Introduction
Hepatocellular carcinoma (HCC) is the most common
form of primary liver cancer in adults and the third leading cause
of cancer-related mortality worldwide (1). HCC occurs most frequently in patients
with chronic liver disease and cirrhosis (2). The common risk factors of HCC include
hepatitis B and C virus infection, heavy consumption of alcohol,
exposure to aflatoxin B1 and non-alcoholic steatohepatitis
(3). Despite significant advances
in diagnosis and treatment, liver resection and transplantation are
considered the only curative options at present (3). To date, no effective treatment
strategies have been developed for patients with advanced HCC,
including the approved therapeutic agent, sorafenib (4), owing to chemoresistance and excessive
cytotoxicity resulting in dismal prognosis (5). Therefore, identification of early and
effective diagnostic markers and therapeutic targets for this
disease remains an urgent unmet medical need.
The discovery of genes involved in tumor progression
has prompted research into various methods of therapeutic
intervention. Based on advances in genome-wide technology in cancer
research, we established a microarray dataset to identify the genes
responsible for dismal prognosis in earlier studies (6–8). In
the present study, transmembrane protein 165 (TMEM165), a Golgi
protein (9), was shown to be
associated with aggressive characteristics of HCC. TMEM165 is a
highly conserved hydrophobic protein of 324 amino acids that
contains 7 transmembrane-spanning domains according to topology
predictions (10). Recent research
has revealed that mutations in TMEM165 were associated with
a rare autosomal recessive disease designated ‘congenital disorders
of glycosylation’ leading to defects in metabolic processes through
impairment of galactosylation and sialylation of N-glycoproteins
(10–14). The majority of congenital disorders
of glycosylation are caused by defects in glycosylation machinery
components, such as SLC35A1, B4GALT1 and MGAT2 (15–17).
In contrast to the glycosylation-governing proteins, TMEM165
maintains Golgi ion homeostasis and vesicular Golgi trafficking
(18). While TMEM165 has been
extensively characterized in association with congenital disorders
of glycosylation, its relationship with cancer remains to be
determined. HCC-related proteins have been identified in multiple
cellular compartments, including the nucleus, cytoplasm,
mitochondria and plasma membrane. However, association of Golgi
proteins with HCC has rarely been documented in the literature to
date. To the best of our knowledge, GP73 (a 73-kDa Golgi
transmembrane protein) is the protein that has been relatively well
documented as a useful serum biomarker for HCC and shown to be
involved in the progression of benign liver disease (17,19,20).
In the present study, we identified a Golgi transmembrane protein,
TMEM165, that is overexpressed in HCC patient tissues.
TMEM165 expression was significantly associated with high
levels of α-fetoprotein as well as presence of macroscopic vascular
and serosal invasion. Consistently, TMEM165 depletion
attenuated invasion activities of HCC cells via decrease in MMP-2
expression. Our findings revealed that TMEM165 could serve
as a novel HCC marker associated with cancer aggressiveness and
presents a potential therapeutic target.
Materials and methods
Patients and tissue samples
HCC tissues were acquired from patients with HCC
subjected to hepatic surgery between September 1992 and December
2004 at Korea Cancer Center Hospital. A total of 88 HCCs used in
the present study included 33 pair-matched HCC and adjacent liver
tissues, and 55 HCC tissues. Normal liver tissues were obtained
from patients with colon or rectal cancer, metastasized into liver,
who underwent surgical resection at Seoul National University
Hospital and Korea Cancer Center Hospital. The adjacent liver
tissues around these metastatic cancers were regarded and used as a
normal liver (n=15) when liver fibrosis as well as HCC were not
observed in the patients. Our study was approved by the
Institutional Review Boards of Korea Cancer Center Hospital and
Seoul National University Hospital. Written informed consent was
either waived by the Institutional Review Board of Korea Cancer
Center Hospital or received from patients at Seoul National
University Hospital, respectively. Clinicopathological information
collected retrospectively was utilized to examine associations
between TMEM165 expression patterns and HCC characteristics.
Fibrosis was graded on a scale of 0–4 (stage 0, not present; stage
1, portal; stage 2, periportal; stage 3, septal; and stage 4,
cirrhosis).
Cell culture and siRNA
transfection
Cell lines were obtained from two institutions. The
cell lines, SNU354, SNU398, SNU449, SNU475, SNU739 and Huh7 were
purchased from the Korean Cell Line Bank (Seoul, South Korea) and
the cell lines, Hep3B, HepG2, BJ, IMR90, WI38, normal Small Airway
(PCS-301-010) and Prostate (PCS-440-010) epithelial cells from the
American Type Culture Collection (ATCC; Manassas, VA, USA). Huh7,
SNU354, SNU398, SNU449, SNU475 and SNU739 cells were cultured in
RPMI-1640 medium (LM 011–01; Welgene, Daegu, Korea) and BJ, IMR90,
WI38, HepG2 and Hep3B were cultured in MEM (LM 007–07; Welgene)
supplemented with 10% (w/v) fetal bovine serum (FBS; JR Scientific,
Inc., Woodland, CA, USA) and 1% (w/v) penicillin/streptomycin
(Welgene) at 37°C. Normal small Airway and Prostate epithelial
cells were cultured in Airway Epithelial Basal media (ATCC PCS
300–030) supplemented with Bronchial Epithelial Cell Growth kit
(ATCC PCS-300-040) and Prostate Epithelial Cell Basal Medium (ATCC
PCS-440-030) supplemented with Prostate Epithelial Cell Growth kit
(ATCC PCS-440-040), respectively. For knockdown of TMEM165,
siRNA transfection was performed using Lipofectamine RNA iMAX
reagent (cat. no. 13778030; Invitrogen; Thermo Fisher Scientific,
Inc., Waltham, MA, USA) according to the manufacturer's
instructions. The following siRNA sequences were used:
TMEM165 siRNA#1, 5′-AGCCAUCAUGGCAAUGCGCUAUU-3′ (sense) and
5′-UAGCGCAUUGCCAUGAUGGCUUU-3′ (antisense); and TMEM165
siRNA#2, 5′-UUGGGUAGGACACCCAAUAUAUU-3′ (sense) and
5′-UAUAUUGGGUGUCCUACCCAAUU-3′ (antisense).
RNA extraction and cDNA synthesis
Total RNA was extracted from cultured cells and
fresh tissues using the RNeasy Mini Kit (cat. no. 74106; Qiagen,
Inc., Valencia, CA, USA) according to the manufacturer's protocol.
The quality and concentration of isolated total RNA were determined
using a NanoDrop ND-1000 spectrophotometer (NanoDrop; Thermo Fisher
Scientific, Inc., Wilmington, DE, USA). RNA samples were stored at
−80°C until gene expression analysis. cDNA was synthesized using
the iScript reverse transcriptase kit (cat. no. 170890; Bio-Rad
Laboratories, Inc., Hercules, CA, USA) in keeping with the
manufacturer's protocol.
Quantitative and semi-quantitative
RT-PCR
Quantitative RT-PCR was performed with the KAPA SYBR
FAST Universal qPCR kit (kit code: KK4301; Kapa Biosystems, Inc.,
Boston, MA, USA) on the CFX96 Real-Time RT-PCR Detection System
(Bio-Rad Laboratories, Inc.). The primer sequences used for
real-time RT-PCR were: TMEM165 forward,
5′-GGCAGTAATTGGAGGAAGAATGATAGC-3′ and reverse,
5′-ACCAGAATCAGGGCTTATAAATAGTGC-3′; and β2-microglobulin forward,
5′-AAGGACTGGTCTTTCTATCTCTTGTA-3′ and reverse,
5′-ACTATCTTGGGGTGTGACAAAGTC-3′. Median Ct values from triplicate
experiments were used for statistical analyses and TMEM165
levels were normalized to median β2-microglobulin expression.
Relative TMEM165 expression in HCC and adjacent liver
tissues, compared to that in normal liver tissues, was analyzed
using the comparative threshold cycle 2−ΔΔCt method.
Semi-quantitative RT-PCR was performed using the Maxime PCR PreMix
kit (cat. no. 25185; iNtRON Biotechnology, Kyungi-do, Korea) and
the following primers: TMEM165 forward,
5′-GGCAGTAATTGGAGGAAGAATGATAGC-3′ and reverse,
5′-ACCAGAATCAGGGCTTATAAATAGTGC-3′; β2-microglobulin forward,
5′-GTGCTCGCGCTACTCTCTCT-3′ and reverse, 5′-CGGCAGGCATACTCATCTTT-3′;
MMP-1 forward, 5′-ACAAAGTTGATGCAGTTTTCA-3′ and reverse,
5′-GAATCCATAAGCCACAAACTT-3′; MMP-2 forward,
5′-ATCTTTGCTGGAGACAAATTC-3′ and reverse,
5′-AACTTCACGCTCTTCAGACTT-3′; MMP-9 forward,
5′-AACTTTGACAGCGACAAGAA-3′ and reverse,
5′-TTGAACAAATACAGCTGGTTC-3′; MMP-13 forward,
5′-CCCAAAATTTTCTACCTCTGA-3′ and reverse,
5′-TTTTGATGATGATGAAACCTG-3′; β-actin forward,
5′-GGACTTCGAGCAAGAGATGG-3′ and reverse, 5′-AGCACTGTGTTGGCGTACAG-3′;
RPS26 forward, 5′-CCAAGGACAAGGCCATTAAG-3′ and reverse,
5′-AGCACCCGCAGGTCTAAATC-3′; and GAPDH forward,
5′-GTCAGTGGTGGACCTGACCT-3′ and reverse, 5′-TGCTGTAGCCAAATTCGTTG-3′.
The optimal PCR cycle for each primer set was determined by
monitoring PCR products harvested at 2-cycle intervals with at
least 3 different cycles. PCR amplification was performed with an
initial 2 min of denaturation at 94°C, followed by cycling a 20 sec
denaturation at 94°C, 10 sec annealing at 55°C and 30 sec extension
at 72°C with a final 5 min extension at 72°C. The final PCR
products were loaded on agarose gels and mRNA levels quantitated
using NIH ImageJ software (http://rsb.info.nih.gov/ij).
Western blot analysis
Total tissue samples were homogenized in lysis
buffer [50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 5 mM EDTA, 1% (v/v)
Nonidet P-40, 0.1% (w/v) sodium dodecyl sulfate, 0.5% (w/v) sodium
deoxycholate] including protease inhibitor cocktail (cat. no.
P3100-010; GenDEPOT, Inc., Barker, TX, USA) and placed on ice for
20 min. After incubation, tissues were centrifuged for 20 min at
13,000 × g at 4°C and the supernatant fractions were collected.
Quantified and sampled proteins were separated on a 12.0% (w/v) SDS
gel and transferred to nitrocellulose membranes (cat. no. 10401396;
Whatman, Maidstone, UK). Next, the membranes were blocked with 5%
(w/v) skim milk in TBS with Tween-20 (TBS-T) buffer [25 mM
Tris-HCl, pH 7.4, 140 mM NaCl, 2.7 mM KCl, 0.05% (w/v) Tween-20]
for 1 h at room temperature, followed by incubation with primary
antibodies against TMEM165 (diluted to 1:1,000; cat. no.
20485–1-AP; ProteinTech Group, Rosemont, IL, USA) and GAPDH
(diluted to 1:2,000; cat. no. sc-25778; Santa Cruz Biotechnology,
Inc., Santa Cruz, CA, USA) for 1 h at room temperature. After
washing 3 times with TBS-T, the membranes were treated with
horseradish peroxidase-conjugated secondary antibody (diluted to
1:3,000; cat. nos. A120-101P and A90-116P; Bethyl Laboratories,
Inc., Montgomery, TX, USA) at room temperature for 1 h. Following 3
further washes with TBS-T, the membranes were reacted with Luminol
chemiluminescent reagent (cat. no. sc-2048; Santa Cruz
Biotechnology, Inc.) according to the manufacturer's protocol. The
intensities of the band were analyzed using the NIH ImageJ software
(National Institutes of Health, Bethesda, MD, USA).
Clonal survival analysis
Huh7 and SNU475 cells were transiently transfected
with control and TMEM165 siRNAs. At 24 h after transfection,
1,000 cells were seeded on a 6-well plate. Cells were fixed with
10% (w/v) formaldehyde and stained with 0.1% (w/v) crystal violet
10 days after seeding. Colonies were counted using digital images
obtained with ImageJ. All experiments were performed in
triplicate.
Invasion analysis
Polycarbonate membrane Transwell inserts (cat. no.
354480; Corning Inc., Corning, NY, USA) were coated with 20 µg/ml
Matrigel. Following transient transfection, Huh7 and SNU475 cells
(2×104) were seeded on the upper compartment of the
chamber in serum-free medium. As a chemoattractant, complete medium
containing 10% (v/v) FBS was placed in the bottom compartment.
After 24 and 48 h of incubation, the cells were fixed, stained with
Hemacolor® solution (Merck KGaA, Darmstadt, Germany) and
counted under a light microscope in 4 different fields. All
experiments were performed in triplicate.
Statistical analysis
Statistical analysis was performed using SPSS v.23.0
software (IBM Corp., Armonk, NY, USA). Receiver operating
characteristic (ROC) curves were generated to assess the optimal
cut-off point for TMEM165 expression. A one-way analysis of
variance (ANOVA) with post hoc Tukey's HSD test was conducted to
compare the expression levels of TMEM165 in normal,
non-tumor and tumor samples. Correlations between TMEM165
expression and clinicopathological parameters were determined using
a 2-sample t-test, Pearson's correlation test, Chi-squared
(χ2) and Fisher's exact tests. For the comparison of
invasion and clonal survival, ANOVA with post hoc Dunnett's test
was performed. The data were considered significant at
P<0.05.
Results
TMEM165 is overexpressed in HCC
To establish whether TMEM165 is involved in the
pathogenesis of HCC, we examined its expression patterns in tumor
and adjacent liver tissue specimens obtained from HCC patients
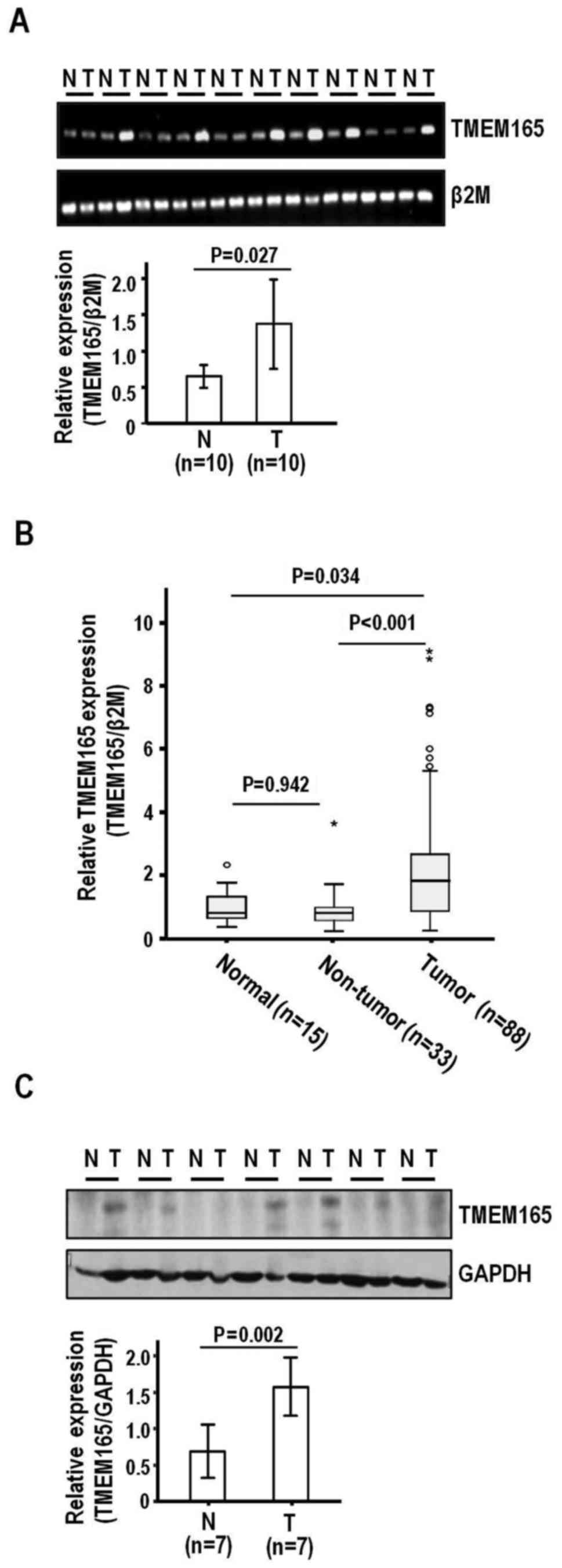
subjected to surgical resection. Semi-quantitative RT-PCR disclosed
higher TMEM165 expression in HCC than adjacent liver tissues
(P=0.027) (Fig. 1A). To confirm
TMEM165 expression, we performed quantitative real-time
RT-PCR on an extended HCC sample set (n=88) (Table I) comprised of background liver
cirrhosis (n=29) and fibrosis (n=3:7:16:24:29 for grades 0:1:2:3:4,
respectively). Similar to semi-quantitative RT-PCR findings,
real-time RT-PCR revealed significantly elevated TMEM165
expression in HCC, compared to adjacent non-tumor liver
(P<0.001) and normal liver tissues (P=0.034) (Fig. 1B). In contrast to tumor tissues,
adjacent liver tissues did not exhibit a significant difference in
TMEM165 expression, compared to normal liver tissues
(P=0.942). When expression levels were adjusted and compared with
those of normal liver tissues, the mean increases in TMEM165
expression in HCC and adjacent liver tissues were 2.40- and
1.02-fold (median, 1.89- and 0.99-fold, respectively). Consistent
with the mRNA expression data, the protein levels were higher in
HCC than adjacent liver tissues (P=0.002) (Fig. 1C). The collective findings clearly
demonstrated TMEM165 overexpression in HCC.
 | Table I.Patient demographics and pathological
data (n=88). |
Table I.
Patient demographics and pathological
data (n=88).
| Variables | Classification | Distribution |
|---|
| Sex | Male : Female | 77 : 11 |
| Age (years) | Year, mean ± SD
(range) | 52.05±10.79
(25–74) |
| Etiology | Hepatitis B :
Hepatitis C | 85 : 03 |
| Total
bilirubin | <1 mg/dl : ≥1
mg/dl | 59 : 29 |
| AFP | <20 ng/dl : ≥20
ng/dl | 38 : 49 |
|
| <50 ng/dl : ≥50
ng/dl | 47 : 40 |
|
| <200 ng/dl :
≥200 ng/dl | 55 : 32 |
| AST | IU/l, mean ± SD
(range) | 51.92±31.07
(15–182) |
| ALT | IU/l, mean ± SD
(range) | 46.25±25.21
(10–130) |
| Child-Pugh
classification | A : B and C | 70 : 07 |
| Tumor size | cm, mean ± SD
(range) | 5.8±3.25
(1.0–24.0) |
| Tumor number | Single :
Multiple | 59 : 17 |
| Macroscopic
vascular invasion | No : Yes | 78 : 5 |
| Microscopic
vascular invasion | No : Yes | 34 : 39 |
| Serosal
invasion | No : Yes | 68 : 12 |
| Edmondson
gradea | І : II : III :
VI | 11 : 48 : 27 :
1 |
| Fibrosis | 0 : 1 : 2 : 3 :
4 | 3 : 7 : 16 : 24 :
29 |
TMEM165 overexpression is clinically
associated with macroscopic vascular invasion, microscopic serosal
invasion, and high α-fetoprotein levels of HCC
Next, we examined the potential association of
TMEM165 mRNA levels with clinicopathological parameters to
ascertain whether overexpression has a clinical impact on HCC.
Variations in TMEM165 expression determined based on
real-time analysis were mostly <2.0-fold between normal and
adjacent liver tissues. To further verify an optimal cut-off point
for TMEM165 expression, we performed ROC analysis using SPSS
software. In the analysis, the clinical parameters, AFP level
(<50 vs. ≥50 ng/ml), macroscopic invasion (no and yes) and
serosal invasion (no and yes) were associated with TMEM165
(P<0.05). The optimal cut-off points of TMEM165
expression for these parameters were 1.98- (P=0.009), 3.03-
(P=0.032), and 2.32-fold (P=0.018), respectively, indicating a
2.0-fold is suitable for comparison with clinicopathological
parameters. Accordingly, we subdivided HCCs into two groups based
on TMEM165 expression employing 2.0-fold as a cut-off point.
Among the 88 tumor tissues examined, 41 (46.6%) exhibited ≥2.0-fold
increase in TMEM165 transcript expression. Consistent with
the ROC analysis, higher α-fetoprotein levels (≥50 ng/ml, P=0.022;
≥200 ng/ml, P=0.024) and presence of macroscopic vascular invasion
(P=0.023) and serosal invasion (P=0.006) were significantly
associated with TMEM165 overexpression (≥2.0-fold) (Table II). In contrast, HCC-based
parameters such as sex (P=0.187), age (P=0.479), aspatate
transaminase (P=0.204), alanine transaminase (P=0.386), Child-Pugh
classification (P=0.273), tumor size (P=0.113), tumor number
(P=0.212), microscopic vascular invasion (P=0.377), Edmondson grade
(P=0.274) and cirrhosis (P=0.334), were not associated with
TMEM165 overexpression. Furthermore, HCCs classified based
on significant parameters (Table
II) exhibited marked differences in TMEM165 expression
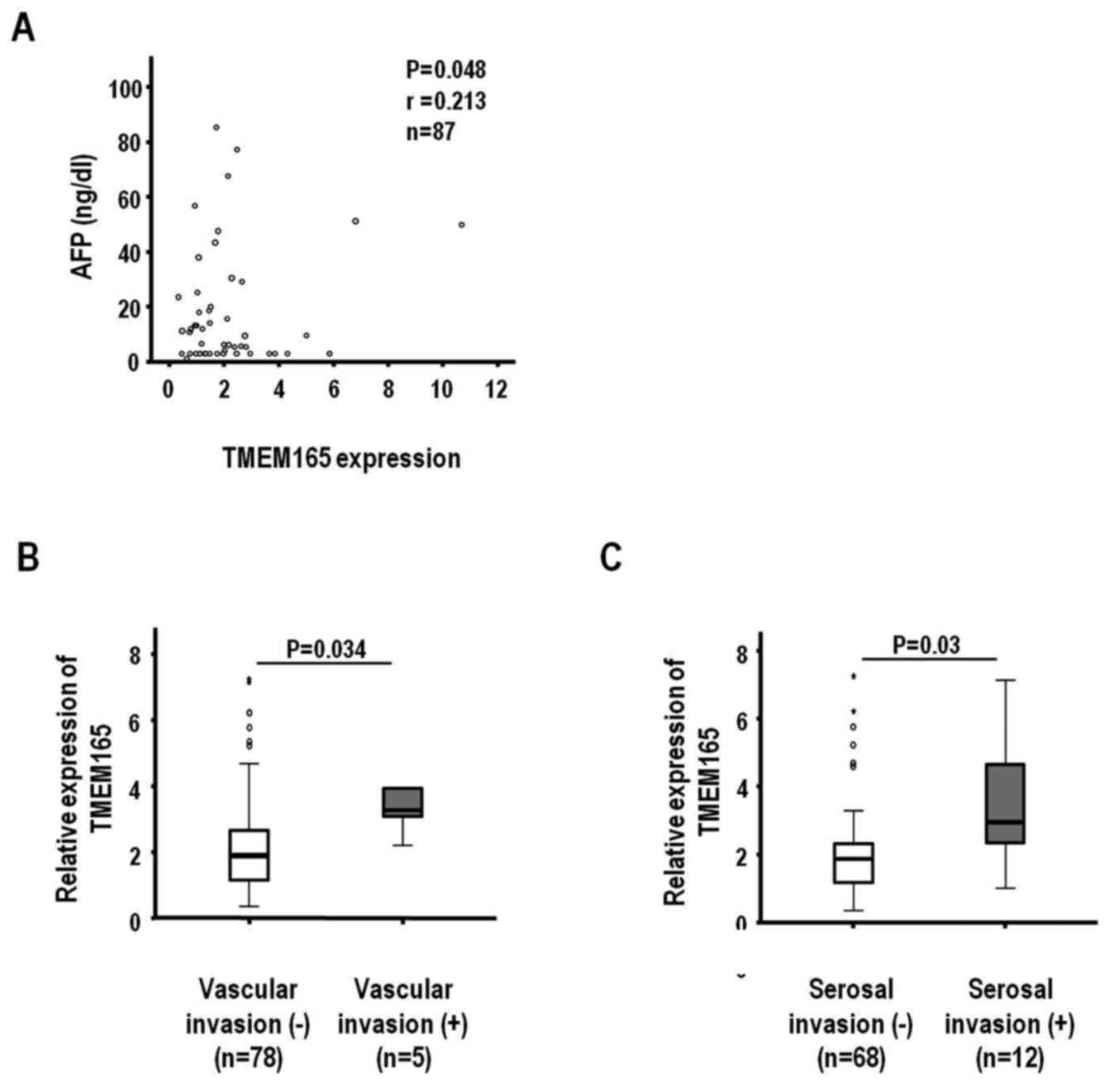
(Fig. 2). Specifically, the
TMEM165 expression level was positively correlated with the
AFP level (Fig. 2A) (n=87, r=0.213
and P=0.048). Additionally, median increases in TMEM165
expression were 1.90- and 3.26-fold according to macroscopic
vascular invasion (no and yes, P=0.034) (Fig. 2B) and 1.87- and 2.95-fold according
to serosal invasion (no and yes, P=0.03), respectively (Fig. 2C). The clinicopathological
association data indicated that TMEM165-associated
parameters influence aggressiveness of HCCs, based on the finding
that overexpression of this protein is clinically associated with
the invasive characteristics of this tumor type.
 | Table II.Correlation between TMEM165
expression and clinicopathological parameters (n=88). |
Table II.
Correlation between TMEM165
expression and clinicopathological parameters (n=88).
|
| TMEM165
expression |
|
|---|
|
|
|
|
|---|
| Variables | <2.0 fold | ≥2.0 fold |
P-valuea |
|---|
| Sex |
|
Male | 43 | 34 | 0.187 |
|
Female | 4 | 7 |
|
| Age (years) |
|
<52 | 20 | 19 | 0.479 |
|
≥52 | 26 | 22 |
|
| AFP (ng/ml) |
|
| 0.149 |
|
<20 | 23 | 15 |
|
|
≥20 | 23 | 26 |
|
| AFP (ng/ml) |
|
<50 | 30 | 17 | 0.022 |
|
≥50 | 16 | 24 |
|
| AFP (ng/ml) |
|
| 0.024 |
|
<200 | 34 | 21 |
|
|
≥200 | 12 | 20 |
|
| AST (U/l) |
|
<40 | 20 | 22 | 0.204 |
|
≥40 | 27 | 19 |
|
| ALT (U/l) |
|
<35 | 17 | 17 | 0.386 |
|
≥35 | 30 | 24 |
|
| Child-Pugh
classification |
| A | 36 | 34 | 0.273 |
|
B,C | 5 | 2 |
|
| Tumor size
(cm) |
|
<5 | 26 | 16 | 0.113 |
| ≥5 | 21 | 24 |
|
| Tumor number |
|
Single | 33 | 26 | 0.212 |
|
Multiple | 7 | 10 |
|
| Macroscopic
vascular invasion |
| No | 43 | 35 | 0.023 |
|
Yes | 0 | 5 |
|
| Microscopic
vascular invasion |
| No | 17 | 17 | 0.377 |
|
Yes | 22 | 17 |
|
| Serosal
invasion |
| No | 41 | 27 | 0.006 |
|
Yes | 2 | 10 |
|
| Edmondson
grade |
|
I,II | 33 | 26 | 0.274 |
| III,
IV | 13 | 15 |
|
| Cirrhosis |
| No | 28 | 22 | 0.334 |
|
Yes | 14 | 15 |
|
TMEM165 depletion decreases invasion
of HCC cells without affecting clonal survival
In view of the clinical association of
TMEM165 overexpression with macroscopic vascular and serosal
invasion, we further investigated the possibility of regulatory
effects on the invasion of HCC cells. Prior to evaluation, we
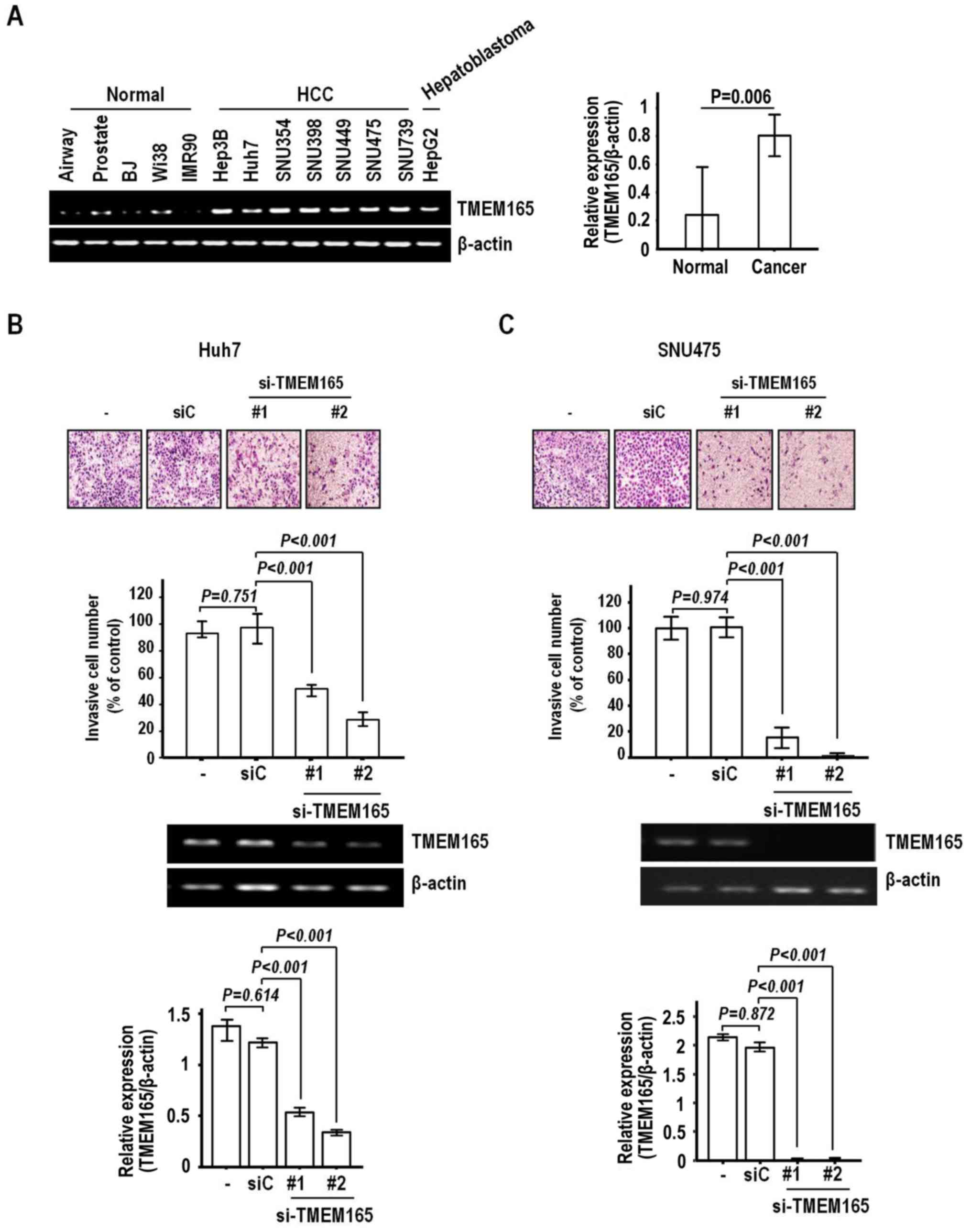
determined the expression patterns of TMEM165 in normal and
cancer cell lines to select suitable cells. For this experiment, we
employed the established HCC cell lines Hep3B, Huh7, SNU354,
SNU398, SNU449, SNU475 and SNU739 and hepatoblastoma cell line
HepG2. Consistent with data from HCC specimens, most cancer cell
lines exhibited greater expression than normal human fibroblast
(BJ, Wi38, IMR90) and epithelial cells, including those of the
Small Airway and Prostate (Fig.
3A).
Next, we examined the effect of TMEM165
depletion on invasion of Huh7 cells that express high levels of
TMEM165. Notably, TMEM165 depletion achieved via
transfection of siRNA induced a significant decrease in the
invasive activity of Huh7 cells (Fig.
3B). Two siRNAs (#1 and #2) recognizing different regions on
the coding sequence induced a noticeable level of suppression of
invasive activity (51.3 and 72.1%, respectively, compared to the
control siRNA). Significantly reduced invasion was additionally
observed in the SNU475 cell line (Fig.
3C). The rates of inhibition of invasion by TMEM165
siRNA #1 and #2 were 88.2 and 95.7%, respectively, clearly
indicating that knockdown of TMEM165 in HCC suppresses
invasive activity to a significant extent. To further confirm this
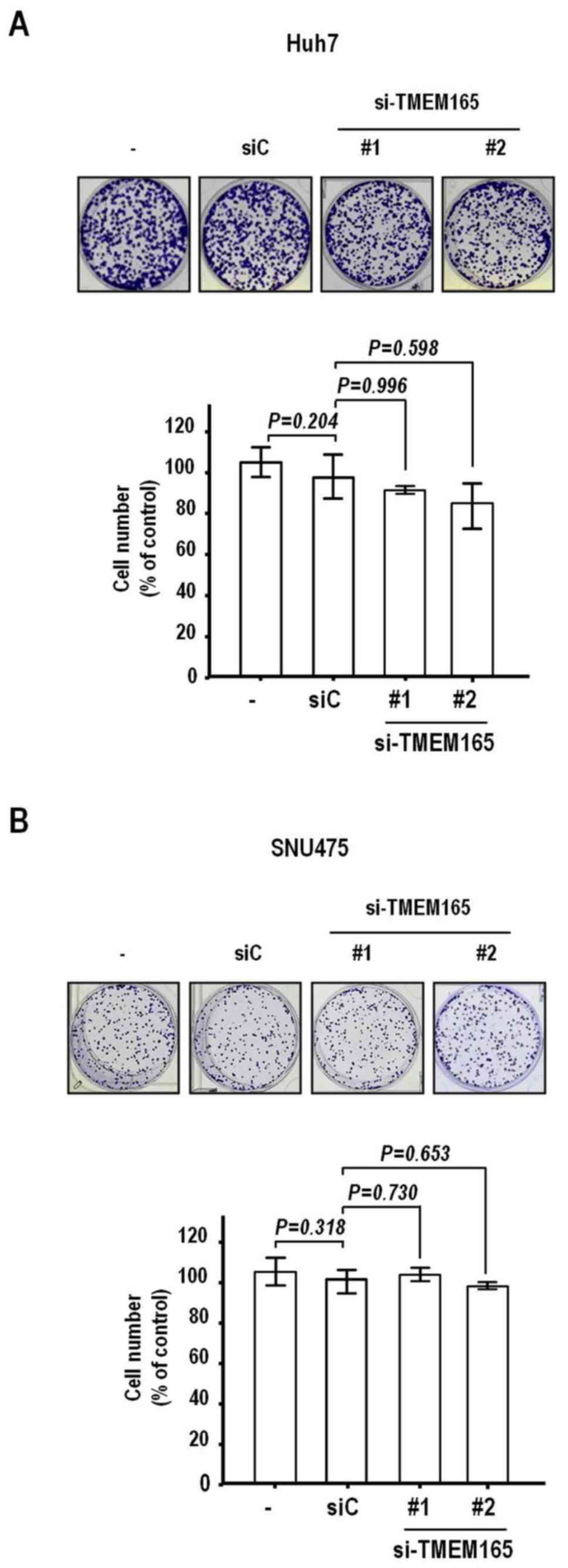
finding, we assessed the effects of TMEM165 depletion on
clonal growth that can detect cell proliferation and death.
Notably, clonal growth was hardly inhibited in both cell lines
after transfection of TMEM165 siRNAs (P>0.1, Fig. 4A and B). Thus, the TMEM165
depletion-mediated suppression of HCC cell invasion activity was
not attributable to decreased clonal survival.
TMEM165 affects matrix
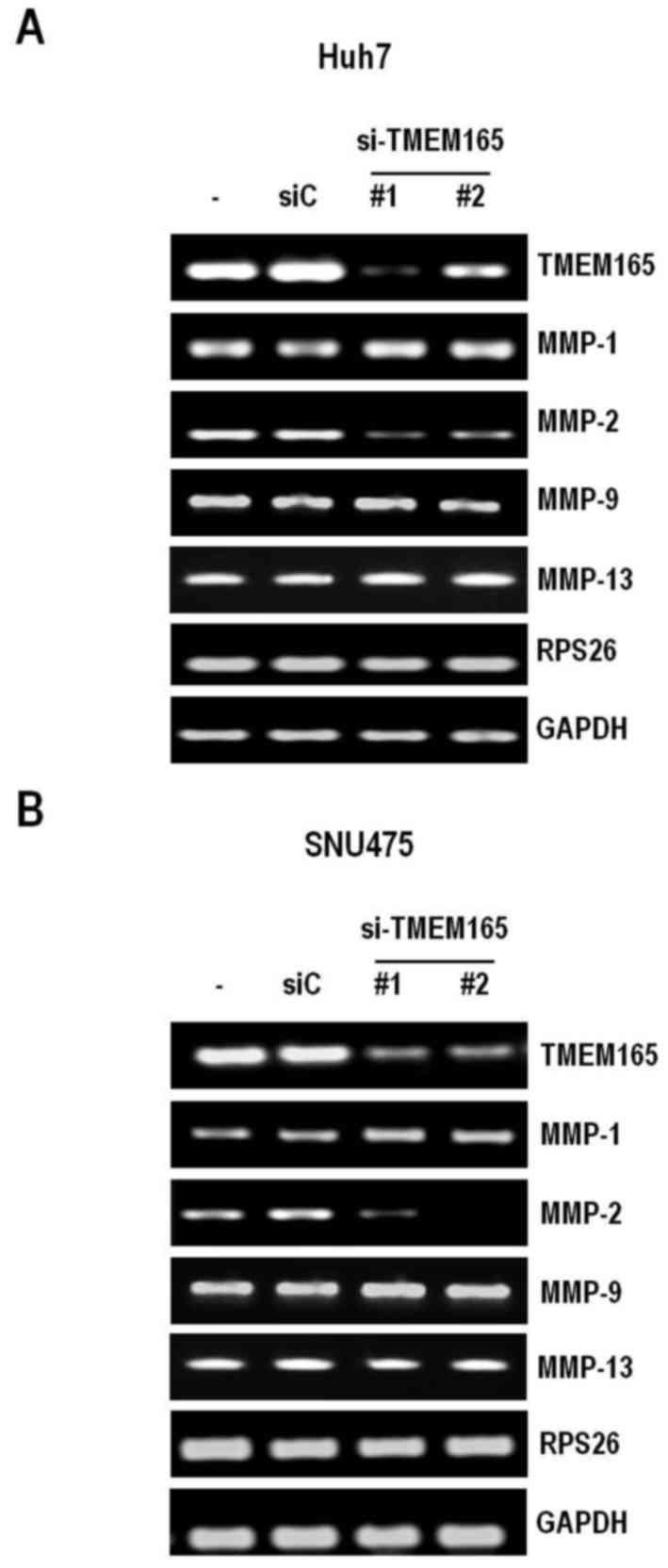
metalloproteinase-2 (MMP-2) expression
To further determine the mechanisms underlying
TMEM165-mediated regulation of invasion, we examined the
potential involvement of MMPs, well-known invasion activators.
Specifically, we assessed the expression levels of MMP-1, −2
and −9 in HCC cells depleted of TMEM165. As expected
from the cell invasion data, MMP-2 levels were severely
decreased under conditions of TMEM165 depletion in both Huh7
and SNU475 cell lines (Fig. 5A and
B). In contrast, MMP-1 and −9 levels were not
affected. To determine whether TMEM165 controls cancer cell
invasion in a similar manner to Golgi phosphoprotein 2, we examined
the expression of MMP-13 that are regulated by Golgi
phosphoprotein 2 (21). Notably,
TMEM165 depletion had no effect on the expression patterns
of this molecule, indicating that the mechanism by which TMEM165
promotes the invasive activity of cancer cells is distinct from
that of Golgi phosphoprotein 2.
Discussion
The TMEM165 protein, mainly characterized in
association with congenital disorders of glycosylation type 2
disease, has recently attracted significant research attention
(13,14,22,23).
Based on cohort studies, 3 different mutations in TMEM165
have been reported to date, specifically, homozygous point mutation
in the deep intronic splice region, homozygous missense mutation
and heterozygous missense mutation, all of which culminate in loss
or decrease in function of the protein (11,14).
Recent in vitro studies have disclosed that TMEM165 is
associated with Golgi homeostasis sensitive to manganese
concentrations (18), indicating
that mutation and loss of function of the protein can trigger
defects in manganese-sensitive Golgi homeostasis. The importance of
Golgi proteins in promoting cancer progression has been highlighted
in previous studies. Golgi phosphoprotein 2 (GP73) is aberrantly
expressed in HCC and knockdown of this gene inhibits cancer cell
invasiveness via altering expression of E-cadherin, N-cadherin and
MMP-13 (24,25). Moreover, Golgi phosphoprotein 3
(GOLPH3) is overexpressed in various cancer types, in turn,
increasing invasion and migration of cancer cells (26–28).
GOLPH3 interacts with phosphatidylinositol-4-phosphate and
myosin18A to facilitate Golgi to plasma membrane trafficking and
regulates the directional migration of cells (29). Despite the recent focus on Golgi
proteins, the specific function of TMEM165 in cancer remains to be
determined.
We previously established an integrative analysis
tool to determine the gene expression patterns between non-tumor
and tumor regions in correlation with clinical outcomes (6). Based on these experiments, we
demonstrated for the first time that the TMEM165 protein is
significantly upregulated in tumor compared to non-tumor tissues of
HCC patients. Notably, overexpression of TMEM165 (using the
2-fold cut-off criterion) was positively correlated with
aggressiveness of HCC characterized by high AFP levels as well as
presence of macroscopic vascular and microscopic serosal invasion.
In keeping with this finding, depletion of TMEM165
significantly reduced invasiveness of human HCC cell lines in
vitro, accompanied by a decrease in expression of MMP-2,
an endopeptidase that degrades the extracellular matrix to
stimulate cancer invasion and metastasis. However, other MMPs
(MMP-1, −9, and −13) were not affected by depletion of
TMEM165. These results support a role of TMEM165 as a key
molecule that directs cancer cell invasion, consequently promoting
aggressiveness. Clonal survival analysis can be effectively used to
detect several phenotypic changes, including proliferation, death
and senescence. TMEM165 depletion decreased cancer cell
invasion without affecting clonal survival, indicative of a
negligible effect on these phenotypes. Inhibition of cellular
proliferation, death or senescence is reported to trigger a
decrease in the invasion rate of cancer cells (30). Therefore, TMEM165 regulation of
cancer cell invasion is credible, since the possibility that
phenotypic changes, such as cellular proliferation, death, and
senescence, can affect cancer cell invasion was eliminated.
Despite the significant correlation between
TMEM165 overexpression and aggressiveness of the tumor
phenotype, TMEM165 was not associated with patient prognosis
in the current study (data not shown). This may be attributable to
the small number of samples used for our analysis. Studies on
larger cohorts are therefore required to validate the expression
patterns of TMEM165 in relation to patient prognosis.
Furthermore, while decreased invasiveness of cancer cells upon
TMEM165 knockdown was evident, the precise mechanisms
require elucidation in future studies. Considering the relationship
between TMEM165 mutation and type II congenital disorder of
glycosylation, it is plausible that its overexpression in HCC is
associated with abnormalities in galactosylation and sialylation of
N-glycoproteins observed in the disease. These protein
modifications influence the invasive activity of cancer cells
(31). Several invasion-related
proteins (CD44, integrin and E-cadherin) have been revealed to be
markedly affected by glycosylation (32–34).
In summary, TMEM165 is both transcriptionally and
translationally overexpressed in HCC and associated with invasive
activity.
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from the
National Research Foundation of Korea (nos. 2012M3A9B6055346,
2017M3A9A8033561 and 2017R1A2B4008805) and the Korea Institute of
Radiological and Medical Sciences (nos. 50531–2018 and 50542–2018)
funded by the Ministry of Science, ICT and Future Planning,
Republic of Korea.
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
KHL, SBK and JH conceived and supervised the study.
KHL, JSL and MYK wrote the manuscript. SBK and ERP reviewed and
edited manuscript. MYK, JSL, ERP, YNS and JYJ prepared material
and/or performed experiment. JSL and MYK performed statistical
analysis. EHC, SHP, CJH, DWC, JJJ, KSS and SBK generated clinical
data. All authors read and approved the manuscript and agree to be
accountable for all aspects of the research in ensuring that the
accuracy or integrity of any part of the work are appropriately
investigated and resolved.
Ethics approval and consent to
participate
The present study was approved by the Institutional
Review Boards of Korea Cancer Center Hospital and Seoul National
University Hospital. Written informed consent was either waived by
the Institutional Review Board of Korea Cancer Center Hospital or
received from patients at Seoul National University Hospital,
respectively.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
HCC
|
human hepatocellular carcinoma
|
|
AFP
|
α-fetoprotein
|
|
AST
|
aspartate aminotransferase
|
|
ALT
|
alanine transaminase
|
|
MMP
|
matrix metalloproteinase
|
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
El-Serag HB: Hepatocellular carcinoma. N
Engl J Med. 365:1118–1127. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bruix J and Sherman M: American
Association for the Study of Liver Diseases: Management of
hepatocellular carcinoma: An update. Hepatology. 53:1020–1022.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Llovet JM, Ricci S, Mazzaferro V, Hilgard
P, Gane E, Blanc JF, de Oliveira AC, Santoro A, Raoul JL, Forner A,
et al: Sorafenib in advanced hepatocellular carcinoma. N Engl J
Med. 359:378–390. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Cheng AL, Kang YK, Chen Z, Tsao CJ, Qin S,
Kim JS, Luo R, Feng J, Ye S, Yang TS, et al: Efficacy and safety of
sorafenib in patients in the Asia-Pacific region with advanced
hepatocellular carcinoma: A phase III randomised, double-blind,
placebo-controlled trial. Lancet Oncol. 10:25–34. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kim BY, Suh KS, Lee JG, Woo SR, Park IC,
Park SH, Han CJ, Kim SB, Jeong SH, Yeom YI, et al: Integrated
analysis of prognostic gene expression profiles from hepatitis B
virus-positive hepatocellular carcinoma and adjacent liver tissue.
Ann Surg Oncol. 19 Suppl 3:S328–S338. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kim BY, Choi DW, Woo SR, Park ER, Lee JG,
Kim SH, Koo I, Park SH, Han CJ, Kim SB, et al:
Recurrence-associated pathways in hepatitis B virus-positive
hepatocellular carcinoma. BMC Genomics. 16:2792015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Park ER, Kim SB, Lee JS, Kim YH, Lee DH,
Cho EH, Park SH, Han CJ, Kim BY, Choi DW, et al: The mitochondrial
hinge protein, UQCRH, is a novel prognostic factor for
hepatocellular carcinoma. Cancer Med. 6:749–760. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Potelle S, Morelle W, Dulary E, Duvet S,
Vicogne D, Spriet C, Krzewinski-Recchi MA, Morsomme P, Jaeken J,
Matthijs G, et al: Glycosylation abnormalities in Gdt1p/TMEM165
deficient cells result from a defect in Golgi manganese
homeostasis. Hum Mol Genet. 25:1489–1500. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Rosnoblet C, Legrand D, Demaegd D,
Hacine-Gherbi H, de Bettignies G, Bammens R, Borrego C, Duvet S,
Morsomme P, Matthijs G, et al: Impact of disease-causing mutations
on TMEM165 subcellular localization, a recently identified protein
involved in CDG-II. Hum Mol Genet. 22:2914–2928. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Foulquier F, Amyere M, Jaeken J, Zeevaert
R, Schollen E, Race V, Bammens R, Morelle W, Rosnoblet C, Legrand
D, et al: TMEM165 deficiency causes a congenital disorder of
glycosylation. Am J Hum Genet. 91:15–26. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bammens R, Mehta N, Race V, Foulquier F,
Jaeken J, Tiemeyer M, Steet R, Matthijs G and Flanagan-Steet H:
Abnormal cartilage development and altered N-glycosylation in
Tmem165-deficient zebrafish mirrors the phenotypes associated with
TMEM165-CDG. Glycobiology. 25:669–682. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Schulte Althoff S, Grüneberg M, Reunert J,
Park JH, Rust S, Mühlhausen C, Wada Y, Santer R and Marquardt T:
TMEM165 deficiency: Postnatal changes in Glycosylation. JIMD Rep.
26:21–29. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Dulary E, Potelle S, Legrand D and
Foulquier F: TMEM165 deficiencies in Congenital Disorders of
Glycosylation type II (CDG-II): Clues and evidences for roles of
the protein in Golgi functions and ion homeostasis. Tissue Cell.
49:150–156. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jaeken J, Schachter H, Carchon H, De Cock
P, Coddeville B and Spik G: Carbohydrate deficient glycoprotein
syndrome type II: A deficiency in Golgi localised
N-acetyl-glucosaminyltransferase II. Arch Dis Child. 71:123–127.
1994. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Peters V, Penzien JM, Reiter G, Körner C,
Hackler R, Assmann B, Fang J, Schaefer JR, Hoffmann GF and
Heidemann PH: Congenital disorder of glycosylation IId (CDG-IId) -
A new entity: Clinical presentation with Dandy-Walker malformation
and myopathy. Neuropediatrics. 33:27–32. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Martinez-Duncker I, Dupré T, Piller V,
Piller F, Candelier JJ, Trichet C, Tchernia G, Oriol R and
Mollicone R: Genetic complementation reveals a novel human
congenital disorder of glycosylation of type II, due to
inactivation of the Golgi CMP-sialic acid transporter. Blood.
105:2671–2676. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Potelle S, Dulary E, Climer L, Duvet S,
Morelle W, Vicogne D, Lebredonchel E, Houdou M, Spriet C,
Krzewinski-Recchi MA, et al: Manganese-induced turnover of TMEM165.
Biochem J. 474:1481–1493. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Marrero JA, Romano PR, Nikolaeva O, Steel
L, Mehta A, Fimmel CJ, Comunale MA, D'Amelio A, Lok AS and Block
TM: GP73, a resident Golgi glycoprotein, is a novel serum marker
for hepatocellular carcinoma. J Hepatol. 43:1007–1012. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wei H, Li B, Zhang R, Hao X, Huang Y, Qiao
Y, Hou J and Li X and Li X: Serum GP73, a marker for evaluating
progression in patients with chronic HBV infections. PLoS One.
8:e538622013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Jin D, Tao J, Li D, Wang Y, Li L, Hu Z,
Zhou Z, Chang X, Qu C and Zhang H: Golgi protein 73 activation of
MMP-13 promotes hepatocellular carcinoma cell invasion. Oncotarget.
6:33523–33533. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Krzewinski-Recchi MA, Potelle S, Mir AM,
Vicogne D, Dulary E, Duvet S, Morelle W, de Bettignies G and
Foulquier F: Evidence for splice transcript variants of TMEM165, a
gene involved in CDG. Biochim Biophys Acta. 1861:737–748. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Morelle W, Potelle S, Witters P, Wong S,
Climer L, Lupashin V, Matthijs G, Gadomski T, Jaeken J, Cassiman D,
et al: Galactose supplementation in patients with TMEM165-CDG
rescues the glycosylation defects. J Clin Endocrinol Metab.
102:1375–1386. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chen X, Wang Y, Tao J, Shi Y, Gai X, Huang
F, Ma Q, Zhou Z, Chen H, Zhang H, et al: mTORC1 up-regulates GP73
to promote proliferation and migration of hepatocellular carcinoma
cells and growth of xenograft tumors in mice. Gastroenterology.
149:741–752. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yang Y, Liu Q, Zhang H, Zhao H, Mao R, Li
Z, Ya S, Jia C and Bao Y: Silencing of GP73 inhibits invasion and
metastasis via suppression of epithelial-mesenchymal transition in
hepatocellular carcinoma. Oncol Rep. 37:1182–1188. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ma Y, Wang X, Wu Y, Sun B, Lv H, Rong F
and Zheng X: Overexpression of GOLPH3 protein is associated with
worse prognosis in patients with epithelial ovarian cancer. Tumour
Biol. 35:11845–11849. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li Q, Ma Y and Xu W: High GOLPH3
expression is associated with poor prognosis and invasion of
hepatocellular carcinoma. Mol Med Rep. 11:4315–4320. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li W, Guo F, Gu M, Wang G, He X, Zhou J,
Peng Y, Wang Z and Wang X: Increased expression of GOLPH3 is
associated with the proliferation of prostate cancer. J Cancer.
6:420–429. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Taft MH, Behrmann E, Munske-Weidemann LC,
Thiel C, Raunser S and Manstein DJ: Functional characterization of
human myosin-18A and its interaction with F-actin and GOLPH3. J
Biol Chem. 288:30029–30041. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Schaeffer D, Somarelli JA, Hanna G, Palmer
GM and Garcia-Blanco MA: Cellular migration and invasion uncoupled:
Increased migration is not an inexorable consequence of
epithelial-to-mesenchymal transition. Mol Cell Biol. 34:3486–3499.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Pinho SS and Reis CA: Glycosylation in
cancer: Mechanisms and clinical implications. Nat Rev Cancer.
15:540–555. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
English NM, Lesley JF and Hyman R:
Site-specific de-N-glycosylation of CD44 can activate hyaluronan
binding, and CD44 activation states show distinct threshold
densities for hyaluronan binding. Cancer Res. 58:3736–3742.
1998.PubMed/NCBI
|
|
33
|
Guo HB, Lee I, Kamar M, Akiyama SK and
Pierce M: Aberrant N-glycosylation of beta1 integrin causes reduced
alpha5beta1 integrin clustering and stimulates cell migration.
Cancer Res. 62:6837–6845. 2002.PubMed/NCBI
|
|
34
|
Pinho SS, Seruca R, Gärtner F, Yamaguchi
Y, Gu J, Taniguchi N and Reis CA: Modulation of E-cadherin function
and dysfunction by N-glycosylation. Cell Mol Life Sci.
68:1011–1020. 2011. View Article : Google Scholar : PubMed/NCBI
|