and Bcl-2 was significantly
repressed, while the protein level of Bax was elevated
significantly in both U87 and U251 cell lines after transfection
with miRNA-320b, demonstrating that miRNA-320b probably played a
crucial role in the regulation of proliferation, apoptosis,
migration and invasion of human glioma cells.
Discussion
A growing number of miRNAs have been demonstrated to
be aberrantly expressed in wide spectrum of human diseases, such as
Parkinson (16), diabetes (17), atherosclerosis (18), immune diseases (19) and especially malignant tumors
(20–24). Recently, dysregulated miRNAs were
identified to be crucial in the regulation of carcinogenesis and
cancer progression (25–27). miR-320b, an important member of
miR-320 family, has been reported to be involved in a variety of
biological processes. Yan et al found that miR-320b level
was significantly lower in prediabetes compared with newly
diagnosed type 2 diabetes (T2D) and normal glucose tolerance (NGT)
subjects (28). Zhang et al
reported that low serum miR-320b expression could act as a novel
indicator of carotid atherosclerosis (29). Moreover, Qin et al provided
evidence that elevating the expression of circulating miRNA-320b
might serve as a biomarker for unexplained recurrent spontaneous
abortion (URSA) (30). However,
limited studies were focused on the role of miRNA-320b as an
important regulator in tumor with its pathological role in glioma
still poorly understood.
In this study, we investigated the function of
miR-320b in glioma, detecting a greatly decreased expression
pattern of miR-320b in glioma tissues compared with normal brain
tissues as well as in both U87 and U251 glioma cell lines in
contrast with NC, which was similar to the results by Tadano et
al and Li et al in colorectal adenoma (31) and nasopharyngeal carcinoma patients
(14). To further identify the
clinical significance of miR-320b, we explored the association
between miR-320b and clinicopathological features of glioma
patients, finding that its expression was tightly related to tumor
clinical stage, with low expression of miR-320b more frequently
exhibited in high degree gliomas. Moreover, we detected the
correlation of miR-320b expression with overall survival,
discovering that glioma patients with decreased expression of
miRNA-320b had a worse prognosis of overall survival. These data
suggested the involvement of miR-320b in the occurrence and
development of glioma. Therefore, functional and pathogenetic
studies were conducted in vitro to elucidate its precise
role in glioma.
Li et al reported that decrease of miRNA-320b
enhanced NPC cell proliferation by targeting TRIAP1 (14). In addition, Wang et al and
Tadano et al discovered that miR-320b could markedly promote
cell proliferation ability of human colorectal cancer cells by
targeting c-Myc and CDK6 both in vitro and in vivo
(15,31). Similar results were presented in our
study, since MTS and colony formation assays revealed that
miRNA-320b mimics significantly suppressed cell proliferation both
in U87 and U251 cell lines compared with NC group.
Dysregulation of apoptosis and cell cycle are
principal element in the transformation and progression of glioma.
Bcl-2 family members, mainly including Bax and Bcl-2, play crucial
roles in the process of cancer cell apoptosis. Cyclin D1, usually
involved in promotion of tumor growth and regulation of the
development of gliomas, is a key regulator of G1/S cell cycle
transition. Furthermore, flow cytometric analysis showed that
overexpression of 320b statistically induced G0/G1 phase arrest and
cell apoptosis, and correspondingly reduced the percentage of S
phase cells by significantly repressing the expression of Cyclin D1
and BCL-2 and upregulating Bax level. Concordant with our results,
Wang et al discovered that miR-320b suppressed cell
proliferation by accumulating of cells in G1 phase both in HCT-116
and SW-480 cells (15). Actually,
the function of miR-320b in the cell cycle and apoptosis was not
clarified in our study, which needs to be elucidated in the
future.
Previous studies have shown that all kinds of
secreted factors are also very important in the promotion of tumor
metastasis. As important secreted proteins MMPs, MMP2 and MMP9 play
prominent roles in invasion and migration in the context of
malignancy. In this study, we observed that miR-320b mimic could
suppress glioma cell migration and invasion through significantly
decreasing the expression of MMP2 and MMP9 in contrast with NC
groups in both U87 and U251 cells. In colorectal cancer, similar
inhibitory effects of miR-320b were observed in SW620 cells and RKO
cells by targeting β-catenin, Neuropilin-1 and Rac-1, resulting in
inhibition of proliferation and invasion by competing with its
homologous microRNA-320a (32).
In conclusion, for the first time, our study
demonstrated that miR-320b was greatly downregulated in glioma
tissues and correlated with poor prognostic outcome of glioma
patients. Furthermore, overexpression of miR-320b by mimics
markedly inhibited proliferation, invasion and migration, as well
as promoted apoptosis of glioma cells. All these findings suggested
that miR-320b played a pivotal role in the regulation of glioma
progression and could probably be used as an underlying prognostic
marker and a novel therapeutic target for gliomas in the
future.
Acknowledgements
This study was supported by The National Natural
Science Foundation of China (no. 81503166).
References
|
1
|
Sathornsumetee S and Rich JN: New
treatment strategies for malignant gliomas. Expert Rev Anticancer
Ther. 6:1087–1104. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Preusser M, Lim M, Hafler DA, Reardon DA
and Sampson JH: Prospects of immune checkpoint modulators in the
treatment of glioblastoma. Nat Rev Neurol. 11:504–514. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Floyd JA, Galperin A and Ratner BD: Drug
encapsulated polymeric microspheres for intracranial tumor therapy:
A review of the literature. Adv Drug Deliv Rev. 91:23–37. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Westphal M and Lamszus K: Circulating
biomarkers for gliomas. Nat Rev Neurol. 11:556–566. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jonas S and Izaurralde E: Towards a
molecular understanding of microRNA-mediated gene silencing. Nat
Rev Genet. 16:421–433. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lu LF, Gasteiger G, Yu IS, Chaudhry A,
Hsin JP, Lu Y, Bos PD, Lin LL, Zawislak CL, Cho S, et al: A Single
miRNA-mRNA interaction affects the immune response in a context-
and cell-type-specific manner. Immunity. 43:52–64. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zheng L, Pu J, Qi T, Qi M, Li D, Xiang X,
Huang K and Tong Q: miRNA-145 targets v-ets erythroblastosis virus
E26 oncogene homolog 1 to suppress the invasion, metastasis, and
angiogenesis of gastric cancer cells. Mol Cancer Res. 11:182–193.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yuan Z, Ding S, Yan M, Zhu X, Liu L, Tan
S, Jin Y, Sun Y, Li Y and Huang T: Variability of miRNA expression
during the differentiation of human embryonic stem cells into
retinal pigment epithelial cells. Gene. 569:239–249. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yu G, Li H, Wang J, Gumireddy K, Li A, Yao
W, Tang K, Xiao W, Hu J, Xiao H, et al: miRNA-34a suppresses cell
proliferation and metastasis by targeting CD44 in human renal
carcinoma cells. J Urol. 192:1229–1237. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hur K, Toiyama Y, Takahashi M, Balaguer F,
Nagasaka T, Koike J, Hemmi H, Koi M, Boland CR and Goel A:
MicroRNA-200c modulates epithelial-to-mesenchymal transition (EMT)
in human colorectal cancer metastasis. Gut. 62:1315–1326. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang H, Yan C, Shi X, Zheng J, Deng L,
Yang L, Yu F, Yang Y and Shao Y: MicroRNA-575 targets BLID to
promote growth and invasion of non-small cell lung cancer cells.
FEBS Lett. 589:805–811. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang Y, Huang W, Ran Y, Xiong Y, Zhong Z,
Fan X, Wang Z and Ye Q: miR-582-5p inhibits proliferation of
hepatocellular carcinoma by targeting CDK1 and AKT3. Tumour Biol.
36:8309–8316. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jiang X, Hu C, Arnovitz S, Bugno J, Yu M,
Zuo Z, Chen P, Huang H, Ulrich B, Gurbuxani S, et al: miR-22 has a
potent anti-tumour role with therapeutic potential in acute myeloid
leukaemia. Nat Commun. 7:114522016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li Y, Tang X, He Q, Yang X, Ren X, Wen X,
Zhang J, Wang Y, Liu N and Ma J: Overexpression of mitochondria
mediator gene TRIAP1 by miR-320b loss is associated with
progression in nasopharyngeal carcinoma. PLoS Genet.
12:e10061832016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang H, Cao F, Li X, Miao HEJ, Xing J and
Fu CG: miR-320b suppresses cell proliferation by targeting c-Myc in
human colorectal cancer cells. BMC Cancer. 15:7482015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Batistela MS, Josviak ND, Sulzbach CD and
de Souza RL: An overview of circulating cell-free microRNAs as
putative biomarkers in Alzheimer's and Parkinson's Diseases. Int J
Neurosci. Jul 20–2016.(Epub ahead of print). PubMed/NCBI
|
|
17
|
Hsu YC, Chang PJ, Ho C, Huang YT, Shih YH,
Wang CJ and Lin CL: Protective effects of miR-29a on diabetic
glomerular dysfunction by modulation of DKK1/Wnt/β-catenin
signaling. Sci Rep. 6:305752016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tana C, Giamberardino MA and Cipollone F:
microRNA profiling in atherosclerosis, diabetes and migraine. Ann
Med. 49:93–105. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bao MH, Li JM, Luo HQ, Tang L, Lv QL, Li
GY and Zhou HH: NF-κB-regulated miR-99a modulates endothelial cell
inflammation. Mediators Inflamm. 2016:53081702016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ke Y, Zhao W, Xiong J and Cao R: miR-149
inhibits non-small-cell lung cancer cells EMT by targeting FOXM1.
Biochem Res Int. 2013:5067312013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cui F, Wang S, Lao I, Zhou C, Kong H,
Bayaxi N, Li J, Chen Q, Zhu T and Zhu H: miR-375 inhibits the
invasion and metastasis of colorectal cancer via targeting SP1 and
regulating EMT-associated genes. Oncol Rep. 36:487–493.
2016.PubMed/NCBI
|
|
22
|
Shi H, Ji Y, Zhang D, Liu Y and Fang P:
MiR-135a inhibits migration and invasion and regulates EMT-related
marker genes by targeting KLF8 in lung cancer cells. Biochem
Biophys Res Commun. 465:125–130. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Babae N, Bourajjaj M, Liu Y, Van Beijnum
JR, Cerisoli F, Scaria PV, Verheul M, Van Berkel MP, Pieters EH,
Van Haastert RJ, et al: Systemic miRNA-7 delivery inhibits tumor
angiogenesis and growth in murine xenograft glioblastoma.
Oncotarget. 5:6687–6700. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yuan J, Xiao G, Peng G, Liu D, Wang Z,
Liao Y, Liu Q, Wu M and Yuan X: miRNA-125a-5p inhibits glioblastoma
cell proliferation and promotes cell differentiation by targeting
TAZ. Biochem Biophys Res Commun. 457:171–176. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Varamo C, Occelli M, Vivenza D, Merlano M
and Nigro Lo C: MicroRNAs role as potential biomarkers and key
regulators in melanoma. Genes Chromosomes Cancer. 56:3–10. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ebrahimi S, Ghasemi F, Hassanian SM,
Shahidsales S, Maftouh M, Akbarzade H, Parizadeh SA, Hassanian SM
and Avan A: Circulating microRNAs as novel potential diagnostic and
prognosis biomarkers in pancreatic cancer. Curr Pharm Des.
22:6444–6450. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kurozumi A, Goto Y, Okato A, Ichikawa T
and Seki N: Aberrantly expressed microRNAs in bladder cancer and
renal cell carcinoma. J Hum Genet. 62:49–56. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yan S, Wang T, Huang S, Di Y, Huang Y, Liu
X, Luo Z, Han W and An B: Differential expression of microRNAs in
plasma of patients with prediabetes and newly diagnosed type 2
diabetes. Acta Diabetol. 53:693–702. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhang R, Qin Y, Zhu G, Li Y and Xue J: Low
serum miR-320b expression as a novel indicator of carotid
atherosclerosis. J Clin Neurosci. 33:252–258. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Qin W, Tang Y, Yang N, Wei X and Wu J:
Potential role of circulating microRNAs as a biomarker for
unexplained recurrent spontaneous abortion. Fertil Steril.
105:1247–1254.e3. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Tadano T, Kakuta Y, Hamada S, Shimodaira
Y, Kuroha M, Kawakami Y, Kimura T, Shiga H, Endo K, Masamune A, et
al: MicroRNA-320 family is downregulated in colorectal adenoma and
affects tumor proliferation by targeting CDK6. World J Gastrointest
Oncol. 8:532–542. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhou J, Zhang M, Huang Y, Feng L, Chen H,
Hu Y, Chen H, Zhang K, Zheng L and Zheng S: MicroRNA-320b promotes
colorectal cancer proliferation and invasion by competing with its
homologous microRNA-320a. Cancer Lett. 356B:669–675. 2015.
View Article : Google Scholar
|