Introduction
Esophageal carcinoma (EC) is a common malignancy
that affects >450,000 individuals worldwide and causes high
morbidity and mortality. Esophageal squamous cell carcinoma (ESCC)
and esophageal adenocarcinoma are the two major histologic types of
EC (1). ESCC is a frequent type of
histopathology in China, especially in Xinjiang and Northwestern
regions, where ESCC in Kazakhs is abnormally higher than that in
other minorities (2). Although
treatment methods have improved, ESCC has become a major public
health concern because of its increasing mortality. Therefore, we
should further explore the molecular mechanisms related to the
pathogenesis of EC to develop effective treatments.
MicroRNAs (miRNAs) are a class of small (21–25
nucleotides) naturally occurring non-coding RNAs. miRNAs can
perform dual functions as a tumor promotor or suppressor by
targeting different mRNAs or dysregulating their expression levels.
Their regulatory mechanisms occur through the posttranscriptional
repression of their target genes by targeting 3′UTR (3). miRNAs are highly and extensively
regulated, and miRNA binding sites can be present in 5′UTR and
within coding sequences (4). miRNAs
are highly conserved in species (5). With tissue specificity, miRNAs are
suitable for early cancer detection and prognosis prediction
(6). Mature miRNAs play important
regulatory roles in cell proliferation, differentiation, and death
(7). Therefore, miRNAs can be used
as biomarkers to diagnose and treat diseases, including ESCC, whose
early recognition is crucial. The role of miRNAs in the
pathological process of ESCC has also been widely examined
(8). For instance, Chen et
al (9) used microarrays to
examine the miRNA expression profiles of 119 paired ESCC tissue
samples and indicated a four-miRNA signature that potentially
predicts patient survival. Takeshita et al performed a miRNA
array by using serum samples from ESCC patients and found that
miR-1246 can be strongly utilized as a novel diagnostic and
prognostic ESCC biomarker (10).
The role of miRNAs has also been demonstrated in the diagnostic and
prognostic uses of circulation in ESCC patients.
Metastasis and proliferation are related to cancer
prognosis, and various steps of metastasis involve several miRNAs
(11). miRNA arrays have been used
to determine the functions of dysregulated miRNAs in cancer.
However, we have yet to reveal the mechanisms on how to effectively
screen key miRNAs in dysregulated genes. First, we aimed to examine
the miRNA microarray platforms in ESCC tissue samples and evaluate
the miRNA expression levels by using an ESCC cell line
subpopulation with high invasive ability and an ESCC cell line
treated with the anticancer drug nedaplatin (NDP). We also
performed qRT-PCR to explore the miRNAs involved in cell invasion
and proliferation. Second, we applied a systems bioinformatics
approach to investigate the biological characteristics of
dysregulated miRNAs in ESCC and to elucidate their molecular
mechanisms in tumor development and invasion. Finally, we selected
relevant functional miRNAs.
Materials and methods
Patients and specimens
ESCC tissues and related non-tumor normal esophageal
tissues were obtained from 8 Chinese-Kazakh ESCC patients
undergoing esophagectomy in 2015 at The First Affiliated Hospital
of the Medical College, Shihezi University. None of these patients
received pre-surgical radiochemotherapy. The excised specimens were
quickly frozen and stored in liquid nitrogen until RNA was
extracted. A pathologist in The First Affiliated Hospital of the
Medical College, Shihezi University provided distinct histological
confirmation of the ESCC diagnosis. The clinicopathological
information of all of the patients is shown in Table I. This study was approved by the
ethics committees of The First Affiliated Hospital of the Medical
College, Shihezi University, and informed consent was obtained from
each patient.
 | Table I.Clinical features of patient with
ESCC. |
Table I.
Clinical features of patient with
ESCC.
| No. | Sex | Age | Locationa | UICC Tb | UICC Nb | TNM staging | Tumor
gradec |
|---|
| 1 | M | 72 | Lt | 4 | 3 | IIIC | M |
| 2 | M | 53 | Lt | 3 | 0 | IIA | M |
| 3 | F | 67 | Mt | 2 | 0 | IIA | M |
| 4 | M | 44 | Mt | 2 | 0 | IIA | P |
| 5 | F | 64 | Mt | 2 | 0 | IIA | W |
| 6 | F | 58 | Lt | 3 | 1 | IIIA | M |
| 7 | F | 72 | UtMt | 3 | 0 | IIB | W |
| 8 | F | 77 | Lt | 2 | 0 | IIA | W |
Agilent miRNA array and computational
analysis
ESCC tissues were transported to Shanghai
Biotechonology Co., Ltd. (Shanghai, China) for miRNA isolation and
microarray assays were performed by using an Agilent Human miRNA
microarray platform (design ID: 38169). Labeling and hybridization
were performed using the protocols. Only the first three pairs of
ESCC tissues (Table I) could be
used for further processes. The slides were scanned using an
Agilent microarray scanner (Agilent Technologies, CA, USA) and
Feature Extraction 10.7. Raw data were normalized using the
quantile algorithm of Gene Spring Software 12.6 (Agilent
Technologies). Student's t-test was conducted to screen
differentially expressed genes. The filtration criteria were as
follows: i) fold change (linear) ≤0.5 or fold change (linear) ≥2
and ii) t-test P<0.05.
Cell lines and cell culture
ECA109 is a human ESCC cell line obtained from
Shihezi University (Xinjiang, China). The cell line was cultured in
Roswell Park Memorial Institute (RPMI)-1640 nutrient mixture (Life
Technologies, Grand Island, NY, USA) supplemented with 10% fetal
bovine serum (FBS) in a humidified atmosphere containing 5%
CO2 at 37°C.
Construction of highly invading cell
models
Highly invading cell models were selected in
accordance with a previously described method (12). Subpopulations from the ECA109 ESCC
cell line were chosen by using a Corning Matrigel™ invasion chamber
(NY, USA) with a 6-well plate and 8.0-µm pores. ECA109
(2×104 cells) was seeded into the wells and suspended in
600 µl of RPMI-1640 containing 10% FBS. After the cells were
cultured at 37°C for 48 h, the chambers were removed. The invasion
ability of the cells that invaded the lower chamber was higher than
that of the cells in other parts of the chamber. After an
appropriate number of cells was set, the next round of selection
was conducted. Among the first-round selections, the sublines in
the lower chambers were nominated as ECA109T1-1, the sublines from
the second and third rounds were, respectively, designated as
ECA109T1-2 and ECA109T1-3. The parental line in the first series
was designated ECA109T1. If subpopulations were passaged >20
times, a new round of selection was performed.
Invasion Transwell assays
The cells were starved with an Opti-MEM (Corning,
NY, USA) medium for 6 h. Then, 106 cells were suspended
in 1 ml of serum-free medium, and 100 µl or 10×104 cells
were plated onto the upper chamber of a 24-well plate with 8.0-µm
pores Corning Matrigel Invasion Chamber and coated with 0.7 mg/ml
Matrigel (BD Biosciences. They were placed in 600 µl of RPMI-1640
medium containing 10% FBS in the lower chamber. The cells were
incubated for 48 h, and those on the lower surface of the filter
were fixed using paraformaldehyde and stained with 0.5% crystal
violet. Five fields across the lower membrane were photographed at
×100 magnification. The cells were then counted using ImageJ
(http://rsbweb.nih.gov/ij/).
Construction of cytotoxicity cell
model and assay
ECA109 was seeded into a 96-well microplate at a
population of 4,000 cells per well and cultured for 12 h in
RPMI-1640 medium with 10% FBS. Then, the cells were treated with 10
µl of 64 mg/l NDP in the Opti-MEM (Corning) medium. The NDP
concentration was compared with the results of Su et al
(13). The cells incubated in the
Opti-MEM medium were set up with the control group and incubated
for 48 h at 37°C. Afterwards, each well was mixed with 10 µl of
CCK-8 (Seabiotech, Shanghai, China) and further incubated for 3 h
at 37°C. Optical density (OD) was detected by using a
spectrophotometer (Bio-Rad, Hercules, CA, USA) at 450 nm. Growth
inhibition was calculated as the percentage of the untreated
controls. To construct the cytotoxicity cell model, we treated the
ECA109 cells with 200 µl of NDP (64 mg/l) for 48 h in RPMI-1640
medium with 10% FBS. We isolated the miRNAs to detect miRNA
expression.
RNA isolation and qRT-PCR for miRNA
detection
miRNAs were isolated from the cells by using a
miRNeasy mini kit (Qiagen, Stanford, CA, USA) according to the
manufacturer's protocol. RNA quality was evaluated at 260–280 nm OD
ratio. Total RNA (2,000 ng) was reverse-transcribed to cDNA by
using a miRcute Plus miRNA first-strand cDNA synthesis kit
(Tiangen, China). miRNA quantitation was performed using a TaqMan
miRNA assay (Applied Biosystems, Carlsbad, CA, USA) in a CFX96
real-time thermal cycler (Bio-Rad). The oligonucleotides used in
this study are listed in Table II.
All of the samples were normalized using U6, and their relative
expression levels were calculated using the 2−∆∆CT
method.
 | Table II.The oligonucleotides used in this
study. |
Table II.
The oligonucleotides used in this
study.
| Namea | Sequence
(5′-3′) |
|---|
| U6 F | TGC GGG TGC TCG CTT
CGG CAG C |
| miR-6125 F | TTA TTG CGG AAG GCG
GAG CGG |
| miR-1285-3p F | GTC TGG GCA ACA AAG
TGA GAC CT |
| miR-21-5p F | GCG GTA GCT TAT CAG
ACT GAT GTT G |
| miR-424-3p F | GCA AAA CGT GAG GCG
CTG CTA T |
| miR-4697-5p F | TAG GGG GCG CAG TCA
CTG AC |
| miR-5194 F | ATG AGG GGT TTG GAA
TGG GAT GG |
| miR-652-5p F | CAA CCC TAG GAG AGG
GTG CC |
| miR-29c-5p F | GTG ACC GAT TTC TCC
TGG TGT TC |
| miR-30a-3p F | GCT TTC AGT CGG ATG
TTT GCA GC |
| miR-378a-3p F | ACT GGA CTT GGA GTC
AGA AGG C |
| miR-378i F | GAC TGG ACT AGG AGT
CAG AAG G |
Chip data
The gene expression omnibus (GEO) database of NCBI
(National Center for Biotechnology Information) is a public
database that stores chip data, next generation sequencing data,
and other high-throughput experimental data (http://www.ncbi.nlm.nih.gov/geo/). The accession
number of the mRNA Chip data is GSE20347, which includes 17 pairs
of ESCC tissues and paired normal adjacent tissues. The chip used
for mRNA analysis is the Affymetrix Human Genome U133A 2.0 Array
(GPL571), which comprises 47,000 transcriptions and variants
covering 38,500 human genes with identified functions.
Identification of differentially
expressed mRNAs
GSE20347 was used as an input into BRB-array tools
(version 4.5.0 Stable Release). The expression data of mRNA were
standardized relative to the reference array, and the median array
was used as a reference with the following filtration criteria: i)
the signal value of a probe should be 1.5-fold of the median
(bidirectional) and the changes should be ≥20% of the total
expression value; ii) the missing value should be ≤50%; iii) if a
number of probes have been used for one gene, only the probe with
the highest IQR is retained. Differential analysis was performed
using the class comparison tool of BRB-array tools. Differential
mRNA was defined as P<0.01, FDR<0.01, and the folds of
expression change were ≥2 or ≤0.5.
miRNA prediction analysis and
interaction network with mRNAs
TargetScan 7.0 and microT-CDS were used to identify
the predicted miRNA targets. If the number of target genes was too
large, we selected the first 600 genes sorted by the cumulative
weighted context score in TargetScan and miTG score in microT.
After intersecting the predictions with GEO differential mRNAs, we
finally obtained the special genes targeted by miRNAs in ESCC. The
interaction network between miRNAs and mRNAs was visualized using
Cytoscape.
Interaction network and functional
annotation of differential miRNAs and mRNAs
Genes were uploaded to Cytoscape software for
further analysis. The BiNGO plugin (14) in Cytoscape was used to complete GO
(Gene Ontology) analysis. Biological process (BP), molecular
function (MF), and cellular component (CC) networks were analyzed.
The following filtration criteria were considered: i) Benjamini and
Hochberg FDR correction for multiple testing correction; ii)
significance level P<0.05; lists were sorted by the corrected
P-value. The first ten GO were selected for further clustering.
MCODE plugin (15) in Cytoscape was
used to find clusters or highly interconnected regions in these
three GO networks. DAVID online software (16) was used to perform Kyoto Encyclopedia
of Genes and Genomes (KEGG) signaling pathway analysis. Options
were set as follows: i) gene count >2; ii) display by Benjamini;
and iii) P<0.05.
Statistical analysis
Each experiment was conducted at least in
triplicate. Data are presented as means ± standard deviation for
each group. One-way ANOVA and Student's t-test in SPSS version 17.0
(Chicago, IL, USA) were carried out. P<0.05 was considered
significant.
Results
miRNA expression profile of ESCC
To determine the effect of miRNAs on the development
and progression of ESCC in Kazakhs, we initially performed an miRNA
microarray analysis (cutoff, P<0.05, >2.0-fold change) with
three paired ESCC and adjacent tissues. Our results showed that six
miRNAs (miR-1286-3p, miR-21-5p, miR-424-3p, miR-4697-5p, miR-5194,
and miR-652-5p) were significantly overexpressed and five miRNAs
(miR-29c-5p, miR-30a-3p, miR-378a-3p, miR-378i, and miR-584-5p)
were weakly expressed in ESCC tissues compared with normal
esophageal tissues (Fig. 1A).
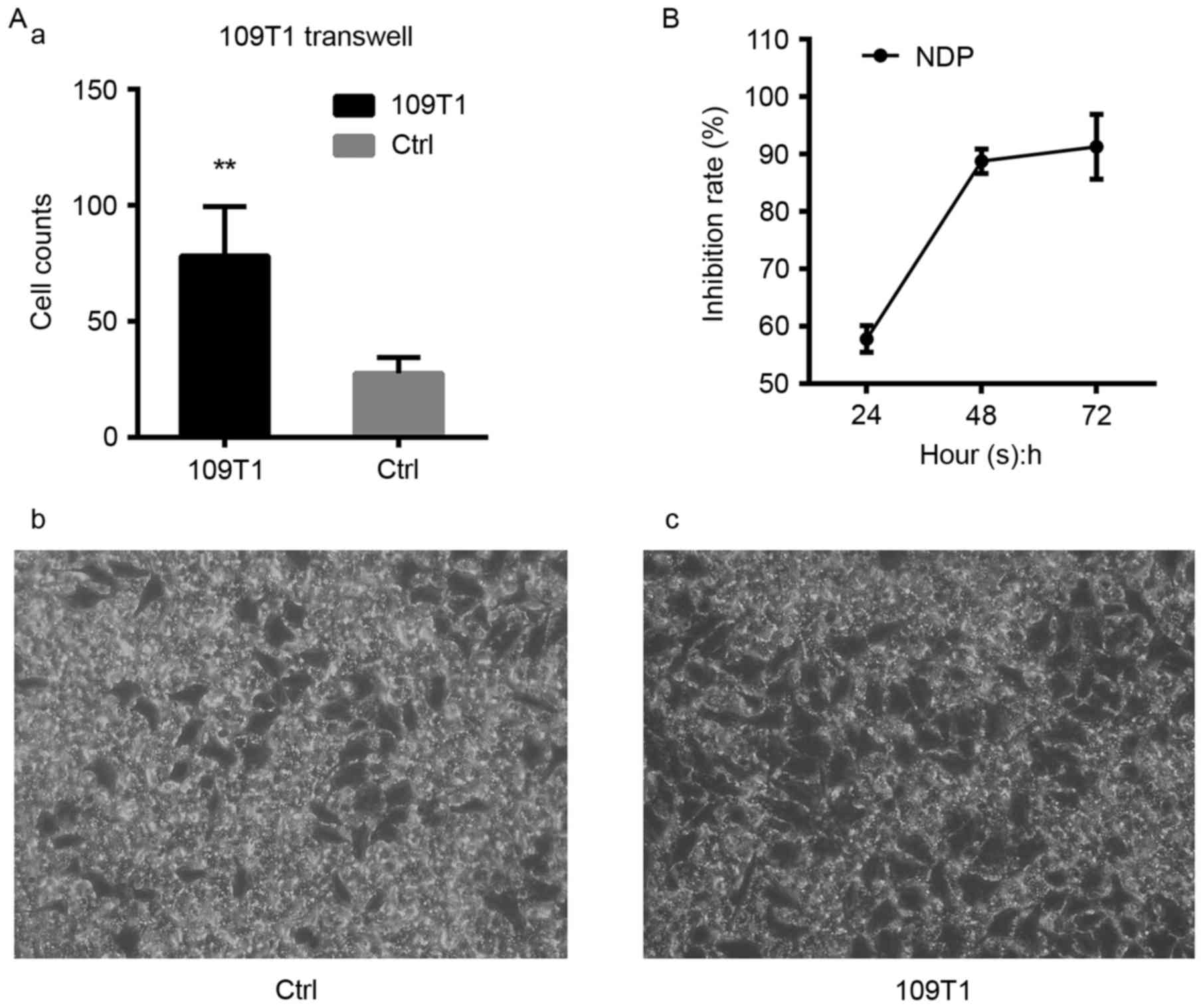
ECA109T1 exhibits stronger
invasiveness and motility than ECA109 cells
After completing one selection cycle for highly
invading cells, we obtained ECA109T1 with high invasiveness. A
Matrigel Transwell invasion assay was conducted to detect the
differences in invasiveness between ECA109T1 and ECA109. Cells of
ECA109T1 group invaded at a rate of 78±9.494 to the lower chamber,
whereas ECA109 group invaded at 27.60±3.076 (P<0.01, n=5). This
finding demonstrated that the invasiveness of ECA109T1 was more
significant than that of ECA109 (Fig.
2A), also indicated that this cell model could be used to
select key miRNAs that participate in tumor metastasis.
NDP-treated ECA109 cell line
suppresses proliferation
The cytotoxicity assay via CCK-8 was carried out to
detect the inhibition rate of ECA109 and ECA109 proliferation by 64
mg/l of NDP at 24, 48, and 72 h. Our results revealed that the
inhibition rate was increased with time (P<0.05) (Fig. 2B). Thus, NDP could suppress cell
proliferation and elicit an antiproliferative effect. The expected
results could not be achieved because of the state of NDP-treated
ECA109 at 72 h. ECA109 treated with 64 mg/l NDP for 48 h was used
to construct the cytotoxicity cell model and further subjected to
RT-qPCR analysis.
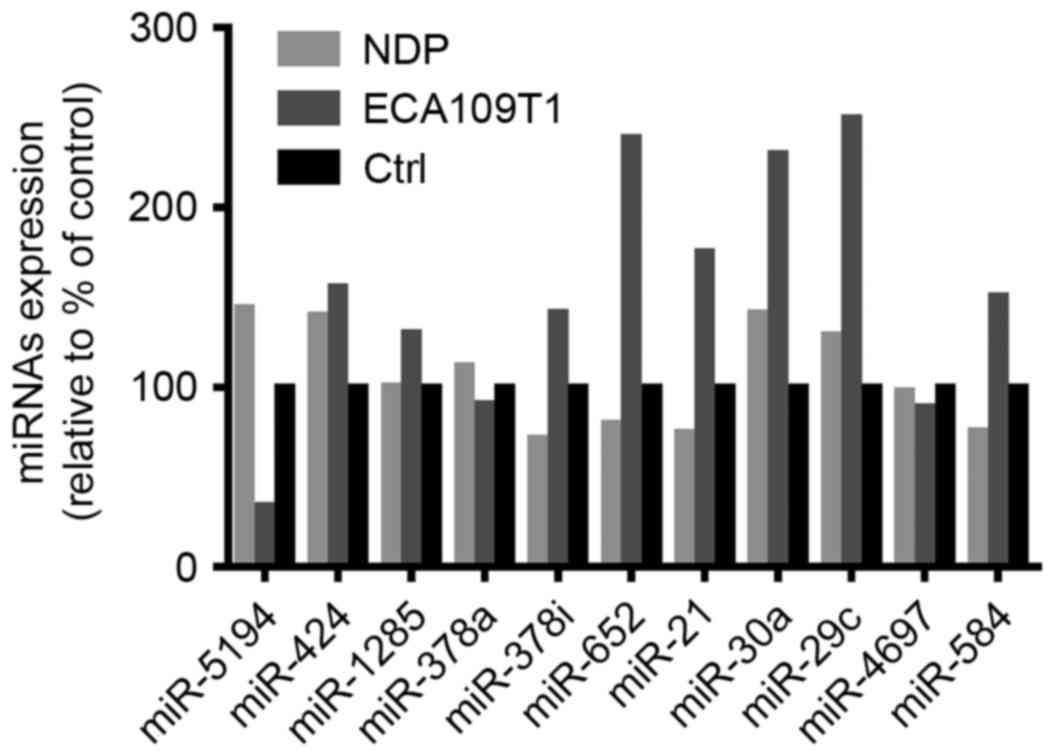
RT-qPCR revealed miRNAs with
consistent expression
To determine the miRNAs that participate in ESCC
cell invasion and proliferation, we performed RT-qPCR detection and
quantitated each miRNA from miRNA profiling within the two cell
models (Fig. 3). After the qRT-PCR
results were compared with the microarray results, an upregulated
gene was released by the microarray, and miR-652-5p and miR-21-5p
were overexpressed in ECA109T1 and suppressed in NDP-treated ECA109
cells. Therefore, miR-652-5p and miR-21-5p were consistently
expressed in the miRNA microarray and cell models of ESCC.
Moreover, these two miRNAs may be critical in ESCC occurrence and
development. These miRNAs were further used for bioinformatics
analyses.
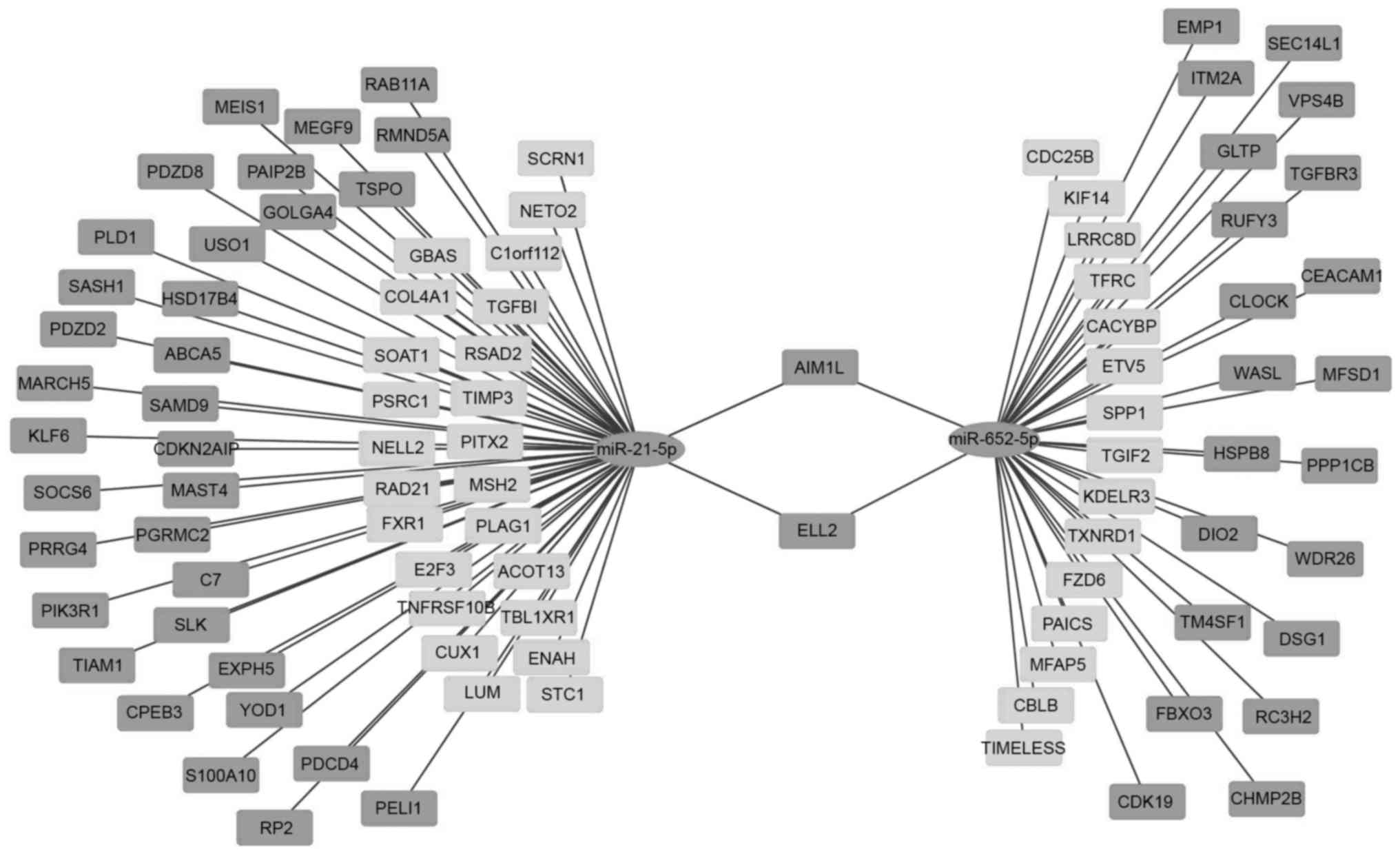
Differentially expressed mRNA
screening and interaction network with miRNAs
A total of 650 target mRNAs were present in
miR-21-5p and 713 targets were found in miR-652-5p. The mRNA
(GSE20347) data were downloaded and integrated in the BRB-array
tools for standardization and filtration. A total of 6,572 probes
were selected for further analysis (Fig. 1B). The class comparison tool of
BRB-array tools was used to analyze chip data. A total of 1,163
mRNAs exhibited differential expression (P<0.01, FDR<0.01,
and the expression fold-change of ≥2 or ≤0.5). Of these mRNAs, 640
were upregulated and 521 were downregulated. The intersected genes
of miR-21-5p were 59 (35 upregulated and 24 downregulated) and
those of miR-652-5p were 38 (23 upregulated and 15 downregulated).
The interaction network of miRNAs and intersected genes was
visualized by using Cytoscape (Fig.
4).
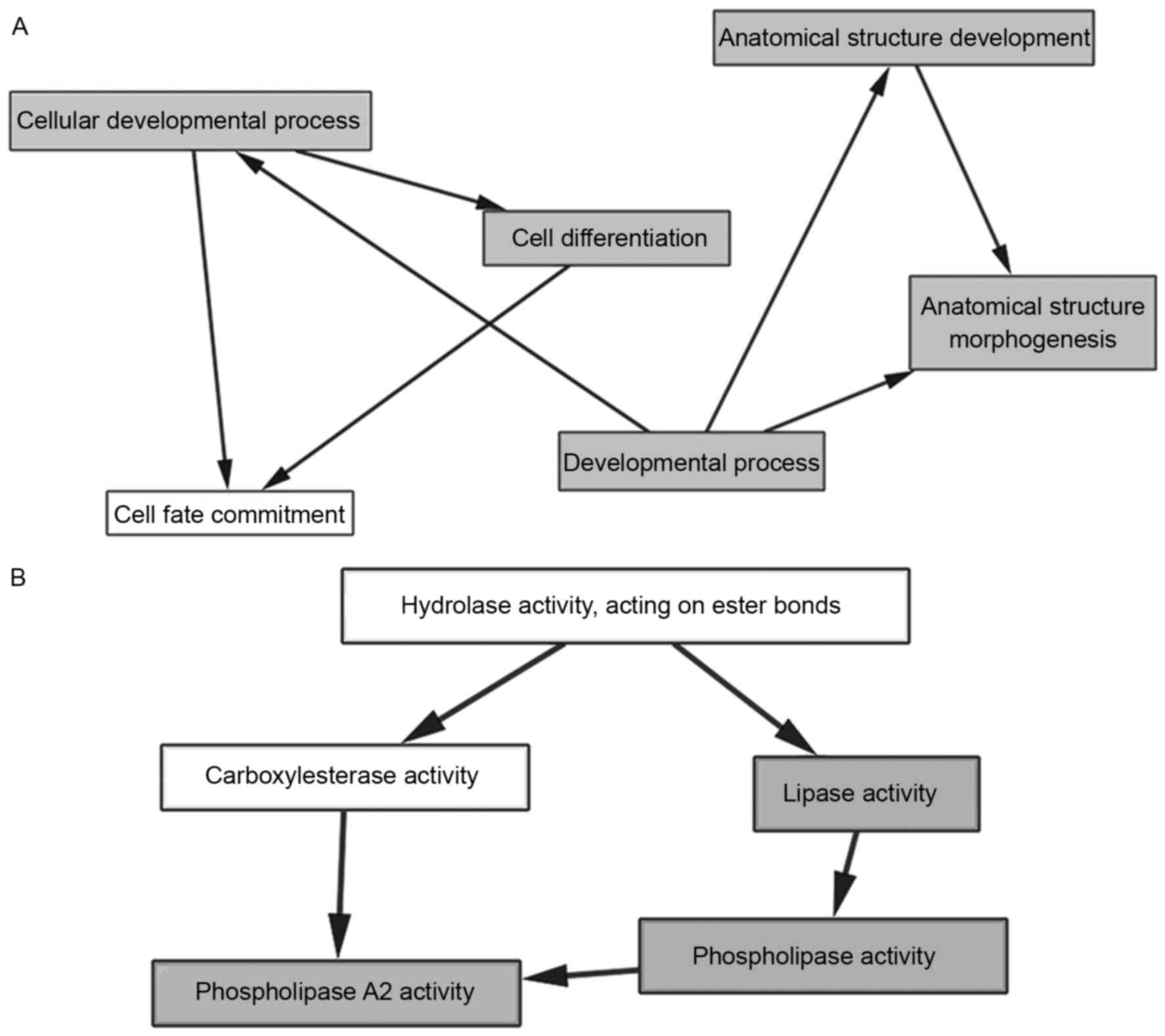
Network analysis of GO, KEGG, and PPI
interactions
GO analysis was performed using the BiNGO plugin in
Cytoscape. The first 10 BP and MF were arranged according to the
P-value of GO and were chosen for further MCODE clustering
analysis. No nodes were displayed in CC if P-value was <0.05.
These results indicated that 8 GO and 5 related genes were
essential for ESCC GO. BP was as follows: i) cellular developmental
process, ii) developmental process, iii) cell differentiation, iv)
anatomical structure development, and v) anatomical structure
morphogenesis (Fig. 5A). MF was as
follows: i) phospholipase activity, ii) lipase activity, and iii)
phospholipase A2 activity (Fig.
5B). The mRNAs targeted by dysregulated miRNAs are listed in
Table III. DAVID KEGG pathway
analysis showed three pathway records: i) endocytosis, ii) pathways
in cancer, and iii) actin cytoskeleton regulation (Table IV).
 | Table III.Key genes in GO clusters. |
Table III.
Key genes in GO clusters.
| miRNA | Upa | Downa |
|---|
| miR-21-5p | PLD1 | MSH2, STC1 |
| miR-652-5p | DSG1 | N/Ab |
 | Table IV.KEGG annotation. |
Table IV.
KEGG annotation.
| Term |
P-valuea | Genes |
|---|
| Endocytosis | 0.005 | PLD1, CBLB, TFRC,
VPS4B, RAB11A, CHMP2B |
| Pathways in
cancer | 0.015 | E2F3, PLD1, CBLB,
COL4A1, MSH2, PIK3R1, FZD6 |
| Regulation of actin
cytoskeleton | 0.044 | ENAH, TIAM1, WASL,
PPP1CB, PIK3R1 |
Discussion
This study aimed to analyze the molecular mechanisms
of invasion and proliferation by considering miRNA expression
profiles, cell models, and bioinformatics. We obtained 11
differential miRNAs from the microarray profiles of Kazakh
ethnicity ESCC and normal adjacent tissues. The ESCC cell line
subpopulation with a high invasive ability and cytotoxicity cell
model treated with NDP was constructed. However, further studies
should be performed to investigate whether these models can
decrease the number of miRNAs. Combined with the profile, miR-21-3p
and miR-652-3p were further analyzed. Bioinformatics was applied to
observe the molecular functions of miRNAs macroscopically. The
target genes of miRNAs were obtained via the prediction databases
and combined with the GEO data of the ESCC mRNA via BRB-array
tools. Cytoscape and its plugin contributed to GO classification
and enrichment analysis. In these two miRNA targets, PLD1, MSH2,
STC1, and DSG1 may be implicated in cancer occurrence and
development.
ESCC in Northwestern China, especially in Xinjiang
where the high-risk Kazakh population resides, has a high motility
(17). First, some behavioral and
environmental risk factors play important roles in ESCC development
among Kazakhs (2). After ~10 years
of national integration, environmental and behavioral factors have
decreased. Thus, gene susceptibility and polymorphism are involved
(18). Second, Xinjiang is
considered an important area for ESCC investigation because of the
presence of multiracial society and high incidence. To determine
special miRNAs with racial specificity, we collected three paired
Kazakh ESCC and adjacent tissue samples for further detection. The
collection of tissue from Kazakhs is challenging because of
religion, concept, and other factors. However, we will consider
additional factors and increase the sample size in our future
studies. Our results revealed that miR-21-3p, miR-652-3p, and their
potential targets may be involved in ESCC invasion and
proliferation.
Bioinformatics approaches are the main patterns used
to analyze microarray profiling results and predict miRNA functions
(19,20). However, bioinformatics depends on
experimental databases, and we found that this approach may easily
eliminate miRNAs that have been discovered with few experimental
bases. In our experiment, the overbalance in the data size of
bioinformatics was related to the databases between miRNAs
discovered earlier and later. False-negative results may increase
when one method is used. Therefore, two different cell models were
constructed in this study. ECA109 is a well-differentiated ESCC
cell line from the middle esophagus of a Chinese patient. ECA109T1
is significantly more invasive than its parent cells. Moreover, the
mice injected with invasive subpopulation selected by Transwell
chambers had faster tumor growth and metastasis (12). NDP is a new platinum derivative
selected from a series of platinum analogs based on its pronounced
preclinical antitumor activity against various solid tumors with
low nephrotoxicity (21). It can be
combined with paclitaxel and other chemotherapeutic agents to
improve treatment effects and reduce adverse effects. In
vitro, NDP affects ESCC proliferation and apoptosis (13). We used NDP to construct a
cytotoxicity cell model and investigate its therapeutic effects.
However, further studies should determine whether the combination
of different chemotherapeutic agents differentiates
microenvironment or molecular mechanisms.
qRT-PCR revealed the genes differentially expressed
in two cell models. After qRT-PCR findings were matched with
microarray results, miR-21-5p and miR-652-5p showed that they may
be important in ESCC invasion and proliferation. miR-21 is an
oncogene in various diseases, and its overexpression in tissues and
serum is associated with poor prognosis (22). Although miR-21 is considered a
potential therapeutic target and biomarker of ESCC (23,24),
the relationship between miR-652-5p and ESCC has yet to be
elucidated. As such, this parameter will be our next focus.
Bioinformatics analysis also demonstrated that dysregulated miRNAs
and their target genes (PLD1, MSH2, STC1, and DSG1) were mostly
involved in cell differentiation, anatomical structure (BP), and
lipase and phospholipase activities (MF). Phospholipase D1 (PLD1)
is a phospholipid-metabolizing enzyme whose overexpression promotes
cell growth and proliferation by activating Akt (25). Zhang et al revealed that the
downregulated PLD1 inhibits the proliferation of gastric carcinoma
cells and this enzyme is targeted by miR-638 (26). Human MutS homolog 2 (MSH2) encodes
the DNA repair protein. In a clinical survival analysis, MSH2
expression is related to metastasis and tumor shape (27) and recognized as a target by miR-21
(28). The reduced desmoglein-1
(DSG1) expression in cervical tumors is associated with survival
and prognosis (29). The
association between miRNAs and these four target genes should be
determined in vivo and in vitro in future molecular
biological studies. Fatty acids are synthetized rapidly in growing
tumors (30). In clinical research,
the pharmacological control of Fas/FasL signaling may improve the
therapeutic efficacy and outcome in ESCC patients receiving
preoperative chemoradiotherapy (31). Lipase and phospholipase activation
may suppress ESCC development. These target genes likely
participate in three pathways, namely, endocytosis, cancer
pathways, actin cytoskeleton regulation, which are mostly related
to cancer. Furthermore, miR-21-5p and miR-652-5p may perform vital
functions in ESCC.
Finally, we successfully selected differentially
expressed miRNA and their potential target genes through cell
modeling and bioinformatics analysis. Our results indicated that
these miRNAs and targets may be used for future therapeutic
investigations. These selection processes may improve the
efficiency of microarray selection. In the future, miRNAs and their
targets should be verified, multiple drugs should be combined, and
other supplements should be administered to improve the conclusion
of this study.
Acknowledgements
The authors would like to acknowledge the help from
First Hospital Affiliated to Medical College of Shihezi University.
This study was supported in part by doctor/scientific grant
2014BB019 from Xinjiang Production and Construction Corps and NFSC
(81260301, 81360358, 81460362) from the National Science Foundation
of China.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zheng S, Vuitton L, Sheyhidin I, Vuitton
DA, Zhang Y and Lu X: Northwestern China: a place to learn more on
oesophageal cancer. Part one: behavioural and environmental risk
factors. Eur J Gastroenterol Hepatol. 22:917–925. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lee RC, Feinbaum RL and Ambros V: The
C. elegans heterochronic gene lin-4 encodes small RNAs with
antisense complementarity to lin-14. Cell. 75:843–854. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tay Y, Zhang J, Thomson AM, Lim B and
Rigoutsos I: MicroRNAs to Nanog, Oct4 and Sox2 coding regions
modulate embryonic stem cell differentiation. Nature.
455:1124–1128. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Stark A, Brennecke J, Bushati N, Russell
RB and Cohen SM: Animal microRNAs confer robustness to gene
expression and have a significant impact on 3′UTR evolution. Cell.
123:1133–1146. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ferracin M, Veronese A and Negrini M:
Micromarkers: miRNAs in cancer diagnosis and prognosis. Expert Rev
Mol Diagn. 10:297–308. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kan T and Meltzer SJ: MicroRNAs in
Barrett's esophagus and esophageal adenocarcinoma. Curr Opin
Pharmacol. 9:727–732. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chen Z, Li J, Tian L, Zhou C, Gao Y, Zhou
F, Shi S, Feng X, Sun N, Yao R, et al: miRNA expression profile
reveals a prognostic signature for esophageal squamous cell
carcinoma. Cancer Lett. 350:34–42. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Takeshita N, Hoshino I, Mori M, Akutsu Y,
Hanari N, Yoneyama Y, Ikeda N, Isozaki Y, Maruyama T, Akanuma N, et
al: Serum microRNA expression profile: miR-1246 as a novel
diagnostic and prognostic biomarker for oesophageal squamous cell
carcinoma. Br J Cancer. 108:644–652. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hurst DR, Edmonds MD and Welch DR:
Metastamir: The field of metastasis-regulatory microRNA is
spreading. Cancer Res. 69:7495–7498. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen YK, Chang WS, Wu IC, Li LH, Yang SF,
Chen JY, Hsu MC, Chen SH, Wu DC, Lee JM, et al: Molecular
characterization of invasive subpopulations from an esophageal
squamous cell carcinoma cell line. Anticancer Res. 30:727–736.
2010.PubMed/NCBI
|
|
13
|
Su XY, Yin HT, Li SY, Huang XE, Tan HY,
Dai HY and Shi FF: Intervention effects of nedaplatin and cisplatin
on proliferation and apoptosis of human tumour cells in vitro.
Asian Pac J Cancer Prev. 13:4531–4536. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Maere S, Heymans K and Kuiper M: BiNGO: A
Cytoscape plugin to assess overrepresentation of gene ontology
categories in biological networks. Bioinformatics. 21:3448–3449.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bader GD and Hogue CWV: An automated
method for finding molecular complexes in large protein interaction
networks. BMC Bioinformatics. 4:22003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Huang W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Cui X, Zhao Z, Liu D, Guo T, Li S, Hu J,
Liu C, Yang L, Cao Y, Jiang J, et al: Inactivation of miR-34a by
aberrant CpG methylation in Kazakh patients with esophageal
carcinoma. J Exp Clin Cancer Res. 33:202014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zheng S, Vuitton L, Sheyhidin I, Vuitton
DA, Zhang Y and Lu X: Northwestern China: a place to learn more on
oesophageal cancer. Part two: gene alterations and polymorphisms.
Eur J Gastroenterol Hepatol. 23:1087–1099. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Johnson CH, Ivanisevic J, Benton HP and
Siuzdak G: Bioinformatics: The next frontier of metabolomics. Anal
Chem. 87:147–156. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhao Y, Zhang X, Zhao Y, Kong D, Qin F,
Sun J and Dong Y: Identification of potential therapeutic target
genes, key miRNAs and mechanisms in acute myeloid leukemia based on
bioinformatics analysis: Med Oncol. 32:1522015.
|
|
21
|
Kameyama Y, Okazaki N, Nakagawa M, Koshida
H, Nakamura M and Gemba M: Nephrotoxicity of a new platinum
compound, 254-S, evaluated with rat kidney cortical slices. Toxicol
Lett. 52:15–24. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Fu W, Pang L, Chen Y, Yang L, Zhu J and
Wei Y: The microRNAs as prognostic biomarkers for survival in
esophageal cancer: A meta-analysis. Scientific World J.
2014:5239792014. View Article : Google Scholar
|
|
23
|
Liu T, Liu Q, Zheng S, Gao X, Lu M, Yang
C, Dai F, Sheyhidin I and Lu X: MicroRNA-21 promotes cell growth
and migration by targeting programmed cell death 4 gene in Kazakh's
esophageal squamous cell carcinoma. Dis Markers. 2014:2328372014.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Komatsu S, Ichikawa D, Takeshita H,
Konishi H, Nagata H, Hirajima S, Kawaguchi T, Arita T, Shiozaki A,
Fujiwara H, et al: Prognostic impact of circulating miR-21 and
miR-375 in plasma of patients with esophageal squamous cell
carcinoma. Expert Opin Biol Ther. 12:(Suppl 1). S53–S59. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Gozgit JM, Pentecost BT, Marconi SA,
Ricketts-Loriaux RSJ, Otis CN and Arcaro KF: PLD1 is overexpressed
in an ER-negative MCF-7 cell line variant and a subset of
phospho-Akt-negative breast carcinomas. Br J Cancer. 97:809–817.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhang J, Bian Z, Zhou J, Song M, Liu Z,
Feng Y, Zhe L, Zhang B, Yin Y and Huang Z: MicroRNA-638 inhibits
cell proliferation by targeting phospholipase D1 in human gastric
carcinoma. Protein Cell. 6:680–688. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Theocharis S, Klijanienko J, Giaginis C,
Rodriguez J, Jouffroy T, Girod A, Point D, Tsourouflis G and
Sastre-Garau X: Expression of DNA repair proteins, MSH2, MLH1 and
MGMT in mobile tongue squamous cell carcinoma: associations with
clinicopathological parameters and patients' survival. J Oral
Pathol Med. 40:218–226. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Cheng JY, Nishi H, Yonekawa C, Takara K
and Isaka K: MicroRNA-21 increases the resistance to CDDP in HeLa
cervical carcinoma cells by targeting PDCD4 and MSH2. FASEB J.
27:(Suppl). 1b5762013.
|
|
29
|
Myklebust MP, Fluge Ø, Immervoll H,
Skarstein A, Balteskard L, Bruland O and Dahl O: Expression of DSG1
and DSC1 are prognostic markers in anal carcinoma patients. Br J
Cancer. 106:756–762. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Medes G, Thomas A and Weinhouse S:
Metabolism of neoplastic tissue. IV. A study of lipid synthesis in
neoplastic tissue slices in vitro. Cancer Res. 13:27–29.
1953.PubMed/NCBI
|
|
31
|
Saigusa S, Tanaka K, Ohi M, Toiyama Y,
Yasuda H, Kitajima T, Okugawa Y, Inoue Y, Mohri Y and Kusunoki M:
Clinical implications of Fas/Fas ligand expression in patients with
esophageal squamous cell carcinoma following neoadjuvant
chemoradiotherapy. Mol Clin Oncol. 3:151–156. 2015.PubMed/NCBI
|