Introduction
Retinoblastoma is an aggressive eye cancer and the
most common intraocular cancer of infancy and childhood (1). Previous studies have indicated that
various vulnerable groups, such as earthquake survivors (2) and rural-to-urban migrants (3), were are in poor health, suffer from
retinoblastoma. In addition, studies on retinoblastoma have led to
the discovery of the first tumor-suppressor gene, RB1, the
loss of which accounts for aberrant cell cycle arrest and the
tumorigenesis of retinoblastoma. Despite research into the
RB family and cell cycle control, the specific pathways that
contribute to the RB loss in tumorigenesis remain largely
unknown. In addition, mice with RB deletion alone were not
retinoblastoma-prone (4),
suggesting the existence of other mechanisms that cooperatively
contributes to the initiation of retinoblastoma.
It is increasingly recognized that microRNAs
(miRNAs) are important regulators of cancer. They are short
non-coding RNAs that post-transcriptionally modulate the expression
of cancer-related genes (5). In
retinoblastoma, a number of miRNAs have been identified to
synergize with loss of the RB family to regulate cell cycles
(6–8). Besides the RB family, the
Runt-related transcription factor 3 (Runx3) gene, another
tumor suppressor in retinoblastoma, was also found to be regulated
by miRNAs. Runx3 is located on the chromosome 1p36 and plays
an important role in mammalian development (9). The loss of Runx3 has been considered
as a prognostic marker for bladder tumor (10), gastric cancer (11) and glioblastoma (12). Previously, the link between miR-106b
and Runx3 has been demonstrated in laryngeal carcinoma
(13). By binding to the
3-untranslated region (3′-UTR) of Runx3, miR-106b
downregulated Runx3 expression, which consequently abolished
the proliferation and invasion of laryngeal carcinoma cells
(13). In retinoblastoma, although
the downregulation of Runx3 has been documented, the
mechanism involved in the regulation of Runx3 has not been
investigated.
Surgery remains the primary treatment for
retinoblastoma to date (14).
However, being an invasive procedure, surgery exposes patients to
substantial risk of losing vision (1). Chemotherapy has also been exploited
for retinoblastoma, however chemotherapy is associated with
potential toxicity and it also increases the risk of secondary
cancers (15,16). Therefore, strategies capable of
modulating retinoblastoma-causative genes are urgently needed to
improve the clinical outcome of retinoblastoma therapy and minimize
harm to the eye.
In the present study, we strived to elucidate the
role of miR-106b in retinoblastoma and investigated the correlation
between miR-106b and Runx3. We revealed that inhibition of
miR-106b led to decreased cell proliferation and migration of human
Y79 retinoblastoma cells. Concurrently, upregulation of
Runx3 was observed. The results shed light on the target of
miR-106b in retinoblastoma, and provide a novel strategy for
inhibiting retinoblastoma progression.
Materials and methods
Cell culture and transfection
Human retinoblastoma Y79 cells were acquired from
the American Type Culture Collection (ATCC; Rockville, MD, USA),
and were maintained by Beina Co. Ltd. (Beijing, China). Dulbecco's
modified Eagles medium (DMEM; Invitrogen, Carlsbad, CA, USA)
containing 5% fetal bovine serum (FBS; HyClone, Logan, UT, USA),
2.0 g/l sodium bicarbonate, 1×105 IU/l penicillin and
100 mg/l streptomycin were used for cell culture. The cells were
cultured in an incubator maintained at 37°C with saturated humidity
and 5% CO2. ASO-miR-106b or ASO-control of 50 nM was
used for transfection with Lipofectamine® 2000
(Invitrogen) according to the manufacturer's recommendations.
Briefly, cells were seeded into 6-well plates at a density of
105 cells/well and cultured overnight. In each well, 100
pmol of ASO-control or ASO-miR-106b was added along with 5 µl
Lipofectamine® 2000. ASO-miR-106b and ASO-control were
prepared by Jiman Phamaceuticals (Shanghai, China).
Cell viability assay
For the cell viability assay, cells of
2.5–3×104 cells/well were first seeded into 96-well
plates, with each well containing 100 µl medium, and treated with
ASO-miR-106b or ASO-control. After culturing for 24, 48 and 72 h,
10 µl Cell Counting Kit-8 (CCK-8) solution was added into the
medium, and incubated for another 4–5 h. The absorbance of each
well at 538 nm was assessed to calculate the cell viability using
the following equation:
Cell
viability=ODuntreated–ODtreatedODuntreated
Transwell cell migration assay
The Transwell apparatus was coated with Matrigel
(both from BD Biosciences, San Jose, CA, USA) of 20–30 µl and
allowed to gel overnight at 37°C. The other side of the membrane
was coated with fibronectin (Invitrogen). Y79 cells
(5×105 cells/ml) of 2,000 µl were added to the chamber
and cultured for 24 h before removing the cells on the membrane.
The membrane on the lower chamber was collected and fixed with
formalin for 30 min, and stained with hematoxylin. Dehydration was
carried out using the standard procedure using ethanol. The
membrane, which was dehydrated with ethanol and xylene, was mounted
onto cover slips, followed by cell counting in 4 fields of view.
The average cell number was calculated.
Apoptosis analysis using flow
cytometry
Cells collected at 48 and 72 h after transfection
were washed with 0.01 mol/l phosphate-buffered saline (PBS), and
centrifuged at 1,500 rpm/m for 5 min. The supernatant was then
discarded. Cell pellets were re-suspected to ensure a cell density
of 1×106 cells/ml. A cell suspension of 500 µl was then
dispensed into a microcentrifuge tube, followed by the addition of
5 µl Annexin V-FITC and 10 µl propidium iodide (PI) (Lianke Biology
Co., Ltd., Hangzhou, China) for staining for 10 min at room
temperature. Flow cytometry was used to detect cells with apoptotic
activity. Experiments were performed in triplicate for each
sample.
RT-PCR
Total RNA of the sample was extracted using a RNA
Extract kit (Promega, Madison, WI, USA). Synthesis of cDNA was
performed using 1 µg purified RNA and a kit acquired from Takara
(Shiga, Japan). RT-PCR was carried out using SYBR-Green mixture
(Takara). Quantification was performed using the 2−ΔΔCt
method. Primers used in the present study were as follows:
5′-GGATTTGGTCGTATTGGGCG-3′ (sense) and 5′-TACTTCTCATGGTTCACAC-3′
(antisense) for GAPDH; 5′-TGCGGCAACACCAGTCGATGG-3′ (sense) and
5′-CCAGTGCAGGGTCCGAGGT-3′ (antisense) for miR-106b;
5′-TGGCAGGCAATGACGA-3′ (sense) and 5′-CAGGGAACGGCTTGGT-3′
(antisense) for Runx3. Primers were synthesized by Sangon
Biotech (Shanghai, China).
Western blot analysis
Cells transfected for 48 and 72 h were lyzed using
RIPA cell lysis buffer (Thermo Fisher Scientific, Inc., Waltham,
MA, USA). The protein concentration was determined using the BCA
assay (Pierce, Rockford, IL, USA). SDS-PAGE was performed using 40
µg of protein. The protein was then transferred onto the
polyvinylidene difluoride (PVDF) membrane, followed by blocking
with 50 g/l non-fat milk for 1.5 h. The goat anti-human
Runx3 antibody (Santa Cruz Biotechnology, Santa Cruz, CA,
USA) was diluted (1:500) in 1% BSA and incubated with the membrane
at 4°C overnight. After washing with TBST (1% Tween-20) 3 times (6
min each time), an HRP-conjugated anti-goat antibody (1:5,000
dilution; Boster Biotechnology Co. Ltd., Wuhan, China) was applied
to the membrane and was incubated for 2 h. Visualization of the
protein band was performed by adding ECL substrates in the dark.
The band intensities of GAPDH were used to normalize the expression
of the Runx3 protein.
Statistical analysis
Statistical analysis was performed using SPSS 19.0
software package (IBM, Chicago, IL, USA). All data were presented
as the mean ± SD. Differences were considered significant when
P<0.05.
Results
Transfection of ASO-miR-106b
downregulates miR-106b expression
To validate the efficacy of ASO-miR-106b in
downregulating miR-106b expression, RT-PCR was performed in Y79
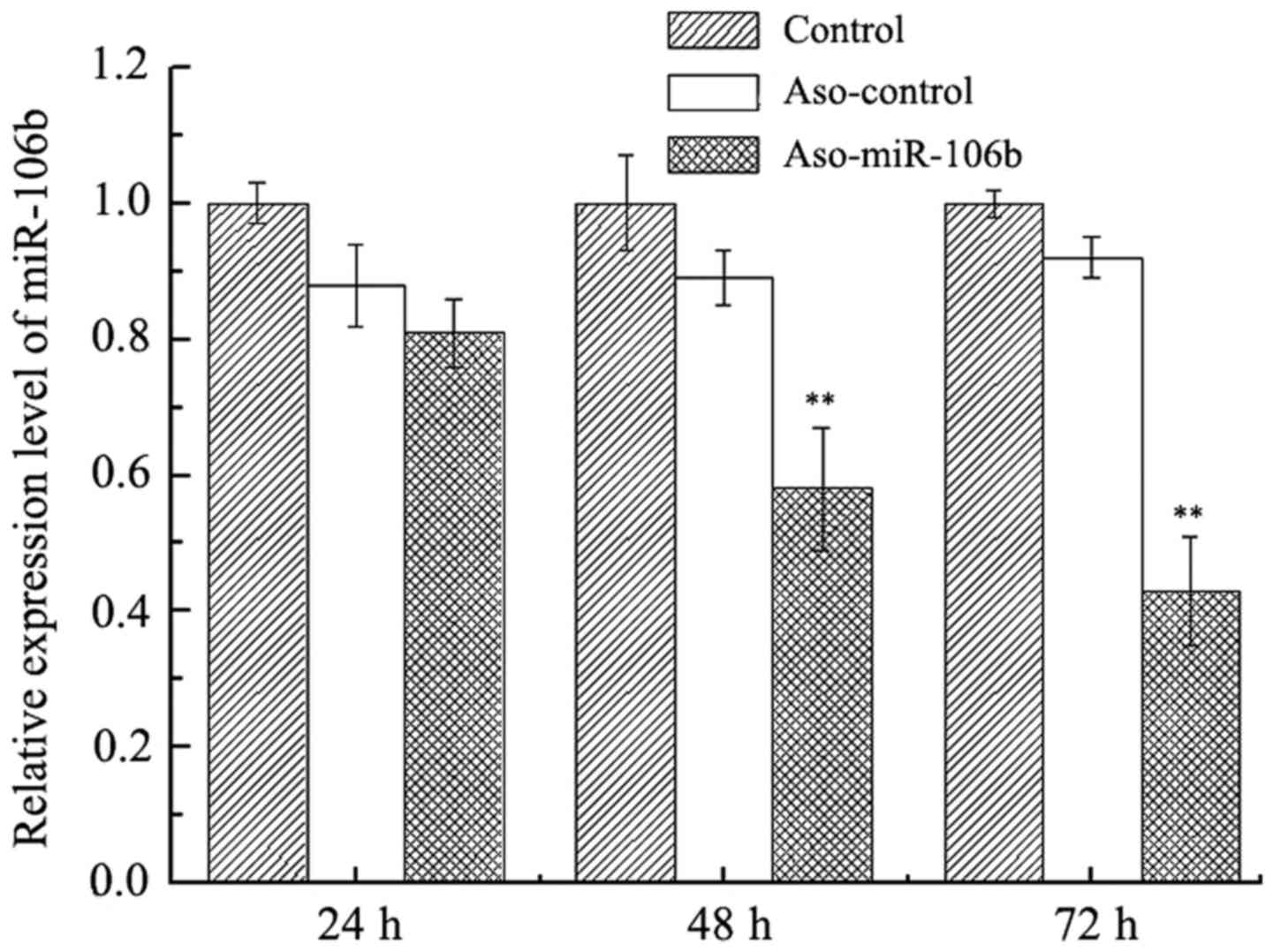
cells transfected with ASO-miR-106b or ASO-control. As shown in
Fig. 1, at 24 h after transfection,
no clear difference in miR-106b levels was seen among all groups.
After 48 and 72 h, a significant downregulation of miR-106b was
observed in cells transfected with miR-106b (P<0.01), suggesting
that ASO-miR-106 effectively inhibited the expression of miR-106.
Conversely, no significant differences were observed in cells
transfected with the ASO-control (P>0.05).
Downregulation of miR-106b decreases
cell viability
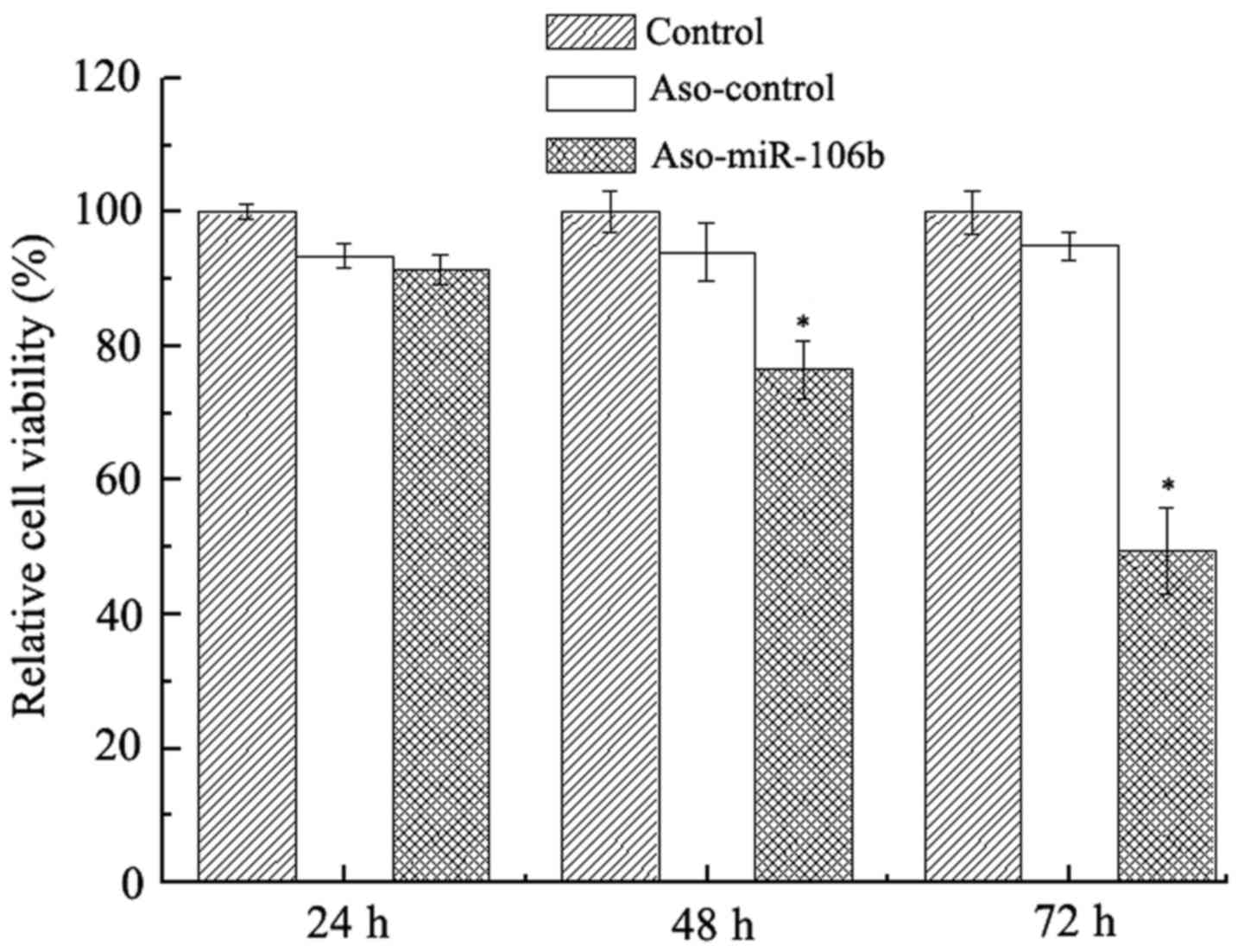
Concomitant with the inhibition of miR-106b
expression, the viability of Y79 was also decreased. As shown in
Fig. 2, while no significant
viability suppression was seen at 24 h after transfection, at 48
and 72 h after transfection, cell viability was significantly
decreased, as revealed by CCK-8 assay (P<0.05).
Downregulation of miR-106b decreases
cell migration
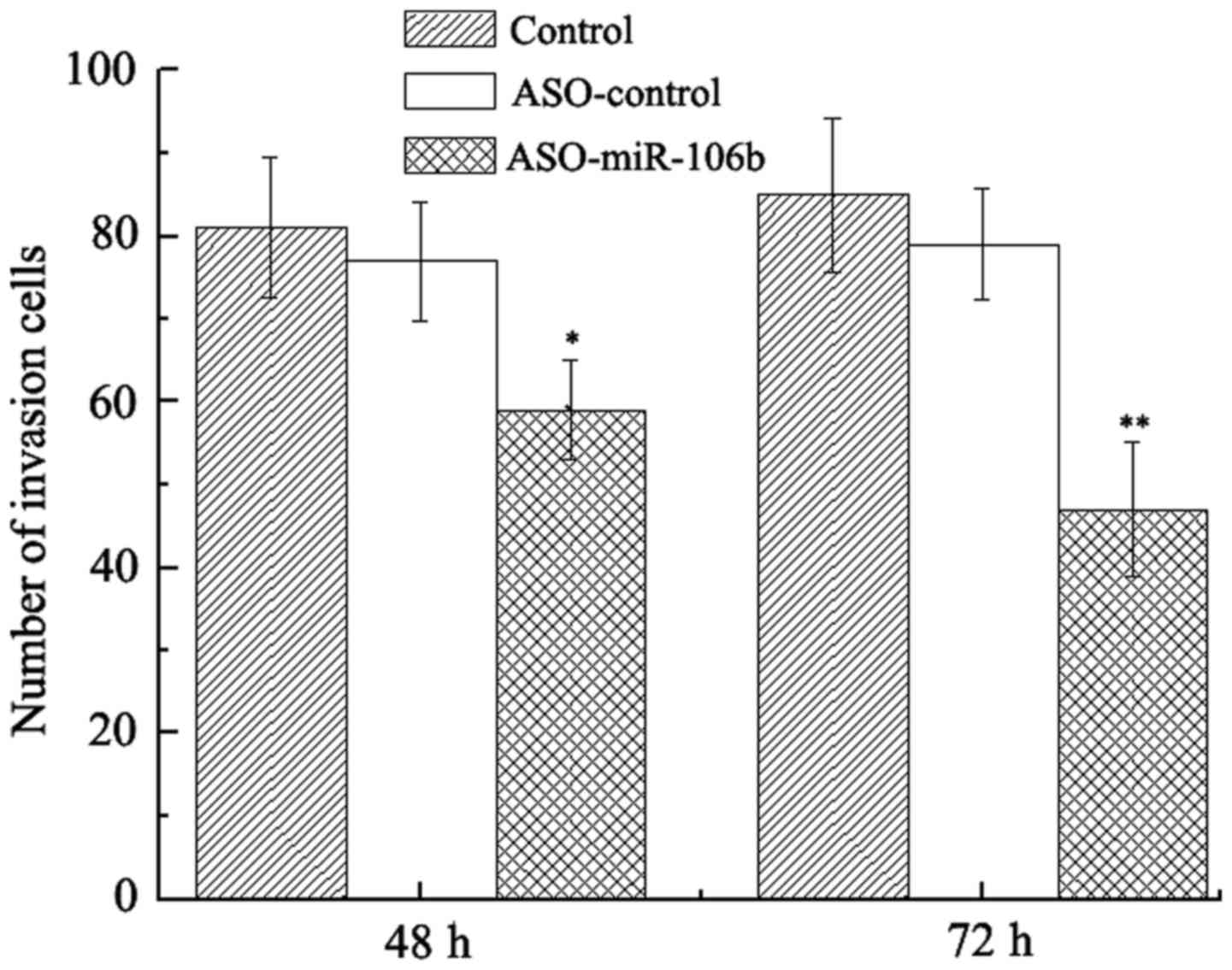
We next examined whether downregulation of miR-106b
decreased the migration of Y79 cells. As expected, at 48 and 72 h
after transfection, the migration of Y79 cells was significantly
decreased as indicated by Transwell assay (P<0.05) (Fig. 3). In contrast, transfection with
ASO-control did not suppress cell migration.
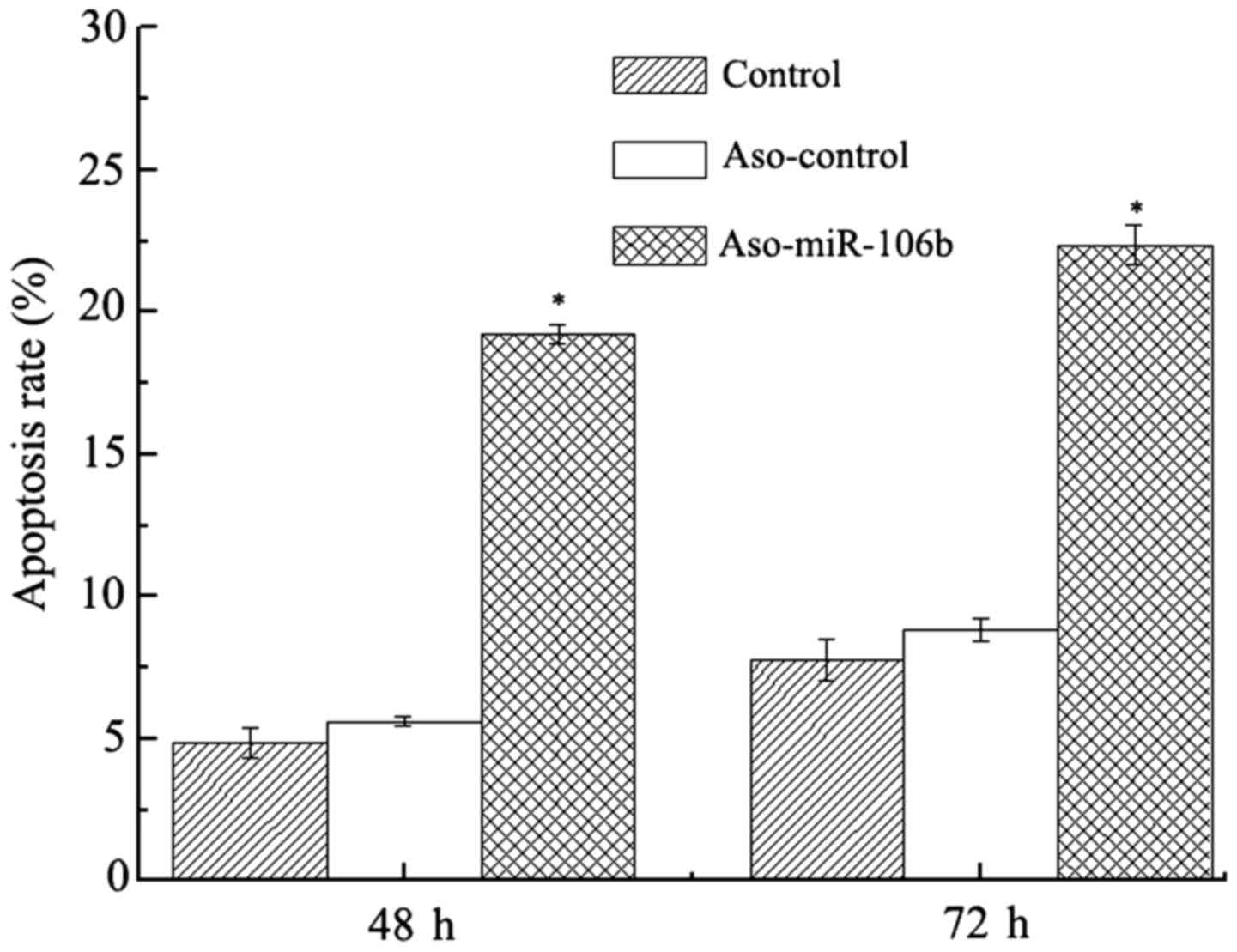
Apoptosis is increased with miR-106
downregulation
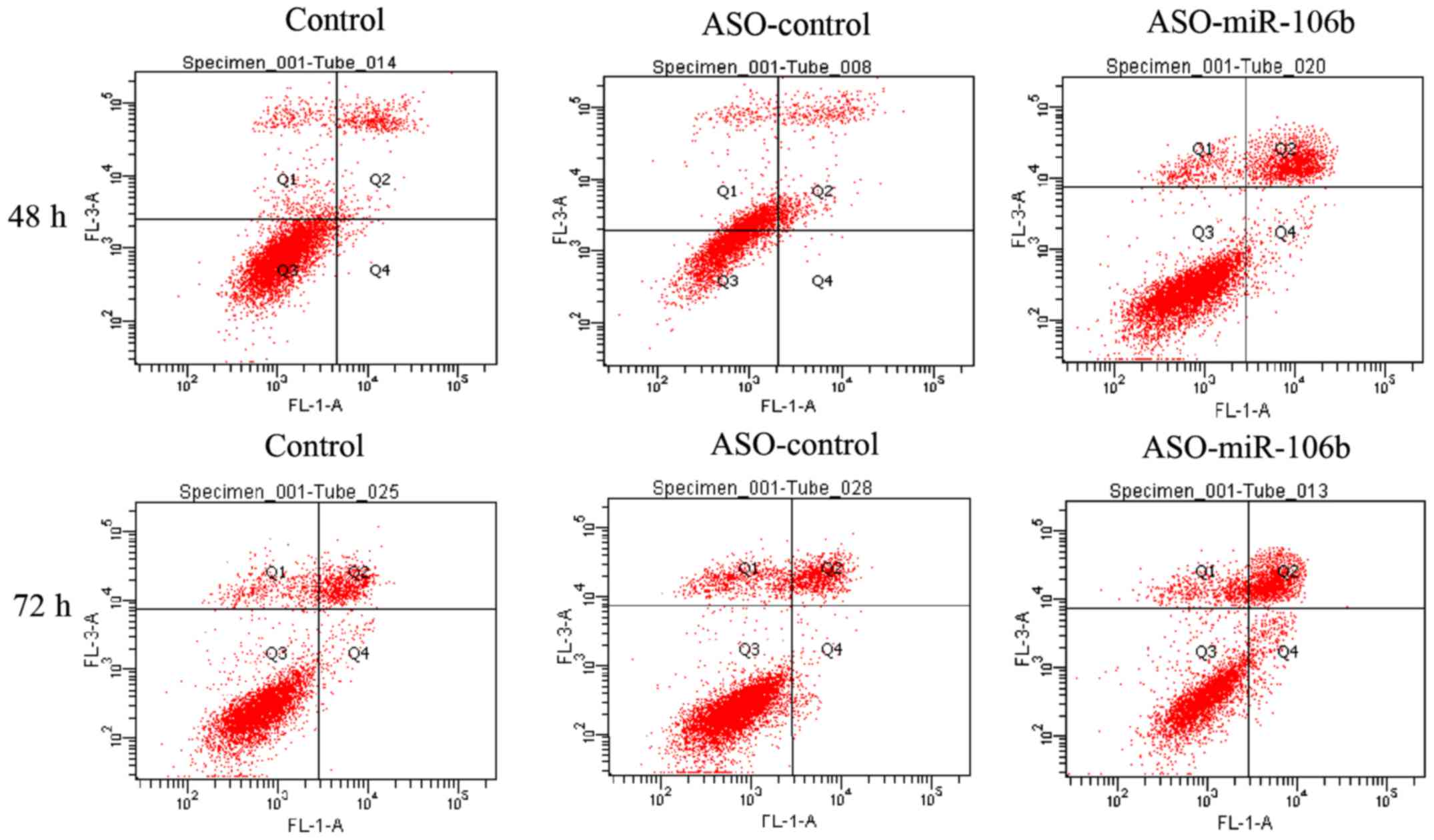
After transfection for 48 and 72 h, the apoptosis in
Y79 cells was evaluated with Annexin-PI staining and flow
cytometry. Consistent with the decreased cell viability and
migration in Y79 cells after ASO-miR-106b transfection, a shift of
cell population toward the Annex+-PI+
population was recorded (Figs. 4
and 5), indicating an increase in
apoptotic activity.
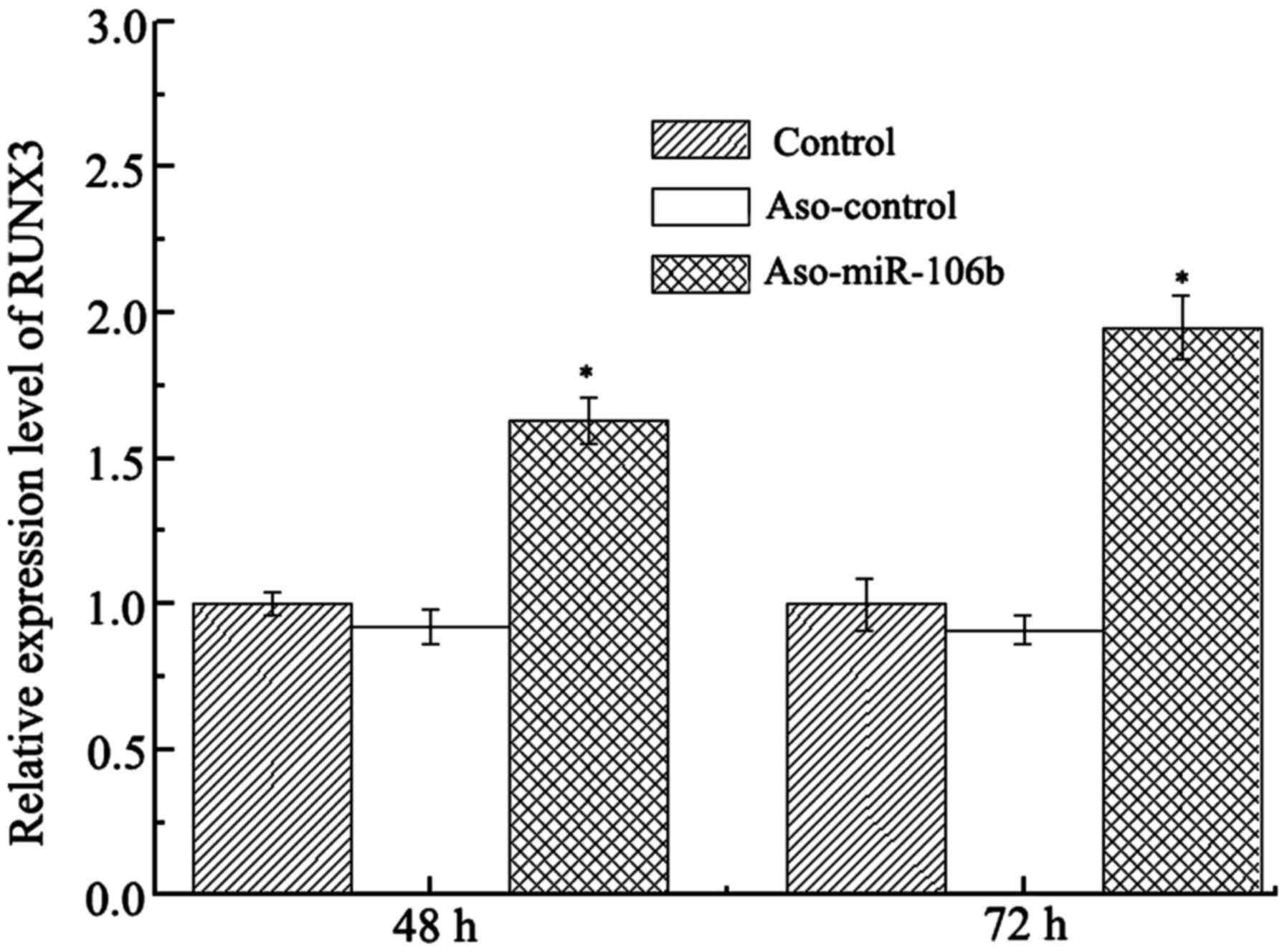
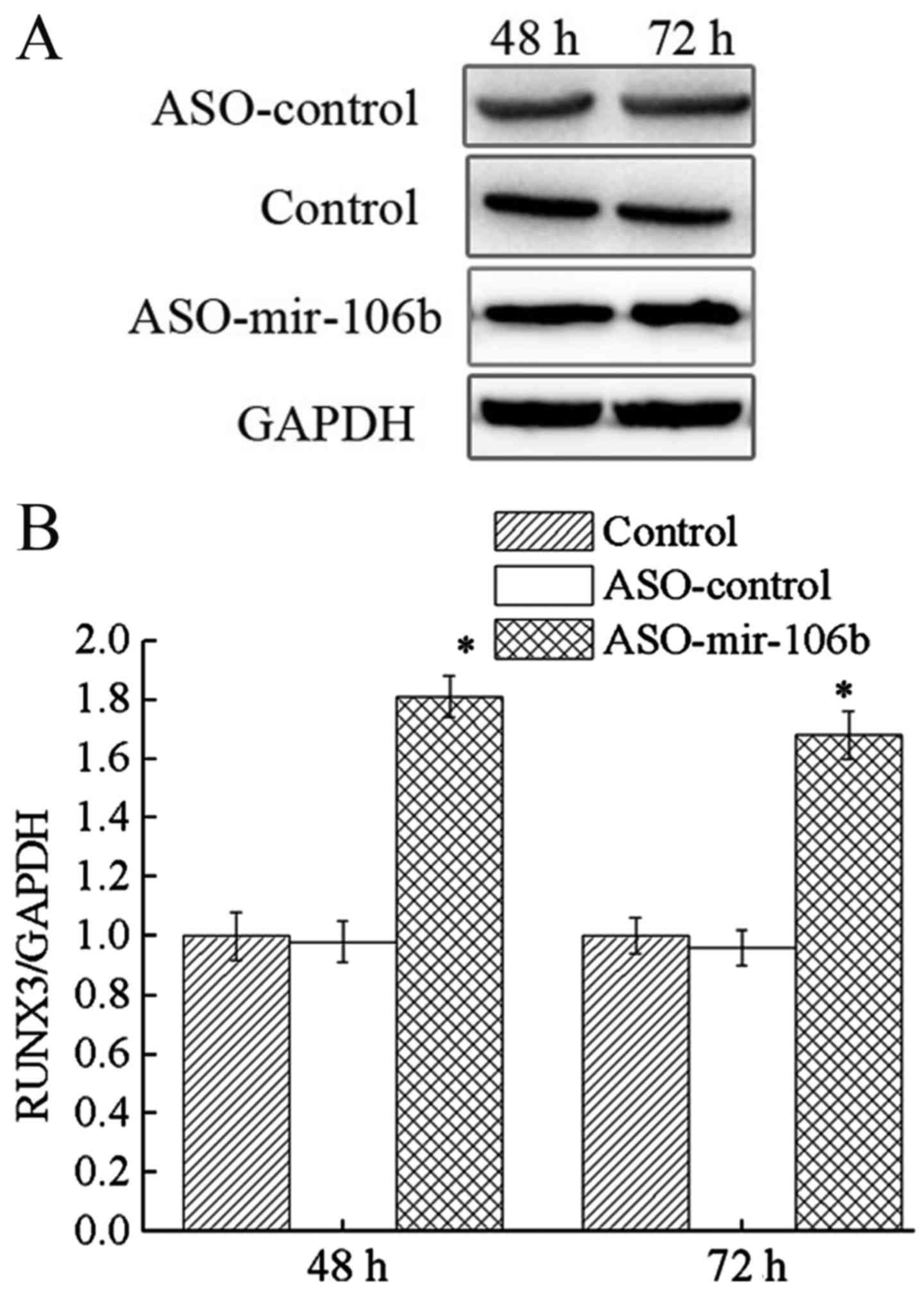
Downregulation of miR-106b increases
Runx3 expression
To explore the target of miR-106b in retinoblastoma,
we analyzed the expression level of Runx3 mRNA and protein
in Y79 cells after ASO-miR-106b transfection. The results revealed
that both Runx3 mRNA and protein expression levels were
upregulated after ASO-miR-106b transfection (P<0.05) (Figs. 6 and 7). Given the role of Runx3 as a tumor
suppressor, this Runx3 upregulation after miR-106b
inhibition is consistent with the diminished cell proliferation and
migration. Therefore, Runx3 acts as a target of miR-106b in the
regulation of retinoblastoma.
Discussion
A vast majority of people suffer from cancer
(17,18). By fine-tuning the expression of
multiple genes post-transcriptionally, miRNAs offer a new paradigm
for re-modulating the gene network wired to the thriving cancer
(19). Numerous miRNA-based
strategies for correcting aberrant gene expression in cancer have
emerged. These miRNAs may assume tumor-promoting or
tumor-inhibiting roles in cancer. For example, given that the loss
of RB1 expression is a hallmark of cancer, miR-106a
inhibition was employed to reverse the downregulation of RB1
(20). In another study, miR-192
was ectopically overexpressed, which inhibited cell proliferation
and induced cell apoptosis in lung cancer cells (21). For retinoblastoma, recent miRNA
microarray analysis yielded a panel of miRNAs as essential
effectors that modulate cancer progression, metastasis and
resistance (22). Nevertheless,
these miRNAs have yet to be utilized as therapeutic targets in
retinoblastoma. Furthermore, despite extensive studies on the
etiology of the disease and genetic diagnosis approaches (23), gene therapies for retinoblastoma are
rarely studied. Therefore, in the present study, we set forth to
explore the therapeutic value of miR-106b in retinoblastoma. The
present study is preceded by much effort in correlating miR-106b
overexpression to the malignant phenotype and resistance of cancer
(24,25). We demonstrated that the inhibition
of miR-106b induced a decrease in Y79 retinoblastoma cell viability
and migration, and induced cell apoptosis. Instead of directly
targeting oncogenes of retinoblastoma, our approach was to inhibit
the expression of miR-106b with antisense oligonucleotides (ASO) to
exert the antitumor effects. Our data potentiated the development
of an in vivo gene therapy strategy that targets miR-106b; a
strategy that lowers the non-specific toxicity commonly observed
with chemotherapy drugs. With the advances in RNA-delivery systems,
we could envision that retinoblastoma therapy based on miR-106b
inhibition may improve the clinical outcome of patients and exempt
them from painful and risky surgery.
Furthermore, we revealed that the antitumor effect
of anti-miR-106b therapy was mediated by Runx3. Similar to
RB1, Runx3 serves as a tumor suppressor and its
methylation is pivotal to the transforming growth factor-β (TGF-β)
pathway. Since its link to epithelial-to-mesenchymal transition and
cancer metastasis, the loss of Runx3 significantly affects
the clinical outcome of cancer patients (26). In spite of efforts in unraveling the
role of Runx3 in cancer progression, very few
Runx3-based therapeutic strategies have been devised.
Previously, ectopic expression of Runx3 was exploited as an
antitumor strategy, whereby the restoration of Runx3 was
shown to drastically suppress tumor growth (26). The findings here revealed that
inhibition of miR-106b is another viable avenue to upregulate
Runx3. This mirrors recent evidence that revealed that
miR-106b significantly promotes TGF-β signaling and cancer
metastasis (25,27). However, the fact that miR-106b is
also an important player in the PTEN/PIK3/AKT pathway (27), WNT pathway (28) and RB family enhances its clinical
value as a target in cancer.
In summary, we demonstrated that anti-miR-106b
therapy effectively inhibited Y79 retinoblastoma cell viability,
proliferation and induced cell apoptosis in vitro.
Runx3 was found to be a target of miR-106b, and the
inhibition of miR-106b upregulated Runx3. Our data is
significant for the development of novel strategies in gene therapy
for retinoblastoma. Further in vivo studies are warranted to
corroborate the role of miR-106b in retinoblastoma, and potentially
apply this strategy to the clinic.
Acknowledgements
The present study received no specific grant from
any funding agency in the public, commercial or non-profit
sectors.
References
|
1
|
Dimaras H, Kimani K, Dimba EA, Gronsdahl
P, White A, Chan HS and Gallie BL: Retinoblastoma. Lancet.
379:1436–1446. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Liang Y and Cao R: Employment assistance
policies of Chinese government play positive roles! The impact of
post-earthquake employment assistance policies on the
health-related quality of life of Chinese earthquake populations.
Soc Indic Res. 120:835–857. 2014. View Article : Google Scholar
|
|
3
|
Liang Y and Guo M: Utilization of health
services and health-related quality of life research of
rural-to-urban migrants in china: A cross-sectional analysis. Soc
Indic Res. 120:277–295. 2015. View Article : Google Scholar
|
|
4
|
Clarke AR, Maandag ER, van Roon M, van der
Lugt NM, van der Valk M, Hooper ML, Berns A and te Riele H:
Requirement for a functional Rb-1 gene in murine development.
Nature. 359:328–330. 1992. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Conkrite K, Sundby M, Mukai S, Thomson JM,
Mu D, Hammond SM and MacPherson D: miR-17~92 cooperates with RB
pathway mutations to promote retinoblastoma. Genes Dev.
25:1734–1745. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Xu Y, Wang K, Gao W, Zhang C, Huang F, Wen
S and Wang B: MicroRNA-106b regulates the tumor suppressor RUNX3 in
laryngeal carcinoma cells. FEBS Lett. 587:3166–3174. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Trompeter HI, Abbad H, Iwaniuk KM, Hafner
M, Renwick N, Tuschl T, Schira J, Müller HW and Wernet P: MicroRNAs
MiR-17, MiR-20a, and MiR-106b act in concert to modulate E2F
activity on cell cycle arrest during neuronal lineage
differentiation of USSC. PLoS One. 6:e161382011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tucker A and Sharpe P: The cutting-edge of
mammalian development; how the embryo makes teeth. Nat Rev Genet.
5:499–508. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kim EJ, Kim YJ, Jeong P, Ha YS, Bae SC and
Kim WJ: Methylation of the RUNX3 promoter as a potential prognostic
marker for bladder tumor. J Urol. 180:1141–1145. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Guo WH, Weng LQ, Ito K, Chen LF, Nakanishi
H, Tatematsu M and Ito Y: Inhibition of growth of mouse gastric
cancer cells by Runx3, a novel tumor suppressor. Oncogene.
21:8351–8355. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mueller W, Nutt CL, Ehrich M,
Riemenschneider MJ, von Deimling A, van den Boom D and Louis DN:
Downregulation of RUNX3 and TES by hypermethylation in
glioblastoma. Oncogene. 26:583–593. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Honavar SG, Shields CL, Shields JA,
Demirci H and Naduvilath TJ: Intraocular surgery after treatment of
retinoblastoma. Arch Ophthalmol. 119:1613–1621. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Errico A: Cancer therapy: Retinoblastoma -
chemotherapy increases the risk of secondary cancer. Nat Rev Clin
Oncol. 11:623. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Smith SJ and Smith BD: Evaluating the risk
of extraocular tumour spread following intravitreal injection
therapy for retinoblastoma: A systematic review. Br J Ophthalmol.
97:1231–1236. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tong AW and Nemunaitis J: Modulation of
miRNA activity in human cancer: A new paradigm for cancer gene
therapy? Cancer Gene Ther. 15:341–355. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liang Y: Satisfaction with economic and
social rights and quality of life in a post-disaster zone in China:
Evidence from earthquake-prone Sichuan. Disaster Med Public Health
Prep. 9:111–118. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Liang Y: Correlations between
health-related quality of life and interpersonal trust: Comparisons
between two generations of Chinese rural-to-urban migrants. Soc
Indic Res. 123:677–700. 2015. View Article : Google Scholar
|
|
19
|
Volinia S, Calin GA, Liu CG, Ambs S,
Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et
al: A microRNA expression signature of human solid tumors defines
cancer gene targets. Proc Natl Acad Sci USA. 103:pp. 2257–2261.
2006; View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Feng S, Cong S, Zhang X, Bao X, Wang W, Li
H, Wang Z, Wang G, Xu J, Du B, et al: MicroRNA-192 targeting
retinoblastoma 1 inhibits cell proliferation and induces cell
apoptosis in lung cancer cells. Nucleic Acids Res. 39:6669–6678.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhao JJ, Yang J, Lin J, Yao N, Zhu Y,
Zheng J, Xu J, Cheng JQ, Lin JY and Ma X: Identification of miRNAs
associated with tumorigenesis of retinoblastoma by miRNA microarray
analysis. Childs Nerv Syst. 25:13–20. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang J, Benavente CA, McEvoy J,
Flores-Otero J, Ding L, Chen X, Ulyanov A, Wu G, Wilson M, Wang J,
et al: A novel retinoblastoma therapy from genomic and epigenetic
analyses. Nature. 481:329–334. 2012.PubMed/NCBI
|
|
23
|
Zheng L, Zhang Y, Liu Y, Zhou M, Lu Y,
Yuan L, Zhang C, Hong M, Wang S and Li X: MiR-106b induces cell
radioresistance via the PTEN/PI3K/AKT pathways and p21 in
colorectal cancer. J Transl Med. 13:2522015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gong C, Qu S, Liu B, Pan S, Jiao Y, Nie Y,
Su F, Liu Q and Song E: MiR-106b expression determines the
proliferation paradox of TGF-β in breast cancer cells. Oncogene.
34:84–93. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wei D, Gong W, Oh SC, Li Q, Kim WD, Wang
L, Le X, Yao J, Wu TT, Huang S, et al: Loss of RUNX3 expression
significantly affects the clinical outcome of gastric cancer
patients and its restoration causes drastic suppression of tumor
growth and metastasis. Cancer Res. 65:4809–4816. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Smith AL, Iwanaga R, Drasin DJ, Micalizzi
DS, Vartuli RL and Ford HL: Abstract A20: The Six1-regulated
miR-106b-25 cluster is a mediator of the tumor promotional effects
of TGF-β signaling in human breast cancer. Cancer Res. 72 Suppl
2:A20. 2012. View Article : Google Scholar
|
|
27
|
Li YL, Wang YH, Zhou H, Yong YH and Cao
YD: miR-106b promoted growth and inhibited apoptosis of
nasopharyngeal carcinoma cells by suppressing the tumor suppressor
PTEN. Int J Clin Exp Pathol. 9:7078–7086. 2016.
|
|
28
|
Malcomson FC, Willis ND, McCallum I, Xie
L, Kelly S, Bradburn M, Belshaw NJ, Johnson IT and Mathers JC:
Differences in the expression of microRNAs implicated in colorectal
carcinogenesis and involved in the WNT signalling pathway in the
macroscopically-normal epithelium of people at higher-risk of
colorectal cancer. Proc Nutr Soc. 74(OCE1): pp. E462015; View Article : Google Scholar
|