Introduction
Prostate cancer (PCa) is one of the most common
cancers of the male reproductive system and the second leading
cause of cancer-related death among men worldwide. In 2015, 41,210
cancer deaths were estimated among males, where 6,801 were from PCa
(1). At present, for patients
diagnosed with clinically localized cancer, effective treatments
for PCa include radical prostatectomy, radiotherapy or
androgen-deprivation therapy. However, approximately 15% of
patients present with local recurrence or develop metastatic tumors
(2). Additionally, some patients
develop castration-resistant prostate cancer (CRPC), for which
treatment is limited and the death rate is high (3,4).
Therefore, it is important to clarify the mechanism of tumor
development and progression to optimize therapeutic strategies for
PCa.
The C-terminal binding proteins (CtBPs) were
initially identified as binding partners of the E1A protein
(5). The CtBP family members CtBP1
and CtBP2 are highly homologous transcriptional corepressors, which
are conserved in both vertebrates and invertebrates (6). Recent studies have observed aberrant
expression of CtBPs in many human malignancies, such as ovarian
cancer, melanoma, breast cancer and esophageal squamous cell
carcinoma (7–10). Growing evidence shows that CtBP2 is
aberrantly upregulated in PCa and its expression is associated with
cancer development and progression (11–13).
Angiogenesis is an essential process in tumor
development, and is an important focus of cancer research and
therapy and may be a target of cancer therapy as the survival and
proliferation of cancer depend on angiogenesis (14). Among the many molecular markers
associated with tumor angiogenesis, the Tel gene has been shown to
play an evolutionarily conserved role in angiogenesis. Tel is
indispensable for the sprouting of human endothelial cells and for
normal development of the Danio rerio blood circulatory
system. Tel orchestrates endothelial sprouting by binding to the
generic co-repressor, CtBP, and forms a Tel-CtBP complex, which
temporally restricts a vascular endothelial growth factor
(VEGF)-mediated pulse of Dll4 expression and thereby directly links
VEGF receptor intracellular signaling and intercellular Notch-Dll4
signaling (15). Thus, we propose
that CtBP2 may play an important role in angiogenesis in PCa.
Follicle stimulating hormone receptor (FSHR) is
thought to play a critical role in reproductive physiology in the
ovary and spermatogenesis by promoting follicular growth and
estrogen synthesis. Stilley et al (16) revealed that FSH is as efficacious as
VEGF in promoting angiogenic processes and stimulating angiogenesis
directly. Yang et al (17)
demonstrated that FSHR is a highly selective tumor vasculature
marker in both primary and metastatic tumors. In this study, we
attempted to integrate and analyze the genome-wide association
study (GWAS) of FSHR and CtBP2, the Cancer Genome Atlas (TCGA) data
and CtBP2 binding data in CistromeMap (18) to explore the mechanism of CtBP2 in
PCa.
Materials and methods
Bioinformatic analysis
First, mRNA expression of CtBP2 and FSHR were
extracted from lymphoblastoid cell lines of 210 HapMap unrelated
individuals at the NCBI's Gene Expression Omnibus website
(http://www.ncbi.nlm.nih.gov/geo/);
series number GSE6536 (19). In
order to find a relationship between CtBP2 and FSHR expression and
the corresponding Genome-wide genotype, the phase III genotype of
these 210 unrelated individuals was downloaded from the HapMap
website. We used Plink software to carry out the association
analysis between CtBP2 and FSHR expression and the corresponding
Genome-wide genotype on Chinese (CHB) and Japanese (JPT)
populations. The annotation of these SNPs was performed in SNPfunc
(http://snpinfo.niehs.nih.gov/snpinfo/snpfunc.htm)
(20). CtBP2 binding data in LNCaP
cell lines was downloaded from CistromeMap database (18). All clinical and mRNA expression
levels (level 3 data, RNA-seq version 2) of PCa patients (PRAD)
were downloaded from TCGA data portal (https://tcga-data.nci.nih.gov/docs/publications/tcga/)
until August 29, 2015. The patients who suffered from other
malignancies or received neoadjuvant therapy were removed.
Cell culture
The prostate carcinoma cell lines LNCaP, DU145 and
PC3 were obtained from the American Type Culture Collection (ATCC
nos. HTB-81™, CRL-1435™, CRL-1740™). All cell lines were cultured
in T-75 flasks to 80% confluence at 37°C in a humidified incubator
containing 5% CO2 (Thermo Fisher Scientific). LNCaP
cells were grown in RPMI-1640 medium, DU145 and PC3 cells in
Dullbeccos modified Eagles medium (DMEM/F12; Hyclone, Logan, UT,
USA) both supplemented with 10% fetal bovine serum (FBS; Thermo
Fisher Scientific, Waltham, MA, USA) and 1% penicillin/streptomycin
(100 U/ml penicillin and 100 µg/ml streptomycin; Hyclone).
RNA interference assay
LNCaP, DU145 and PC3 cells were transfected with
various siRNAs targting CtBP2 (catalog nos.: CtBP2-Homo-451,
CtBP2-Homo-778, CtBP2-Homo-1021), and negative siRNA with a random
sequence was used as a control (Suzhou Genepharma Co., Ltd.,
Suzhou, China). The cells were seeded into 6-well plates at a
density of 5×105 cells/well, and cultured at 37°C with
5% CO2 until the cells reached 80% confluency.
Lipofectamine 2000 reagent (Thermo Fisher Scientific, Inc.) was
used for the transfections according to the manufacturer's
instructions. The siRNA sequences are shown in Table I.
 | Table I.Sequences of CtBP2-siRNA and negative
siRNA. |
Table I.
Sequences of CtBP2-siRNA and negative
siRNA.
| siRNA | Sequence, 5–3 |
|---|
|
CtBP2-siRNA-451 |
|
|
Sense |
GACAGCGAUUGGACAGAAUTT |
|
Antisense |
AUUCUGUCCAAUCGCUGUCTT |
|
CtBP2-siRNA-778 |
|
|
Sense |
GAAUUGCCGUGUGCAACAUTT |
|
Antisense |
AUGUUGCACACGGCAAUUCTT |
|
CtBP2-siRNA-1021 |
|
|
Sense |
CCUUUGGAUUCAGCGUCAUTT |
|
Antisense |
AUGACGCUGAAUCCAAAGGTT |
| Negative siRNA |
|
|
Sense |
UUCUCCGAACGUGUCACGUTT |
|
Antisense |
ACGUGACACGUUCGGAGAATT |
CtBP2 overexpression
LNCaP, DU145 and PC3 cells were cultured in 6-well
plates and transiently transfected with 2 µg of cDNA coding CtBP2,
and the vector pcDNA3.1 was cloned into the cells. Lipofectamine
2000 reagent was used for transfection according to the
manufacturer's protocol. The DNA was diluted in OptiMEM
(Invitrogen, Life Technologies Co., Carlsbad, CA, USA) combined
with Lipofectamine 2000, and incubated for 20 min at room
temperature. After incubation, the complex was added to the culture
medium of each cell line, and cultured at 37°C with 5%
CO2.
RT-PCR
The expression of CtBP2 was analyzed after
transfection. The primers for CtBP2 were as follows:
5′-ACTGTGGCCTTCTGTGACGC-3′ forward and 5′-CTGGTGAGGGTGATGGTGTG-3′
reverse. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used
as a normalization control and was also amplified, and the primers
were: 5′-TCATGAAGTGTGACGTGGACATC-3′ forward and
5′-CAGGAGGAGCAATGATCTTGATCT-3′ reverse. The PCR reaction conditions
were as follows: denaturation at 98°C for 3 min; 32 cycles of
denaturation at 98°C for 10 sec; annealing at 60°C for 30 sec; and
extension at 72°C for 1 min. The final extension was performed at
72°C for 5 min.
The real-time PCR reaction was performed in
biological triplicates using the Applied Biosystems StepOne Plus
real-time PCR system (Applied Biosystems, USA). Relative expression
values were calculated according to the 2−ΔΔCt method.
2−ΔΔCt represents the times ratio of target gene in the
experimental group and the control group, and the formula was as
follows: ΔΔCt = ΔCtexperimental group - ΔCtcontrol
group, where ΔCt = Cttarget gene -
CtGAPDH.
Western blot analysis
The protein levels of CtBP2 after transfection with
CtBP2-siRNA or pcDNA3.1 and the negative control were determined
using the Wes™ (ProteinSimple, San Jose, CA, USA) system. Wes
12–230 kDa Master kit with split buffer (ProteinSimple) was
selected for western analysis. Simple western analysis was carried
out at room temperature under instrument default settings. The
digital images were analyzed with Compass software (ProteinSimple)
on Wes.
Flow cytometry
LNCaP, DU145 and PC3 cells were collected 48 h after
transfection, and cell density was adjusted to 1×106/ml.
Cell suspension (0.5 ml) was placed in a centrifuge tube, and 1.25
µl of Annexin V-FITC (Nanjing KeyGEN Biotech Development Co., Ltd.)
was added to the centrifuge tube. The whole system was carried out
in a dark room at room temperature for 15 min, and centrifuged at
1,000 × g for 5 min. After the supernatant was discarded, the cells
were gently treated with 0.5 ml of cold-binding buffer to
re-suspend. Ten microliters of propidium iodide (PI) was used and
flow cytometry (BD Biosciences, Franklin Lakes, NJ, USA) was
immediately used for detection analysis.
Statistical analysis
Statistical analysis comparisons between groups were
evaluated by using independent-samples t-test in SPSS 22.0
statistical software (SPSS Inc., Chicago, IL, USA). P<0.05 was
indicative of statistical significance. All values are expressed as
mean ± SEM.
Results
CtBP2 function mining by bioinformatic
analysis
We used an online tool (http://bioinformatics.psb.ugent.be/webtools/Venn/) to
generate custom Venn/Euler diagrams for 4 datasets, including the
genes of two datasets associated between CtBP2 and FSHR expression
and HapMap genome-wide genotype, one dataset of binding with CtBP2
in CistromeMap database (18), and
one for the significant difference of mRNA expression between PCa
and adjacent tissues from TCGA. Overlapping genes (1,448) were
found in these 4 datasets (Fig.
1A). Pathway enrichment was performed using DAVID (https://david.ncifcrf.gov/) (21), and 32 pathways were found to be
enriched (Fig. 1B). Among these
enriched pathways, the top 6 pathways were calcium signaling
pathway, arrthythmogenic right ventricular cardiomyopathy (ARVC),
axon guidance, focal adhesion and vascular smooth muscle
contraction (VSMC) pathway. We further investigated 5 genes (CtBP2,
FSHR, VEGFA, FHL2 and SMAD3) in the 1,448 overlapping genes which
were closely correlated with angiogenesis (15–17,22–24).
The results showed that there were significant differences between
the expression of these 5 genes in the TCGA between PCa and normal
adjacent tissues (Fig. 1C).
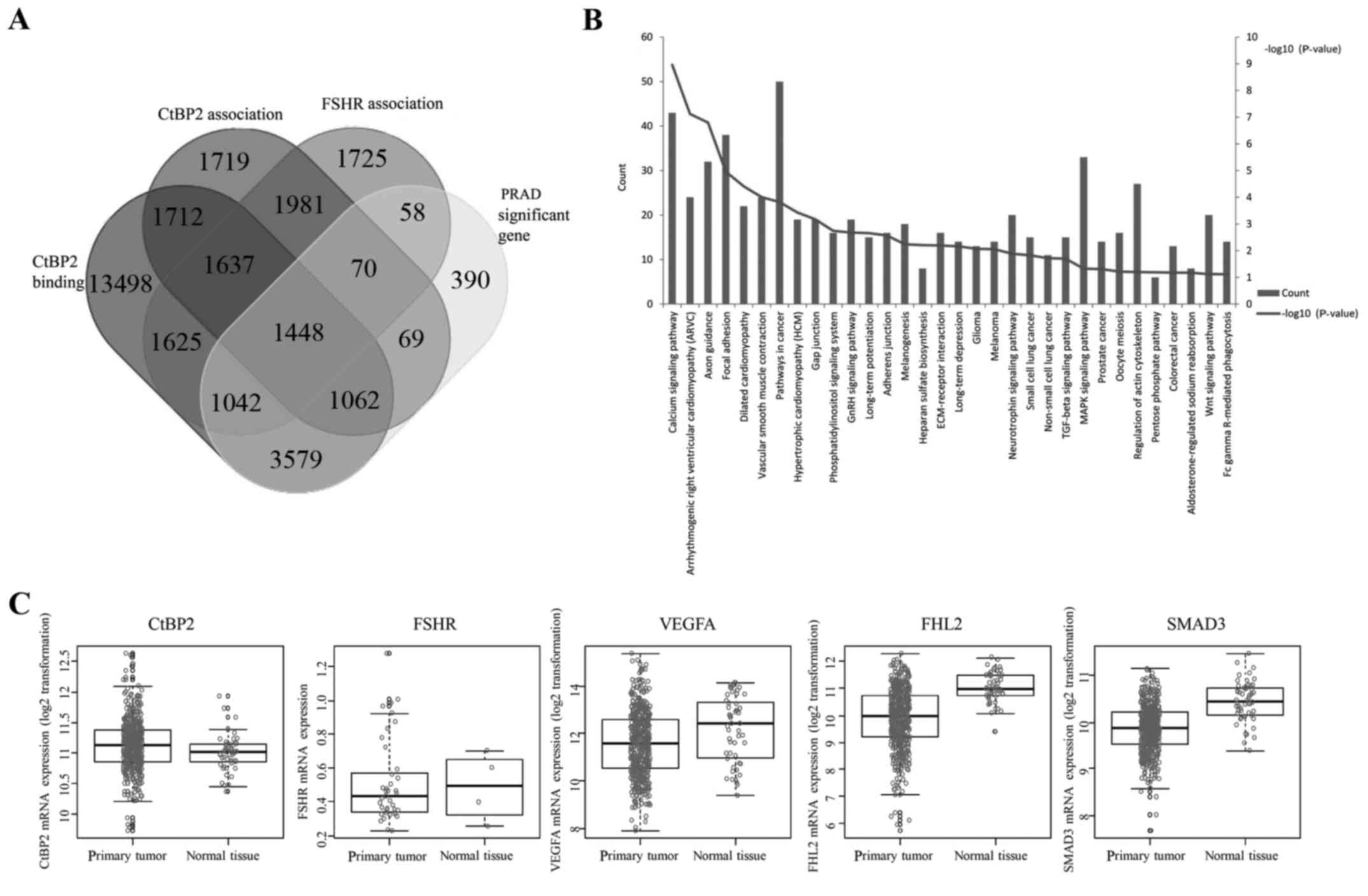 | Figure 1.CtBP2 function mining by
bioinformatic analysis. (A) Custom Venn/Euler diagrams for 4
datasets. Two datasets were the genes associated between CtBP2 or
FSHR expression and HapMap genome-wide genotype, one dataset was
the genes binding with CtBP2 in CistromeMap database (21), and one was the significant
difference in mRNA expression between prostate cancer tissue and
adjacent tissue from TCGA (PRAD). (B) Thirty-two pathways enriched
in DAVID. (C) CtBP2, FSHR, VEGFA, FHL2 and SMAD3 expression between
prostate cancer and adjacent tissues from TCGA. CtBP, C-terminal
binding protein; FSHR, follicle stimulating hormone receptor; VEGF,
vascular endothelial growth factor; FHL2, four and a half LIM
domains 2; SMAD3, SMAD family member 3. |
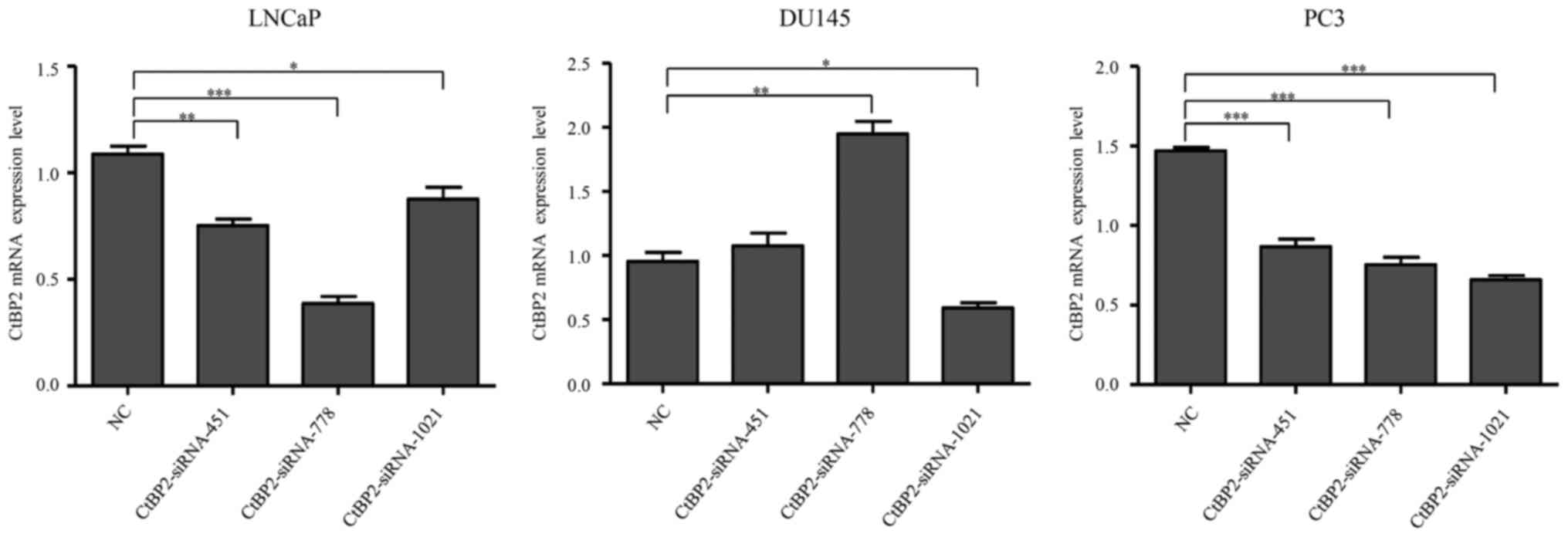
Knockdown of the CtBP2 gene using
siRNAs in the PCa cell lines
In order to ascertain the potential function of
CtBP2 in PCa, we uses siRNAs to interfere with endogenous CtBP2
gene expression (gene ID, 1488). Overall, 3 siRNAs were transfected
into LNCaP, DU145 and PC-3 cells, respectively, with transfection
of the negative siRNA as control. Twenty-four hours after
transfection of the siRNAs, CtBP2 mRNA expression was detected by
RT-PCR. The results showed that, compared to the NC group,
CtBP2-siRNA-451, CtBP2-siRNA-778 and CtBP2-siRNA-1021 significantly
decreased CtBP2 expression in the LNCaP and PC-3 cells, while
CtBP2-siRNA-451 and CtBP2-siRNA-778 increased CtBP2 expression in
the DU145 cell lines, and only CtBP2-siRNA-1021 decreased CtBP2 in
all cell lines. Therefore, CtBP2-siRNA-1021 was selected for the
subsequent experiments (Fig.
2).
CtBP2 expression in LNCaP, DU145 and
PC-3 cells after knockdown and overexpression
CtBP2-siRNA-1021 was transfected into the LNCaP,
DU145 and PC-3 cells, with negative siRNA used as the control.
Twenty-four hours after transfection of CtBP2-siRNA-1021, CtBP2
expression was detected by qRT-PCR and the Wes system. Except for
the Wes system of CtBP2 in the LNCaP cells which did not achieve a
significant difference, RT-PCR and the Wes system results in the
other cell lines demonstrated a obvious decrease in CtBP2 compared
to the NC group (Fig. 3A and B).
Meanwhile, overexpression of CtBP2 in the LNCaP, DU145 and PC-3
cells with the expression plasmid pcDNA3.1 showed opposite results.
Except for CtBP2 in the LNCaP cells as detected by the Wes system,
RT-PCR and Wes system results in other cell lines demonstrated a
notable increase in CtBP2 compared to the NC group (Fig. 3C and D).
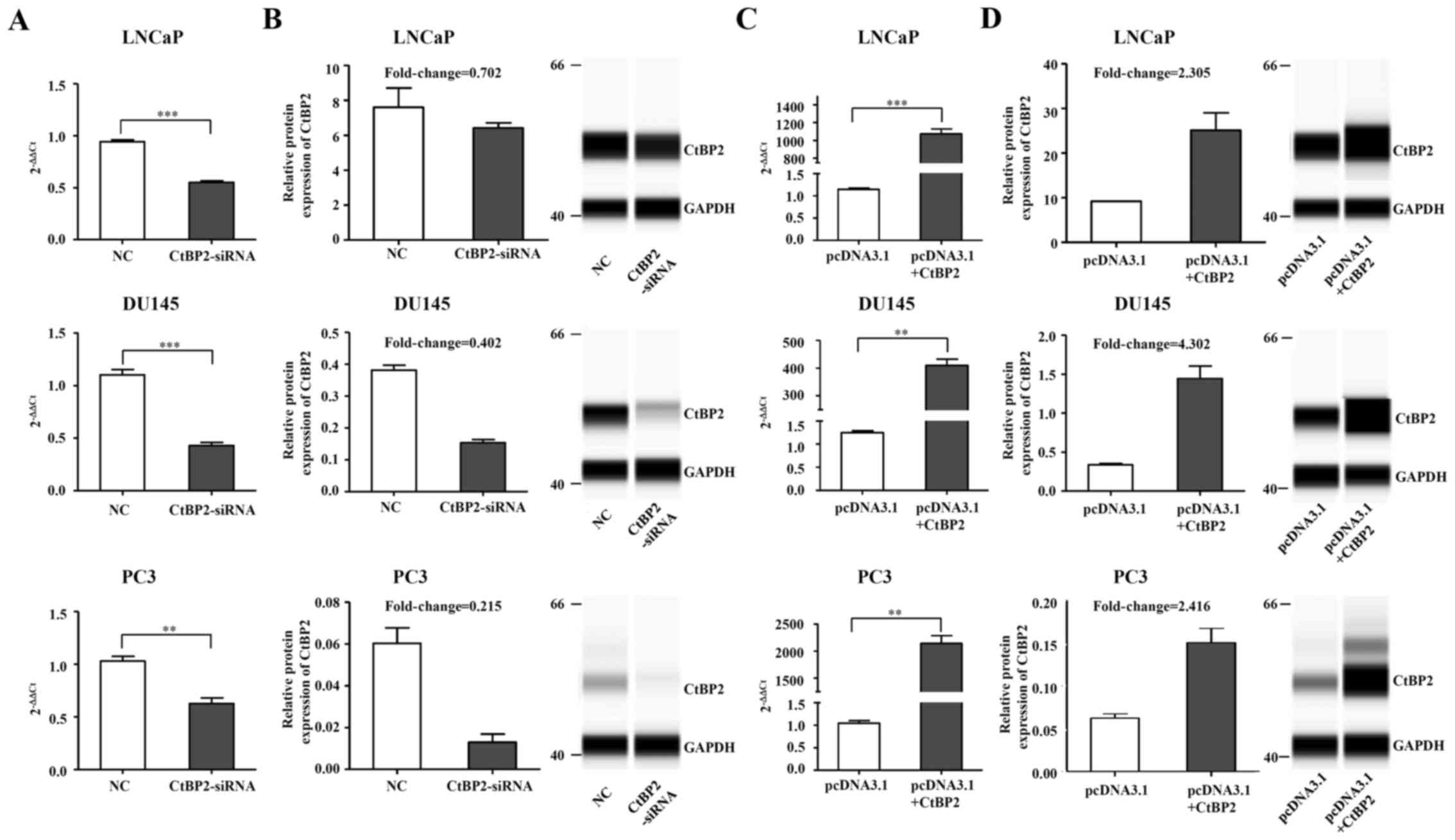 | Figure 3.CtBP2 expression in LNCaP, DU145 and
PC-3 cells after knockdown and overexpression. (A) LNCaP, DU145 and
PC3 cells were transfected with CtBP2-siRNA-1021 and negative siRNA
as control. CtBP2 mRNA expression was confirmed by RT-PCR. (B)
Knockdown of CtBP2 protein expression was detected by Wes system in
the LNCaP, DU145 and PC3 cells. GAPDH was included as a
normalization control. (C) LNCaP, DU145 and PC3 cells were
transfected with pcDNA3.1 containing CtBP2 and empty vector as
control. RT-PCR was used to detect CtBP2 mRNA expression. (D)
Overexpression of CtBP2 protein was confirmed by Wes system in the
LNCaP, DU145 and PC3 cells. The data are expressed as mean ± SEM.
*P<0.05, **P<0.01, ***P<0.001. NC, group transfected with
negative siRNA; CtBP2-siRNA, group transfected with
CtBP2-siRNA-1021; pcDNA3.1, group transfected with empty vector;
pcDNA3.1+CtBP2, group transfected with pcDNA3.1 containing CtBP2.
CtBP, C-terminal binding protein. |
Expression levels of CtBP2, FSHR,
VEGFA, FHL2 and SMAD3 in PCa cells
qPCR was used to detect the expression of CtBP2,
FSHR, VEGFA, FHL2 and SMAD3 after transfection of CtBP2-siRNA-1021
into the LNCaP, DU145 and PC-3 cells (Table II). In the LNCaP cells, FSHR, VEGFA
and FHL2 mRNA levels were significantly decreased in the
CtBP2-siRNA group when compared with levels in the NC group
(p<0.001). In the DU145 cells, FSHR, VEGFA and SMAD3 showed a
significant decrease between the CtBP2-siRNA group and the NC group
(p<0.05). In the PC-3 cells, VEGFA and FHL2 showed an obviously
significant difference (p≤0.001) (Fig.
4A).
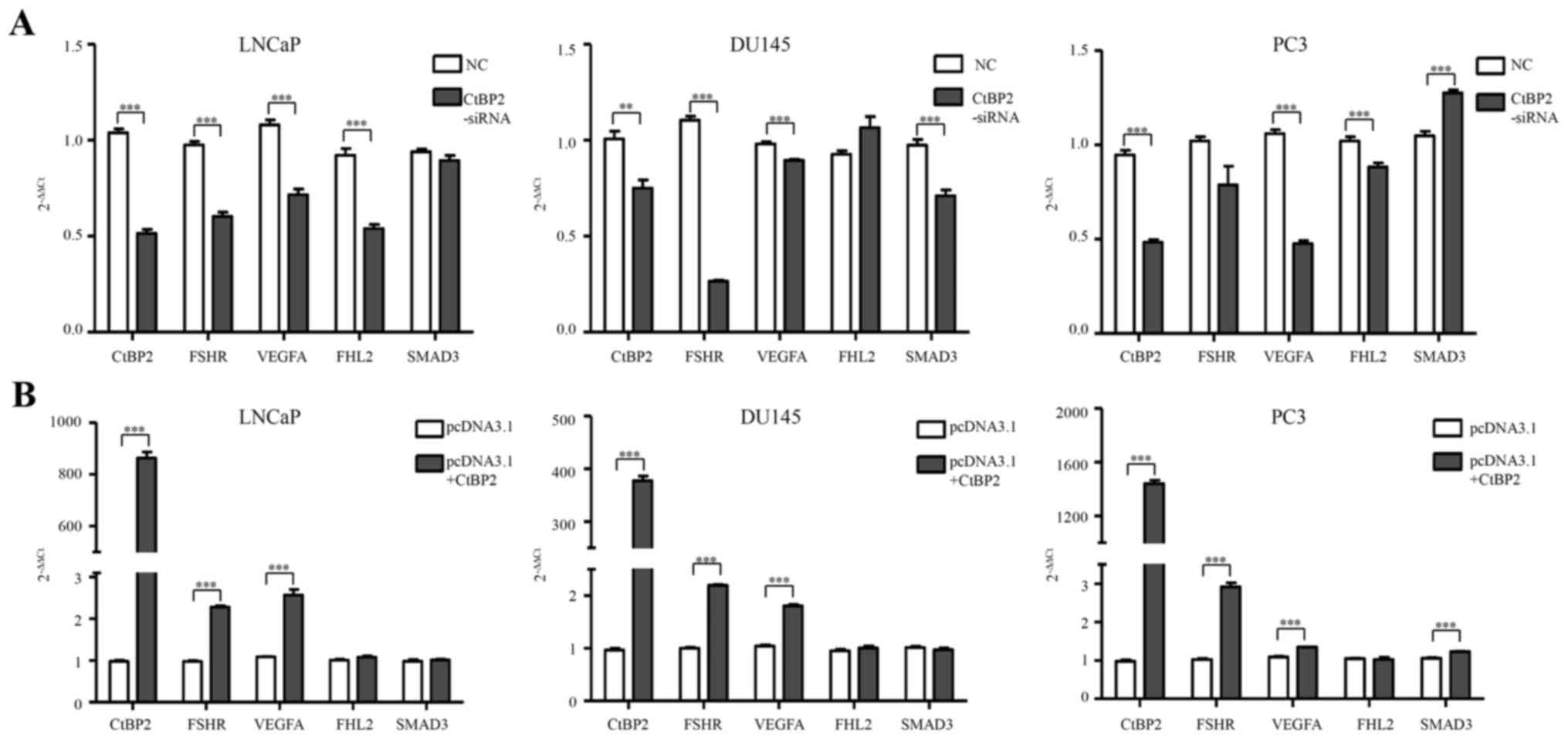 | Figure 4.Expression levels of CtBP2, FSHR,
VEGFA, FHL2 and SMAD3 in PCa cells. (A) qPCR was used to detect the
expression of CtBP2, FSHR, VEGFA, FHL2 and SMAD3 after transfection
with CtBP2-siRNA-1021 and negative siRNA in the LNCaP, DU145 and
PC3 cells. (B) qPCR was used to detect the expression of CtBP2,
FSHR, VEGFA, FHL2 and SMAD3 after transfection with pcDNA3.1
containing CtBP2 and empty vector in LNCaP, DU145 and PC3 cells.
Data are expressed as mean ± SEM. *P<0.05, **P<0.01,
***P<0.001. NC, group transfected with negative siRNA;
CtBP2-siRNA, group transfected with CtBP2-siRNA-1021; pcDNA3.1,
group transfected with empty vector; pcDNA3.1+CtBP2, group
transfected with pcDNA3.1 containing CtBP, C-terminal binding
protein; FSHR, follicle stimulating hormone receptor; VEGF,
vascular endothelial growth factor; FHL2, four and a half LIM
domains 2; SMAD3, SMAD family member 3. PCa, prostate cancer. |
 | Table II.Genes and their primer sequences from
qPCR. |
Table II.
Genes and their primer sequences from
qPCR.
| Genes | Primers |
|---|
| CtBP2 | F:
ACTGTGGCCTTCTGTGACGC |
|
| R:
CTGGTGAGGGTGATGGTGTG |
| FSHR | F:
AGCCTCTGGACCAGTCATTC |
|
| R:
ACAGCAATGGCTGGGATAGG |
| VEGFA | F:
TCAGCGCAGCTACTGCCATC |
|
| R:
ACACTCCAGGCCCTCGTCATTG |
| FHL2 | F:
CAGACTGCTATTCCAACGAG |
|
| R:
TGGCAGATGAAGCAGGTCTC |
| SMAD3 | F:
TACCAGAGAGTAGAGACACCAG |
|
| R:
ATGGAATGGCTGTAGTCGTC |
| IL-8 | F:
TTGGCAGCCTTCCTGATTTC |
|
| R:
ATTTCTGTGTTGGCGCAGTG |
| ATR2 | F:
ATTGCTTCAGCCAGCGTCAG |
|
| R:
TAGCTGGCAAACTGGCCAAG |
| MMP-9 | F:
ACCACGGCCAACTACGACAC |
|
| R:
GTGCAGGCGGAGTAGGATTG |
| CCND1 | F:
ATGGAACACCAGCTCCTGTG |
|
| R:
TCAGATGTCCACGTCCCGCA |
After pcDNA3.1-CtBP2 transfection, FSHR and VEGFA
mRNA levels were significantly increased in the LNCaP cells
compared to levels in the pcDNA3.1+CtBP2 and pcDNA3.1 groups
(p<0.001). A significant difference in FSHR and VEGFA mRNA
expression was noted in the DU145 cells (p<0.001). FSHR, VEGFA
and SMAD3 mRNA expression also showed a significant difference
(p<0.001) in the PC-3 cells (Fig.
4B).
Taken together, interference with the expression of
CtBP2 effectively affected the expression of FSHR and VEGFA. This
indicates that CtBP2 may play an important role in
angiogenesis.
Cell apoptosis of LNCaP, DU145 and
PC-3 cells
Forty-eight hours after transfection, flow cytometry
was used to determine the cell apoptotic rate. CtBP2-siRNA-1021
promoted cell apoptosis in the LNCaP, DU145 and PC-3 cells
(Fig. 5A). There was a significant
increased in the cell apoptosis rate in the CtBP2-siRNA group
compared to that in the NC group in the LNCaP and DU145 cells
(t=18.377, p=0.024 and t=15.080, p=0.006, respectively). However,
there was no significant difference in the cell apoptosis rate
between the CtBP2-siRNA group and the NC group in the PC-3 cells
(p=0.250) (Fig. 5B).
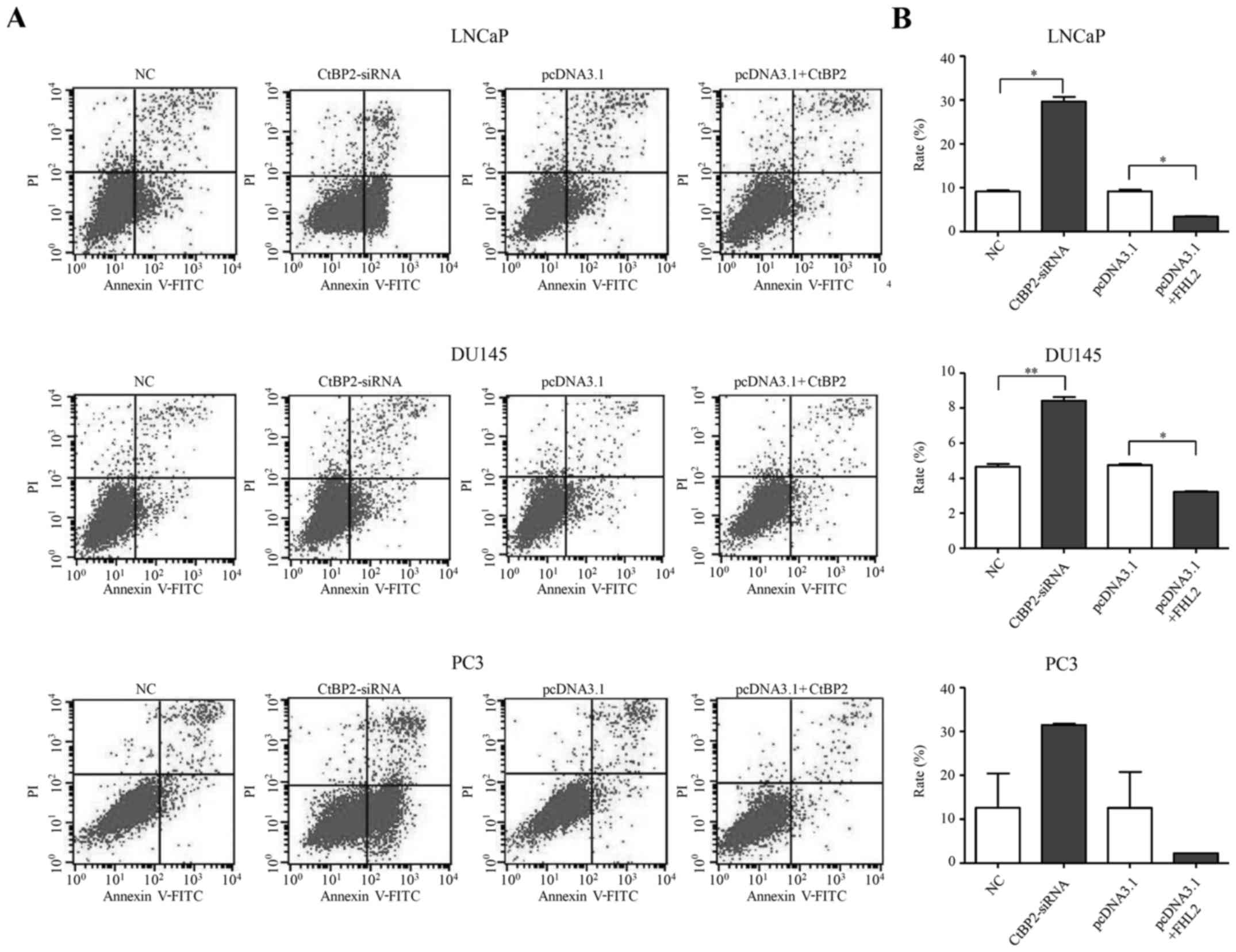 | Figure 5.Cell apoptosis of LNCaP, DU145 and
PC-3 cells. (A) The ratio of apoptosis after transfection with
CtBP2-siRNA-1021 or pcDNA3.1 containing CtBP2 in LNCaP, DU145 and
PC3 cells. (B) Comparison of the ratio of apoptosis after
transfection with CtBP2-siRNA-1021 or pcDNA3.1 containing CtBP2 in
LNCaP, DU145 and PC3 cells. Data are expressed as the mean ± SEM.
*P<0.05, **P<0.01, ***P<0.001. NC, group transfected with
negative siRNA; CtBP2-siRNA, group transfected with
CtBP2-siRNA-1021; pcDNA3.1, group transfected with empty vector;
pcDNA3.1+CtBP2, group transfected with pcDNA3.1 containing CtBP2.
CtBP, C-terminal binding protein. |
CtBP2 overexpression inhibited the apoptosis levels
in the LNCaP, DU145 and PC-3 cells 48 h after transfection
(Fig. 5A). There was a significant
decreased in the cell apoptosis rate between the pcDNA3.1+CtBP2
group and the pcDNA3.1 group in the LNCaP and DU145 cells
(t=16.423, p=0.035 and t=15.080, p=0.006, respectively). There was
also no significant difference between the cell apoptosis rate in
the pcDNA3.1+CtBP2 group and the pcDNA3.1 group in the PC-3 cells
(p=0.427) (Fig. 5B).
Expression of downstream signaling
markers in LNCaP, DU145 and PC3 cells
To identify the biological consequences of CtBP2
interference, we next investigated the expression of subsequent
downstream markers including IL-8, AT2R, CCND1 and MMP9. After
knockdown and overexpression of CtBP2 in LNCaP, DU145 and PC-3
cells, qPCR was used to detect the expression of IL-8, AT2R, CCND1
and MMP9. The primers of CtBP2-associated downstream genes are
shown in Table II. IL-8, AT2R,
CCND1 and MMP9 were significantly decreased between the CtBP2-siRNA
and the NC group in the LNCaP, DU145 and PC-3 cells (Fig. 6A). In regards to pcDNA3.1 containing
CtBP2, IL-8, AT2R, CCND1 and MMP9 showed a significant increase in
the LNCaP and PC-3 cells compared with the pcDNA3.1 group.
Additionally, IL-8, AT2R and MMP9 were significantly increased in
the DU145 cells compared with that in the pcDNA3.1 group (Fig. 6B).
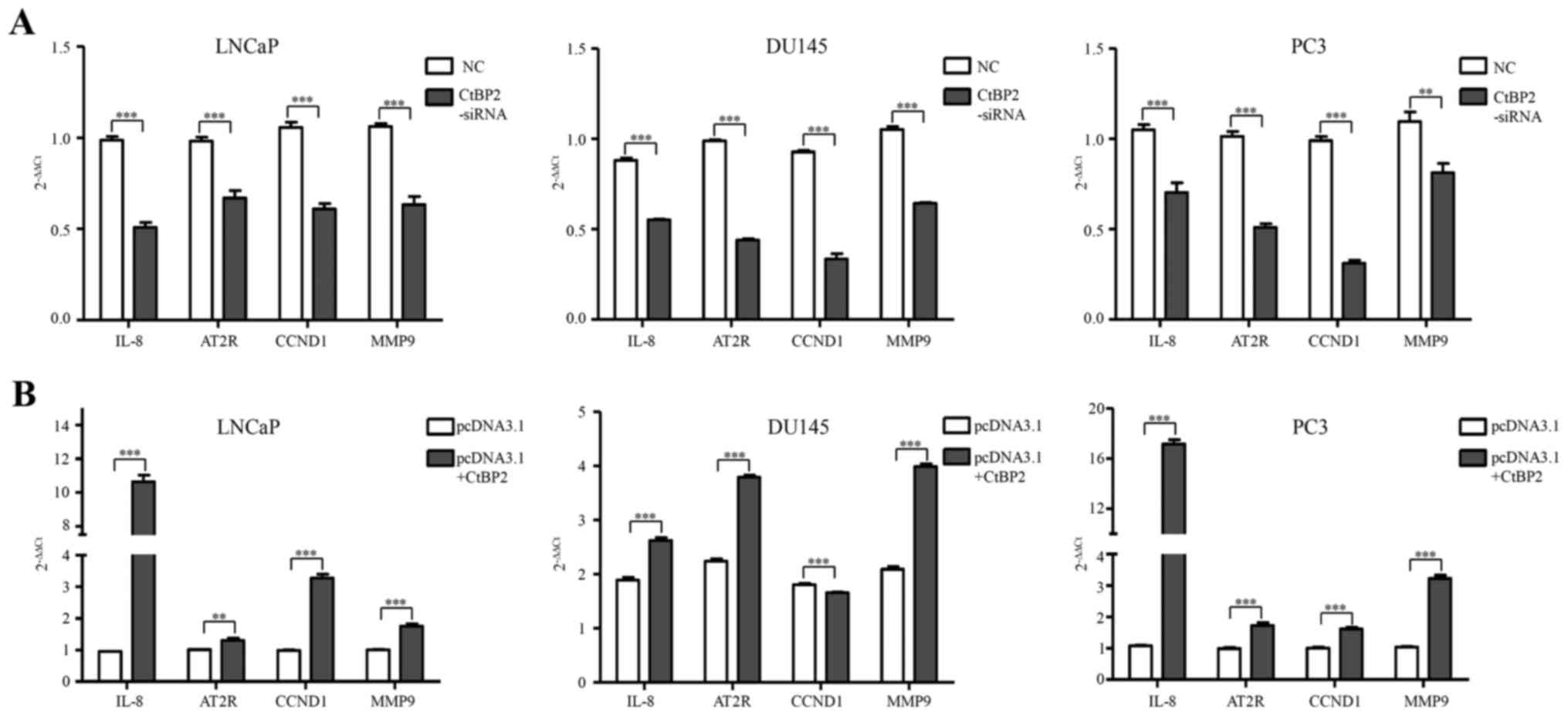 | Figure 6.Expression of downstream signaling
markers in LNCaP, DU145 and PC3 cells. (A) siRNA knockdown of CtBP2
expression inhibited downstream signaling in LNCaP, DU145 and PC3
cells. qPCR was used to detect the expression of IL-8, AT2R, CCND1
and MMP9. (B) CtBP2 overexpression promoted downstream signaling in
LNCaP, DU145 and PC3 cells. qPCR was used to detect the expression
of IL-8, AT2R, CCND1 and MMP9. Data are expressed as mean ± SEM.
*P<0.05, **P<0.01, ***P<0.001. NC, group transfected with
negative siRNA; CtBP2-siRNA, group transfected with
CtBP2-siRNA-1021; pcDNA3.1, group transfected with empty vector;
pcDNA3.1+CtBP2, group transfected with pcDNA3.1 containing CtBP2.
CtBP, C-terminal binding protein; IL-8, interleukin 8; AT2R,
angiotensin II receptor type 2; CCND1, cyclin D1; MMP9, matrix
metallopeptidase 9. |
Discussion
CtBP2 is a transcriptional co-repressor which has
been observed to mediate the repression of the tumor-suppressor
gene product, p16(INK4A) (10).
Takayama et al (13)
demonstrated that CtBP2 modulated the androgen receptor to promote
PCa cell proliferation through c-Myc signaling and also promotes
PCa progression (11). Aberrant
expression of CtBP2 has been found to be associated with
tumorigenesis, tumor progression and poor prognosis of PCa
(12,26). In this study, we provide evidence
that CtBP2 is closely related with angiogenesis in PCa, and its
expression change significantly affects the apoptosis of PCa cells
(LNCaP, DU145 and PC-3) and the downstream markers IL-8, AT2R and
MMP9.
Formation of functional vasculature is an essential
requirement for most physiological processes and the growth of
mammalian tissues. Angiogenesis, the formation of new blood vessels
from pre-existing vasculature, is important for tumor development
and progression (27). Our
bioinformatic analysis indicated that the top 6 pathways of
CtBP2-related genes were correlated with angiogenesis. VEGF and its
receptors VEGFR1/VEGFR2 play major roles in controlling
angiogenesis, including vascularization of solid tumors.
VEGF-induced neoangiogenesis is mediated by NAADP and two-pore
channel-2-dependent Ca2+ signaling (27). Arrhythogenic ARVC and VSMC pathways
are closely related with the vascular system. The principal
mechanisms that regulate the contractile state of VSMCs are changes
in the concentration of cytosolic Ca2+
([Ca2+]c) (28). Axon guidance represents a key stage
in the formation of neuronal network. Meucci et al (29) identified that Axon guidance and
focal adhesion pathways regulate cell motility, tumor invasion and
angiogenesis leading to less aggressive tumors in older age
patients. Dilated cardiomyopathy pathway is also reported to be
related to angiogenesis (30).
The 5 overlapping genes (CtBP2, FSHR, VEGFA, FHL2
and SMAD3) obtained by bioinformatic analysis showed that there was
a significant expression difference in these genes between PCa and
normal adjacent tissues. Following knockdown and overexpression of
CtBP2, the expression of FSHR and VEGFA were notably affected in
all cell lines. In most of the experiments, the expression of FHL2
and SMAD3 was also markedly affected by CtBP2 interference. VEGFA
can induce proliferation and migration of vascular endothelial
cells, and is essential for both physiological and pathological
angiogenesis (31). FSH is as
efficatious as VEGF which is a well-characterized angiogenic factor
in promoting angiogenic processes, and is a highly selective tumor
vasculature marker, which is abundant in both primary and
metastatic tumors (16,17). FHL2 serves a repressor function in
cardiomyocytes through its ability to inhibit ERK1/2
transcriptional coupling (23).
SMAD3 is essential for signaling of transforming growth factor β
(TGF-β) pathway. A role for TGF-β in modulating VEGF release has
been proposed in several cell types, such as glioblastoma (24). These results indicate that CtBP2
plays an important role in angiogenesis.
The association between CtBP2 overexpression and
tumorigenesis and poor clinical outcome and tumor progression of
PCa has been reported (11,12). In this study, we further
demonstrated that the alteration in CtBP2 expression obviously
affected the apoptosis of PCa cells (LNCaP, DU145 and PC3). Zhang
et al (25) revealed that
suppressed expression of CtBP2 in PCa PC3 cells markedly inhibited
cell proliferation by inducing apoptosis in vitro. In this
study, we further confirmed that inhibition of CtBP2 induced
apoptosis in PCa cells (LNCaP, DU145 and PC3). On the contrary,
upregulation of CtBP2 inhibited the apoptosis of PCa cells. A more
recent study demonstrated a critical function for CtBPs in the
transcriptional repression of pro-apoptotic genes such as Bax,
Puma, Bik, and Noxa (32).
In conclusion, CtBP2 expression is associated with
PCa development and progression. We integrated and analyzed 4
datasets from CtBP2 and FSHR GWAS, CtBP2 binding data and the
significant difference in PCa mRNA from the TCGA database, and
performed pathway enrichment. We revealed that the top 6 pathways
were closely related with angiogenesis. CtBP2 interference
indicated that its expression alteration also affected the
expression of VEGFA, FSHR, FHL2 and SMAD3 which are closely related
with angiogenesis. In addition, CtBP2 markedly affected the
apoptosis of PCa cells in vitro, and impacted the expression
of IL-8, AT2R, CCND1 and MMP9 which are associated with cancer
progression. These results highlight the association between CtBP2
and angiogenesis in PCa and indicate that CtBP2 may be a potential
therapeutic target for PCa. The molecular mechanisms underlying the
regulation of angiogenesis by CtBP2 warrant further in vitro
and in vivo investigation.
Acknowledgements
This research was supported by grants from the Anhui
Natural Science Foundation (no. 1408085MH181), Anhui Educational
Departmental Function (KJ2012 Z181), National Natural Science
Foundation of China (nos. 81272853 and 81472414), and the Guangxi
Natural Science Foundation (nos. 2015GXNSFBB139008 and
2014GXNSFBA118201).
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
DiBlasio CJ, Malcolm JB, Hammett J, Wan
JY, Aleman MA, Patterson AL, Wake RW and Derweesh IH: Survival
outcomes in men receiving androgen-deprivation therapy as primary
or salvage treatment for localized or advanced prostate cancer:
20-year single-centre experience. BJU Int. 104:1208–1214. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ramsay AK, McCracken SR, Soofi M, Fleming
J, Yu AX, Ahmad I, Morland R, Machesky L, Nixon C, Edwards DR, et
al: ERK5 signalling in prostate cancer promotes an invasive
phenotype. Br J Cancer. 104:664–672. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Maruyama Y, Miyazaki T, Ikeda K, Okumura
T, Sato W, Horie-Inoue K, Okamoto K, Takeda S and Inoue S: Short
hairpin RNA library-based functional screening identified ribosomal
protein L31 that modulates prostate cancer cell growth via p53
pathway. PLoS One. 9:e1087432014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Turner J and Crossley M: The CtBP family:
Enigmatic and enzymatic transcriptional co-repressors. BioEssays.
23:683–690. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chinnadurai G: CtBP, an unconventional
transcriptional corepressor in development and oncogenesis. Mol
Cell. 9:213–224. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
May T, Yang J, Shoni M, Liu S, He H, Gali
R, Ng SK, Crum C, Berkowitz RS and Ng SW: BRCA1 expression is
epigenetically repressed in sporadic ovarian cancer cells by
overexpression of C-terminal binding protein 2. Neoplasia.
15:600–608. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Deng H, Liu J, Deng Y, Han G, Shellman YG,
Robinson SE, Tentler JJ, Robinson WA, Norris DA, Wang XJ, et al:
CtBP1 is expressed in melanoma and represses the transcription of
p16INK4a and Brca1. J Invest Dermatol. 133:1294–1301.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Di LJ, Byun JS, Wong MM, Wakano C, Taylor
T, Bilke S, Baek S, Hunter K, Yang H, Lee M, et al: Genome-wide
profiles of CtBP link metabolism with genome stability and
epithelial reprogramming in breast cancer. Nat Commun. 4:14492013.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Guan C, Shi H, Wang H, Zhang J, Ni W, Chen
B, Hou S, Yang X, Shen A and Ni R: CtBP2 contributes to malignant
development of human esophageal squamous cell carcinoma by
regulation of p16INK4A. J Cell Biochem. 114:1343–1354.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Debiais-Delpech C, Godet J, Pedretti N,
Bernard FX, Irani J, Cathelineau X, Cussenot O and Fromont G:
Expression patterns of candidate susceptibility genes HNF1β and
CtBP2 in prostate cancer: Association with tumor progression. Urol
Oncol. 32:426–432. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang C, Li S, Qiao B, Yang K, Liu R, Ma
B, Liu Y, Zhang Z and Xu Y: CtBP2 overexpression is associated with
tumorigenesis and poor clinical outcome of prostate cancer. Arch
Med Sci. 11:1318–1323. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Takayama K, Suzuki T, Fujimura T, Urano T,
Takahashi S, Homma Y and Inoue S: CtBP2 modulates the androgen
receptor to promote prostate cancer progression. Cancer Res.
74:6542–6553. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ferrara N and Kerbel RS: Angiogenesis as a
therapeutic target. Nature. 438:967–974. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Roukens MG, Alloul-Ramdhani M, Baan B,
Kobayashi K, Peterson-Maduro J, van Dam H, Schulte-Merker S and
Baker DA: Control of endothelial sprouting by a Tel-CtBP complex.
Nat Cell Biol. 12:933–942. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Stilley JA, Guan R, Duffy DM and Segaloff
DL: Signaling through FSH receptors on human umbilical vein
endothelial cells promotes angiogenesis. J Clin Endocrinol Metab.
99:E813–E820. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yang D, Feng L, Dougherty CA, Luker KE,
Chen D, Cauble MA, Holl Banaszak MM, Luker GD, Ross BD, Liu Z, et
al: In vivo targeting of metastatic breast cancer via tumor
vasculature-specific nano-graphene oxide. Biomaterials.
104:361–371. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Qin B, Zhou M, Ge Y, Taing L, Liu T, Wang
Q, Wang S, Chen J, Shen L, Duan X, et al: CistromeMap: A
knowledgebase and web server for ChIP-Seq and DNase-Seq studies in
mouse and human. Bioinformatics. 28:1411–1412. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Stranger BE, Forrest MS, Dunning M, Ingle
CE, Beazley C, Thorne N, Redon R, Bird CP, de Grassi A, Lee C, et
al: Relative impact of nucleotide and copy number variation on gene
expression phenotypes. Science. 315:848–853. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang P, Dai M, Xuan W, McEachin RC,
Jackson AU, Scott LJ, Athey B, Watson SJ and Meng F: SNP Function
Portal: A web database for exploring the function implication of
SNP alleles. Bioinformatics. 22:e523–e529. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Dennis G Jr, Sherman BT, Hosack DA, Yang
J, Gao W, Lane HC and Lempicki RA: DAVID: Database for Annotation,
Visualization, and Integrated Discovery. Genome Biol. 4:32003.
View Article : Google Scholar
|
|
22
|
Joyal JS, Sun Y, Gantner ML, Shao Z, Evans
LP, Saba N, Fredrick T, Burnim S, Kim JS, Patel G, et al:
Corrigendum: Retinal lipid and glucose metabolism dictates
angiogenesis through the lipid sensor Ffar1. Nat Med. 22:6922016.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Purcell NH, Darwis D, Bueno OF, Müller JM,
Schüle R and Molkentin JD: Extracellular signal-regulated kinase 2
interacts with and is negatively regulated by the LIM-only protein
FHL2 in cardiomyocytes. Mol Cell Biol. 24:1081–1095. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Seystahl K, Tritschler I, Szabo E,
Tabatabai G and Weller M: Differential regulation of TGF-β-induced,
ALK-5-mediated VEGF release by SMAD2/3 versus SMAD1/5/8 signaling
in glioblastoma. Neuro Oncol. 17:254–265. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang C, Gao C, Xu Y and Zhang Z: CtBP2
could promote prostate cancer cell proliferation through c-Myc
signaling. Gene. 546:73–79. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chung AS and Ferrara N: Developmental and
pathological angiogenesis. Annu Rev Cell Dev Biol. 27:563–584.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Favia A, Desideri M, Gambara G, D'Alessio
A, Ruas M, Esposito B, Del Bufalo D, Parrington J, Ziparo E,
Palombi F, et al: VEGF-induced neoangiogenesis is mediated by NAADP
and two-pore channel-2-dependent Ca2+ signaling. Proc
Natl Acad Sci USA. 111:E4706–E4715. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Akata T: Cellular and molecular mechanisms
regulating vascular tone. Part 2: Regulatory mechanisms modulating
Ca2+ mobilization and/or myofilament Ca2+
sensitivity in vascular smooth muscle cells. J Anesth. 21:232–242.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Meucci S, Keilholz U, Tinhofer I and Ebner
OA: Mutational load and mutational patterns in relation to age in
head and neck cancer. Oncotarget. 7:69188–69199. 2016.PubMed/NCBI
|
|
30
|
Yue X, Lin X, Yang T, Yang X, Yi X, Jiang
X, Li X, Li T, Guo J, Dai Y, et al: Rnd3/RhoE modulates
hypoxia-inducible factor 1α/vascular endothelial growth factor
signaling by stabilizing hypoxia-inducible factor 1α and regulates
responsive cardiac angiogenesis. Hypertension. 67:597–605.
2016.PubMed/NCBI
|
|
31
|
Aryal B, Shimizu T, Kadono J, Furoi A,
Komokata T, Inoue M, Ikeda S, Fukukura Y, Nakamura M, Yamakuchi M,
et al: A switch in the dynamics of intra-platelet VEGF-A from
cancer to the later phase of liver regeneration after partial
hepatectomy in humans. PLoS One. 11:e01504462016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Stankiewicz TR, Gray JJ, Winter AN and
Linseman DA: C-terminal binding proteins: Central players in
development and disease. Biomol Concepts. 5:489–511. 2014.
View Article : Google Scholar : PubMed/NCBI
|