Mesenchymal stem cells (MSCs) are multipotent stem
cells that have attracted a great interest for their noteworthy
multilineage differentiation potential (1,2) and
hypoimmunogenic features (3,4). All
these properties of MSCs have led to their application in
regenerative medicine (5,6). Furthermore, due to their tumor-homing
ability (7), MSCs are currently
used as cell-based delivery systems of therapeutic proteins for
cancer treatment (8,9). In particular, through the tumor-homing
ability of MSCs, the localized production of a specific therapeutic
protein is more helpful than the systemic use of a recombinant
protein considering both the effective in situ concentration
of the molecule and the reduction of unwanted systemic actions. To
this end, pro-apoptotic molecules have been linked to MSCs to
counteract tumor growth and a particular interest is evident toward
TRAIL (10–13), a cytotoxic protein inducing
apoptosis mostly in tumor cells, upon binding to the death
domain-containing receptor 4 (DR4) and 5 (DR5). The activity of
TRAIL can be modulated following binding with two membrane-bound
decoy receptors, namely DcR1 and DcR2, which lacking functional
death domains, confer TRAIL resistance to expressing cells
(14).
However, clinical studies based on the use of a
recombinant soluble form of TRAIL, consisting of a non-covalently
assembled homotrimer, as a whole, did not demonstrate therapeutic
efficacy (15,16). Over the past decades, many
recombinant versions of TRAIL have been generated to enhance its
pharmacokinetics and/or antitumor activity (17). To date, it is evident that at least
a hexavalent organization of TRAIL molecules bypass the
pharmacokinetic problems, however not the trimeric form (18). In contrast, in order to manage the
insufficient pharmacokinetic properties, several studies have
examined the practice of in situ production of a standard
soluble TRAIL molecule by different adult stem cells (19–21).
Furthermore, two studies have reported the antitumor activity of
human genetically modified MSCs expressing antibodies in a diabody
format (22,23). Recently, a MSC line, stably
producing TRAIL which is activated in a xenotransplantation tumor
model, has been generated (13).
Although the main known source of MSCs is the bone marrow, a wide
variety of MSCs have been recognized in dental tissues such as
pulp, periodontal ligament and apical papilla, exhibiting several
multilineage potencies including osteogenic, adipogenic and
neurogenic (24–30), besides the expected odontogenic
(31). In our previous studies, we
demonstrated the stem cell properties of dental tissues such as
pulp, follicle and bud, as well as their ability to differentiate
into osteoblasts (32–38). According to our findings,
differentiated dental pulp stem cells (DPSCs) express high levels
of TRAIL. Therefore, we hypothesized that DPSCs could provide,
through the production of TRAIL, an effective anticancer
therapeutic method. Based on these parameters and considering the
pro-apoptotic TRAIL effect, we investigated whether DPSCs
differentiated into osteoblasts, expressing high TRAIL levels were
capable to affect tumor cell viability.
Third molar teeth were obtained from 20 healthy
young donors, who gave their written informed consent. The study
was approved by the Institutional Review Board of the Department of
Dental Science and Surgery-Unit of Periodontology, University of
Bari. The dental pulps were dissected, gently washed with
phosphate-buffered saline (PBS), reduced to small pieces and
digested enzymatically with 3 mg/ml type I collagenase and 4 mg/ml
dispase (Gibco; Thermo Fischer Scientific, Uxbridge, UK) in
agitation for 1 h at 37°C. To obtain single cell suspensions, the
digested solutions were filtered through a 70-µm BD Falcon strainer
(Falcon; BD Biosciences, Sunnyvale, CA, USA). Single cell
suspensions, centrifuged at 1,300 rpm, were seeded at
5×103 cells/cm2 in mesenchymal stem cell
culture medium supplemented with 10% fetal bovine serum (FBS), 100
U/ml penicillin-G, 100 µg/ml streptomycin (Gibco; Thermo Fischer
Scientific) at 37°C, in 5% CO2, replacing the medium
every three days until cells reached confluence. The cells were
then trypsinized and seeded into appropriate common culture dishes
for characterization and experiments, that provide an efficacious
substrate for DPSC adhesion, proliferation and differentiation
(39). For the induction of
osteogenic differentiation, the cells were seeded at a density of
3×103 cells/cm2 in α-MEM supplemented with 2%
FBS, 10−8 M dexamethasone and 50 µg/ml ascorbic acid
(35). For some experiments,
osteogenic differentiated DPSCs were co-cultured with
1×103/cm2 H929 cells (from the ATCC,
Rockville, MD, USA) with or without anti-TRAIL neutralizing
monoclonal antibody (mouse; cat. no. MAB375; 500 ng/ml; R&D
Systems, Minneapolis, MN, USA).
Cell viability was evaluated by the
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
assay. DPSCs were cultured in 96-well tissue-culture plates and
some of them were selected for the time-point 0 (t0),
while the others were differentiated with 50 µg/ml ascorbic acid
and dexamethasone (10−8 M) for 20 days (t20).
Both t0 and t20 cultures were treated with
rh-TRAIL (10–500 ng/ml, TRAIL/TNFSF10; R&D Systems) for 48 h.
The cell viability was assessed by adding 0.5 mg/ml MTT to the
culture medium followed by a 4-h incubation at 37°C in a humidified
5% CO2 atmosphere. To stop the reaction, 150 µl of 0.04
N HCl in absolute isopropanol, was added and the optical density
(OD) was read at 570 nm through an automatic plate reader (550
Microplate Reader; Bio-Rad Laboratories Inc., Hercules, CA, USA).
The obtained values were normalized to cells in control
conditions.
Total cell lysates were obtained from cultures
ceased at different time-points. Briefly, at the indicated
time-points, lysis buffer [50 mmol/l Tris-HCl (pH 8.0), 150 mmol/l
NaCl, 5 mmol/l ethylenediaminetetraacetic acid, 1% NP40 and 1
mmol/l phenylmethyl sulfonyl fluoride] was added to the cell
monolayer and the lysates were recovered after incubation on ice
for 30 min. The proteins were separated by SDS-PAGE gel and
transferred onto nitrocellulose membranes (Hybond; Amersham
Pharmacia, London, UK) and the blots were probed with the
appropriate antibodies (Abs): Mouse caspase-3 (1:500; cat. no.
9662; Cell Signaling Technology, Danvers, MA, USA) and mouse
anti-β-actin monoclonal Abs (1:1,000; Chemicon International Inc.;
EMD Millipore, Billerica, MA, USA), rabbit anti-DR5 (1:200; cat.
no. ab47179; Abcam, Cambridge, UK), anti-DcR2 (1:200; cat. no.
ab2019; Abcam), anti-caspase-8 (1:500; cat. no. 552038; BD
Biosciences, San Diego, CA, USA), anti-cFLIP (1:500; cat. no. 8510;
Cell Signaling Technology) and anti-XIAP (1:500; cat. no. 3B6; Cell
Signaling Technology) polyclonal Abs. Specific reactions with the
appropriate fluorescent-dye-conjugated secondary Ab (1:10,000;
IRDye 800 CW goat anti rabbit IgG or IRDye 800 CW goat anti mouse
IgG; LI-COR Biosciences GmbH, Bad Homburg, Germany), were revealed
with the LI-COR Odyssey Infrared Imaging System (LI-COR
Biosciences, Lincoln, NE, USA).
Statistical analysis was performed using Student's
t-test with the SPSS 22 (SPSS X/PC) software (SPSS, Inc., Chicago,
IL, USA). A value of P<0.05 was considered to indicate
statistically significant differences.
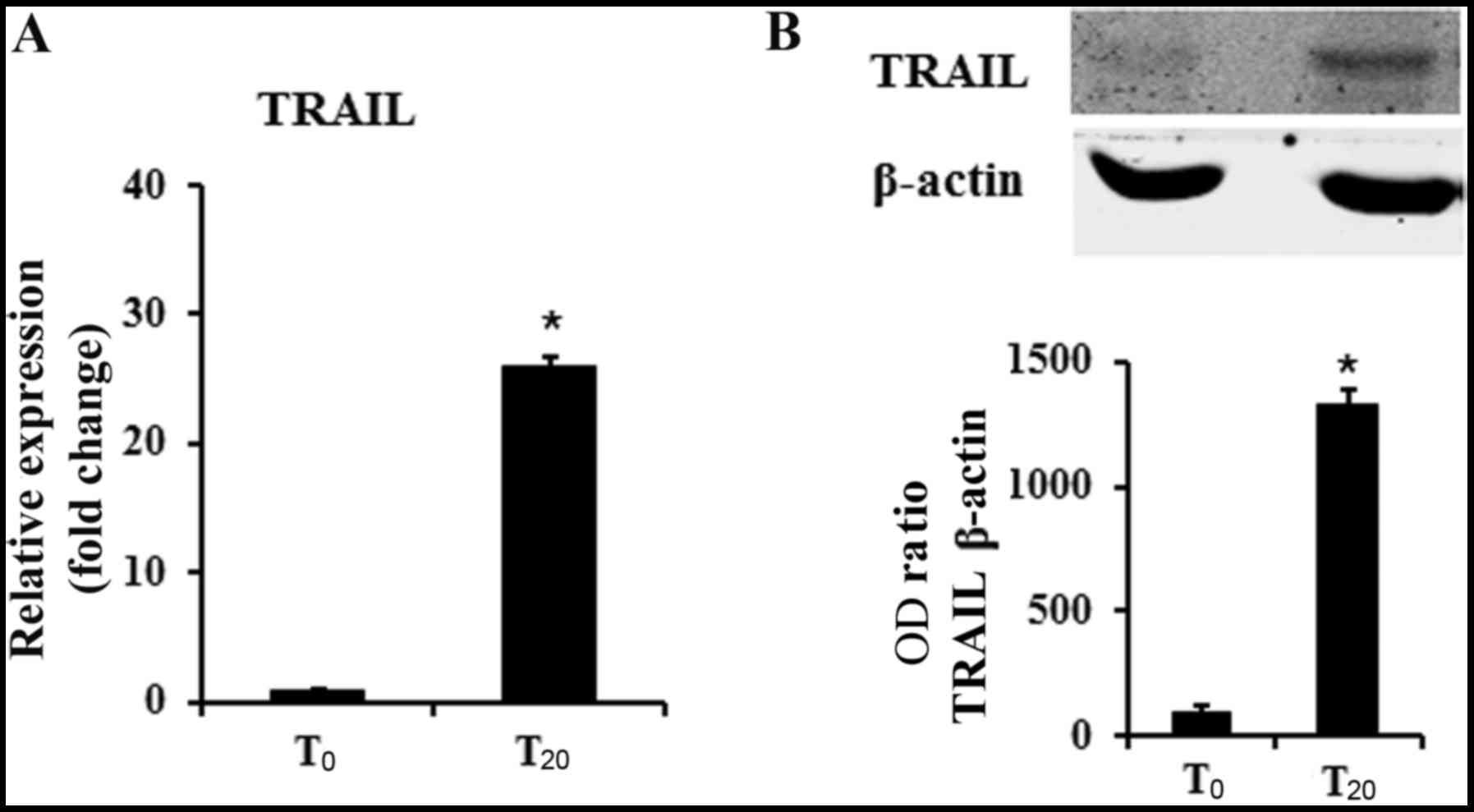
We previously demonstrated that DPSCs cultured in
osteogenic medium displayed an osteoblastic phenotype (32,33).
It is also known from the literature that MSCs produced TRAIL.
These findings prompted us to evaluate the expression of TRAIL in
undifferentiated DPSCs (t0) and in cultures
differentiated for 20 days in osteogenic conditions
(t20). We found that undifferentiated DPSCs already
expressed TRAIL, however, in the cells cultured for 20 days in
osteogenic medium, TRAIL mRNA levels reached a 25-fold increase
(Fig. 1A). These results were also
supported by western blotting indicating a 15-fold increase of
TRAIL in differentiated DPSCs in respect to undifferentiated cells
(Fig. 1B).
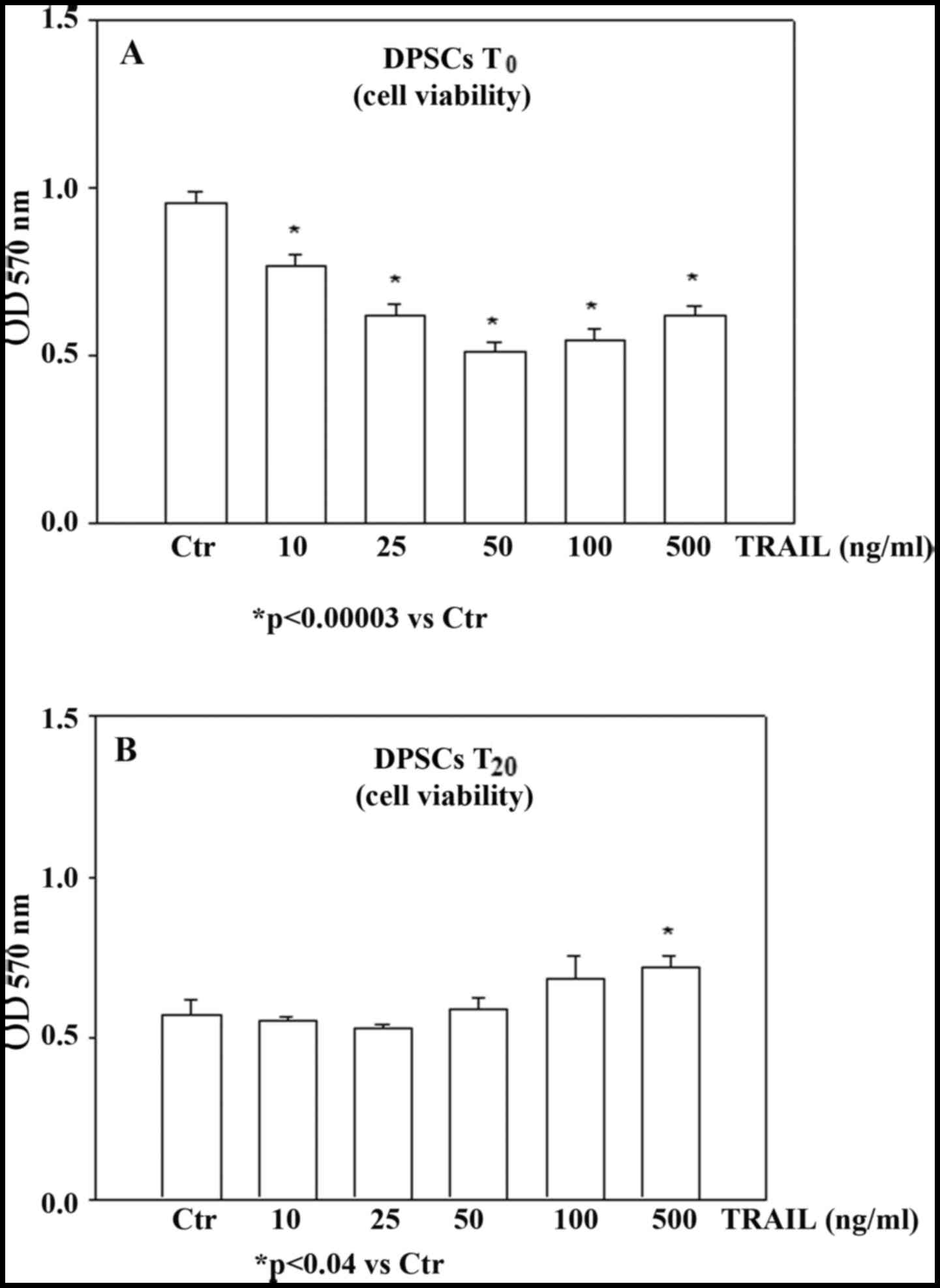
DPSC sensitivity to TRAIL-apoptotic effect was
investigated by analyzing cell viability through an MTT assay in
undifferentiated (t0) and differentiated
(t20) DPSCs in the presence of TRAIL. Undifferentiated
and differentiated DPSCs were first characterized for their
osteoblastic parameters (alkaline phosphatase, osteopontin and
osteocalcin) exhibiting a weak expression at t0 and a
significant increase at t20 (data not shown). The cells
in both conditions were treated with increasing concentrations of
rh-TRAIL (ranging from 10 to 500 ng/ml) for 48 h and their
viability was determined in both TRAIL-treated and untreated cells
as a control. As observed in Fig.
2A, the viability of t0-DPSCs was reduced by TRAIL
in a dose-dependent manner. In detail, when undifferentiated cells
were treated with 10 ng/ml rh-TRAIL for 48 h their viability was
significantly reduced compared to untreated cultures. Treatment
with TRAIL at 25 ng/m further decreased the viability of
t0-DPSCs, while the maximum decrease was observed at 50
ng/ml TRAIL and no additional reduction was observed in the
presence of higher concentrations of the cytokine. Unexpectedly, we
observed that TRAIL did not induce any effect on cell viability on
differentiated DPSCs even at a dose of 50 ng/ml TRAIL (Fig. 2B), thus demonstrating that DPSCs
T20, were resistant to TRAIL-induced apoptosis.
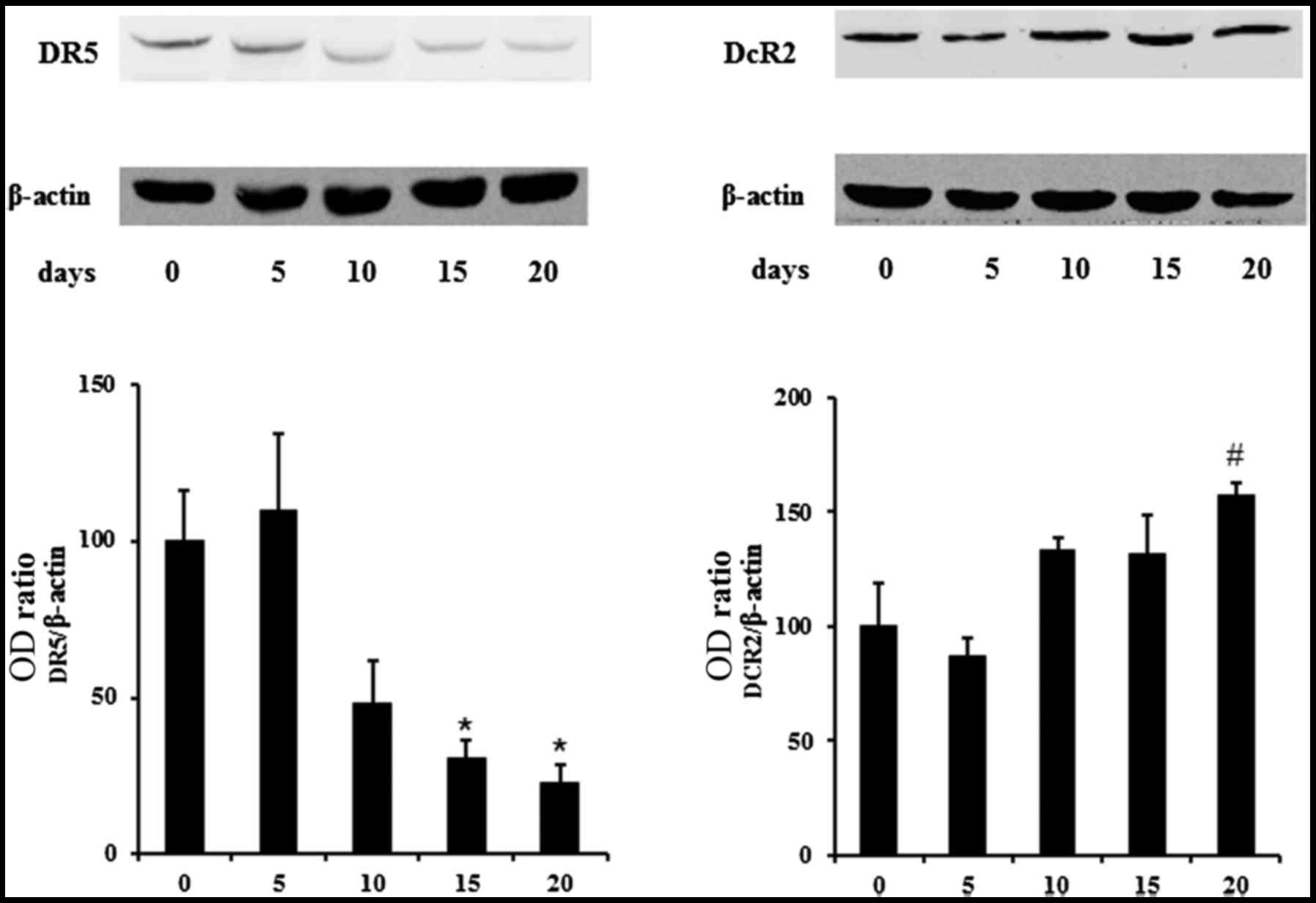
On the base of the aforementioned results, we
explored the possibility that the progression of the osteoblastic
differentiation could affect the expression of death and decoy
TRAIL receptors in DPSCs. Using western blotting we found that
DPSCs constitutively expressed all TRAIL receptors both in
undifferentiated conditions and during their differentiation. In
particular, the expression of DR5, was high in undifferentiated
cells and progressively decreased during the differentiation
process, reaching the lowest level in t20-DPSCs
(Fig. 3). Additionally, the
expression of the decoy receptor DcR2 was low in
t0-DPSCs and increased ~20% following 20 days of
osteoblasic differentiation. The expression of DR4 and DcR1 was not
modified (data not shown). Consequently, the ratio between decoy-
and death-TRAIL receptors shifted in favor of the death-TRAIL
receptors in the undifferentiated DPSCs thus, increasing their
sensitivity to TRAIL apoptotic effect. By contrast, the ratio
shifted in favor of the decoy receptors during the DPSC osteogenic
differentiation. Collectively, these results indicated the absence
of the TRAIL effect on the viability of osteogenic differentiated
DPSCs (Fig. 2).
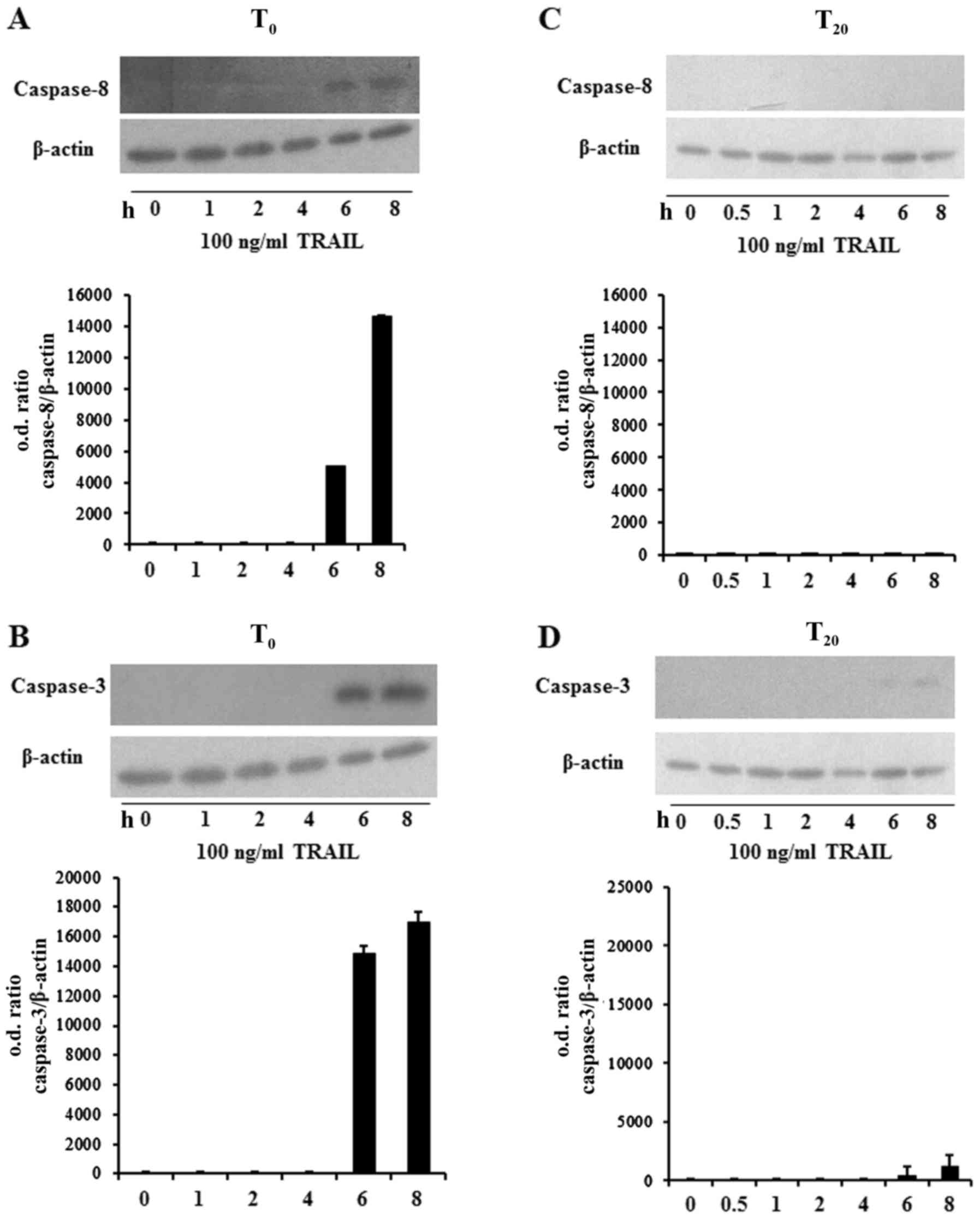
Based on the above described findings we evaluated
the activation of caspase-8 and −3 in TRAIL-treated
undifferentiated and differentiated DPSCs. It is known that
caspase-8 is the early caspase activated during TRAIL-induced
apoptosis in different cell types (41,42).
Using western blot analysis, we demonstrated the activation of
caspase-8 in undifferentiated DPSCs treated with 100 ng/ml TRAIL
from 1 up to 8 h. In particular, caspase activation was evident
after 6 h of TRAIL treatment (Fig.
4A). It is also well-known that activated caspase-8 cleaves
caspase-3. Thus, we explored whether TRAIL treatment induced the
cleavage of caspase-3. As depicted in Fig. 4B, the p17 cleaved form of caspase-3
was found in undifferentiated DPSCs following 6 and 8 h of
TRAIL-treatment exposure. According to the MTT assay, we observed
that TRAIL failed to induce caspase-8 and −3 activation in
differentiated DPSCs (Fig. 4C and
D).
The reported diverse TRAIL sensitivity of the
undifferentiated and differentiated DPSCs prompted us to evaluate
the different levels of the intracellular anti-apoptotic molecules
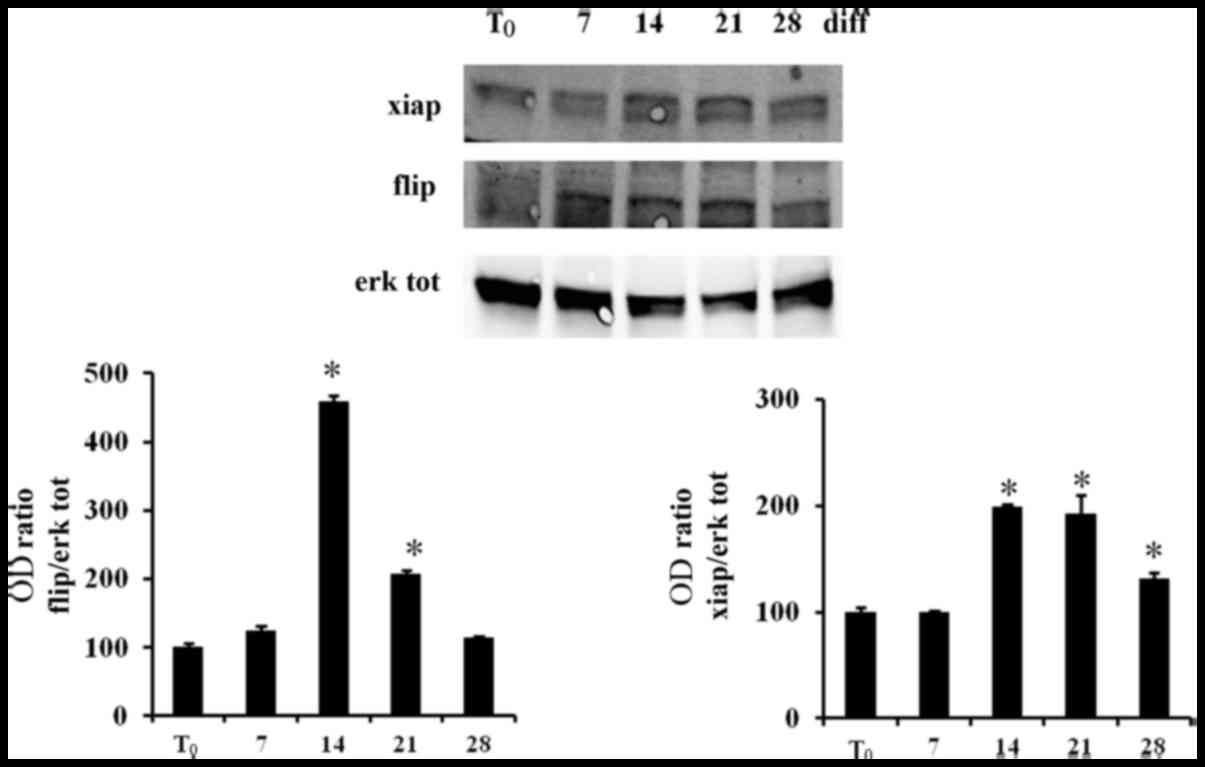
cFLIP and XIAP during the DPSC differentiation. Using western
blotting we observed that the expression of cFLIP and XIAP
increased during the osteoblastic differentiation of DPSCs and the
lowest levels of XIAP and cFLIP were demonstrated in
undifferentiated DPSCs (Fig. 5).
Thus, our findings indicated that differentiated DPSCs were further
preserved from TRAIL pro-apoptotic effect through the increase of
the intracellular inhibitors of caspases cFLIP and XIAP (43,44).
The above-reported high expression of TRAIL in
differentiated DPSCs led us to hypothesize that these cells could
be a vehicle of the pro-apoptotic agent for cancer cells and we
tested this hypothesis on the human myeloma cell line H929.
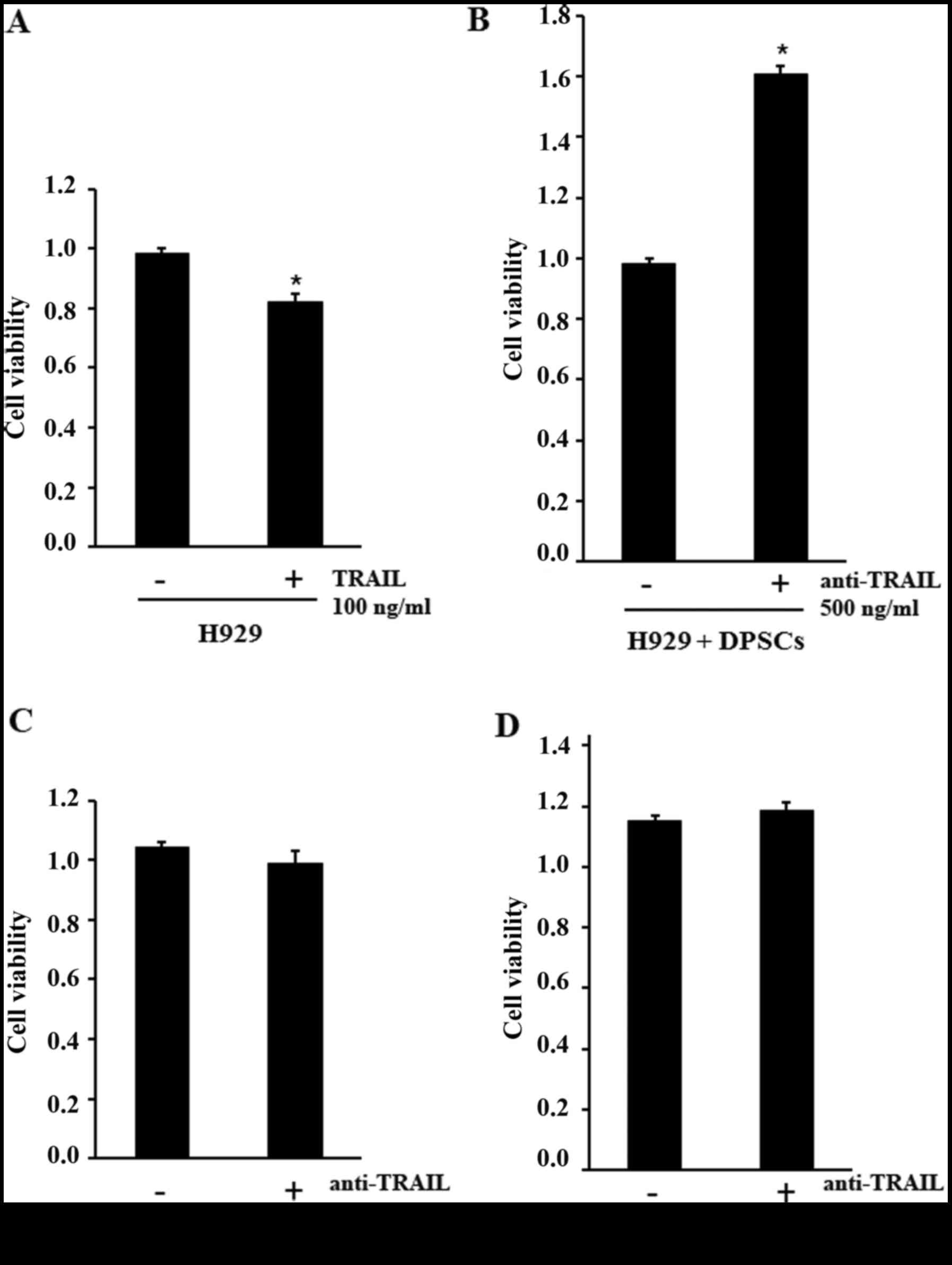
Firstly, we verified the sensitivity of H929 cells to the
TRAIL-apoptotic effect, stimulating the human myeloma cell line
with 100 ng/ml TRAIL and assessing the cell viability with an MTT
assay. The results indicated a significant decrease of H929 cell
viability following treatment, demonstrating that the cells were
sensitive to the TRAIL-apoptotic effect (Fig. 6A). Subsequently, in order to
demonstrate that osteoblastic differentiated DPSCs producing TRAIL
can induce apoptosis of tumor cells, we co-cultured osteoblastic
differentiated DPSCs with H929, with or without a neutralizing
anti-TRAIL antibody. Our results indicated that anti-TRAIL antibody
treatment exhibited a significant increase of cell viability in the
co-culture, which should be related to the neutralization of TRAIL
produced by differentiated DPSCs (Fig.
6B). Anti-TRAIL antibody did not affect cell viability of H929
cells and differentiated DPSCs cultured alone (Fig. 6C and D). Notably, cell-cell contact
was fundamental for this interaction; in fact the media from
osteoblastic differentiated DPSCs did not affect the viability of
H929 cells (data not shown).
In the present study, we demonstrated that the
expression of TRAIL increased during the osteoblastic
differentiation of DPSCs and in parallel, that differentiated DPSCs
lost their sensitivity to TRAIL-induced apoptosis. These two
properties of DPSCs led us to use these cells as a TRAIL-vehicle to
induce an apoptotic effect on H929 myeloma cell line in
vitro.
In detail, we found that only undifferentiated DPSCs
were sensitive to TRAIL-induced apoptosis in a time- and
dose-dependent manner, while DPSCs cultured in osteogenic
conditions were resistant. We demonstrated that the diverse
responses of undifferentiated and differentiated DPSCs to TRAIL
were related to the different expression of TRAIL-receptors DR5 and
DCR2 during osteoblastic differentiation. DR5 is a death
domain-containing receptor, required for the induction of cell
death (55–58), while DCR2 is a decoy factor able to
counteract the pro-apoptotic action of TRAIL (59,60).
Our results indicated that undifferentiated DPSCs expressed higher
levels of DR5 and lower levels of DcR2 compared to differentiated
DPSCs. Thus, in undifferentiated DPSCs the ratio of TRAIL-receptors
moved towards the death receptors, leading these cells to be more
sensitive to the pro-apoptotic action of TRAIL. Conversely, in
differentiated DPSCs the receptor ratio shifted to the decoy
receptors, resulting in defense from TRAIL-mediated apoptosis. We
demonstrated that cell viability in DPSCs at t0 was
negatively affected by TRAIL treatment, while DPSCs at
t20 were protected from death. However, we hypothesized
that cell-resistance or -sensitivity to TRAIL-mediated apoptosis
was not only determined by the balance between the expression of
its death and decoy receptors, but could be partially related to
the levels of certain intracellular anti-apoptotic molecules, such
as c-FLIP and XIAP (61–64). Supporting the observation of
different TRAIL sensitivity between t0- and
t20-DPSCs, we found low levels of the anti-apoptotic
factors c-FLIP and XIAP in t0-DPSCs, that increased
during osteogenic differentiation.
Furthermore, according to the shifted ratio of
receptors towards death receptors and low levels of cFLIP and XIAP
in undifferentiated DPSCs, we demonstrated, in these cells, after
the TRAIL administration, the immediate activation of caspase-8
that in turn resulted in the executioner caspase-3 cleavage
starting at 6 h of incubation. These events represented the crucial
intracellular steps in the initiation of the apoptotic pathway
activated by TRAIL in other cells (65–67).
Notably, the stimulation of TRAIL failed to activate this pathway
in t20-DPSCs, since caspase-8 and −3 were almost
unexpressed. The latter finding was in agreement with the
resistance to TRAIL-mediated cell death exhibited by these cells in
osteogenic conditions and with the expression of receptors and
anti-apoptotic molecules. The obtained results led us to
hypothesize on the possible role of differentiated DPSCs as a
vehicle of TRAIL to cancer cells and prompted us to test this
hypothesis on cancer cells. We used the human myeloma cell line
H929 to test our hypothesis due to its non-adherent behavior in
culture. Previous studies have tested the use of MSCs as a vehicle
of TRAIL demonstrating the ability to induce either in vitro
apoptosis in several cancer cell lines (11), or in vivo remission of colon
tumor and sarcomas established in nude mice (10,12).
Furthermore, the localized action of TRAIL delivered by MSCs
appeared to bypass the resistance of cancer cells, such as breast
and colorectal, to soluble TRAIL (68,69).
More recently, a stable MSC line expressing a highly bioactive form
of TRAIL was generated and demonstrated a significant tumor
regression in an in vivo Colo205 mouse xenograft tumor model
(13). The advantage of our model
is that DPSCs differentiated in osteogenic conditions, expressed
high levels of endogenous TRAIL, thus this system did not require
any transfection technology and the cells were resistant to
TRAIL-mediated apoptosis. Our results indicated that the cell
viability of human myeloma H929 cells was affected by the
TRAIL-apoptotic effect (Fig. 6A).
Notably when the H929 cells were co-cultured with DPSCs and treated
with anti-TRAIL neutralizing antibody, they recovered high levels
of cell viability, with a complete rescue of the apoptotic effect
(Fig. 6A). In conclusion, our
results revealed that differentiated DPSCs expressed high levels of
TRAIL and were not sensitive to its pro-apoptotic effect, thus they
may be an optimal carrier of this antitumor agent in cancer cells.
Furthermore, the results demonstrated that this effect completely
depended on TRAIL. When TRAIL, produced by DPSCs co-cultured with
myeloma cells, was neutralized using blocking antibodies, an
increase of H929 cell viability was obtained.
Notably, a recent well-designed study indicated that
osteoblasts could counteract leukemia progression in mice, while
osteoblast impairment promoted the disease. The authors concluded
that osteoblasts could be a therapeutic target in akute leukemia,
although the basic mechanism of this interesting finding has not
yet been described (70). This
study is aligned with our results and it is intriguing to
hypothesize that TRAIL expressed by osteoblasts mediates this
effect. Collectively, these emerging results suggested that
osteoblasts and osteogenic differentiated MSCs could have a
potential therapeutic role in hindering tumor progression.
The authors would like to thank Pasqua Bellocci for
her technical support. The author A. Di Benedetto has received
funding from the ‘Fondo di Sviluppo e Coesione 2007–2013, APQ
Ricerca Regione Puglia ‘Programma regionale a sostegno della
specializzazione intelligente e della sostenibilità sociale ed
ambientale-Future In Research’.
The authors declare that they have no competing
interests.
|
1
|
Aslan H, Zilberman Y, Kandel L, Liebergall
M, Oskouian RJ, Gazit D and Gazit Z: Osteogenic differentiation of
noncultured immunoisolated bone marrow-derived CD105+
cells. Stem Cells. 24:1728–1737. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Muraglia A, Cancedda R and Quarto R:
Clonal mesenchymal progenitors from human bone marrow differentiate
in vitro according to a hierarchical model. J Cell Sci.
113:1161–1166. 2000.PubMed/NCBI
|
|
3
|
Davies LC, Heldring N, Kadri N and Le
Blanc K: Mesenchymal stromal cell secretion of programmed death-1
ligands regulates T cell mediated immunosuppression. Stem Cells.
35:766–776. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Madec AM, Mallone R, Afonso G, Abou Mrad
E, Mesnier A, Eljaafari A and Thivolet C: Mesenchymal stem cells
protect NOD mice from diabetes by inducing regulatory T cells.
Diabetologia. 52:1391–1399. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Horwitz EM, Prockop DJ, Fitzpatrick LA,
Koo WW, Gordon PL, Neel M, Sussman M, Orchard P, Marx JC, Pyeritz
RE and Brenner MK: Transplantability and therapeutic effects of
bone marrow-derived mesenchymal cells in children with osteogenesis
imperfecta. Nat Med. 5:309–313. 1999. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Scarano A, Crincoli V, Di Benedetto A,
Cozzolino V, Lorusso F, Podaliri Vulpiani M, Grano M, Kalemaj Z,
Mori G and Grassi FR: Bone regeneration induced by bone porcine
block with bone marrow stromal stem cells in a minipig model of
mandibular ‘Critical Size’. Stem Cells Int. 2017:90828692017.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hagenhoff A, Bruns CJ, Zhao Y, von
Luttichau I, Niess H, Spitzweg C and Nelson PJ: Harnessing
mesenchymal stem cell homing as an anticancer therapy. Expert Opin
Biol Ther. 16:1079–1092. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Niess H, von Einem JC, Thomas MN, Michl M,
Angele MK, Huss R, Günther C, Nelson PJ, Bruns CJ and Heinemann V:
Treatment of advanced gastrointestinal tumors with genetically
modified autologous mesenchymal stromal cells (TREAT-ME1): Study
protocol of a phase I/II clinical trial. BMC Cancer. 15:2372015.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Nowakowski A, Drela K, Rozycka J, Janowski
M and Lukomska B: Engineered mesenchymal stem cells as an
anti-cancer trojan horse. Stem Cells Dev. Sep 7–2016.(Epub ahead of
print). View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yu R, Deedigan L, Albarenque SM, Mohr A
and Zwacka RM: Delivery of sTRAIL variants by MSCs in combination
with cytotoxic drug treatment leads to p53-independent enhanced
antitumor effects. Cell Death Dis. 4:e5032013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yuan Z, Kolluri KK, Sage EK, Gowers KH and
Janes SM: Mesenchymal stromal cell delivery of full-length tumor
necrosis factor-related apoptosis-inducing ligand is superior to
soluble type for cancer therapy. Cytotherapy. 17:885–896. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Grisendi G, Spano C, D'Souza N, Rasini V,
Veronesi E, Prapa M, Petrachi T, Piccinno S, Rossignoli F, Burns
JS, et al: Mesenchymal progenitors expressing TRAIL induce
apoptosis in sarcomas. Stem Cells. 33:859–869. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Marini I, Siegemund M, Hutt M, Kontermann
RE and Pfizenmaier K: Antitumor activity of a mesenchymal stem cell
line stably secreting a tumor-targeted TNF-related
apoptosis-inducing ligand fusion protein. Front Immunol. 8:5362017.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
von Karstedt S, Montinaro A and Walczak H:
Exploring the TRAILs less travelled: TRAIL in cancer biology and
therapy. Nat Rev Cancer. 17:352–366. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Herbst RS, Eckhardt SG, Kurzrock R,
Ebbinghaus S, O'Dwyer PJ, Gordon MS, Novotny W, Goldwasser MA,
Tohnya TM, Lum BL, et al: Phase I dose-escalation study of
recombinant human Apo2L/TRAIL, a dual proapoptotic receptor
agonist, in patients with advanced cancer. J Clin Oncol.
28:2839–2846. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Soria JC, Smit E, Khayat D, Besse B, Yang
X, Hsu CP, Reese D, Wiezorek J and Blackhall F: Phase 1b study of
dulanermin (recombinant human Apo2L/TRAIL) in combination with
paclitaxel, carboplatin, and bevacizumab in patients with advanced
non-squamous non-small-cell lung cancer. J Clin Oncol.
28:1527–1533. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ganten TM, Koschny R, Sykora J,
Schulze-Bergkamen H, Büchler P, Haas TL, Schader MB, Untergasser A,
Stremmel W and Walczak H: Preclinical differentiation between
apparently safe and potentially hepatotoxic applications of TRAIL
either alone or in combination with chemotherapeutic drugs. Clin
Cancer Res. 12:2640–2646. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Siegemund M, Seifert O, Zarani M, Džinić
T, De Leo V, Göttsch D, Münkel S, Hutt M, Pfizenmaier K and
Kontermann RE: An optimized antibody-single-chain TRAIL fusion
protein for cancer therapy. MAbs. 8:879–891. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Guiho R, Biteau K, Grisendi G, Taurelle J,
Chatelais M, Gantier M, Heymann D, Dominici M and Redini F: TRAIL
delivered by mesenchymal stromal/stem cells counteracts tumor
development in orthotopic Ewing sarcoma models. Int J Cancer.
139:2802–2811. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yan C, Song X, Yu W, Wei F, Li H, Lv M,
Zhang X and Ren X: Human umbilical cord mesenchymal stem cells
delivering sTRAIL home to lung cancer mediated by MCP-1/CCR2 axis
and exhibit antitumor effects. Tumour Biol. 37:8425–8435. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lathrop MJ, Sage EK, Macura SL, Brooks EM,
Cruz F, Bonenfant NR, Sokocevic D, MacPherson MB, Beuschel SL,
Dunaway CW, et al: Antitumor effects of TRAIL-expressing
mesenchymal stromal cells in a mouse xenograft model of human
mesothelioma. Cancer Gene Ther. 22:44–54. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Compte M, Cuesta AM, Sánchez-Martin D,
Alonso-Camino V, Vicario JL, Sanz L and Alvarez-Vallina L: Tumor
immunotherapy using gene-modified human mesenchymal stem cells
loaded into synthetic extracellular matrix scaffolds. Stem Cells.
27:753–760. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang X, Yang Y, Zhang L, Lu Y, Zhang Q,
Fan D, Zhang Y, Zhang Y, Ye Z and Xiong D: Mesenchymal stromal
cells as vehicles of tetravalent bispecific Tandab (CD3/CD19) for
the treatment of B cell lymphoma combined with IDO pathway
inhibitor D-1-methyl-tryptophan. J Hematol Oncol. 10:562017.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gronthos S, Mankani M, Brahim J, Robey PG
and Shi S: Postnatal human dental pulp stem cells (DPSCs) in vitro
and in vivo. Proc Natl Acad Sci USA. 97:pp. 13625–13630. 2000;
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Seo BM, Miura M, Gronthos S, Bartold PM,
Batouli S, Brahim J, Young M, Robey PG, Wang CY and Shi S:
Investigation of multipotent postnatal stem cells from human
periodontal ligament. Lancet. 364:149–155. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sonoyama W, Liu Y, Fang D, Yamaza T, Seo
BM, Zhang C, Liu H, Gronthos S, Wang CY, Wang S and Shi S:
Mesenchymal stem cell-mediated functional tooth regeneration in
swine. PLoS One. 1:e792006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Pisciotta A, Carnevale G, Meloni S, Riccio
M, De Biasi S, Gibellini L, Ferrari A, Bruzzesi G and De Pol A:
Human dental pulp stem cells (hDPSCs): Isolation, enrichment and
comparative differentiation of two sub-populations. BMC Dev Biol.
15:142015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Nagatomo K, Komaki M, Sekiya I, Sakaguchi
Y, Noguchi K, Oda S, Muneta T and Ishikawa I: Stem cell properties
of human periodontal ligament cells. J Periodontal Res. 41:303–310.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Marrelli M, Paduano F and Tatullo M: Human
periapical cyst-mesenchymal stem cells differentiate into neuronal
cells. J Dent Res. 94:843–852. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Giorgini E, Conti C, Ferraris P, Sabbatini
S, Tosi G, Centonze M, Grano M and Moric G: FT-IR microscopic
analysis on human dental pulp stem cells. Vibrational Spectroscopy.
57:30–34. 2011.
|
|
31
|
Tatullo M, Falisi G, Amantea M, Rastelli
C, Paduano F and Marrelli M: Dental pulp stem cells and human
periapical cyst mesenchymal stem cells in bone tissue regeneration:
Comparison of basal and osteogenic differentiated gene expression
of a newly discovered mesenchymal stem cell lineage. J Biol Regul
Homeost Agents. 29:713–718. 2015.PubMed/NCBI
|
|
32
|
Mori G, Brunetti G, Oranger A, Carbone C,
Ballini A, Lo Muzio L, Colucci S, Mori C, Grassi FR and Grano M:
Dental pulp stem cells: Osteogenic differentiation and gene
expression. Ann N Y Acad Sci. 1237:47–52. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Mori G, Centonze M, Brunetti G, Ballini A,
Oranger A, Mori C, Lo Muzio L, Tetè S, Ciccolella F, Colucci S, et
al: Osteogenic properties of human dental pulp stem cells. J Biol
Regul Homeost Agents. 24:167–175. 2010.PubMed/NCBI
|
|
34
|
Mori G, Ballini A, Carbone C, Oranger A,
Brunetti G, Di Benedetto A, Rapone B, Cantore S, Di Comite M,
Colucci S, et al: Osteogenic differentiation of dental follicle
stem cells. Int J Med Sci. 9:480–487. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Di Benedetto A, Carbone C and Mori G:
Dental pulp stem cells isolation and osteogenic differentiation: A
good promise for tissue engineering. Methods Mol Biol.
1210:117–130. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Di Benedetto A, Brunetti G, Posa F,
Ballini A, Grassi FR, Colaianni G, Colucci S, Rossi E,
Cavalcanti-Adam EA, Lo Muzio L, et al: Osteogenic differentiation
of mesenchymal stem cells from dental bud: Role of integrins and
cadherins. Stem Cell Res. 15:618–628. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Posa F, Di Benedetto A, Colaianni G,
Cavalcanti-Adam EA, Brunetti G, Porro C, Trotta T, Grano M and Mori
G: Vitamin D effects on osteoblastic differentiation of mesenchymal
stem cells from dental tissues. Stem Cells Int. 2016:91508192016.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Di Benedetto A, Posa F, Carbone C, Cantore
S, Brunetti G, Centonze M, Grano M, Lo Muzio L, Cavalcanti-Adam EA
and Mori G: NURR1 downregulation favors osteoblastic
differentiation of MSCs. Stem Cells Int. 2017:76170482017.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Tatullo M, Marrelli M, Falisi G, Rastelli
C, Palmieri F, Gargari M, Zavan B, Paduano F and Benagiano V:
Mechanical influence of tissue culture plates and extracellular
matrix on mesenchymal stem cell behavior: A topical review. Int J
Immunopathol Pharmacol. 29:3–8. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Pfaffl MW: A new mathematical model for
relative quantification in real-time RT-PCR. Nucleic Acids Res.
29:e452001. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Peter ME, Scaffidi C, Medema JP, Kischkel
F and Krammer PH: The death receptors. Results Probl Cell Differ.
23:25–63. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Medema JP, Scaffidi C, Kischkel FC,
Shevchenko A, Mann M, Krammer PH and Peter ME: FLICE is activated
by association with the CD95 death-inducing signaling complex
(DISC). EMBO J. 16:2794–2804. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Golks A, Brenner D, Fritsch C, Krammer PH
and Lavrik IN: c-FLIPR, a new regulator of death receptor-induced
apoptosis. J Biol Chem. 280:14507–14513. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Deveraux QL, Takahashi R, Salvesen GS and
Reed JC: X-linked IAP is a direct inhibitor of cell-death
proteases. Nature. 388:300–304. 1997. View
Article : Google Scholar : PubMed/NCBI
|
|
45
|
Wiley SR, Schooley K, Smolak PJ, Din WS,
Huang CP, Nicholl JK, Sutherland GR, Smith TD, Rauch C, Smith CA,
et al: Identification and characterization of a new member of the
TNF family that induces apoptosis. Immunity. 3:673–682. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Pitti RM, Marsters SA, Ruppert S, Donahue
CJ, Moore A and Ashkenazi A: Induction of apoptosis by Apo-2
ligand, a new member of the tumor necrosis factor cytokine family.
J Biol Chem. 271:12687–12690. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Ashkenazi A, Pai RC, Fong S, Leung S,
Lawrence DA, Marsters SA, Blackie C, Chang L, McMurtrey AE, Hebert
A, et al: Safety and antitumor activity of recombinant soluble Apo2
ligand. J Clin Invest. 104:155–162. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Walczak H, Miller RE, Ariail K, Gliniak B,
Griffith TS, Kubin M, Chin W, Jones J, Woodward A, Le T, et al:
Tumoricidal activity of tumor necrosis factor-related
apoptosis-inducing ligand in vivo. Nat Med. 5:157–163. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Tinhofer I, Biedermann R, Krismer M,
Crazzolara R and Greil R: A role of TRAIL in killing osteoblasts by
myeloma cells. FASEB J. 20:759–761. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Brunetti G, Oranger A, Carbone C, Mori G,
Sardone FR, Mori C, Celi M, Faienza MF, Tarantino U, Zallone A, et
al: Osteoblasts display different responsiveness to TRAIL-induced
apoptosis during their differentiation process. Cell Biochem
Biophys. 67:1127–1136. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Roux S, Lambert-Comeau P, Saint-Pierre C,
Lépine M, Sawan B and Parent JL: Death receptors, Fas and TRAIL
receptors, are involved in human osteoclast apoptosis. Biochem
Biophys Res Commun. 333:42–50. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Colucci S, Brunetti G, Cantatore FP,
Oranger A, Mori G, Pignataro P, Tamma R, Grassi FR, Zallone A and
Grano M: The death receptor DR5 is involved in TRAIL-mediated human
osteoclast apoptosis. Apoptosis. 12:1623–1632. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Mori G, Brunetti G, Colucci S, Oranger A,
Ciccolella F, Sardone F, Pignataro P, Mori C, Karapanou V, Ballini
A, et al: Osteoblast apoptosis in periodontal disease: Role of
TNF-related apoptosis-inducing ligand. Int J Immunopathol
Pharmacol. 22:95–103. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Mori G, Brunetti G, Colucci S, Ciccolella
F, Coricciati M, Pignataro P, Oranger A, Ballini A, Farronato D,
Mastrangelo F, et al: Alteration of activity and survival of
osteoblasts obtained from human periodontitis patients: Role of
TRAIL. J Biol Regul Homeost Agents. 21:105–114. 2007.PubMed/NCBI
|
|
55
|
Pan G, O'Rourke K, Chinnaiyan AM, Gentz R,
Ebner R, Ni J and Dixit VM: The receptor for the cytotoxic ligand
TRAIL. Science. 276:111–113. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Walczak H, Degli-Esposti MA, Johnson RS,
Smolak PJ, Waugh JY, Boiani N, Timour MS, Gerhart MJ, Schooley KA,
Smith CA, et al: TRAIL-R2: A novel apoptosis-mediating receptor for
TRAIL. EMBO J. 16:5386–5397. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Wu GS, Burns TF, McDonald ER III, Jiang W,
Meng R, Krantz ID, Kao G, Gan DD, Zhou JY, Muschel R, et al:
KILLER/DR5 is a DNA damage-inducible p53-regulated death receptor
gene. Nat Genet. 17:141–143. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Schneider P, Thome M, Burns K, Bodmer JL,
Hofmann K, Kataoka T, Holler N and Tschopp J: TRAIL receptors 1
(DR4) and 2 (DR5) signal FADD-dependent apoptosis and activate
NF-kappaB. Immunity. 7:831–836. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Marsters SA, Sheridan JP, Pitti RM, Huang
A, Skubatch M, Baldwin D, Yuan J, Gurney A, Goddard AD, Godowski P
and Ashkenazi A: A novel receptor for Apo2L/TRAIL contains a
truncated death domain. Curr Biol. 7:1003–1006. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Degli-Esposti MA, Dougall WC, Smolak PJ,
Waugh JY, Smith CA and Goodwin RG: The novel receptor TRAIL-R4
induces NF-kappaB and protects against TRAIL-mediated apoptosis,
yet retains an incomplete death domain. Immunity. 7:813–820. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Safa AR: c-FLIP, a master anti-apoptotic
regulator. Exp Oncol. 34:176–184. 2012.PubMed/NCBI
|
|
62
|
Chawla-Sarkar M, Bae SI, Reu FJ, Jacobs
BS, Lindner DJ and Borden EC: Downregulation of Bcl-2, FLIP or IAPs
(XIAP and survivin) by siRNAs sensitizes resistant melanoma cells
to Apo2L/TRAIL-induced apoptosis. Cell Death Differ. 11:915–923.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Lee TJ, Lee JT, Park JW and Kwon TK:
Acquired TRAIL resistance in human breast cancer cells are caused
by the sustained cFLIP(L) and XIAP protein levels and ERK
activation. Biochem Biophys Res Commun. 351:1024–1030. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Deveraux QL, Leo E, Stennicke HR, Welsh K,
Salvesen GS and Reed JC: Cleavage of human inhibitor of apoptosis
protein XIAP results in fragments with distinct specificities for
caspases. EMBO J. 18:5242–5251. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Sprick MR, Rieser E, Stahl H, Grosse-Wilde
A, Weigand MA and Walczak H: Caspase-10 is recruited to and
activated at the native TRAIL and CD95 death-inducing signalling
complexes in a FADD-dependent manner but can not functionally
substitute caspase-8. EMBO J. 21:4520–4530. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Nagata S: Apoptosis by death factor. Cell.
88:355–365. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Ashkenazi A and Dixit VM: Death receptors:
Signaling and modulation. Science. 281:1305–1308. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Mueller LP, Luetzkendorf J, Widder M,
Nerger K, Caysa H and Mueller T: TRAIL-transduced multipotent
mesenchymal stromal cells (TRAIL-MSC) overcome TRAIL resistance in
selected CRC cell lines in vitro and in vivo. Cancer Gene Ther.
18:229–239. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Lin T, Huang X, Gu J, Zhang L, Roth JA,
Xiong M, Curley SA, Yu Y, Hunt KK and Fang B: Long-term tumor-free
survival from treatment with the GFP-TRAIL fusion gene expressed
from the hTERT promoter in breast cancer cells. Oncogene.
21:8020–8028. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Krevvata M, Silva BC, Manavalan JS,
Galan-Diez M, Kode A, Matthews BG, Park D, Zhang CA, Galili N,
Nickolas TL, et al: Inhibition of leukemia cell engraftment and
disease progression in mice by osteoblasts. Blood. 124:2834–2846.
2014. View Article : Google Scholar : PubMed/NCBI
|