Introduction
Breast cancer is one of the most common malignant
diseases among women both in developed and developing countries,
accounting for 29% of new cancer cases and 14% of cancer-associated
deaths among American women (1). In
China, the incidence of breast cancer is lower than in America, but
during the past decade, the incidence has been increasing markedly,
especially in rural areas (2).
Therefore, more effective methods for breast cancer diagnosis and
treatments are urgently needed.
MicroRNAs (miRNAs) are endogenous non-coding RNAs
that function as gene regulators mainly by binding to the 3′UTR of
their target mRNAs, inducing mRNAs degradation or translation
repression (3). Biological
processes, including cell proliferation, differentiation, apoptosis
and senescence are controlled by miRNAs. Aberrant expression of
miRNAs is associated with a variety of cancers, such as lung
cancer, gastric cancer, and breast cancer (4–6). Let-7
was originally discovered in Caenorhabditis elegans,
controlling the timing of stem-cell division and differentiation
(7). Subsequently, let-7 and its
family members have been found playing important roles in tumor
suppression. Akao et al reported that let-7 might suppress
the growth of human colon cancer cells and reduced oncogene
RAS and c-myc expression (8). Esquela-Kerscher et al found
that let-7 could inhibit the growth of lung cancer cell lines, as
well as the growth of lung cancer cell xenografts in mice (9). Let-7c-5p, a member of let-7 family, is
downregulated in prostate cancer, overexpression of let-7c-5p
suppresses androgen receptor expression and leads to inhibition of
prostate cancer cell proliferation (10). Nwaeburu et al found that
forced expression of let-7c-5p by quercetin could activate Numbl
expression and resulted in suppression of pancreatic cancer
progression (11). Some other
studies suggest that let-7c-5p functions as a anti-oncogene in
human non-small cell lung cancer, hepatocellular carcinoma and
colorectal cancer (12–14).
Although let-7c-5p has been studied in several
cancers, its function and molecular mechanism in breast cancer
remain to be further identified. Here, we measured let-7c-5p
expression in breast cancer tissues and corresponding adjacent
tissues, and examined the effects of let-7c-5p on human breast
cancer cell proliferation and apoptosis. Moreover, for the first
time, we found that ERCC6 was a target of let-7c-5p in
breast cancer.
Materials and methods
Tissues collection
Nine paraffin-embedded breast cancer and
corresponding adjacent tissue samples were collected from Zhejiang
Cancer Hospital (Hangzhou, China). This study was approved by the
ethics committee of the Zhejiang Sci-Tech University (Hangzhou,
China) and Zhejiang Cancer Hospital. All the samples were stored at
4°C until RNA extraction.
RNA extraction and qRT-PCR
Total RNA was extracted from paraffin-embedded
tissues using Recover All™ Total Nucleic Acid Isolation (Ambion,
Austin, TX, USA) following the manufacturer's instructions. RNA
concentrations were measured by the NanoDrop ND-2000
spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). RNA
reverse transcription was performed with 100 ng of total RNA.
Expression of let-7c-5p, ERCC6 and internal control GAPDH were
detected using SYBR Premix Ex Taq™ II (Takara, Dalian, China).
qRT-PCR was performed on the Applied Biosystems 7500 real-time PCR
system (Applied Biosystems, Foster City, CA, USA). Relative
expression was calculated using the 2−∆∆Ct method, the
expression of let-7c-5p and ERCC6 was normalized with GAPDH.
Primers for qRT-PCR are listed in Table
I.
 | Table I.Primers for qRT-PCR. |
Table I.
Primers for qRT-PCR.
| Primer | Sequence
(5′-3′) |
|---|
| let-7c-5p |
GTCGTATCCAGTGCAGGGTCCGAGG |
| stem-loop |
TATTCGCACTGGATACGACAACCAT |
| let-7c-5p | F:
GAGGTAGTAGGTTGTATGGTTG |
|
| R:
GCAGGGTCCGAGGTATTC |
| ERCC6 | F:
CAATAGTCTGCCTCCCCACCCC |
|
| R:
CAACTTCTCGTTCCTCAACACATC |
| GAPDH | F:
TGCCAAATATGATGACATCAAGAA |
|
| R:
GGAGTGGGTGTCGCTGTTG |
Cell culture and transfection
The MCF-7 human breast cancer cell line was obtained
from Zhejiang Sci-Tech University, cultured in DMEM-high glucose
medium supplemented with 10% fetal bovine serum (Gibco, Grand
Island, NY, USA), at 37°C in a 5% CO2 humidified
incubator (HF90, Heal Force, Hong Kong). Mimics for miR-NC
(5′-CAGUACUUUUGUGUAGUACAA-3′) and let-7c-5p
(5′-UGAGGUAGUAGGUUGUAUGGUU-3′) were purchased from GenePharma
(Shanghai, China). Reverse transfection was performed in this study
using GeneTran™ III High Efficiency Transfection Reagent (Biomiga,
Inc., San Diego, CA, USA) according to the manufacturer's
recommendations.
MTT assay
MCF-7 cells were seeded in 96-well plates at
approximately 4000 cells per well and transfected with miR-NC or
let-7c-5p mimics. MTT (Sigma, St. Louis, MO, USA) was used to
examine the effects of let-7c-5p on cell proliferation at 24, 48
and 72 h post-transfection. At each time, 20 µl MTT solution was
added to each well, and incubated at 37°C for 4 h, the formazan
produced by viable cells was dissolved in 150 µl dimethylsulfoxide
(DMSO). The cell numbers were evaluated by reading the absorbance
at 490 nm (15).
Cell apoptosis assay
Cells were seeded in 6-well plates at approximately
3×105 cells per well and transfected with miR-NC or
let-7c-5p mimics. All cells were harvested at 24 h
post-transfection, washed with cold PBS for twice, stained with
FITC and PI using BD Pharmingen FITC Annexin V™ apoptosis detection
kit I (BD Biosciences, San Jose, CA, USA) following the
manufacturer's instructions. Flow cytometry analysis was finished
on BD Accuri C6 flow cytometer with C6 software.
Dual luciferase reporter assay
The potential target genes of let-7c-5p were
predicted using the algorithms of TargetScan (http://www.targetscan.org/vert_71/), PicTar
(http://pictar.mdc-berlin.de/) and
starBase (http://starbase.sysu.edu.cn/). ERCC6 was taken as a
potential target gene of let-7c-5p. There was only one potential
complementary site for let-7c-5p in the 3′UTR of ERCC6 mRNA, and
the wild-type 3′UTR fragment containing putative binding site was
amplified by PCR and cloned into the XbaI and NotI
sites downstream of Renilla luciferase vector pRL-TK
(Promega, Beijing, China), the recombinant plasmid was named
pRL-ERCC6-wt. The putative let-7c-5p binding site was deleted by
overlap-extension PCR, and the recombinant plasmid was named
pRL-ERCC6-mut. Primers are listed in Table II, constructs were verified by
sequencing.
 | Table II.Primers for construction of
recombinant plasmids. |
Table II.
Primers for construction of
recombinant plasmids.
| Primer | Sequence
(5′-3′) |
|---|
| Wild-type | F:
ACAACATTGCTTCCTA |
| 3′UTR | R:
TCAATCCAAGTATTTTCTCC |
| Mutant | F:
ACAACATTGCTTCCTA |
| 3′UTR-1 | R:
AAGTTTTAATTCACATCATGCAAACAA |
| Mutant | F:
TGTTTGCATGATGTGAATTACAACTT |
| 3′UTR-2 | R:
TCAATCCAAGTATTTTCTCC |
For dual luciferase reporter assay, cells were
seeded in 24-well plates in triplicate and cotransfected with the
firefly luciferase vector pGL3 and recombinant plasmid pRL-ERCC6-wt
or pRL-ERCC6-mut, together with mimics for either miR-NC or
let-7c-5p at a final concentration of 20 nM. After 48 h, luciferase
activities were measured using the dual luciferase reporter assay
system (Promega). Renilla luciferase activity was normalized
with firefly luciferase activity.
Western blotting
Cells were harvested at 48 h post-transfection and
lysed in RIPA buffer on ice. Total proteins were separated by 10%
SDS-PAGE, electroblotted onto PVDF membranes. Membranes were
blocked with 5% non-fat milk powder for 2 h at room temperature and
incubated overnight at 4°C with a polyclonal antibody: anti-ERCC6
antibody (1:2000 dilution; Abcam, Cambridge, MA, USA) or anti-GAPDH
antibody (1:2000 dilution; HuaBio, China). After washing three
times with TBS-T, membranes were incubated with a secondary
antibody anti-rabbit HRP-conjugate (1:2000 dilution; HuaBio) for 2
h at room temperature (16).
Antibody detection was performed with ECL Western Blotting
Substrate (Solarbio, Beijing, China), and photos were taken using
the Tanon 5500 imaging system (Tanon, Shanghai, China).
Statistical analysis
Statistical analysis was performed using SPSS
software version 17.0 (SPSS, Inc., Chicago, IL, USA). All data were
presented as mean ± standard deviation (SD). Statistical
significance was tested by Student's t-test. P<0.05 was
considered to indicate a statistically significant difference.
Results
Reduced expression of let-7c-5p in
breast cancer tissues
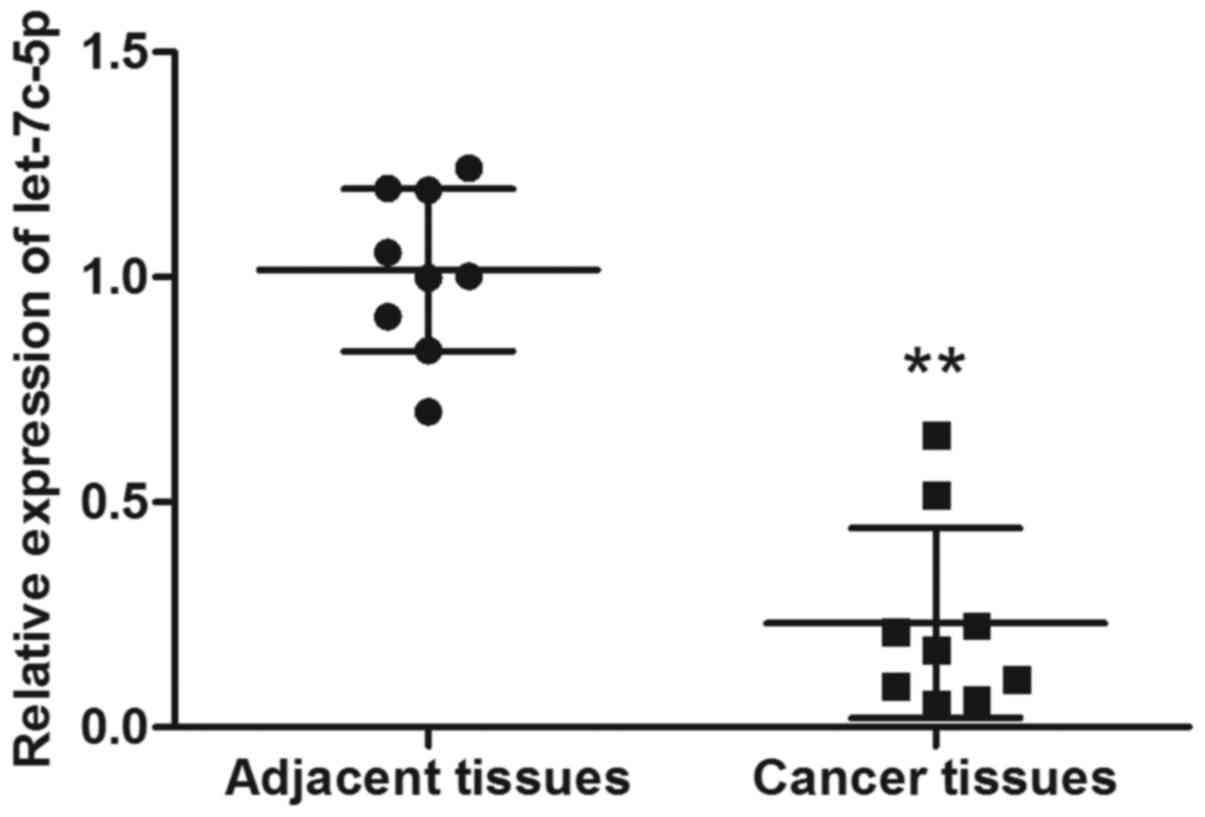
As shown in Fig. 1,
expression of let-7c-5p was significantly decreased in the nine
breast cancer tissues, indicating that let-7c-5p could potentially
serve as a biomarker for breast cancer.
Overexpression of let-7c-5p inhibits
MCF-7 cell proliferation and induces cell apoptosis
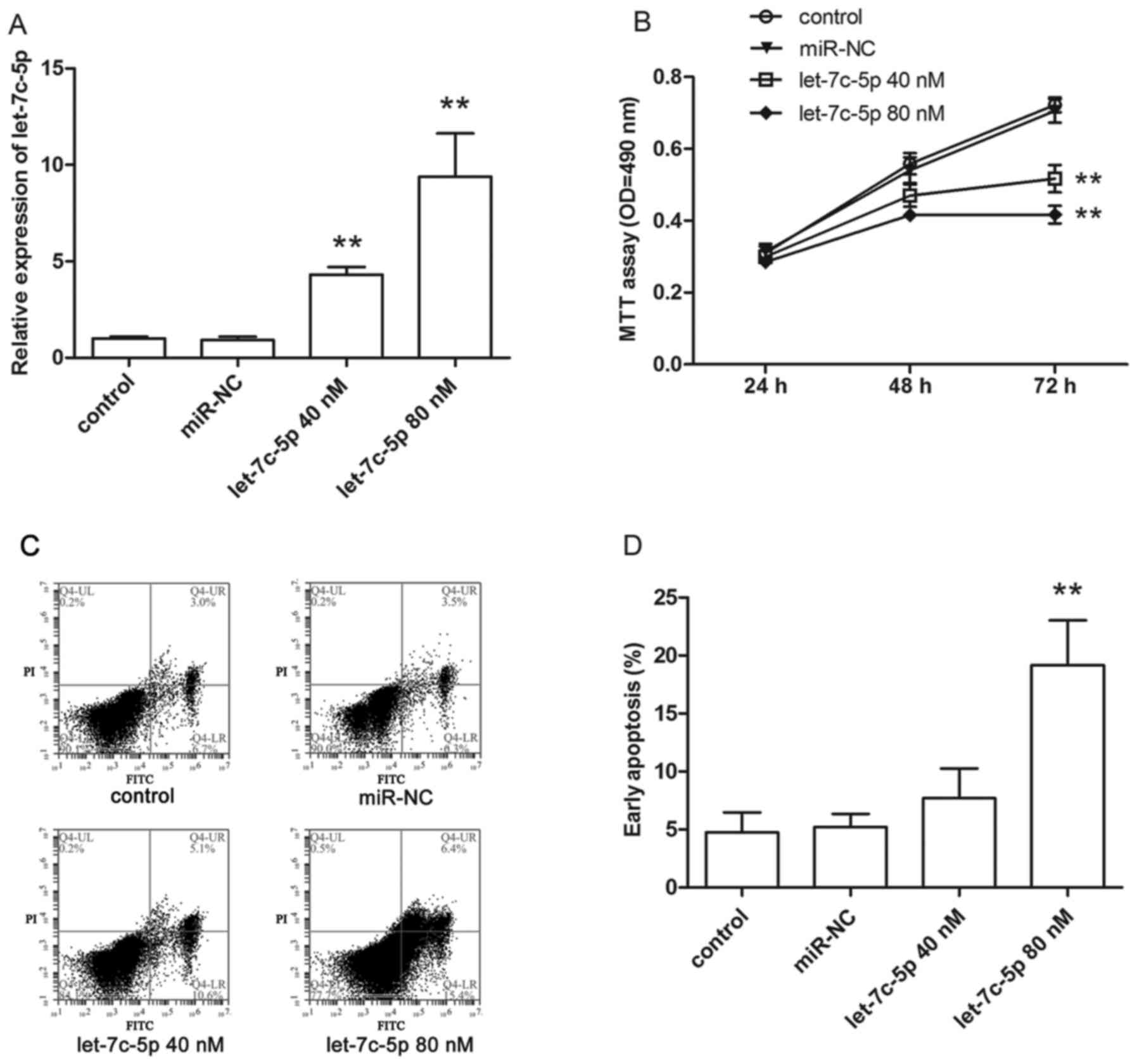
To determine the transfection efficiency, cells were
harvested after transfection with 40 or 80 nM let-7c-5p mimics for
24 h. Total RNA was isolated from cells using the traditional
TRIzol method. qRT-PCR was performed to measure the expression of
let-7c-5p. As shown in Fig. 2A,
let-7c-5p expression was enhanced significantly by let-7c-5p
mimics.
To examine the effects of let-7c-5p on MCF-7 cell
proliferation in vitro, MTT assay was performed at 24, 48
and 72 h post-transfection, the cell growth curve showed that the
proliferation ability of transfected cells was reduced remarkably
compared to control group cells (Fig.
2B).
Additionally, we explored whether cell apoptosis
could be induced by let-7c-5p. After transfection for 24 h, cells
were harvested, and cell apoptosis was detected by flow cytometer.
As shown in Fig. 2C, early stage
apoptotic cells are presented in the lower right quadrant, a higher
rate of apoptosis was observed in transfected cells, especially in
those transfected with 80 nM let-7c-5p mimics (Fig. 2D).
ERCC6 is a direct target gene of
let-7c-5p in breast cancer
TargetScan, PicTar and starBase were used to predict
the target genes of let-7c-5p, and the intersection of the
prediction results from the three algorithms showed that there were
41 candidate genes with at least one binding site of let-7c-5p
(Table III). Of these genes,
ITGB3 and MAP4K3 have been verified as targets of
let-7c-5p in human non-small cell lung cancer. In this experiment,
we aimed to find a new target of let-7c-5p in breast cancer.
ERCC6, which has been reported to be associated with cancer
risk, was selected as a putative target gene.
 | Table III.Potential target genes of let-7c-5p
predicted by TargetScan, PicTar and starBase. |
Table III.
Potential target genes of let-7c-5p
predicted by TargetScan, PicTar and starBase.
| Potential target
genes | Position |
|---|
| ADRB2 |
chr5:148207936–148207943 |
| AHCTF1 |
chr1:247003542–247003549 |
| APBB3 |
chr5:139937904–139937911 |
| BACH1 |
chr21:30717135–30717142 |
| CD200R1 |
chr3:112641981–112641988 |
| CDV3 |
chr3:133307221–133307228 |
| CLDN12 |
chr7:90043275–90043282 |
| COIL |
chr17:55016284–55016291 |
| COL1A1 |
chr17:48262068–48262074 |
| DMD |
chrX:31138095–31138102 |
| E2F6 |
chr2:11585579–11585586 |
| ERCC6 |
chr10:50666729–50666736 |
| FGD6 |
chr12:95474757–95474764 |
| FNIP1 |
chr5:130979161–130979168 |
| FRAS1 |
chr4:79462429–79462436 |
| GAN |
chr16:81412340–81412347 |
| GNPTAB |
chr12:102140912–102140919 |
| GOLT1B |
chr12:21670278–21670285 |
| HAND1 |
chr5:153854615–153854622 |
| INTS2 |
chr17:59943299–59943306 |
| IRS2 |
chr13:110408191–110408198 |
| ITGB3 |
chr17:45388633–45388639 |
| KLHDC8B |
chr3:49213851–49213858 |
| LRIG2 |
chr1:113666778–113666785 |
| LRIG3 |
chr12:59266314–59266321 |
| MAP4K3 |
chr2:39476696–39476703 |
| MED8 |
chr1:43850492–43850499 |
| MLL2 |
chr12:49414882–49414889 |
| NAP1L1 |
chr12:76441001–76441008 |
| NLK |
chr17:26522778–26522784 |
| PRPF38B |
chr1:109243302–109243309 |
| RANBP2 |
chr2:109401057–109401064 |
| RNF20 |
chr9:104324767–104324773 |
| RRM2 |
chr2:10269649–10269656 |
| SEMA4C |
chr2:97526289–97526296 |
| SLC20A1 |
chr2:113421361–113421367 |
| SLC35D2 |
chr9:99083376–99083383 |
| TMEM2 |
chr9:74298636–74298643 |
| WDR37 |
chr10:1176192–1176199 |
| ZCCHC3 |
chr20:280315–280321 |
| ZFYVE26 |
chr14:68214907–68214914 |
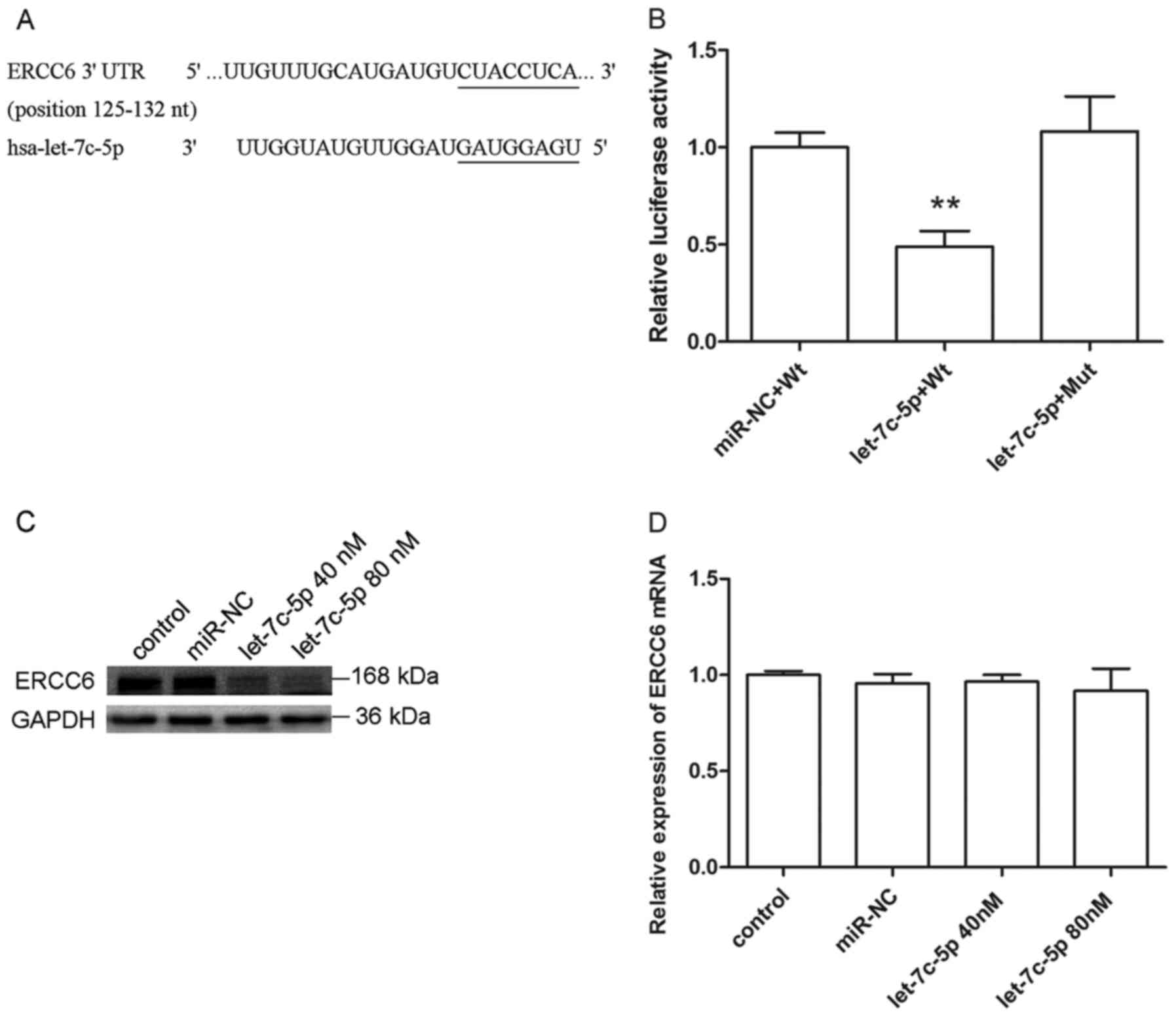
The let-7c-5p binding sites in 3′UTR of ERCC6
mRNA were predicted by TargetScan, as a result, there was only one
potential complementary sequence for let-7c-5p at nt 125–132
(Fig. 3A). To validate whether
let-7c-5p targets the 3′UTR of ERCC6 mRNA, we used the
firefly luciferase vector pGL3 as an internal control plasmid, and
co-transfected pGL3 and recombinant plasmid pRL-ERCC6-wt or
pRL-ERCC6-mut along with miR-NC or let-7c-5p mimics into MCF-7
cells. Luciferase activities were measured after co-transfection
for 48 h, as shown in Fig. 3B, dual
luciferase reporter assay demonstrated that the Renilla
luciferase activity of pRL-ERCC6-wt, which contained the wild-type
3′UTR fragment of ERCC6, was decreased by approximately 50%
by let-7c-5p, compared to the miR-NC group. In addition, the
luciferase activity of pRL-ERCC6-mut, in which the predicted
binding site has been deleted, was not suppressed significantly.
These results showed that ERCC6 was a direct target gene for
let-7c-5p in breast cancer.
To better understand the effects of let-7c-5p on
ERCC6, we evaluated the ERCC6 mRNA and protein expression
level in MCF-7 cells after treatment with let-7c-5p mimics for 48
h. As shown in Fig. 3C and D,
protein expression of ERCC6 was clearly inhibited by let-7c-5p, but
the mRNA expression of ERCC6 did not change significantly.
These findings suggested that let-7c-5p regulated ERCC6 mainly
through suppression of ERCC6 protein accumulation, but not
affecting the mRNA level.
Discussion
A large number of studies suggest that miRNAs are
involved in multiple human cancers, some are oncogenes, like
miR-183, which is over-expressed in synovial sarcoma,
rhabdomyosarcoma and colon cancer, targets the tumor suppressor
gene EGR1 and contributes to cell migration in these tumor
types (17). Some other miRNAs may
act as tumor suppressor genes, such as miR-100, which is found to
reduce colon cancer cell motility and growth by targeting Lgr5
in vitro (18). MiRNA let-7c-5p
acts as a cancer suppressor in different ways, such as preventing
early cancer progression through repressing HMGA2 expression
(19), inhibiting migration and
invasion of human non-small cell lung cancer and colorectal cancer
(13,14), and inducing cell apoptosis and
disrupting cell cycle in human hepatocellular carcinoma cells
(12). As for breast cancer,
let-7c-5p is downregulated in both tissues and serum of the
patients, and the expression of let-7c-5p is affected by
postmenopausal status (20). Higher
expression level of let-7c-5p is reported to be correlated with
better clinical prognosis of patients with estrogen
receptor-positive breast cancer (21), thus we hypothesize that let-7c-5p
may play a suppressive part in breast cancer.
In this study, we also confirmed that let-7c-5p
expression was decreased in breast cancer tissues compared with
that of matched adjacent tissues, moreover, we found that
overexpression of let-7c-5p could inhibit breast cancer cell
proliferation and induce cell apoptosis significantly, these
observations authenticated the hypothesis that let-7c-5p acted as a
tumor suppressor in breast cancer. Furthermore, we found that
let-7c-5p could bind to the 3′UTR of ERCC6 mRNA. However,
miRNAs regulate their target mRNAs in different ways, if the miRNAs
have perfect or near-perfect complementarity to the 3′UTR of target
mRNAs, the target mRNAs will be destroyed by miRNA, in this
situation, both the mRNA and protein levels will be affected. On
the other hand, the miRNAs have partial complementary sites in the
3′UTR of target mRNAs, the inhibition of protein accumulation will
be direct, but the mRNA level may not be affected significantly
(22,23). Herein, let-7c-5p has partial
complementarity to the 3′UTR of ERCC6 mRNA, as a result,
ERCC6 protein expression was suppressed but ERCC6 mRNA
expression was not inhibited obviously by let-7c-5p in MCF-7
cells.
ERCC6, also known as CSB, is a chromatin remodeling
factor. The protein encoded by ERCC6 gene is a DNA-binding
protein, which has important activity in nucleotide excision repair
(24). It has been identified that
mutations in ERCC6 gene are associated with the rare disease
Cockayne syndrome type B (25). In
recent years, ERCC6 is also showed to be involved in
cancers. Ma et al unraveled some genetic variants of
ERCC6 jointly taking part in the lung cancer development
among Chinese people (26). Chiu
et al and Liu et al also reported that ERCC6
polymorphisms were associated with the increased risk of gastric
cancer and oral cancer (27,28).
Chromosomal instability is observed in most cancers
including breast cancer. In cancer cells with high levels of
chromosomal instability and replication stress, DNA damage occurs
more frequently, and accumulation of DNA damage results in cell
malfunction and cell apoptosis (29,30).
So more DNA repair mechanisms need to be activated to correct the
damage and maintain the activity of cancer cells (31,32).
In the current study, we find that suppression of ERCC6 protein
expression by let-7c-5p is related with reduced growth ability of
breast cancer cells and higher rate of apoptosis, indicating ERCC6
may be involved in DNA damage repair in breast cancer and reduced
ERCC6 protein level leads to DNA damage accumulation.
Many clinical anticancer drugs target DNA directly
(33), such as cisplatin, which
interferes with DNA replication and induces DNA damage by binding
to DNA and forming DNA-cisplatin adducts. However, cisplatin
resistance is a serious problem in cancer treatment, and activation
of DNA repair mechanism is one of the main mechanisms involved in
cisplatin resistance (34). The
expression of excision repair cross-complementation group 1
(ERCC1), which participates in nucleotide excision repair, can be
induced by cisplatin treatment, and increased excision of
DNA-cisplatin adducts contributes to cisplatin resistance (35). As a DNA-binding protein, ERCC6 is
also likely to be associated with anticancer drugs resistance via
the nucleotide excision repair. Nevertheless, this potential
mechanism of ERCC6 has not been verified, thus more studies on
ERCC6 are still needed.
In conclusion, our study suggested that let-7c-5p
was a crucial contributor to cell apoptosis and inhibition of cell
proliferation in breast cancer, partially through targeting ERCC6.
These findings may provide a new potential strategy for breast
cancer treatment.
Acknowledgements
This study was supported by the Natural Science
Foundation of Zhejiang Province (LY15C050002 and LQ17H160013). This
study was also supported by the National Natural Science Foundation
of China (81402100), the Foundation of Health and Family Planning
Commission of Jiangsu Province (Q201408) and the Social Development
Foundation of Zhejiang (SH2016031, SH2014026).
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Filipowicz W, Bhattacharyya SN and
Sonenberg N: Mechanisms of post-transcriptional regulation by
microRNAs: Are the answers in sight? Nat Rev Genet. 9:102–114.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chen LT, Xu SD, Xu H, Zhang JF, Ning JF
and Wang SF: MicroRNA-378 is associated with non-small cell lung
cancer brain metastasis by promoting cell migration, invasion and
tumor angiogenesis. Med Oncol. 29:1673–1680. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Guo B, Liu L, Yao J, Ma R, Chang D, Li Z,
Song T and Huang C: miR-338-3p suppresses gastric cancer
progression through a PTEN-AKT axis by targeting P-REX2a. Mol
Cancer Res. 12:313–321. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Shi Y, Luo X, Li P, Tan J, Wang X, Xiang T
and Ren G: miR-7-5p suppresses cell proliferation and induces
apoptosis of breast cancer cells mainly by targeting REGγ. Cancer
Lett. 358:27–36. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Roush S and Slack FJ: The let-7 family of
microRNAs. Trends Cell Biol. 18:505–516. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Akao Y, Nakagawa Y and Naoe T: let-7
microRNA functions as a potential growth suppressor in human colon
cancer cells. Biol Pharm Bull. 29:903–906. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Esquela-Kerscher A, Trang P, Wiggins JF,
Patrawala L, Cheng A, Ford L, Weidhaas JB, Brown D, Bader AG and
Slack FJ: The let-7 microRNA reduces tumor growth in mouse models
of lung cancer. Cell Cycle. 7:759–764. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Nadiminty N, Tummala R, Lou W, Zhu Y,
Zhang J, Chen X, White RW eVere, Kung HJ, Evans CP and Gao AC:
MicroRNA let-7c suppresses androgen receptor expression and
activity via regulation of Myc expression in prostate cancer cells.
J Biol Chem. 287:1527–1537. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Nwaeburu CC, Bauer N, Zhao Z, Abukiwan A,
Gladkich J, Benner A and Herr I: Up-regulation of microRNA let-7c
by quercetin inhibits pancreatic cancer progression by activation
of Numbl. Oncotarget. 7:58367–58380. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhu X, Wu L, Yao J, Jiang H, Wang Q, Yang
Z and Wu F: MicroRNA let-7c inhibits cell proliferation and induces
cell cycle arrest by targeting CDC25A in human hepatocellular
carcinoma. PLoS One. 10:e01242662015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Han HB, Gu J, Zuo HJ, Chen ZG, Zhao W, Li
M, Ji DB, Lu YY and Zhang ZQ: Let-7c functions as a metastasis
suppressor by targeting MMP11 and PBX3 in colorectal cancer. J
Pathol. 226:544–555. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhao B, Han H, Chen J, Zhang Z, Li S, Fang
F, Zheng Q, Ma Y, Zhang J, Wu N, et al: MicroRNA let-7c inhibits
migration and invasion of human non-small cell lung cancer by
targeting ITGB3 and MAP4K3. Cancer Lett. 342:43–51. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Mao Y, Wu S, Zhao R and Deng Q: MiR-205
promotes proliferation, migration and invasion of nasopharyngeal
carcinoma cells by activation of AKT signalling. J Int Med Res.
44:231–240. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wu Q, Jin H, Yang Z, Luo G, Lu Y, Li K,
Ren G, Su T, Pan Y, Feng B, et al: MiR-150 promotes gastric cancer
proliferation by negatively regulating the pro-apoptotic gene EGR2.
Biochem Biophys Res Commun. 392:340–345. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sarver AL, Li L and Subramanian S:
MicroRNA miR-183 functions as an oncogene by targeting the
transcription factor EGR1 and promoting tumor cell migration.
Cancer Res. 70:9570–9580. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhou MK, Liu XJ, Zhao ZG and Cheng YM:
MicroRNA-100 functions as a tumor suppressor by inhibiting Lgr5
expression in colon cancer cells. Mol Med Rep. 11:2947–2952.
2015.PubMed/NCBI
|
|
19
|
Park SM, Shell S, Radjabi AR, Schickel R,
Feig C, Boyerinas B, Dinulescu DM, Lengyel E and Peter ME: Let-7
prevents early cancer progression by suppressing expression of the
embryonic gene HMGA2. Cell Cycle. 6:2585–2590. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li XX, Gao SY, Wang PY, Zhou X, Li YJ, Yu
Y, Yan YF, Zhang HH, Lv CJ, Zhou HH, et al: Reduced expression
levels of let-7c in human breast cancer patients. Oncol Lett.
9:1207–1212. 2015.PubMed/NCBI
|
|
21
|
Sun X, Xu C, Tang SC, Wang J, Wang H, Wang
P, Du N, Qin S, Li G, Xu S, et al: Let-7c blocks estrogen-activated
Wnt signaling in induction of self-renewal of breast cancer stem
cells. Cancer Gene Ther. 23:83–89. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Pillai RS, Bhattacharyya SN and Filipowicz
W: Repression of protein synthesis by miRNAs: How many mechanisms?
Trends Cell Biol. 17:118–126. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Selby CP and Sancar A: Human
transcription-repair coupling factor CSB/ERCC6 is a DNA-stimulated
ATPase but is not a helicase and does not disrupt the ternary
transcription complex of stalled RNA polymerase II. J Biol Chem.
272:1885–1890. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ghai SJ, Shago M, Shroff M and Yoon G:
Cockayne syndrome caused by paternally inherited 5 Mb deletion of
10q11.2 and a frameshift mutation of ERCC6. Eur J Med Genet.
54:272–276. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ma H, Hu Z, Wang H, Jin G, Wang Y, Sun W,
Chen D, Tian T, Jin L, Wei Q, et al: ERCC6/CSB gene polymorphisms
and lung cancer risk. Cancer Lett. 273:172–176. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chiu CF, Tsai MH, Tseng HC, Wang CL, Tsai
FJ, Lin CC and Bau DT: A novel single nucleotide polymorphism in
ERCC6 gene is associated with oral cancer susceptibility in
Taiwanese patients. Oral Oncol. 44:582–586. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Liu JW, He CY, Sun LP, Xu Q, Xing CZ and
Yuan Y: The DNA repair gene ERCC6 rs1917799 polymorphism is
associated with gastric cancer risk in Chinese. Asian Pac J Cancer
Prev. 14:6103–6108. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Leung HW, Wu CH, Lin CH and Lee HZ:
Luteolin induced DNA damage leading to human lung squamous
carcinoma CH27 cell apoptosis. Eur J Pharmacol. 508:77–83. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kaina B: DNA damage-triggered apoptosis:
Critical role of DNA repair, double-strand breaks, cell
proliferation and signaling. Biochem Pharmacol. 66:1547–1554. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Minocherhomji S, Ying S, Bjerregaard VA,
Bursomanno S, Aleliunaite A, Wu W, Mankouri HW, Shen H, Liu Y and
Hickson ID: Replication stress activates DNA repair synthesis in
mitosis. Nature. 528:286–290. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Burrell RA, McClelland SE, Endesfelder D,
Groth P, Weller MC, Shaikh N, Domingo E, Kanu N, Dewhurst SM,
Gronroos E, et al: Replication stress links structural and
numerical cancer chromosomal instability. Nature. 494:492–496.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kasyanenko N: DNA as a target for
anticancer drugs based on the coordination compounds of metals.
FEBS J. 280:89. 2013.
|
|
34
|
O'Grady S, Finn SP, Cuffe S, Richard DJ,
O'Byrne KJ and Barr MP: The role of DNA repair pathways in
cisplatin resistant lung cancer. Cancer Treat Rev. 40:1161–1170.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhu H, Luo H, Zhang W, Shen Z, Hu X and
Zhu X: Molecular mechanisms of cisplatin resistance in cervical
cancer. Drug Des Devel Ther. 10:1885–1895. 2016. View Article : Google Scholar : PubMed/NCBI
|