Introduction
Rhomboid genes encode the most distributed and
evolutionarily conserved polytopic intramembrane proteins (1). This superfamily comprises active
intramembrane serine proteases that can activate or transactivate
the epidermal growth factor receptor (EGFR) signaling pathway and a
group of non-catalytic members with diverse cellular functions
(1). The human genome contains 14
rhomboid genes that can be grouped into five rhomboid proteases
(RHBDL1/2/3/4 and PARL) and nine pseudoproteases
(iRhom1/2, Derlin1/2/3, RHBDD2/3, UBAC2 and TMEM115)
(2). Previous studies have
demonstrated that rhomboid proteases and pseudoproteases are
involved in several cellular processes such as cell proliferation,
apoptosis, endoplasmic reticulum (ER) stress and EGFR activation.
Rhomboids have also been associated with human diseases, such as
neurodegenerative conditions, as well as cancer (2).
RHBDD2 pseudoprotease has been found overexpressed
in advanced-stage breast and colorectal cancers (3–6).
Although the particular RHBDD2 function has not yet been
assessed, its expression has been associated with breast cancer
cell migration, proliferation, and cellular response to ER stress
(7). A previous study has suggested
the existence of two human RHBDD2 transcriptional variants
which are differentiated by the alternative splicing of exon II in
the mature mRNA (4). The
RHBDD2 mRNA variant 1 (RHBDD2-1) encodes a protein
known as isoform A, while RHBDD2 variant 2 (RHBDD2-2)
encodes a shorter protein called isoform B (4).
It is known that a subtle balance between splicing
variants is crucial to cellular homeostasis, while the unbalanced
expression of splicing variants contributes to cancer development
(8). The expression pattern of
specific variants of numerous genes (eg. BRCA1/2 genes) is
altered during oncogenesis giving the cell an adaptive phenotype to
the changing tumor environment (9,10). The
switch between isoforms is regulated by different genetics and
environmental factors finally determining the tumor phenotype
(11,12).
In the present study, we analyzed the expression of
the RHBDD2 splicing variants in neoplastic and normal breast
samples, as well as in breast cancer cell lines under nutritional
stress conditions. In addition, we determined the RHBDD2 protein
subcellular localization in breast cancer cells.
Materials and methods
Breast tissue samples
Breast tissue specimens were obtained from different
hospitals and medical centres associated with the School of Medical
Sciences of the National University of La Plata. Twenty-three
breast primary tumor samples (all invasive ductal carcinomas), as
well as 12 normal mammary gland samples obtained from aesthetic
mammoplasties were studied. All samples were obtained from female
patients ranging from 28 to 89 years old, during the period from
February 2011 to October 2015.
Breast cancer cell line culture and
glucose starvation (GS) assay
The human MCF7, T47D and MDA-MB-231 breast cancer
cell lines were purchased from the ATCC® Bioresource
Center (Manassas, VA, USA). Cells were cultured in Dulbecco's
modified Eagle's medium (DMEM) (D7777; Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany) with 10% fetal bovine serum (FBS; Natocor,
Villa Carlos Paz, Argentina), 100 U/ml penicillin and 100 µg/ml
streptomycin (P0781; Sigma-Aldrich; Merck KGaA) at 37°C in
humidified atmosphere with 5% CO2. MCF7 and T47D cell
lines were also cultured under glucose starvation (GS) conditions.
Briefly, cells were cultured on a 6-well plate to 70% confluence in
complete DMEM (D7777) with glucose as described above. Then, the
medium was replaced by incomplete DMEM medium (D5030;
Sigma-Aldrich; Merck KGaA) without glucose and FBS, supplemented
with 100 U/ml penicillin and 100 µg/ml streptomycin. Cells were
maintained under GS condition for 3, 6, 9 and 12 h. Control cells
were cultured in complete DMEM. At each time point, the cells were
harvested and total protein and RNA were isolated.
Protein and total RNA were isolated using TRI
Reagent™ solution (Thermo Fisher Scientific, Inc., Waltham, MA,
USA) according to the manufacturer's protocol. The SuperScript™
Reverse Transcriptase kit (Thermo Fisher Scientific, Inc.) was used
for cDNA synthesis according to the manufacturer's protocol.
Gene expression analysis
Total RHBDD2 and RHBDD2-2 (splicing
variant 2) expression levels were analyzed by RT-qPCR using the
StepOne™ Real-Time PCR System and associated Software v2.3 (Thermo
Fisher Scientific, Inc.). In addition, genes involved in cell
stress response were evaluated: DDIT3 mRNA expression was
measured as reference for cell GS response (13), while HSPA5 and CALR
were evaluated for ER cell stress (14,15).
Gene expression levels were calculated by the 2−ΔCq
method (16), using as reference
rRNA18S. The SYBR™ Select Master Mix (Thermo Fisher
Scientific, Inc.) was used for RT-qPCR reaction solution, according
to manufacturer's protocol. The following primers were used:
total RHBDD2 (Fw: 5′-ggtgtttggcatggttgtg-3′, Rv:
5′-cgatggaatagcagtaggtga-3′); RHBDD2-2 (Fw:
5′-attacagcagaggagactgg-3′, Rv: 5′-gatgtaggttaccagcctgt-3′);
DDIT3 (Fw: 5′-agccaaaatcagagctggaa-3′ and Rv:
5′-tggatcagtctggaaaagca-3); HSPA5 (Fw:
5′-cacagtggtgcctaccaaga-3′ and Rv: 5′-tgt ctt ttg tca ggg gtc
ttt-3′); CALR (Fw: 5′-aca acc ccg agt att ctc cc-3′ and Rv:
5′-tgt caa aga tgg tgc cag ac-3′) and rRNA18S (Fw: 5′-gta
acc cgtt gaa ccc catt-3′, Rv: 5′-cca tcc aat cgg tag tag cg-3′).
RT-PCR thermal profile was as follows: 5 min at 95°C, 40 cycles of
40 sec at 95°C-30 sec at 55°C (for all primer pairs)-30 sec at
72°C, and a final cycle at 95°C for 1 min-55°C for 30 sec and 96°C
for 30 sec was added.
RHBDD2 isoform analysis in breast
cancer cell lines and tissue samples
The RHBDD2 splicing variants and protein
isoform expression were evaluated in breast cancer cell lines and
breast normal and tumor samples selected for showing RHBDD2
mRNA expression. Total RHBDD2 and RHBDD2-2 mRNA
expression were analyzed by RT-qPCR, using the primers mentioned
above. We also evaluated by direct sequencing the RHBDD2
splicing variant RT-PCR products obtained with the primer pair: Fw:
5′-tgaagtccgaggccctt-3′ (complementary to exon I) and Rv:
5′-caaagcgccagatgatgata-3′ (complementary to exon III).
The RHBDD2 protein isoforms were detected by
SDS-PAGE followed by Western-blot analysis. Total protein was
isolated from human cell lines and breast samples with TRI Reagent™
Solution (Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. The primary antibody: rabbit anti-RHBDD2
(1:1,000; cat. no. TA306891; Origene, Rockville, MD, USA) was
incubated overnight at 4°C. The antibody target sequence is located
at the C-terminal domain of both RHBDD2 isoforms. Then, the
secondary antibody, goat anti-rabbit IgG-HRP conjugate (1:2,000;
cat. no. P0448; Dako Denmark, Glostrup, Hovedstaden, DK) was
incubated for 3 h at 4°C. The estimated molecular weight for RHBDD2
isoform A is 39.21 kDa (364 aa), and for RHBDD2 isoform B is 23.64
kDa (223 aa). ACTB was used as a loading reference (42 kDa). It was
detected using the primary antibody: mouse anti-β-actin-HRP
conjugate (1:5,000; cat. no. ab173838; Abcam, Cambridge, MA, USA)
incubated for 3 h at 4°C. Protein bands were visualized by
chemiluminescence reaction on radiographic plates using the EasySee
Western Blot kit (DW101-01; TransBionovo, BJ, CN). The relative
expression of the RHBDD2 isoforms was determined by density band
analysis with ImageJ software (https://imagej.nih.gov/ij/).
RHBDD2 subcellular localization by
confocal immunofluorescence
The RHBDD2 subcellular localization was determined
by fluorescence immunocytochemistry in MCF7, T47D and MDA-MB-231
cells. Cells were grown on a 100-mm2 cover glass to 70%
confluence and fixed with 4% formaldehyde or cold acetone.
Initially, the cell membrane was permeabilized with 0.01% Triton,
incubating for 10 min at room temperature. Then, the cells were
incubated overnight at 4°C with the primary antibodies: rabbit
anti-RHBDD2 (1:400); mouse anti-CANX (1:50; cat. no. MA3-027;
Thermo Fisher Scientific, Inc.) for ER detection (17); mouse anti-GalNacT3 (culture
supernatant donated by Professor Ulla Mandel, Copenhagen Center for
Glycomics, University of Copenhagen) for Golgi apparatus detection
(18); and mouse anti-Ykt6p v-SNARE
(E-2) (1:50; cat. no. sc-365732; Santa Cruz Biotechnology, Dallas,
TX, USA) for transport vesicle detection (19). Later, cells were incubated for 2 h
at 4°C with the secondary antibodies: goat anti-rabbit IgG-Cy3
conjugate (1:200; cat. no. 711165152; Jackson ImmunoResearch
Laboratories, Inc., West Grove, PA, USA) and goat anti-mouse
IgG-biotin conjugate (1:100; cat. no. BA9200; Vector Laboratories,
Inc., Burlingame, CA, USA). Then, the cells were incubated with
streptavidin-FITC conjugated-(1:10,000; cat. no. SA10002; Thermo
Fisher Scientific, Inc.) for 30 min at room temperature. Finally,
cells were visualized using the Confocal FluoView™ 1000
immunofluorescence microscope. Images were acquired at red and
green fluorescence signal channels with the associated FluoView
Software (Olympus Latin America, Miami, FL, USA). Colocalization
analysis was performed with the JaCoP application on ImageJ
software. The Pearson's (R) correlation coefficient was calculated
for colocalization quantification (20).
Statistical analysis
RT-qPCR experiments were performed in triplicate for
each data point. Data were analyzed using the Student's t-test or
one-way analysis of variance (ANOVA). Data are expressed as the
means ± 2 standard deviations (SDs) of the sample. All tests were
two-tailed, and the level of statistical significance was set at
P≤0.05. Statistical analysis was performed with R Software
(https://www.r-project.org/).
In silico analysis of RHBDD2 splicing
variants in breast carcinomas
To further investigate the relevance of both
RHBDD2 splicing variants in human breast carcinomas,
RHBDD2 transcript variant profiles were analyzed employing
The Cancer Genome Atlas-Breast Cancer (TCGA-BRCA) RNA-Seq dataset
obtained from the UCSC Xena TOIL RNA-seq recompute resource.
Briefly, RHBDD2-1 (ENST00000006777.10) and
RHBDD2-2 (ENST00000428119.1) expression levels were
evaluated in 1,092 primary invasive breast carcinomas.
Seven-hundred and twenty-nine cases out of 1,092 showed high
expression levels of RHBDD2-1 (Var 1), RHBDD2-2 (Var
2) or both variants that were subsequently grouped according to
their intrinsic subtypes, proliferative and Risk-Of-Recurrence
scores (ROR-P). Intrinsic subtypes and their derivate scores
(proliferation and ROR-P) were determined using the 50-gene (PAM50)
predictor bioclassifier R script (21).
Results
The RHBDD2 gene encodes two splicing
variants differentially expressed in tumor and normal tissues
RHBDD2 gene sequence analysis allowed the
identification of 5 exons and 4 introns with two putative
transcripts differentiated by the alternative splicing of exon II
in the mature mRNA (4). The
RHBDD2 mRNA variant 1 (RHBDD2-1) is a transcript of
1,756 nt encoded by the exons I, III, IV and V. The RHBDD2-2
variant is a transcript of 1,878 nt encoded by the exons I, II,
III, IV and V.
The expression pattern of the RHBDD2 splicing
variants was evaluated on breast cancer cell lines and on known
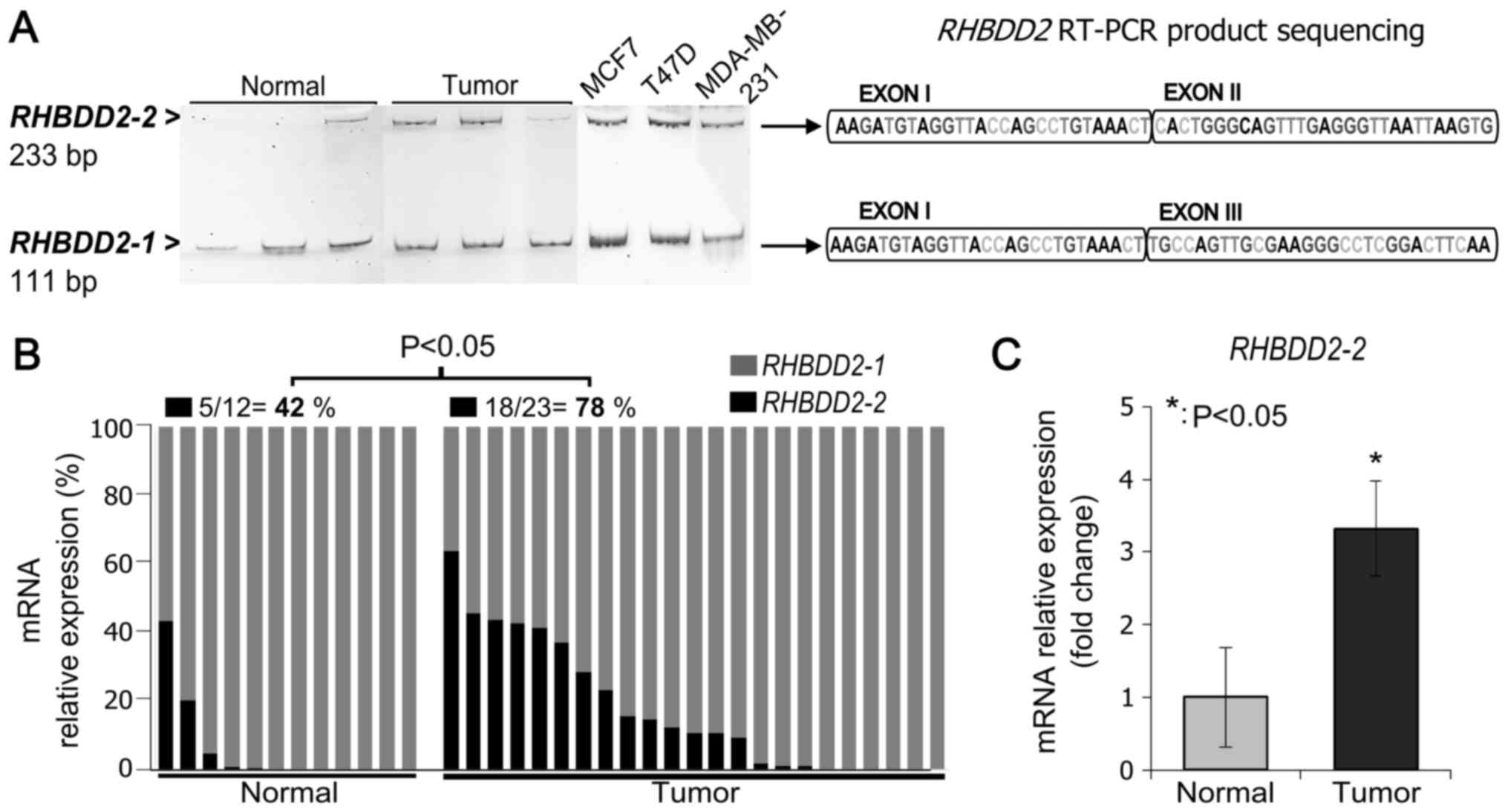
RHBDD2-positive breast tumor and normal samples by RT-PCR.
We used a primer pair designed to detect both mRNA variants; the
forward primer spanned exon I and the reverse primer spanned exon
III. The expected PCR products for the RHBDD2-1 and
RHBDD2-2 variants (111 bp and 233 bp, respectively) were
detected on the analyzed samples. The identity of the amplification
products was confirmed by PCR amplicon sequencing (Fig. 1A).
In addition, RHBDD2 splicing variants
expression were analyzed by RT-qPCR in normal and tumor samples
(Fig. 1B). RHBDD2-1
expression was detected in all normal and tumor samples. While,
RHBDD2-2 expression was more frequently detected in tumors
(78%, 18 out of 23) than in normal tissues (42%, 5 out of 12)
(P<0.05, Fig. 1B). Moreover, a
significant increment in the RHBDD2-2 expression level was
detected in tumors in respect to normal breast samples (P<0.05,
Fig. 1C).
RHBDD2–2 expression is associated with
poor patient prognostic factors
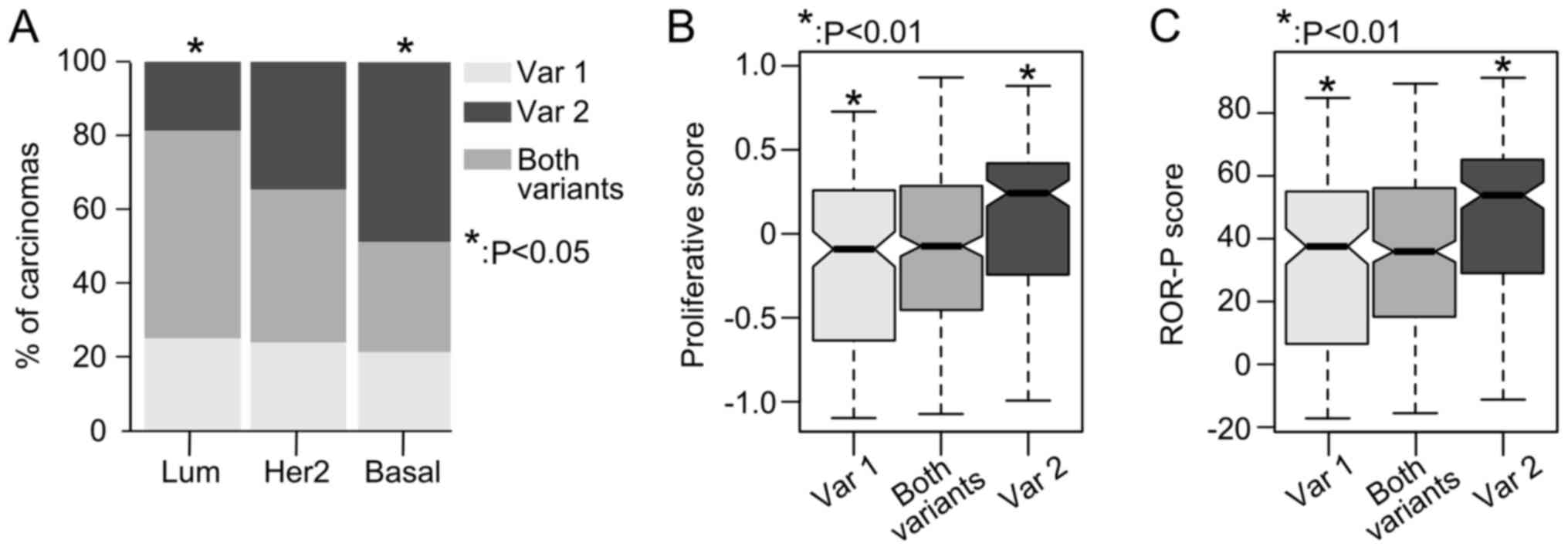
The expression profiles of RHBDD2 splicing
variants were analyzed in a 729 breast cancer samples, obtained
from the TCGA-BRCA RNA-Seq database.
RHBDD2-2 variant expression was found to be
significantly more frequent in basal-like breast carcinomas (49% of
cases) in respect to luminal-like tumors (19% of cases, P<0.05),
but not in respect to Her2-enriched tumors (P>0.05) (Fig. 2A). In addition, patients with
primary breast carcinomas that expressed the RHBDD2-2
variant had an increased proliferative and ROR-P scores compared
with tumors expressing the RHBDD2-1 variant (P<0.01,
Fig. 2B and C).
RHBDD2-2 is translated to protein and
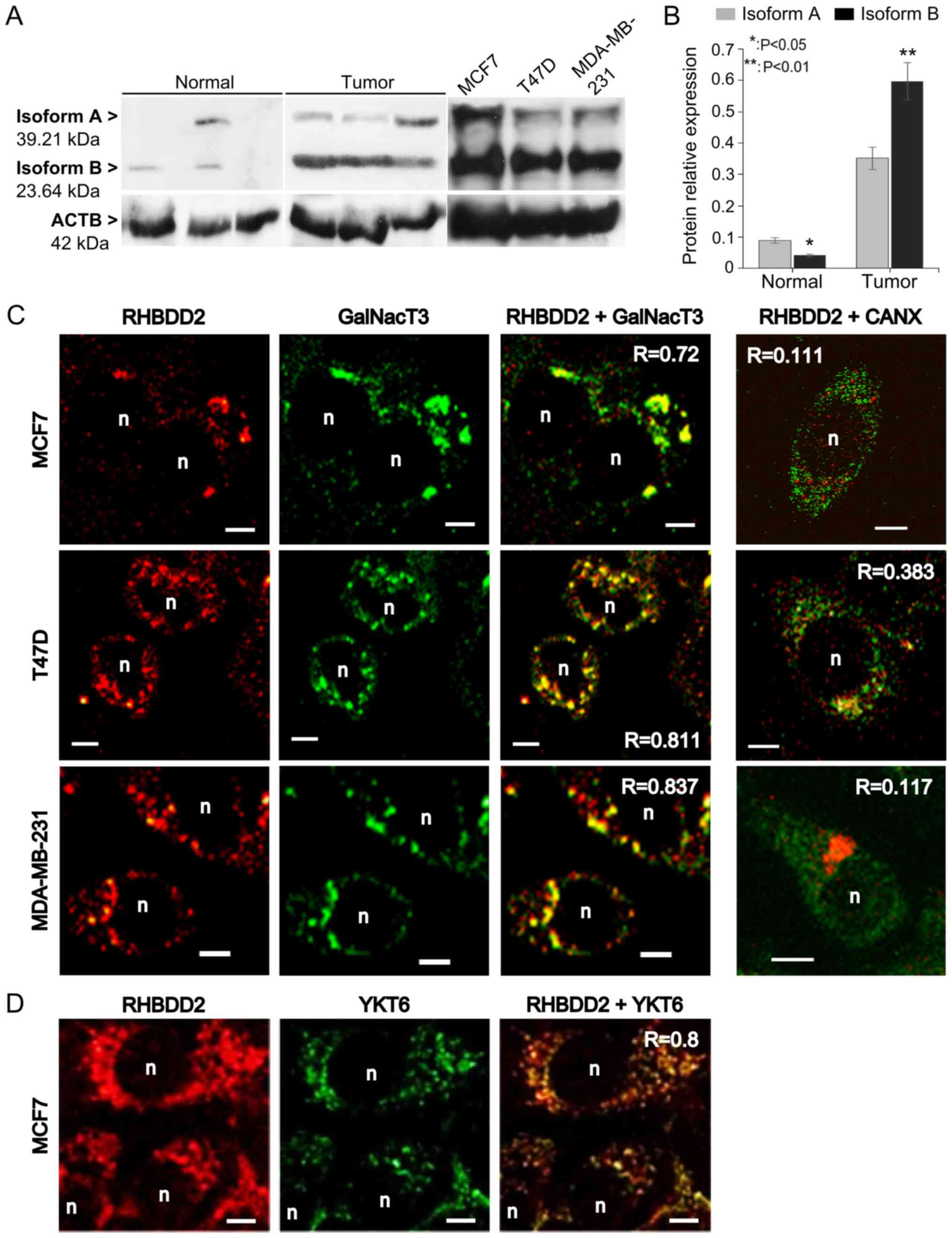
overexpressed in breast cancer cells
In order to confirm RHBDD2-2 expression at the
protein level, total protein from breast cancer cell lines, normal
and breast tumor samples was analyzed by western blot. An
anti-RHBDD2 antibody against a C-terminus aa sequence present in
both RHBDD2 isoforms was used. The RHBDD2-1 mRNA variant
encodes a 364 aa protein known as isoform A (39.21 kDa). The
RHBDD2-2 variant encodes a 223 aa protein we called isoform
B protein (23.64 kDa) with a shorter N-terminus, translated from an
internal AUG codon in exon III.
Both RHBDD2 protein isoforms were detected: isoform
A at 39 kDa and isoform B at 24 kDa. ACTB was used as reference for
relative protein isoform amount estimation (Fig. 3A). As expected, a significant
overexpression of total RHBDD2 protein was detected in the tumor
samples (P<0.01). Individual RHBDD2 isoform analysis detected a
statistically significant increment of RHBDD2 isoform B expression
in respect to isoform A in the tumoral samples (P<0.01). In
contrast, isoform B expression was lower than isoform A in the
normal samples (P<0.05, Fig.
3B).
RHBDD2 subcellular localization
RHBDD2 subcellular localization was determined by
confocal immunofluorescence microscopy in the human breast cancer
cell lines MCF7, T47D and MDA-MB-231. Quantification of
colocalization analysis revealed a positive correlation between
RHBDD2 and the Golgi apparatus marker GalNacT3 (R>0.7, Fig. 3C), also the ER-Golgi/intra-Golgi
transport vesicle marker Ytk6 v-SNARE (R=0.8, Fig. 3D), but not the ER marker CANX
(R<0.4, Fig. 3C).
Glucose starvation triggers RHBDD2
variant 2 expression in breast cancer cells
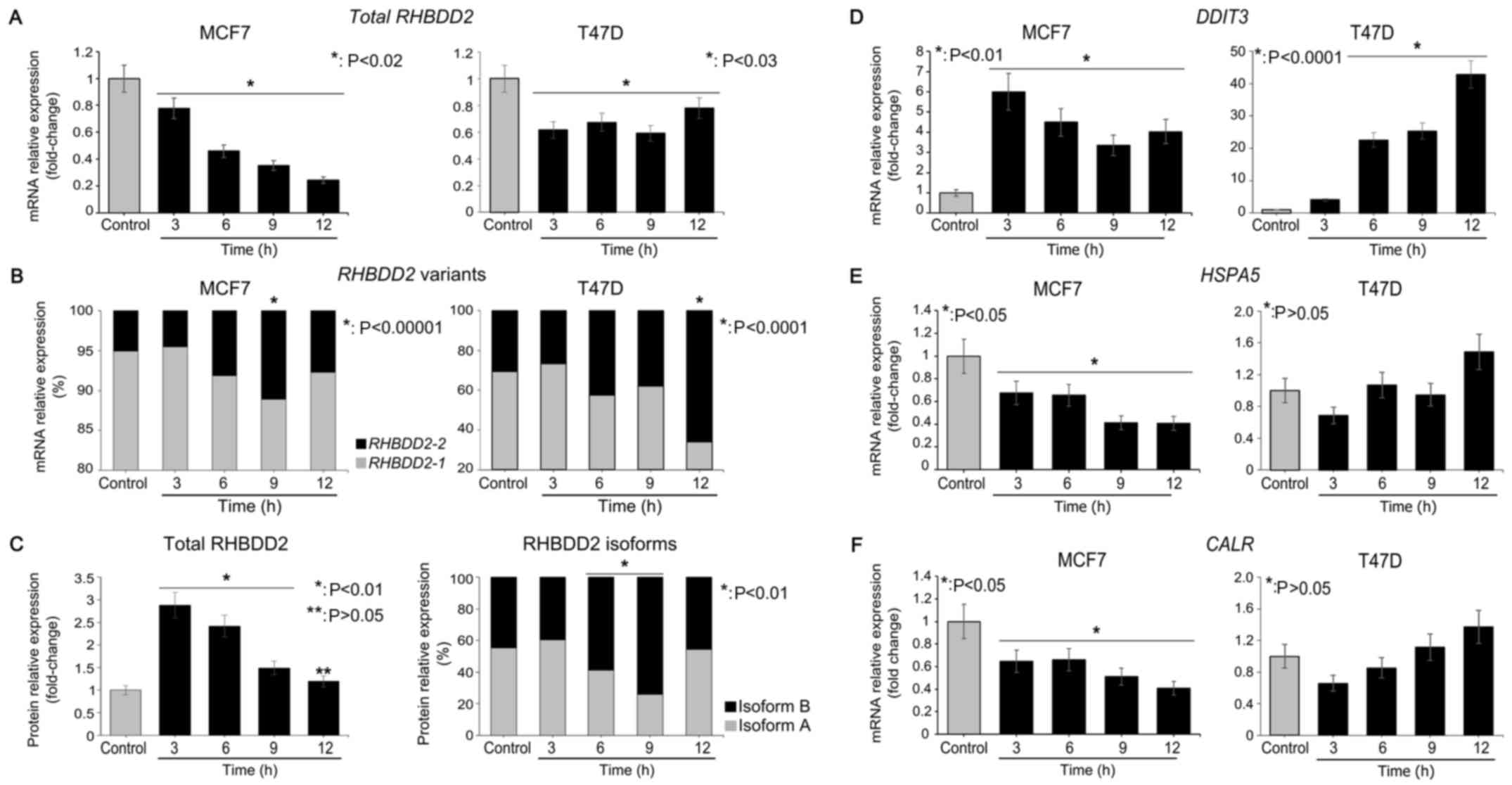
Total RHBDD2 and RHBDD2-2 mRNA levels
were evaluated in the MCF7 and T47D cells under GS conditions from
3 to 12 h by RT-qPCR. A significant decrease in total RHBDD2
mRNA expression was detected under GS conditions in the MCF7
(P<0.02) and T47D (P<0.03) cell lines (Fig. 4A). However, a statistically
significant induction of RHBDD2-2 expression was observed at
6 h of GS treatment in both cancer cell lines (P<0.01). The
RHBDD2-2 highest expression levels were detected at 9 and 12
h of GS treatment in the MCF7 (P<0.00001) and T47D (P<0.0001)
cells respectively (Fig. 4B). In
addition, a concomitant significant RHBDD2-1 decrease was
observed (P<0.03, Fig. 4B),
indicating a switch from RHBDD2 variant 1 to RHBDD2 variant 2 in
response to GS. The cellular response to GS was confirmed by
DDIT3 mRNA level analysis (13). As expected, a significant increment
of DDIT3 expression was detected under GS in the MCF7
(P<0.01) and T47D (P<0.0001) cells (Fig. 4D). In addition, CALR and
HSPA5 mRNA levels were analyzed as ER cell stress response
biomarkers (14,15). These data suggest that GS treatment
does not induce ER cell stress response in both cellular models
(Fig. 4E and F).
Furthermore, expression of RHBDD2 protein isoforms
was also analyzed in MCF7 cells. A significant increment of total
RHBDD2 protein was detected at 3, 6 and 9 h of GS respect to
controls (P<0.01, Fig. 4C). In
addition, a significant increment of isoform B expression was
detected at 6 and 9 h of GS in respect to the control cells
(P<0.01), while a significant reduction of isoform A was
observed (P<0.01, Fig. 4C).
Discussion
The RHBDD2 gene belongs to the Rhomboid
transmembrane protein superfamily. Among the rhomboid proteases and
pseudoproteases members, RHBDD2 is classified as a
pseudoprotease because of the loss of the catalytic site (1). Although the RHBDD2 cell
function is unknown, its aberrant expression has been associated
with malignant diseases (2).
RHBDD2 overexpression was initially described by Abba et
al in patients with advanced breast cancer as a consequence of
gene amplification events (3).
Recently, we described a significant association between
RHBDD2 overexpression among breast carcinomas with
low/negative progesterone receptor expression (6). In the present study, we demonstrated
that RHBDD2 mRNA variants and their coding proteins can be
differentially detected in breast tumor and normal specimens. As
previously described (3,4,6), total
RHBDD2 (mRNA and protein) was found to be upregulated in
breast carcinomas. However, individual splicing variant analysis
allowed us to detect a significant increment in RHBDD2-2
variant expression in breast carcinomas in respect to normal
tissues. More importantly, we identified an increased expression of
isoform B in respect to isoform A in tumor samples, extending our
previous knowledge of RHBDD2 expression in this malignant disease.
Furthermore, in-silico analysis of TCGA-BRCA RNA-Seq data
showed a significantly increased expression of the RHBDD2-2
variant in the basal-like breast cancer subtype. This subtype is
characterized by an aggressive phenotype and does not respond to
hormonal therapy but to chemotherapy with anthracycline and taxane
(22). The RHBDD2-2 variant
was also associated with the highest cell proliferation and ROR-P
scores indicating a worse outcome for patients with this variant
expression in respect to RHBDD2-1.
Sustained tumor growth requires the capability to
deal with a nutrient-deprived microenvironment before neovascular
development. Glucose deprivation has diverse effects in cancer cell
lines depending on lineage differences and the presence of
mutations in the several pathways it may trigger (23). Besides the alteration in glucose
metabolism and cancer-related pathways (eg. mTOR and AKT), glucose
starvation may trigger changes in RNA processing, protein delivery
and Golgi physiology (24,25). In the present study, we evaluated
the RHBDD2 response to nutritional stress by glucose
deprivation based on its previously described association with the
modulation of cellular stress conditions (5,7). A
decrease in total RHBDD2 mRNA expression level was detected
in response to GS. However, we observed a significant increase in
RHBDD2-2 expression concomitant with a reduction in
RHBDD2-1 mRNA levels. Under normal glucose conditions
RHBDD2-1 was the most expressed variant. In agreement with
these data, RHBDD2 protein isoform B significantly increased its
expression in response to glucose deprivation in contrast to the
isoform A, whose expression was reduced. The mechanisms that could
modulate mRNA splicing in response to glucose starvation in in
vitro models have not been completely determined. Regulation of
mRNA splicing by nutritional factors has been described in
glucose-6-phosphate dehydrogenase (G6PD) expression
(24). Starvation inhibits
G6PD splicing by decreasing the rate of intron removal,
leading to a decrease in mature mRNA (26). Glucose starvation-induced splicing
regulatory events have also been described in tumor cell lines. In
murine ovarian carcinoma cells, VEGF mRNA variant expression
and stability are affected by glucose starvation (27).
mRNA sequence analysis indicated that protein
isoform A is translated from an AUG codon in the RHBDD2-1
exon I, while isoform B should be translated from an internal AUG
codon located in the RHBDD2-2 exon III. Two well-known
stress-related regulatory mechanisms could be driving the
translation process of isoform B: the internal ribosome entry sites
(IRES) and the upstream open reading frames (uORFs) (28). IRES elements are specialized RNA
regulatory sequences governing cap-independent translation
initiation from internal AUGs that are translated during cellular
stress when cap-dependent translation is compromised (29). uORFs are sequences defined by an
initiation codon in frame with a termination codon located upstream
or downstream to the main AUG. uORFs has been described to modulate
the expression of stress-related mRNAs such as CHOP, ATF4/5
and GADD34 (30).
Importantly, RHBDD2 mRNA sequence analysis using the IRESite
resource (http://iresite.org/) allow us to
identify IRES elements in the 5′UTR region of the RHBDD2-2
variant. These regulatory sequences could be modulating the
translational process of the RHBDD2 isoform B under different
cellular stress conditions.
Using confocal microscopy, we were able to
corroborate the localization of the RHBDD2 proteins at the Golgi
apparatus of human breast cancer cells, as was previously
determined in mouse-derived cell lines (31,32).
We also identified the RHBDD2 proteins associated with V-SNARE
transport vesicles involved in different Golgi trafficking
processes. Overall, we proposed that a switch in RHBDD2
splicing variants and its protein products in the Golgi and
associated transport vesicles may be related to a pro-survival
signaling pathway initiated under the stressful tumor
microenvironment conditions. Nevertheless, further studies should
be conducted in other cancer cell lines and in vivo models
in order to corroborate our findings and also to elucidate the role
of the RHBDD2-2 variant in breast cancer progression.
Acknowledgements
The authors thank to Professor Ulla Mandel for
antibody donation.
Funding
The present study was supported by the National
Agency of Scientific and Technological Promotion (PICT-2015-0149)
and The National Cancer Institute of Argentina (INC-MSAL).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
MCA and RC conceived and designed the study. RC,
MER, AG, VF and SP performed the experiments. RC, MCA and EL
performed the statistical and bioinformatic analysis. MER, MIL, MVC
collect the samples. RC and MCA wrote the paper. RC, MCA, MER and
MVC reviewed and edited the manuscript. All authors read and
approved the manuscript and agree to be accountable for all aspects
of the research in ensuring that the accuracy or integrity of any
part of the work are appropriately investigated and resolved.
Ethics approval and consent to
participate
All procedures performed in studies involving human
participants were in accordance with the ethical standards of the
Bioethics Committee of the School of Medical Sciences (COBIMED)
from the National University of La Plata, Protocol N°
0800–017399/13–000 and with the 1964 Helsinki Declaration and its
later amendments or comparable ethical standards. Informed consent
was obtained from all individual participants included in the
study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Freeman M: The rhomboid-like superfamily:
Molecular mechanisms and biological roles. Annu Rev Cell Dev Biol.
30:235–254. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bergbold N and Lemberg MK: Emerging role
of rhomboid family proteins in mammalian biology and disease.
Biochim Biophys Acta. 1828:2840–2848. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Abba MC, Sun H, Hawkins KA, Drake JA, Hu
Y, Nunez MI, Gaddis S, Shi T, Horvath S, Sahin A and Aldaz CM:
Breast cancer molecular signatures as determined by SAGE:
Correlation with lymph node status. Mol Cancer Res. 5:881–890.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Abba MC, Lacunza E, Nunez MI, Colussi A,
Isla-Larrain M, Segal-Eiras A, Croce MV and Aldaz CM: Rhomboid
domain containing 2 (RHBDD2): A novel cancer-related gene
over-expressed in breast cancer. Biochim Biophys Acta.
1792:988–997. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lacunza E, Canzoneri R, Rabassa ME,
Zwenger A, Segal-Eiras A, Croce MV and Abba MC: RHBDD2: A
5-fluorouracil responsive gene overexpressed in the advanced stages
of colorectal cancer. Tumour Biol. 33:2393–2399. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Canzoneri R, Lacunza E, Isla Larrain M,
Croce MV and Abba MC: Rhomboid family gene expression profiling in
breast normal tissue and tumor samples. Tumour Biol. 35:1451–1458.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lacunza E, Rabassa ME, Canzoneri R,
Pellon-Maison M, Croce MV, Aldaz CM and Abba MC: Identification of
signaling pathways modulated by RHBDD2 in breast cancer cells: A
link to the unfolded protein response. Cell Stress Chaperones.
19:379–388. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Biamonti G, Catillo M, Pignataro D,
Montecucco A and Ghigna C: The alternative splicing side of cancer.
Semin Cell Dev Biol. 32:30–36. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Venables JP: Aberrant and alternative
splicing in cancer. Cancer Res. 64:7647–7654. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ghigna C, Valacca C and Biamonti G:
Alternative splicing and tumor progression. Curr Genomics.
9:556–570. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
David CJ and Manley JL: Alternative
pre-mRNA splicing regulation in cancer: Pathways and programs
unhinged. Genes Dev. 24:2343–2364. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Oltean S and Bates DO: Hallmarks of
alternative splicing in cancer. Oncogene. 33:5311–5318. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Carlson SG, Fawcett TW, Bartlett JD,
Bernier M and Holbrook NJ: Regulation of the C/EPB-related gene
gadd153 by glucose deprivation. Mol Cell Biol. 13:4736–4744. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lee AS: The ER chaperone and signalling
regulator GRP78/BiP as a monitor of endoplasmic reticulum stress.
Methods. 35:373–381. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Michalak M, Groenendyk J, Szabo E, Gold LI
and Opas M: Calreticulin, a multi-process calcium-buffering
chaperone of the endoplasmic reticulum. Biochem J. 417:651–666.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2−ΔΔCT method. Methods.
25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bergeron JJ, Brenner MB, Thomas DY and
Williams DB: Calnexin: A membrane-bound chaperone of the
endoplasmic reticulum. Trends Biochem Sci. 19:124–128. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Röttger S, White J, Wandall HH, Olivo JC,
Stark A, Bennett EP, Whitehouse C, Berger EG, Clausen H and Nilsson
T: Localization of three human polypeptide GalNAc-transferases in
HeLa cells suggests initiation of O-linked glycosylation throughout
the Golgi apparatus. J Cell Sci. 111:45–60. 1998.PubMed/NCBI
|
|
19
|
Zhang T and Hong W: Ykt6 forms a SNARE
complex with syntaxin 5, GS28, and Bet1 and participates in a late
stage in endoplasmic reticulum-Golgi transport. J Biol Chem.
276:27480–27487. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bolte S and Cordelières FP: A guided tour
into subcellular colocalization analysis in light microscopy. J
Microsc. 224:213–232. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Parker JS, Mullins M, Cheang MC, Leung S,
Voduc D, Vickery T, Davies S, Fauron C, He X, Hu Z, et al:
Supervised risk predictor of breast cancer based on intrinsic
subtypes. J Clin Oncol. 27:1160–1167. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Dai X, Li T, Bai Z, Yang Y, Liu X, Zhan J
and Shi B: Breast cancer intrinsic subtype classification, clinical
use and future trends. Am J Cancer Res. 5:2929–2943.
2015.PubMed/NCBI
|
|
23
|
He N, Kim N, Jeong E, Lu Y, Mills GB and
Yoon S: Glucose starvation induces mutation and lineage-dependent
adaptive responses in a large collection of cancer cell lines. Int
J Oncol. 48:67–72. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Amir-Ahmady B and Salati LM: Regulation of
the processing of glucose-6-phosphate dehydrogenase mRNA by
nutritional status. J Biol Chem. 276:10514–10523. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hicks SW and Machamer CE: Golgi structure
in stress sensing and apoptosis. Biochim Biophys Acta.
1744:406–414. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Cyphert TJ, Suchanek AL, Griffith BN and
Salati LM: Starvation actively inhibits splicing of
glucose-6-phosphate dehydrogenase mRNA via a bifunctional ESE/ESS
element bound by hnRNP K. Biochim Biophys Acta. 1829:905–915. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang L, Conejo-Garcia JR, Yang N, Huang
W, Mohamed-Hadley A, Yao W, Benencia F and Coukos G: Different
effects of glucose starvation on expression and stability of VEGF
mRNA isoforms in murine ovarian cancer cells. Biochem Biophys Res
Commun. 292:860–868. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Spriggs KA, Bushell M and Willis AE:
Translational regulation of gene expression during conditions of
cell stress. Mol Cell. 40:228–237. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Martínez-Salas E, Piñeiro D and Fernández
N: Alternative mechanisms to initiate translation in eukaryotic
mRNAs. Comp Funct Genomics. 2012:3915462012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Barbosa C, Peixeiro I and Romão L: Gene
expression regulation by upstream open reading frames and human
disease. PLoS Genet. 9:e10035292013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ahmedli NB, Gribanova Y, Njoku CC, Naidu
A, Young A, Mendoza E, Yamashita CK, Ozgül RK, Johnson JE, Fox DA
and Farber DB: Dynamics of the rhomboid-like protein RHBDD2
expression in mouse retina and involvement of its human ortholog in
retinitis pigmentosa. J Biol Chem. 288:9742–9754. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ferretti VA, Canzoneri R, Barbeito CG,
Croce MV, Abba MC and Lacunza E: Spatiotemporal expression of
Rhomboid domain containing 2 (Rhbdd2) during rat development. Acta
Histochem. 117:635–641. 2015. View Article : Google Scholar : PubMed/NCBI
|