Introduction
Hepatocellular carcinoma (HCC) is one of the most
common malignant tumors. It is the fifth most common cancer and
ranks third in cancer-related deaths. In developing countries the
incidence rate is even higher (1),
and in developed countries the incidence rate is gradually
increasing (2). Due to higher and
earlier metastasis, HCC has a very poor prognosis with a 5-year
survival of only ~3–5% (3,4). Several options are available for HCC
therapy, such as local treatment, surgical resection, targeted
biological therapy, systemic chemotherapy and liver
transplantation. However, the cure rate and overall survival have
been unsatisfactory. Many cancer patients are unable to undergo
operational therapy surgery due to poor liver function resulted
resulting from chronic liver diseases (5). Therefore, there is an urgent need for
early diagnosis and treatment of HCC. Identification of HCC-related
genes is essential to have a better understating understanding of
the pathogenic mechanisms in order to develop new diagnostic and
therapeutic strategies.
Chromatin assembly factor 1(CAF-1) is a trimer
histone chaperone located in the nucleus, which mediates chromatin
assembly after DNA replication and repair (6). CAF-1 is a heterotrimeric complex
composed of three subunits p150, p60 and p48. It recruits histones
H3 and H4 to DNA to facilitate nucleosome assembly, participates in
the regulation of DNA repair and epigenetic changes in embryonic
stem cellsas well as cell proliferation. It has been revealed to be
an important histone chaperone in eukaryotic cells such as yeast
and multicellular organisms (7).
CAF1B is a p60 subunit of CAF-1, which is positively related to
cell proliferation, and is located in the nucleolus during
interphase and in the site of DNA replication in the S phase
(8). It has been proposed as a
clinical marker to distinguish quiescent from proliferating cells
(9). High expression of CAF-1 has
been revealed to be associated with the invasion and metastasis of
oral squamous cell carcinoma (10).
High expression of CAF1B was demonstrated to be an indicator of
poor prognosis in neuroblastoma (11). In the DNA synthesis phase CAF-1 is
reversibly phosphorylated by Cyclin/Cdk2 (12). These findings indicated that the
expression level of CAF-1 as well as CAF1B is closely related to
tumor metastasis, invasion and poor prognosis (11,13).
Numerous studies have shown that CAF-1 controls specific chromatin
recombination in the S phase to promote cell cycle progression and
influences cell proliferation and apoptosis. It has been
demonstrated that CAF-1 is essential for differentiation and
proliferation of higher eukaryotic animal cells. Recently, CAF-1,
particularly CHAF1B, was proposed as a new marker for tumor
proliferation and prognosis and overexpression of CAF-1/p60 was
used as a clinical marker for malignant tumor progression in some
human malignant tumors (14,15).
Previous studies have confirmed that CHAF1B is
expressed in HCC cell lines Hep3B, HUH-7, HepG2 and SMMC-7721.
Analysis of the expression data in the TCGA database revealed that
the gene had higher mRNA abundance in a large number of HCC tissues
than in the adjacent tissues. Therefore, we speculated that CHAF1B
may be related to the biological behavior of HCC. In the present
study, we knocked down the CHAF1B gene in HUH-7 cells using an RNAi
lentiviral vector to investigate its effect on proliferation,
apoptosis and the cell cycle of the cells. The findings would
provide insight into the role of the gene in HCC, help in the
identification of new signaling molecules for HCC research and
validate its potential in HCC diagnosis and therapy.
Materials and methods
Cell line, reagents and equipment
Human HCC cell line HUH-7 was purchased from
Yingniurui Biological Co. (Wuxi, Jiangsu, China). DMEM high glucose
medium was obtained from Sijiqing Biotech (Hangzhou, China) (batch
no. KGM12800S-500), DMSO was purchased from Amresco (Solon, OH,
USA) (batch no. 302A0325), 0.25% trypsin (containing EDTA) was
purchased from Kaiji Biotech (Jiangsu, China) (batch no. 20160818),
penicillin and streptomycin mix solution and GoldView I nucleic
acid stain were obtained from Beijing Solarbio Science &
Technology Co., Ltd. (Beijing, China) (batch no. 20160909), fetal
bovine serum (FBS) was purchased from BI Biotech (Beijing, China)
(batch no. 1552680) and Transwell cells were obtained from Falcon
(Atlanta, GA, USA). GoldStar Taq MasterMix and UltraSYBR Mixture
were products from CWBiotech (Beijing, China). Primers were
synthesized at General Biotech (Beijing, China). The
ultra-sensitive chemiluminescence imaging system
(ChemiDoc™XRS+) and real-time PCR instrument CFX
Connect™ were purchased by Bio-Rad Laboratories, Inc.
(Hercules, CA, USA).
Experimental grouping
The cells were divided into the control that did not
receive any treatment, the empty-vector group that was transfected
with a vector without an insert andthe CHAF1B group that was
transfected with CHAF1B.
Construction of lentivirus for CHAF1B
knockdown
Interference sequences (upstream
5′-CACCGCCACTTAGAAGATGTGTATGCGAACATACACATCTTCTAAGTGGC and
downstream 5′-AAAAGCCACTTAGAAGATGTGTATGTTCGCATACACATCTTCTAAGTGGC)
for CHAF1B were designed and synthesized and ligated to lentivirus
vector pDS019_pL_shRNA_F. The lentivirus containing vector
pDS019-PL-shRNA-GFP-homo-CHAF1B was packed and verified using a
fluorescence microscope. The virus titer was determined using the
TCID50 method and its expression was confirmed using
western blot and PCR analyses. The virus was then amplified and
purified using CsC1 gradient centrifugation. Cells without any
treatment were used as the blank control, while the empty vector
was used as a control.
Cell invasion and migration
assays
Prior to the assessment of the invasion ability, the
cells were infected with the CHAF1B-knockdown virus or control for
24 h. Then 5×105 cells in serum-free medium were placed
into the upper chamber of an insert (8 µm pore size; BD Biosciences
(San Jose, CA, USA). After 48 h of incubation, the cells remaining
on the upper membrane were removed with cotton wool, whereas the
cells that had migrated or invaded through the membrane were
stained with 0.1% crystal violet in 25% methanol/PBS, imaged and
counted under an inverted microscope (Life Technologies; Thermo
Fisher Scientific, Inc., Waltham, MA, USA). The experiments were
independently repeated three times.
For the assessment of migration ability, an in
vitro scratch assay was used as previously reported (16).
Colony-formation ability assay
Cells (0.4 ml) were inoculated into culture medium
in the wells of a 96-plate (400 cells/well) in a total volume of 3
ml and cultured for 2 to 3 weeks. The medium was refreshed every 3
days. Once the colonies became visible, they were fixed in
methanol, and then washed and stained with Giemsa staining solution
for 30 min. The number of colonies were counted and the
colony-forming ability was calculated. The experiments were
repeated three times independently.
Flow cytometry
Cells were washed with pre-chilled PBS three times
and fixed in 1 ml 70% pre-chilled ethanol overnight at 4°C and
washed again with PBS three times. The cells were re-suspended in 1
ml PI/Triton X-100 staining solution containing 0.2 mg RNase A for
15 min at 37°C and then analyzed by flow cytometry. Cells
(2×104) were assayed at each cell cycle.
Expression profiling and
annotation
Total RNA was extracted using TRIzol reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) and an RNeasy Kit
(Qiagen GmbH, Hilden, Germany). The first strands of cDNA were
synthesized using a cDNA synthesis kit (Invitrogen, Thermo Fisher
Scientific, Inc.) and labelled using a monochromatic DNA labeling
kit (Roche NimbleGen, Inc., Madison, WI, USA). The labeled cDNA was
hybridized to GeneChip Human Gene 2.0 ST Array (Affymetrix; Thermo
Fisher Scientific, Inc.) using a hybridization kit (Roche
NimbleGen, Inc.) and the signals were scanned, with the GenePix
4000B chip scanner. The data were normalized using NimbleScan
software (v. 2.0) and analyzed for differentially expressed genes
using Agilent GeneSpring GX software (v. 11.5.1). Pathway analysis
was performed against the latest KEGG (Kyoto Encyclopedia of Genes
and Genomes) database.
Real-time PCR
Total RNA was extracted from the cells using TRIzol
reagent according to the supplier's instructions, reversely
transcripted into cDNA and used for PCR assays using primers for
CHAF1B as shown in Table I. PCR was
carried out in a total volume of 20 µl containing 1 µl of diluted
and pre-amplified cDNA, 12.5 µl of 2X UltraSYBR Mixture and 1 µl of
each fluorescence TaqMan probe using primers listed in Table I. The cycling conditions were 50°C
for 2 min, 95°C for 3 min followed by 40 cycles, each one
consisting of 15 sec at 95°C and 30 sec at 50.6°C. The samples were
run in triplicate and the mean value was calculated for each
case.
 | Table I.Primers for RT-PCR. |
Table I.
Primers for RT-PCR.
| Gene | Primer
sequence |
|---|
| DNAJB1 | F
3′-GATGGCTCTGATGTCATTTATC-5′ |
|
| R
3′-GCCTTCTCCAGGAACTTTT-5′ |
| SBNO1 | F
3′-ATCCAGTGTTACTCCTCCTG-5′ |
|
| R
3′-TGCTATCGTCCTTCCTTT-5′ |
| BLM | F
3′-TGAATCCAGAAACCAGCAC-5′ |
|
| R
3′-AAGCAGTTCGTTCCCACA-5′ |
| PSMB6 | F
3′-CTGGGAAAGCCGAGAAGT-5′ |
|
| R
3′-ATGCGGTCGTGAATAGGT-5′ |
| SLC30A7 | F
3′-TAGGCTTGATTTCCGACTC-5′ |
|
| R
3′-CCGCTCTAACATACCCATA-5′ |
| DDX3X | F
3′-GGAGCGAATGCGTAAGGT-5′ |
|
| R
3′-TTAGGGTGCGAAATGCTG-5′ |
| TWF2 | F
3-′AGGTTGTGATTGAGGACGAG-5′ |
|
| R
3′-GGAGTTATCAGGCGACCAG-5′ |
| SMC3 | F
3′-TGTGATGAACCTCCTTGA-5′ |
|
| R
3′-TCTGAGAATCTGGTGCTG-5′ |
| TMEM259 | F
3′-CGAGACGCCCACCAAAGT-5 |
|
| R
3′-CATCGTAGCCCAGGAACTCAT-5′ |
| β-actin | F
3′-CCTGTATGCCTCTGGTCG-5′ |
|
| R
3′-GGCGTAACCCTCGTAGAT-5′ |
The data were managed using the Applied Biosystems
software RQ Manager v. 1.2.1 (Applied Biosystems, Foster City, CA,
USA). The relative expression was calculated using the comparative
Ct method and by obtaining the fold change value
(2−ΔΔCt) according to a previously described protocol
(17).
Western blot analysis
Cells were lysed using RIPA buffer (50 mM Tris, pH
7.2; 150 mM NaCl; 1% Triton X-100; and 0.1% SDS) containing
protease (1:100; Roche Diagnostics, Indianapolis, IN, USA) and
phosphatase (1:100; Sigma-Aldrich, St. Louis, MO, USA) inhibitors.
The protein concentrations were determined using a bicinchoninic
acid assay (Pierce; Thermo Fisher Scientific, Inc.). Proteins (60
µg) were separated by SDS-PAGE and transferred (Bio-Rad
Laboratories, Inc.) to PVDF membranes (EMD Millipore, Billerica,
MA, USA). The membranes were incubated with rabbit polyclonal
antibodies specific for SMC3 (1:2,000; cat. no. ab9263) and β-actin
(1:1,000; cat. no. ab8266; both from Abcam, Cambridge, MA, USA),
PSMB6 (1:2,000; cat. no. A4053) TWF2 (1:2,000; cat. no. A5860) BLM
(1:2,000; cat. no. A6535) and SLC30A7 (1:2,000; cat. no. A5172; all
from ABclonal Biotech Co., Ltd., Woburn, MA, USA). The expression
levels of these three proteins were standardized to human α-actin
using a mouse polyclonal anti-α-actin antibody (1:1,000; cat. no.
MAB1501R; EMD Millipore). Primary antibodies were detected using
goat anti-rabbit or goat anti-mouse horseradish peroxidase
(HRP)-conjugated secondary antibodies (1:1,000; AP189P; EMD
Millipore). Immunoreactive bands were visualized using the
ultra-sensitive chemiluminescence imaging system (ChemiDocXRS+)
according to the manufacturer's instructions, and then quantified
by densitometry using a ChemiGenius Gel Bio Imaging System
(Syngene, Frederick, MD, USA).
Construction of HCC nude mouse
models
Infected and control HUH-7 cells were digested,
washed three times with PBS and re-suspended in PBS. Cells (200 µl;
3×106/mouse) were subcutaneously injected into nude
mice. The animals were scarified three weeks later and the tumor
tissues were collected for analysis.
H&E staining
Slides were dewaxed, dehydrated and stained with
hematoxylin staining solution for 5 min. The stained slides were
soaked in hydrochloric acid alcohol for 30 sec, washed twice with
PBS, briefly soaked in 0.5% ammonia for 1 to 2 min. After being
washed with PBS, the slides were counterstained with eosin for 2
min, washed 2 times with PBS, dehydrated, sealed with neutral
balsam and images were captured under a E400 Nikon microscope at
×400 magnification (Nikon, Tokyo, Japan).
Statistical analysis
Statistical analyses were performed using SPSS 20
software (SPSS, Inc., Chicago, IL, USA). All experiments were
repeated at least three times and performed in triplicate. The
means were compared using the Student's t-test or one-way ANOVA
with the corresponding post hoc test. P≤0.05 was considered to
indicate a statistically significant result.
Results
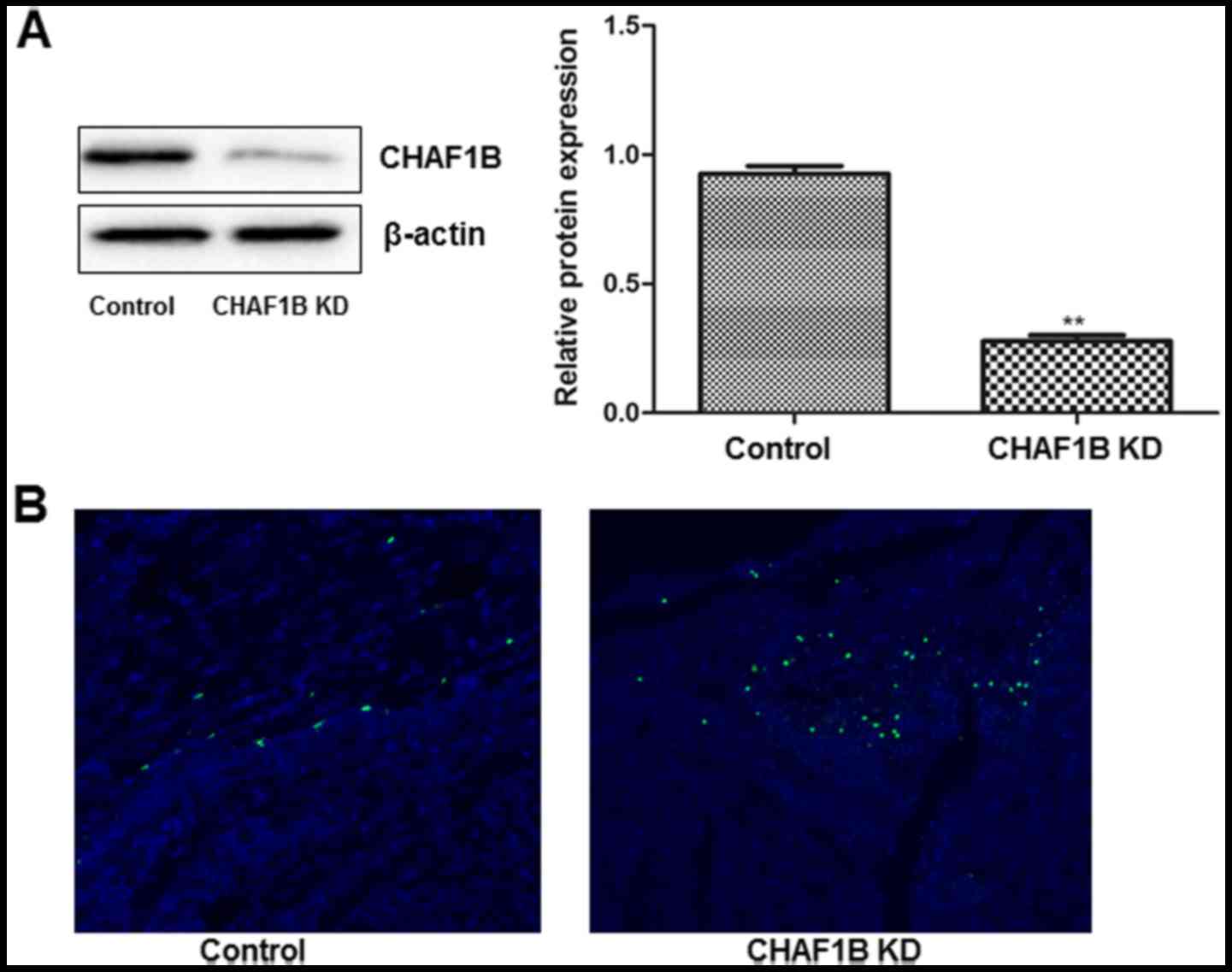
Transfection of HUH-7 cells with
CHAF1B-silencing lentivirus
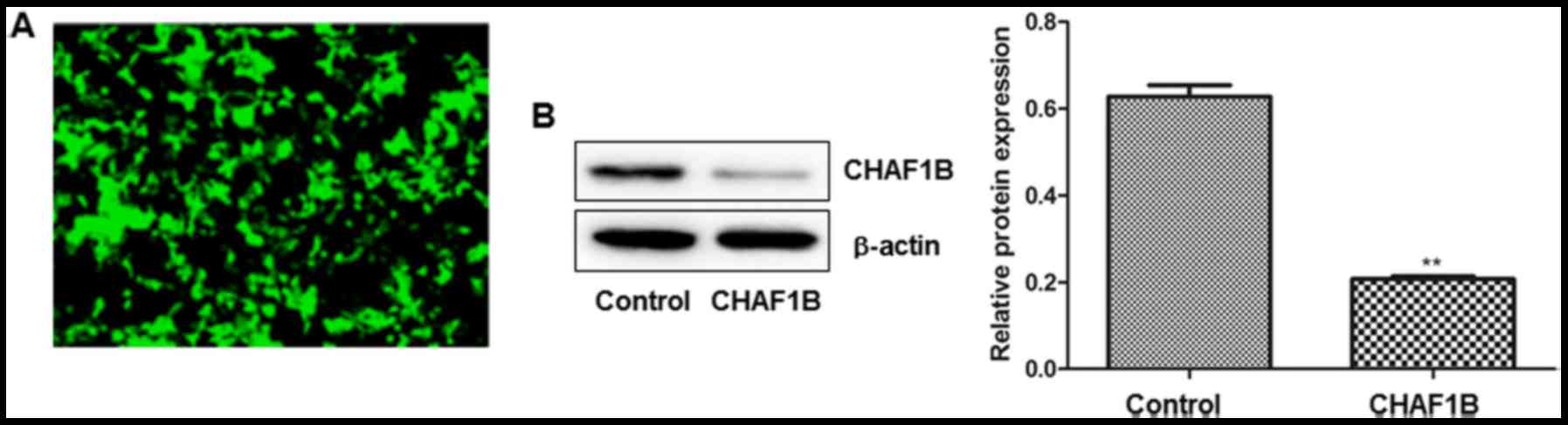
We examined the transfection efficiency by observing
the intensity of GFP fluorescence 24 h after the transfection.
Observations revealed that >95% of cells were fluorescent after
the cells were transfected with a lentivirus harboring CHAF1B-shRNA
(Fig. 1A). No fluorescence was
observed in the untransfected cells (data not shown). Western blot
analysis revealed that CHAF1B was significantly downregulated at
the protein level (Fig. 1B).
Invasion ability of HUH-7 cells after
CHAF1B knockdown
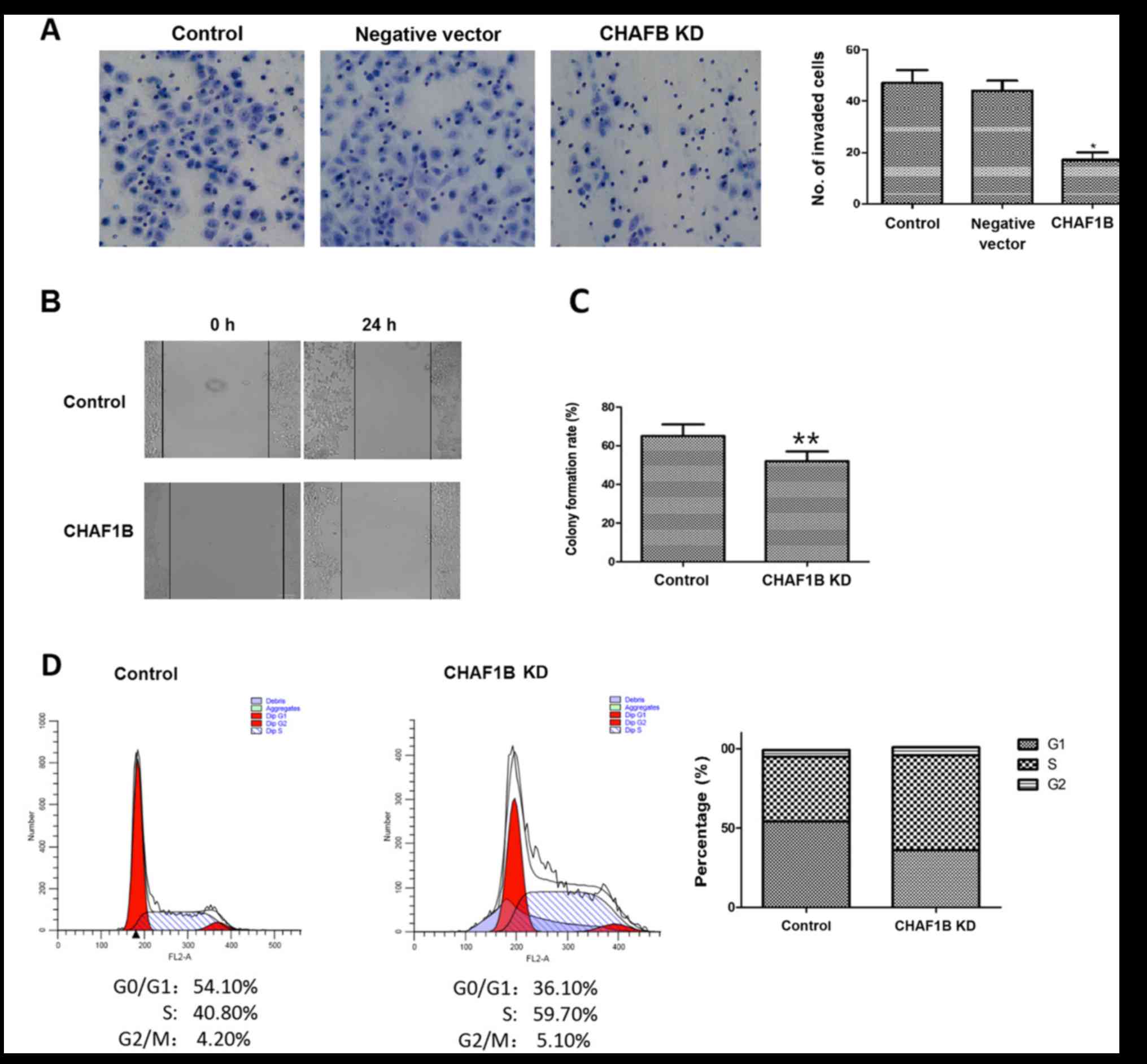
We then examined the invasion ability of HUH-7 cells
after the knockdown of the CHAF1B gene. The number of invaded cells
was significantly lower after knockdown as compared to the control
(16.33 vs. 42.13, P<0.01), while the negative vector did not
change the invasion ability (Fig.
2A), indicating that the downregulation of CHAF1B expression
reduces the invasion ability of HUH-7 cells, suggesting that it may
be used as a new approach for HCC therapy.
Migration and colony-forming ability
of HUH-7 cells after CHAF1B knockdown
We next investigated the effect of CHAF1B knockdown
on migration and colony-forming ability of HUH-7 cells. The scratch
assays revealed that compared with the control, CHAF1B knockdown
significantly reduced the migration distance of HUH-7 cells
(98.6±3.29 µm vs. 41.2±2.59 µm, P<0.01) (Fig. 2B). We also compared the colony
formation ability as a measure of tumor-forming ability. The
results revealed that CHAF1B knockdown significantly reduced the
colony formation rate as compared with the control (Fig. 2C, P<0.01). These findings
revealed that silencing of CHAF1B may reduce the tumorigenic
ability of HUH-7 cells and inhibit tumor occurrence and
development.
Effect of CHAF1B knockdown on the cell
cycle of HUH-7 cells
Flow cytometric analysis revealed that there were
significantly less HUH-7 cells in the G0/G1
phase and more in the S phase after CHAF1B knockdown (36.10 and
59.7 vs. 54.10 and 40.8%). However, the percentage of cells at the
G2/M phase were similar (4.20 vs. 5.10%), indicating that knockdown
of the CHAF1B gene reduced the number of cells in the G1 phase and
increased the cells in the S phase, resulting in changes in the
distribution of cells in different phases (Fig. 2D).
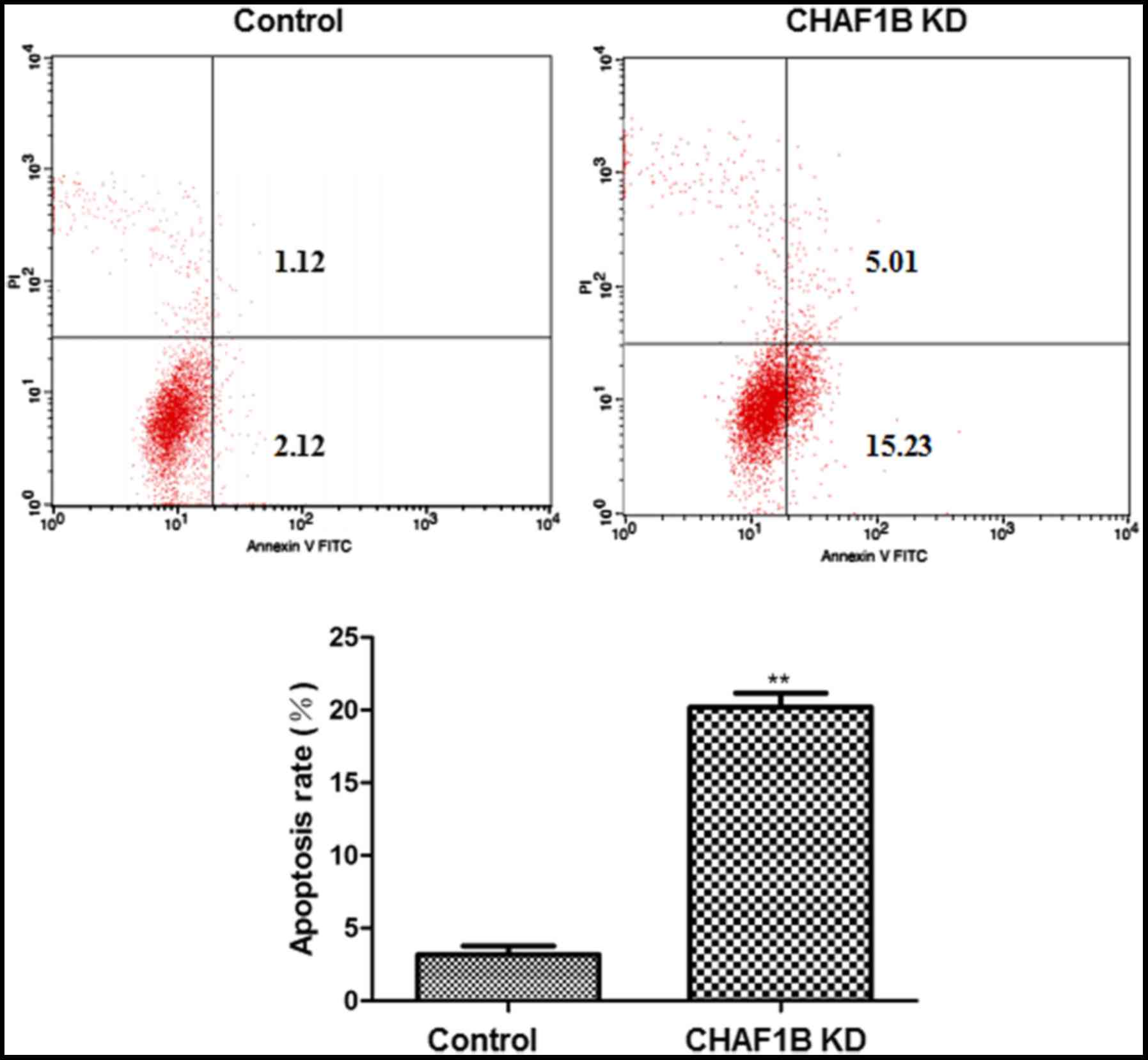
Effect of CHAF1B knockdown on the
apoptosis of HUH-7 cells
Flow cytometric analysis revealed that following
CHAF1B knockdown, HUH-7 cells had a significantly higher apoptosis
rate as compared with the control (Fig.
3).
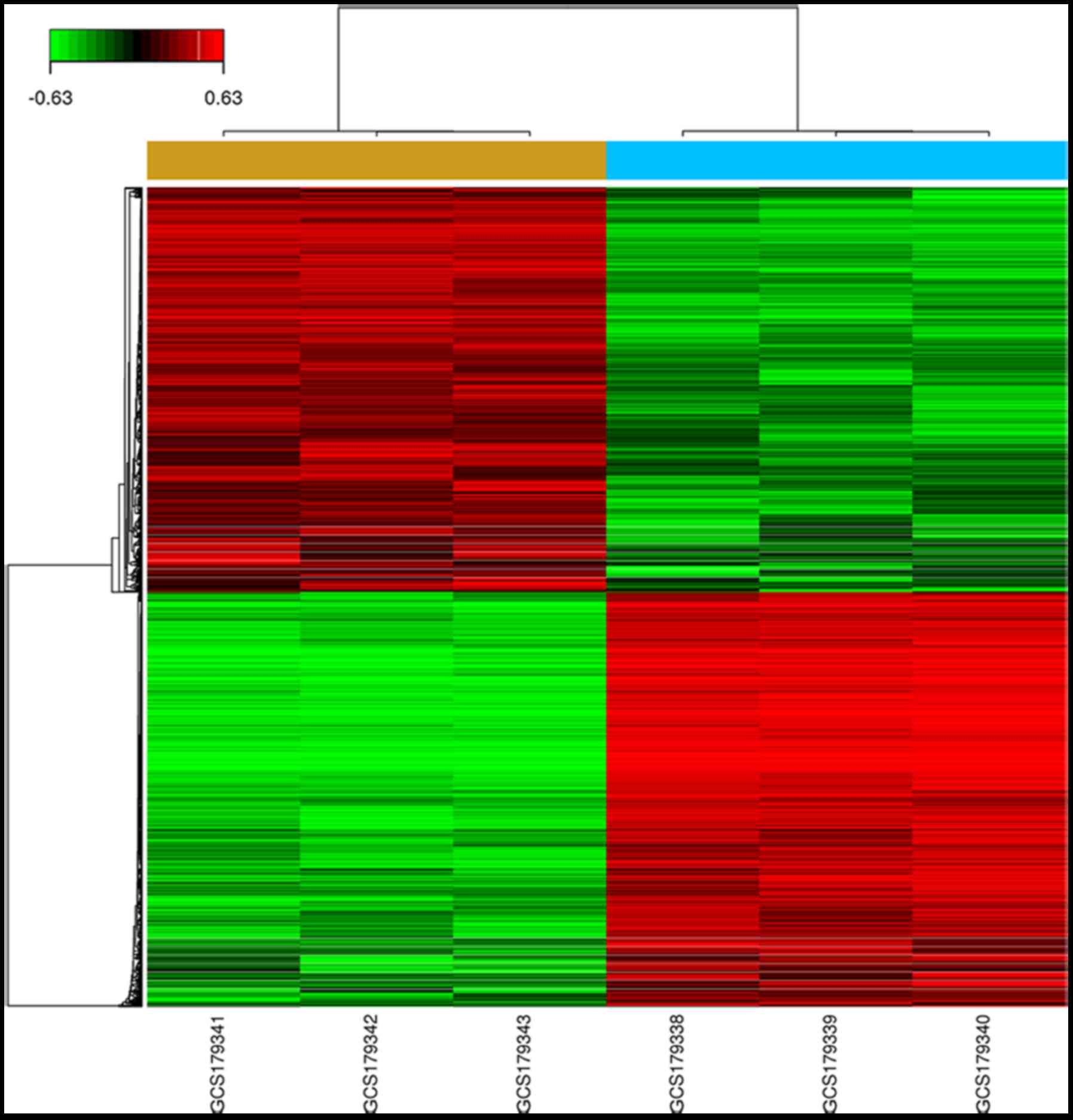
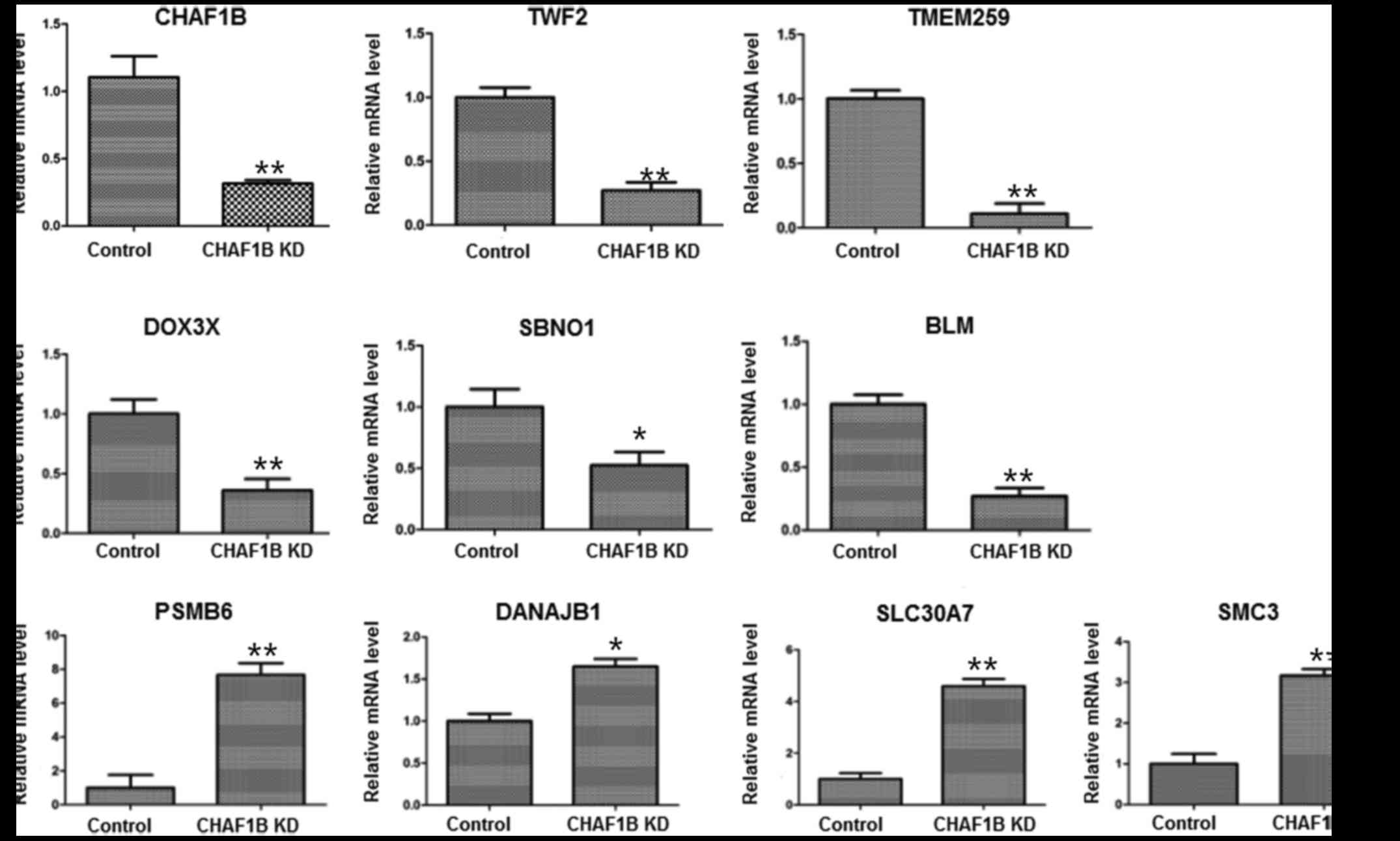
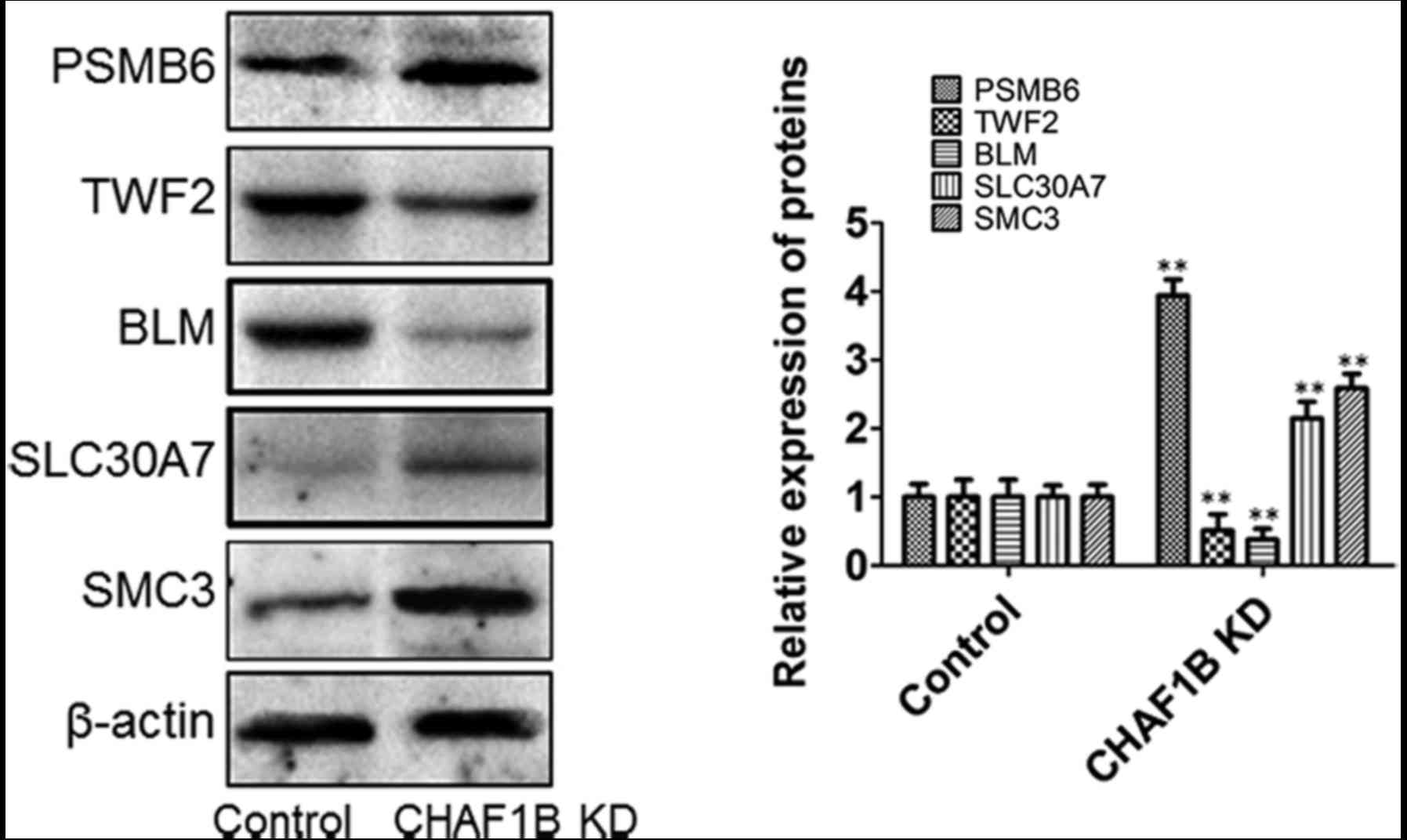
Expression of downstream genes in HUH-7 cells after
CHAF1B knockdown. We profiled the expression of genes using the
gene chip analysis. Several genes were found differentially
expressed in the knockdown cells as compared with the control
(Fig. 4), among them, the
expression levels of PSMB6, SLC30A7, SMC3, TWF2 and BLM were the
most markedlyaltered. RT-PCR results revealed that the mRNA levels
of TWF2, TMEM259, DDX3X, SBNO1 and BLM were downregulated while the
levels of PSMB6, DNAJB1, SLC30A7 and SMC3 were upregulated after
CHAF1B knockdown (Fig. 5). Western
blot analyses revealed that the protein levels of PSMB6, SLC30A7
and SMC3 were significantly increased and the levels of BLM and
TWF2 were significantly decreased following CHAF1B knockdown
(Fig. 6).
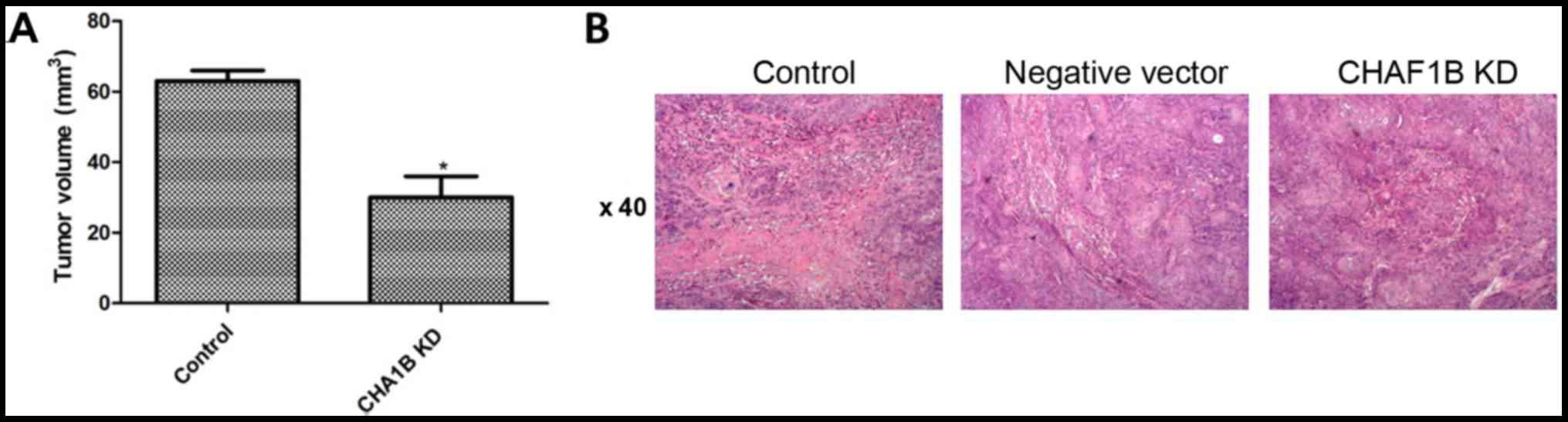
Effect of CHAF1B knockdown on tumor
growth
We then compared the growth of tumors in the mouse
models prepared using CHAF1B-knockdown HUH-7 cells. Twenty-eight
days after the injections, the volume and tumor formation rate were
similar between the control and the empty vector. However, these
figures were significantly lower in mice injected with CHAF1B
knockdown-cells as compared with the control (Fig. 7A). H&E staining further
confirmed that tumors were present in rats in all groups (Fig. 7B). Western blot analyses revealed
that CHAF1B was significantly lower after CHAF1B-knockdown
(Fig. 8A) and TUNEL assay revealed
that there were more apoptotic cells in the CHAF1B-knockdown HUH-7
cells (Fig. 8B). These findings
demonstrated that CHAF1B knockdown reduced the tumorigenicity of
HUH-7 cells.
Discussion
Chromatin assembly factors (CAFs) have subunits of
different sizes (150 kDa, CAF-1-p150 or CHAF1a), (60 kDa, CAF-1-p60
or CHAF1B) and (48 kDa, CAF-1-p48 or RbAp48, p48) (18). Verreault et al (19) purified the complex and revealed that
CHAF1B can reposition nucleosomes, and change the position and
structure of nucleosomes to regulate the level of chromatin during
DNA replication, transcription, repair and recombination (20). They determined that transcriptional
factors (TFs) and CAFs bind to specific sites in the promoter to
change the position and structure of nucleosomes and sensitize
chromatin to ribozymes. The synthesis of DNA is the basis for
nucleosome assembly and plays a key role in maintaining genome
stability. During the replication of DNA, the synthesis of CAF-1 in
histones H3 and H4 is turned off directly in many tissues, and the
large subunit (CAF-1-P150, CHAF1a) and small subunit (RbAp48, P48)
of CAF-1 have a variety of functions, for instance, as histone
chaperones (21).
In recent years, CAF1B has been demonstrated to be
highly expressed in a variety of malignant tumors, and has been
identified as a marker of poor prognosis in cancer patients
(7,10). The expression level of CHAF1B has
been demonstrated to be related to tumor invasiveness (11). It has been revealed that from radial
growth in the early stage to pituitary growth, the expression level
of CAF1B is significantly elevated, and the level also increases
during the transition from benign nevus to malignant melanoma
(21). Polo et al revealed
that in breast, cervical and endometrial cancer as well as in renal
cell carcinoma, higher expression of CAF-1 was related to
histological grading and may be used as an independent factor of
poor prognosis (22). Our
experimental results also revealed that knockdown of the CHAF1B
gene reduced the invasiveness, migration and colony-forming ability
of HCC cells, further confirming that CHAF1B is involved in HCC
biology.
CHAF1B may play a role in predicting therapeutic
efficacy and monitoring drug response (22). Inhibition of this protein in
invasive tumors can lead to tumor cell death and therefore it may
become a new therapeutic target (23). CAF-1 is a p48 complex composed of
three subunits p150, p60, and p48 (21,24–26).
Its compositions and functions are highly conserved in genome
replication (27). The primary role
of CAF-1 in DNA replication is to facilitate the first step in
assembling nucleosomes to the newly synthesized DNA (9,28,29).
At any time in the cell cycle, DNA damage could lead to the
initiation of DNA damage response (DDR). In eukaryotic tissues, DDR
coordinates DNA damage repair and repair of altered chromatin
(30). For example, CAF-1 may
recruit a large number of damage-repairing proteins to the damaged
chromatin regions as histone chaperones and/or the protein platform
to participate in DNA and chromatin damage repair (9,31–33).
Recent studies also demonstrated that CAF-1 may serve as a protein
platform of epigenetics to inhibit chromatin markers or signal
transduction of active histone, which are important for normal cell
activities (34,35). In addition, numerous studies have
revealed that CAF-1 promoted cell cycle progression by adjusting
the specific chromatin reorganization in the S phase to affect cell
proliferation (34).
In addition, CAF-1 has been demonstrated to be
associated with human diseases. The main function of the p48
subunit is to support the role of retinoblastoma protein RbAp48 in
the inhibition of cell growth. p150 is directly related to
proliferating cell nuclear antigen (PCNA), and actively
participates in the DNA repair process during reproduction
(28). The CAF1B subunits are
highly expressed in breast, oral, tongue, prostate and salivary
gland cancer, as well as in skin melanoma and other malignant
tumors and downregulation of p150 in tongue cancer and head and
neck cancer is an indicator for poor prognosis (36). The present study also demonstrated
that the knockdown of the CHAF1B gene significantly inhibited cell
proliferation and promoted apoptosis.
Collectively, CAF1B elevation was associated with
poor prognosis in HCC, and related to tumor stage, grade and
distant metastasis. Therefore, understanding the expression level
of CAF1B is helpful for the diagnosis and treatment of HCC,
although more studies are warranted to elucidate the specific
mechanism. Our study demonstrated that downregulation of CHAF1B
inhibited the proliferation of HCC cells and induced apoptosis.
These findings provided insight into the involvement of the CHAF1B
gene in tumor signal transduction pathways and indicates the
possible use of the gene for clinical diagnosis and targeted
treatment.
Acknowledgements
This study was supported bythe Department of Science
and Technology of Jiangxi Province (grant no. 20152ACG70020).
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Niu J, Lin Y, Guo Z, Niu M and Su C: The
epidemiological investigation on the risk factors of hepatocellular
carcinoma: A case-control study in southeast china. Medicine.
95:e27582010. View Article : Google Scholar
|
|
2
|
El-Serag HB and Mason AC: Rising incidence
of hepatocellular carcinoma in the United States. N Engl J Med.
340:745–750. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Llovet JM, Burroughs A and Bruix J:
Hepatocellular carcinoma. Lancet. 362:1907–1917. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yang XD, Pan LH, Wang L, Ke Y, Cao J, Yang
C, Zhong JH, Luo W, Guo J and Li LQ: Systematic review of single
large and/or multinodular hepatocellular carcinoma: Surgical
resection improves survival. Asian Pac J Cancer Prev. 16:5541–5547.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lee CW, Tsai HI, Sung CM, Chen CW, Huang
SW, Jeng WJ, Wu TH, Chan KM, Yu MC, Lee WC and Chen MF: Risk
factors for early mortality after hepatectomy for hepatocellular
carcinoma. Medicine. 95:e50282016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
De Koning L, Corpet A, Haber JE and
Almouzni G: Histone chaperones: An escort network regulating
histone traffic. Nat Struct Mol Biol. 14:997–1007. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Xu M, Jia Y, Liu Z, Ding L, Tian R, Gu H,
Wang Y, Zhang H, Tu K and Liu Q: Chromatin assembly factor 1,
subunit A (P150) facilitates cell proliferation in human
hepatocellular carcinoma. Onco Targets Ther. 9:4023–4035. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yu Z, Liu J, Deng WM and Jiao R: Histone
chaperone CAF-1: Essential roles in multi-cellular organism
development. Cell Mol Life Sci. 72:327–337. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Polo SE, Roche D and Almouzni G: New
histone incorporation marks sites of UV repair in human cells.
Cell. 127:481–493. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Mascolo M, Ilardi G, Merolla F, Russo D,
Vecchione ML, de Rosa G and Staibano S: Tissue microarray-based
evaluation of chromatin assembly factor-1 (CAF-1)/p60 as tumour
prognostic marker. Int J Mol Sci. 13:11044–11062. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Staibano S, Mascolo M, Rocco A, Lo Muzio
L, Ilardi G, Siano M, Pannone G, Vecchione ML, Nugnes L, Califano
L, et al: The proliferation marker chromatin assembly factor-1 is
of clinical value in predicting the biological behaviour of
salivary gland tumours. Oncol Rep. 25:13–22. 2011.PubMed/NCBI
|
|
12
|
Jeffery DC, Kakusho N, You Z, Gharib M,
Wyse B, Drury E, Weinreich M, Thibault P, Verreault A, Masai H and
Yankulov K: CDC28 phosphorylates Cac1p and regulates the
association of chromatin assembly factor I with chromatin. Cell
Cycle. 14:74–85. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wu Z, Cui F, Yu F, Peng X, Jiang T, Chen
D, Lu S, Tang H and Peng Z: Up-regulation of CHAF1A, a poor
prognostic factor, facilitates cell proliferation of colon cancer.
Biochem Biophys Res Commun. 449:208–215. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Mascolo M, Ayala F, Ilardi G, Balato A and
Lembo S: Chromatin assembly factor-1/p60 overexpression: A
potential index of psoriasis severity. Eur J Dermatol. 24:509–511.
2014.PubMed/NCBI
|
|
15
|
Sarkar D, Leung EY, Baguley BC, Finlay GJ
and Askarian-Amiri ME: Epigenetic regulation in human melanoma:
Past and future. Epigenetics. 10:103–121. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liang CC, Park AY and Guan JL: In vitro
scratch assay: A convenient and inexpensive method for analysis of
cell migration in vitro. Nat Protoc. 2:329–333. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2−ΔΔCT method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Volk A and Crispino JD: The role of the
chromatin assembly complex (CAF-1) and its p60 subunit (CHAF1b) in
homeostasis and disease. Biochim Biophys Acta. 1849:979–986. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Verreault N, Da Costa D, Marchand A,
Ireland K, Banack H, Dritsa M and Khalife S: PTSD following
childbirth: Prospective study of incidence and risk factors
incanadian women. J PsychosomRes. 73:257–266. 2012. View Article : Google Scholar
|
|
20
|
Cheloufi S, Elling U, Hopfgartner B, Jung
YL, Murn J, Ninova M, Hubmann M, Badeaux AI, Ang Euong C, Tenen D,
et al: The histone chaperone CAF-1 safeguards somatic cell
identity. Nature. 528:218–224. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kaufman PD, Kobayashi R, Kessler N and
Stillman B: The p150 and p60 subunits of chromatin assembly factor
I: A molecular link between newly synthesized histones and DNA
replication. Cell. 81:1105–1114. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Polo SE, Theocharis SE, Grandin L,
Gambotti L, Antoni G, Savignoni A, Asselain B, Patsouris E and
Almouzni G: Clinical significance and prognostic value of chromatin
assembly factor-1 overexpression in human solid tumours.
Histopathology. 57:716–724. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Mascolo M, Ilardi G, Romano MF, Celetti A,
Siano M, Romano S, Luise C, Merolla F, Rocco A, Vecchione ML, et
al: Overexpression of chromatin assembly factor-1 p60,
poly(ADP-ribose) polymerase 1 and nestin predicts metastasizing
behaviour of oral cancer. Histopathology. 61:1089–1105. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kim D, Setiaputra D, Jung T, Chung J,
Leitner A, Yoon J, Aebersold R, Hebert H, Yip CK and Song JJ:
Molecular architecture of yeast chromatin assembly factor 1. Sci
Rep. 6:267022016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Smith S and Stillman B: Purification and
characterization of CAF-I, a human cell factor required for
chromatin assembly during DNA replication in vitro. Cell. 58:15–25.
1989. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhou H, Madden BJ, Muddiman DC and Zhang
Z: Chromatin assembly factor 1 interacts with histone H3 methylated
at lysine 79 in the processes of epigenetic silencing and DNA
repair. Biochemistry. 45:2852–2861. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Shibahara K and Stillman B:
Replication-dependent marking of DNA by PCNA facilitates
CAF-1-coupled inheritance of chromatin. Cell. 96:575–585. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Moggs JG, Grandi P, Quivy JP, Jónsson ZO,
Hübscher U, Becker PB and Almouzni G: A CAF-1-PCNA-mediated
chromatin assembly pathway triggered by sensing DNA damage. Mol
Cell Biol. 20:1206–1218. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Krude T: Chromatin assembly factor 1
(CAF-1) colocalizes with replication foci in HeLa cell nuclei. Exp
Cell Res. 220:304–311. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li GM: New insights and challenges in
mismatch repair: Getting over the chromatin hurdle. DNA Repair.
19:48–54. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Adam S, Polo SE and Almouzni G:
Transcription recovery after DNA damage requires chromatin priming
by the H3.3 histone chaperone HIRA. Cell. 155:94–106. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Baldeyron C, Soria G, Roche D, Cook AJ and
Almouzni G: HP1alpha recruitment to DNA damage by p150CAF-1
promotes homologous recombination repair. J Cell Biol. 193:81–95.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yu Z, Wu H, Chen H, Wang R, Liang X, Liu
J, Li C, Deng WM and Jiao R: CAF-1 promotes notch signaling through
epigenetic control of target gene expression during drosophila
development. Development. 140:3635–3644. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Huang H, Yu Z, Zhang S, Liang X, Chen J,
Li C, Ma J and Jiao R: Drosophila CAF-1 regulates HP1-mediated
epigenetic silencing and pericentric heterochromatin stability. J
Cell Sci. 123:2853–2861. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wen P, Quan Z and Xi R: The biological
function of the WD40 repeat-containing protein p55/Caf1 in
drosophila. Dev Dyn. 241:455–464. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Staibano S, Mignogna C, Lo Muzio L,
Mascolo M, Salvatore G, Di Benedetto M, Califano L, Rubini C and De
Rosa G: Chromatin assembly factor-1 (CAF-1)-mediated regulation of
cell proliferation and DNA repair: A link with the biological
behaviour of squamous cell carcinoma of the tongue? Histopathology.
50:911–919. 2007. View Article : Google Scholar : PubMed/NCBI
|