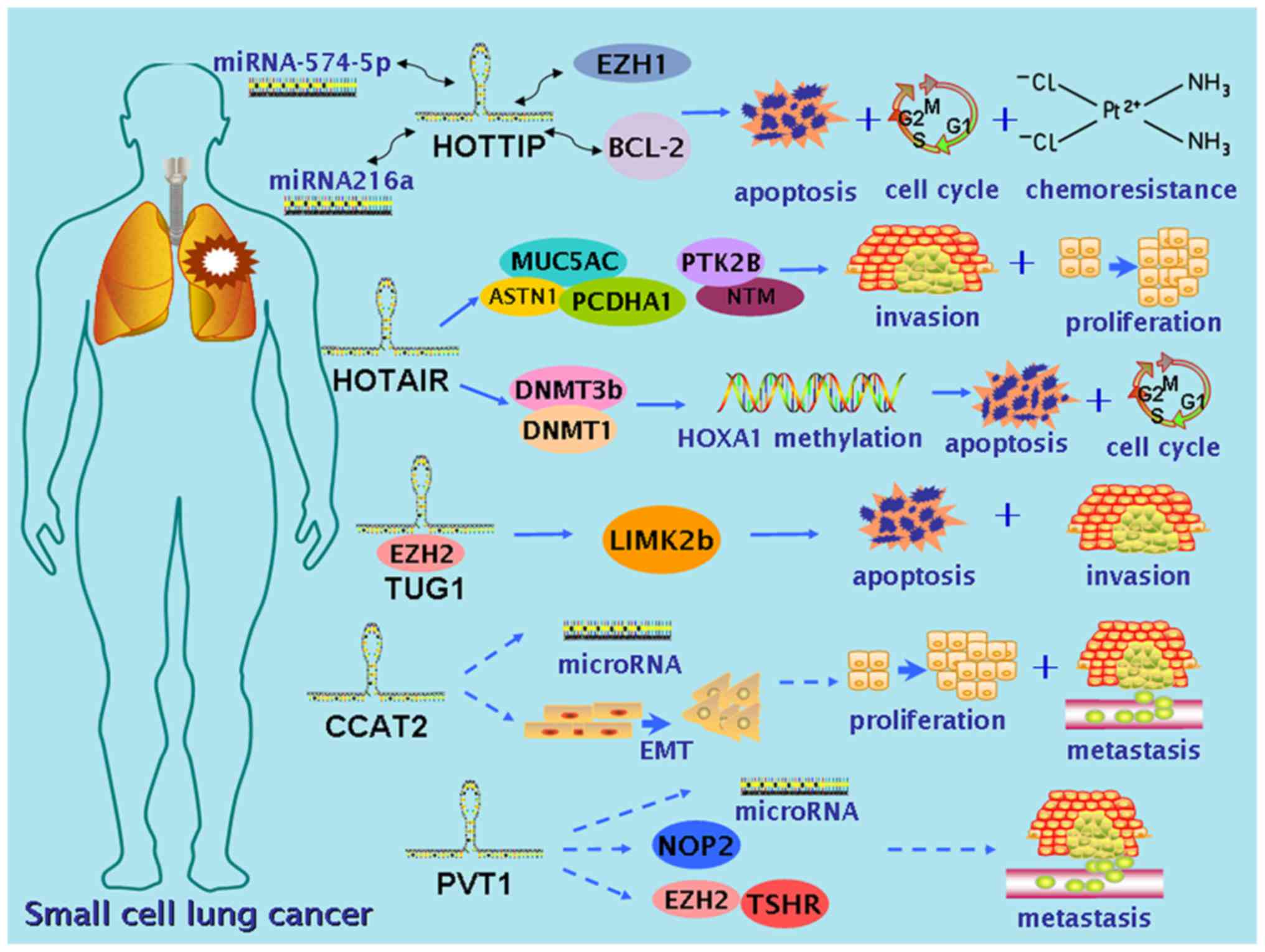
|
1
|
van Meerbeeck JP, Fennell DA and De
Ruysscher DK: Small-cell lung cancer. Lancet. 378:1741–1755. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Barnes H, See K, Barnett S and Manser R:
Surgery for limited-stage small-cell lung cancer. Cochrane Database
Syst Rev. 4:CD0119172017.PubMed/NCBI
|
|
3
|
Sabari JK, Lok BH, Laird JH, Poirier JT
and Rudin CM: Unravelling the biology of SCLC: Implications for
therapy. Nat Rev Clin Oncol. 14:549–561. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Pillai RN and Owonikoko TK: Small cell
lung cancer: Therapies and targets. Semin Oncol. 41:133–142. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Byers LA and Rudin CM: Small cell lung
cancer: Where do we go from here? Cancer. 121:664–672. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Dolly SO, Collins DC, Sundar R, Popat S
and Yap TA: Advances in the development of molecularly targeted
agents in non-small-cell lung cancer. Drugs. 77:813–827. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Seeber A, Leitner C, Philipp-Abbrederis K,
Spizzo G and Kocher F: What's new in small cell lung
cancer-extensive disease? An overview on advances of systemic
treatment in 2016. Future Oncol. 13:1427–1435. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Djebali S, Davis CA, Merkel A, Dobin A,
Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F,
et al: Landscape of transcription in human cells. Nature.
489:101–108. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
George J, Lim JS, Jang SJ, Cun Y, Ozretić
L, Kong G, Leenders F, Lu X, Fernández-Cuesta L, Bosco G, et al:
Comprehensive genomic profiles of small cell lung cancer. Nature.
524:47–53. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Pleasance ED, Stephens PJ, O'Meara S,
McBride DJ, Meynert A, Jones D, Lin ML, Beare D, Lau KW, Greenman
C, et al: A small-cell lung cancer genome with complex signatures
of tobacco exposure. Nature. 463:184–190. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kung JT, Colognori D and Lee JT: Long
noncoding RNAs: Past, present, and future. Genetics. 193:651–669.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ulitsky I and Bartel DP: LincRNAs:
Genomics, evolution, and mechanisms. Cell. 154:26–46. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gibb EA, Brown CJ and Lam WL: The
functional role of long non-coding RNA in human carcinomas. Mol
Cancer. 10:382011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Huang J, Peng J and Guo L: Non-coding RNA:
A new tool for the diagnosis, prognosis, and therapy of small cell
lung cancer. J Thorac Oncol. 10:28–37. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kwok ZH and Tay Y: Long noncoding RNAs:
Lincs between human health and disease. Biochem Soc Trans.
45:805–812. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ohno S: So much ‘junk’ DNA in our genome.
Brookhaven Symp Biol. 23:366–370. 1972.PubMed/NCBI
|
|
17
|
Spornraft M, Kirchner B, Pfaffl MW and
Riedmaier I: Comparison of the miRNome and piRNome of bovine blood
and plasma by small RNA sequencing. Biotechnol Lett. 37:1165–1176.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Busch H, Reddy R, Rothblum L and Choi YC:
SnRNAs, SnRNPs, and RNA processing. Annu Rev Biochem. 51:617–654.
1982. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ota T, Suzuki Y, Nishikawa T, Otsuki T,
Sugiyama T, Irie R, Wakamatsu A, Hayashi K, Sato H, Nagai K, et al:
Complete sequencing and characterization of 21,243 full-length
human cDNAs. Nat Genet. 36:40–45. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bertone P, Stolc V, Royce TE, Rozowsky JS,
Urban AE, Zhu X, Rinn JL, Tongprasit W, Samanta M, Weissman S, et
al: Global identification of human transcribed sequences with
genome tiling arrays. Science. 306:2242–2246. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Okazaki Y, Furuno M, Kasukawa T, Adachi J,
Bono H, Kondo S, Nikaido I, Osato N, Saito R, Suzuki H, et al:
Analysis of the mouse transcriptome based on functional annotation
of 60,770 full-length cDNAs. Nature. 420:563–573. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hon CC, Ramilowski JA, Harshbarger J,
Bertin N, Rackham OJ, Gough J, Denisenko E, Schmeier S, Poulsen TM,
Severin J, et al: An atlas of human long non-coding RNAs with
accurate 5′ ends. Nature. 543:199–204. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hombach S and Kretz M: Non-coding RNAs:
Classification, biology and functioning. Adv Exp Med Biol.
937:3–17. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Schmitt AM and Chang HY: Long noncoding
RNAs in cancer pathways. Cancer Cell. 29:452–463. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang KC and Chang HY: Molecular mechanisms
of long noncoding RNAs. Mol Cell. 43:904–914. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang KC, Helms JA and Chang HY:
Regeneration, repair and remembering identity: The three Rs of
Hox gene expression. Trends Cell Biol. 19:268–275. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Rinn JL, Kertesz M, Wang JK, Squazzo SL,
Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et
al: Functional demarcation of active and silent chromatin domains
in human HOX loci by noncoding RNAs. Cell. 129:1311–1323.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hung T, Wang Y, Lin MF, Koegel AK, Kotake
Y, Grant GD, Horlings HM, Shah N, Umbricht C, Wang P, et al:
Extensive and coordinated transcription of noncoding RNAs within
cell-cycle promoters. Nat Genet. 43:621–629. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Plath K, Mlynarczyk-Evans S, Nusinow DA
and Panning B: Xist RNA and the mechanism of X chromosome
inactivation. Annu Rev Genet. 36:233–278. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lee JT: The X as model for RNA's niche in
epigenomic regulation. Cold Spring Harb Perspect Biol.
2:a0037492010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wutz A, Rasmussen TP and Jaenisch R:
Chromosomal silencing and localization are mediated by different
domains of Xist RNA. Nat Genet. 30:167–174. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Gupta RA, Shah N, Wang KC, Kim J, Horlings
HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhao J, Ohsumi TK, Kung JT, Ogawa Y, Grau
DJ, Sarma K, Song JJ, Kingston RE, Borowsky M and Lee JT:
Genome-wide identification of polycomb-associated RNAs by RIP-seq.
Mol Cell. 40:939–953. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Khalil AM, Guttman M, Huarte M, Garber M,
Raj A, Morales Rivea D, Thomas K, Presser A, Bernstein BE, van
Oudenaarden A, et al: Many human large intergenic noncoding RNAs
associate with chromatin-modifying complexes and affect gene
expression. Proc Natl Acad Sci USA. 106:11667–11672. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Spitale RC, Tsai MC and Chang HY: RNA
templating the epigenome: Long noncoding RNAs as molecular
scaffolds. Epigenetics. 6:539–543. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Collins K: Physiological assembly and
activity of human telomerase complexes. Mech Ageing Dev. 129:91–98.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Chen JL and Greider CW: Telomerase RNA
structure and function: Implications for dyskeratosis congenita.
Trends Biochem Sci. 29:183–192. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Moretti F, Thermann R and Hentze MW:
Mechanism of translational regulation by miR-2 from sites in the 5′
untranslated region or the open reading frame. RNA. 16:2493–2502.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ørom UA, Nielsen FC and Lund AH:
MicroRNA-10a binds the 5′UTR of ribosomal protein mRNAs and
enhances their translation. Mol Cell. 30:460–471. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Hu X, Feng Y, Zhang D, Zhao SD, Hu Z,
Greshock J, Zhang Y, Yang L, Zhong X, Wang LP, et al: A functional
genomic approach identifies FAL1 as an oncogenic long
noncoding RNA that associates with BMI1 and represses p21
expression in cancer. Cancer Cell. 26:344–357. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yin Y, Yan P, Lu J, Song G, Zhu Y, Li Z,
Zhao Y, Shen B, Huang X, Zhu H, et al: Opposing roles for the
lncRNA Haunt and its genomic locus in regulating HOXA
gene activation during embryonic stem cell differentiation. Cell
Stem Cell. 16:504–516. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Cesana M, Cacchiarelli D, Legnini I,
Santini T, Sthandier O, Chinappi M, Tramontano A and Bozzoni I: A
long noncoding RNA controls muscle differentiation by functioning
as a competing endogenous RNA. Cell. 147:358–369. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Tripathi V, Ellis JD, Shen Z, Song DY, Pan
Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, et al: The
nuclear-retained noncoding RNA MALAT1 regulates alternative
splicing by modulating SR splicing factor phosphorylation. Mol
Cell. 39:925–938. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ,
Tao QF, Liu F, Pan W, Wang TT, Zhou CC, et al: A long noncoding RNA
activated by TGF-beta promotes the invasion-metastasis cascade in
hepatocellular carcinoma. Cancer Cell. 25:666–681. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Steck E, Boeuf S, Gabler J, Werth N,
Schnatzer P, Diederichs S and Richter W: Regulation of H19 and its
encoded microRNA-675 in osteoarthritis and under anabolic and
catabolic in vitro conditions. J Mol Med. 90:1185–1195. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Houseley J, Rubbi L, Grunstein M,
Tollervey D and Vogelauer M: A ncRNA modulates histone modification
and mRNA induction in the yeast GAL gene cluster. Mol Cell.
32:685–695. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Hainer SJ, Gu W, Carone BR, Landry BD,
Rando OJ, Mello CC and Fazzio TG: Suppression of pervasive
noncoding transcription in embryonic stem cells by esBAF. Genes
Dev. 29:362–378. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Berghoff EG, Clark MF, Chen S, Cajigas I,
Leib DE and Kohtz JD: Evf2 (Dlx6as) lncRNA regulates
ultraconserved enhancer methylation and the differential
transcriptional control of adjacent genes. Development.
140:4407–4416. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Geisler S and Coller J: RNA in unexpected
places: Long non-coding RNA functions in diverse cellular contexts.
Nat Rev Mol Cell Biol. 14:699–712. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Chen G, Wang Z, Wang D, Qiu C, Liu M, Chen
X, Zhang Q, Yan G and Cui Q: LncRNADisease: A database for
long-non-coding RNA-associated diseases. Nucleic Acids Res.
41:D983–D986. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Lian Y, Cai Z, Gong H, Xue S, Wu D and
Wang K: HOTTIP: A critical oncogenic long non-coding RNA in human
cancers. Mol biosyst. 12:3247–3253. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Wang KC, Yang YW, Liu B, Sanyal A,
Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta
RA, et al: A long noncoding RNA maintains active chromatin to
coordinate homeotic gene expression. Nature. 472:120–124. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Chang S, Liu J, Guo S, He S, Qiu G, Lu J,
Wang J, Fan L, Zhao W and Che X: HOTTIP and HOXA13 are oncogenes
associated with gastric cancer progression. Oncol Rep.
35:3577–3585. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Quagliata L, Matter MS, Piscuoglio S,
Arabi L, Ruiz C, Procino A, Kovac M, Moretti F, Makowska Z,
Boldanova T, et al: Long noncoding RNA HOTTIP/HOXA13 expression is
associated with disease progression and predicts outcome in
hepatocellular carcinoma patients. Hepatology. 59:911–923. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Li Z, Zhao X, Zhou Y, Liu Y, Zhou Q, Ye H,
Wang Y, Zeng J, Song Y, Gao W, et al: The long non-coding RNA
HOTTIP promotes progression and gemcitabine resistance by
regulating HOXA13 in pancreatic cancer. J Transl Med. 13:842015.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Sun Y, Zhou Y, Bai Y, Wang Q, Bao J, Luo
Y, Guo Y and Guo L: A long non-coding RNA HOTTIP expression is
associated with disease progression and predicts outcome in small
cell lung cancer patients. Mol Cancer. 16:1622017. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Sun Y, Hu B, Wang Q, Ye M, Qiu Q, Zhou Y,
Zeng F, Zhang X, Guo Y and Guo L: Long non-coding RNA HOTTIP
promotes BCL-2 expression and induces chemoresistance in small cell
lung cancer by sponging miR-216a. Cell Death Dis. 9:852018.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Bhan A, Soleimani M and Mandal SS: Long
noncoding RNA and cancer: A new paradigm. Cancer Res. 77:3965–3981.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Tsai MC, Manor O, Wan Y, Mosammaparast N,
Wang JK, Lan F, Shi Y, Segal E and Chang HY: Long noncoding RNA as
modular scaffold of histone modification complexes. Science.
329:689–693. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Sun G, Alzayady K, Stewart R, Ye P, Yang
S, Li W and Shi Y: Histone demethylase LSD1 regulates neural stem
cell proliferation. Mol Cell Biol. 30:1997–2005. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: Insights into functions. Nat Rev Genet.
10:155–159. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Tsai MC, Spitale RC and Chang HY: Long
intergenic noncoding RNAs: New links in cancer progression. Cancer
Res. 71:3–7. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Alves Pádua C, Fonseca AS, Muys BR, de
Barros E Lima, Bueno R, Bürger MC, de Souza JE, Valente V, Zago MA
and Silva WA Jr: Brief report: The lincRNA Hotair is required for
epithelial-to-mesenchymal transition and stemness maintenance of
cancer cell lines. Stem Cells. 31:2827–2832. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Yoon JH, Abdelmohsen K, Kim J, Yang X,
Martindale JL, Tominaga-Yamanaka K, White EJ, Orjalo AV, Rinn JL,
Kreft SG, et al: Scaffold function of long non-coding RNA
HOTAIR in protein ubiquitination. Nat Commun. 4:29392013.
View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Ono H, Motoi N, Nagano H, Miyauchi E,
Ushijima M, Matsuura M, Okumura S, Nishio M, Hirose T, Inase N, et
al: Long noncoding RNA HOTAIR is relevant to cellular
proliferation, invasiveness, and clinical relapse in small-cell
lung cancer. Cancer Med. 3:632–642. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Fang S, Gao H, Tong Y, Yang J, Tang R, Niu
Y, Li M and Guo L: Long noncoding RNA-HOTAIR affects
chemoresistance by regulating HOXA1 methylation in small cell lung
cancer cells. Lab Invest. 96:60–68. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Young TL, Matsuda T and Cepko CL: The
noncoding RNA taurine upregulated gene 1 is required for
differentiation of the murine retina. Curr Biol. 15:501–512. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Huang MD, Chen WM, Qi FZ, Sun M, Xu TP, Ma
P and Shu YQ: Long non-coding RNA TUG1 is up-regulated in
hepatocellular carcinoma and promotes cell growth and apoptosis by
epigenetically silencing of KLF2. Mol Cancer. 14:1652015.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Ma B, Li M, Zhang L, Huang M, Lei JB, Fu
GH, Liu CX, Lai QW, Chen QQ and Wang YL: Upregulation of long
non-coding RNA TUG1 correlates with poor prognosis and disease
status in osteosarcoma. Tumour Biol. 37:4445–4455. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Liu Q, Sun S, Yu W, Jiang J, Zhuo F, Qiu
G, Xu S and Jiang X: Altered expression of long non-coding RNAs
during genotoxic stress-induced cell death in human glioma cells. J
Neurooncol. 122:283–292. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Han Y, Liu Y, Gui Y and Cai Z: Long
intergenic non-coding RNA TUG1 is overexpressed in urothelial
carcinoma of the bladder. J Surg Oncol. 107:555–559. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Xu Y, Wang J, Qiu M and Xu L, Li M, Jiang
F, Yin R and Xu L: Upregulation of the long noncoding RNA TUG1
promotes proliferation and migration of esophageal squamous cell
carcinoma. Tumour Biol. 36:1643–1651. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Zhang E, He X, Yin D, Han L, Qiu M, Xu T,
Xia R, Xu L, Yin R and De W: Increased expression of long noncoding
RNA TUG1 predicts a poor prognosis of gastric cancer and regulates
cell proliferation by epigenetically silencing of p57. Cell Death
Dis. 7:e21092016. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Yang L, Lin C, Liu W, Zhang J, Ohgi KA,
Grinstein JD, Dorrestein PC and Rosenfeld MG: ncRNA- and Pc2
methylation-dependent gene relocation between nuclear structures
mediates gene activation programs. Cell. 147:773–788. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Niu Y, Ma F, Huang W, Fang S, Li M, Wei T
and Guo L: Long non-coding RNA TUG1 is involved in cell growth and
chemoresistance of small cell lung cancer by regulating LIMK2b via
EZH2. Mol Cancer. 16:52017. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Ling H, Spizzo R, Atlasi Y, Nicoloso M,
Shimizu M, Redis RS, Nishida N, Gafà R, Song J, Guo Z, et al:
CCAT2, a novel noncoding RNA mapping to 8q24, underlies
metastatic progression and chromosomal instability in colon cancer.
Genome Res. 23:1446–1461. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Gong WJ, Yin JY, Li XP, Fang C, Xiao D,
Zhang W, Zhou HH, Li X and Liu ZQ: Association of
well-characterized lung cancer lncRNA polymorphisms with lung
cancer susceptibility and platinum-based chemotherapy response.
Tumour Biol. 37:8349–8358. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Xin Y, Li Z, Zheng H, Chan MTV and Ka Kei
Wu W: CCAT2: A novel oncogenic long non-coding RNA in human
cancers. Cell Prolif. 50:2017. View Article : Google Scholar
|
|
80
|
Redis RS, Sieuwerts AM, Look MP, Tudoran
O, Ivan C, Spizzo R, Zhang X, de Weerd V, Shimizu M, Ling H, et al:
CCAT2, a novel long non-coding RNA in breast cancer: Expression
study and clinical correlations. Oncotarget. 4:1748–1762. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Cai Y, He J and Zhang D: Long noncoding
RNA CCAT2 promotes breast tumor growth by regulating the Wnt
signaling pathway. Onco Targets Ther. 8:2657–2664. 2015.PubMed/NCBI
|
|
82
|
Huang S, Qing C, Huang Z and Zhu Y: The
long non-coding RNA CCAT2 is up-regulated in ovarian cancer and
associated with poor prognosis. Diagn Pathol. 11:492016. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Zheng J, Zhao S, He X, Zheng Z, Bai W,
Duan Y, Cheng S, Wang J, Liu X and Zhang G: The up-regulation of
long non-coding RNA CCAT2 indicates a poor prognosis for
prostate cancer and promotes metastasis by affecting
epithelial-mesenchymal transition. Biochem Biophys Res Commun.
480:508–514. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Wang CY, Hua L, Yao KH, Chen JT, Zhang JJ
and Hu JH: Long non-coding RNA CCAT2 is up-regulated in gastric
cancer and associated with poor prognosis. Int J Clin Exp Pathol.
8:779–785. 2015.PubMed/NCBI
|
|
85
|
Chen S, Wu H, Lv N, Wang H, Wang Y, Tang
Q, Shao H and Sun C: LncRNA CCAT2 predicts poor prognosis
and regulates growth and metastasis in small cell lung cancer.
Biomed Pharmacother. 82:583–588. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Webb E, Adams JM and Cory S: Variant
(6;15) translocation in a murine plasmacytoma occurs near an
immunoglobulin kappa gene but far from the myc oncogene. Nature.
312:777–779. 1984. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Cory S, Graham M, Webb E, Corcoran L and
Adams JM: Variant (6;15) translocations in murine plasmacytomas
involve a chromosome 15 locus at least 72 kb from the c-myc
oncogene. EMBO J. 4:675–681. 1985.PubMed/NCBI
|
|
88
|
Colombo T, Farina L, Macino G and Paci P:
PVT1: A rising star among oncogenic long noncoding RNAs. Biomed Res
Int. 2015:3042082015. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Ding J, Li D, Gong M, Wang J, Huang X, Wu
T and Wang C: Expression and clinical significance of the long
non-coding RNA PVT1 in human gastric cancer. Onco Targets Ther.
7:1625–1630. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Wang F, Yuan JH, Wang SB, Yang F, Yuan SX,
Ye C, Yang N, Zhou WP, Li WL, Li W, et al: Oncofetal long noncoding
RNA PVT1 promotes proliferation and stem cell-like property of
hepatocellular carcinoma cells by stabilizing NOP2. Hepatology.
60:1278–1290. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Zhou Q, Chen J, Feng J and Wang J: Long
noncoding RNA PVT1 modulates thyroid cancer cell proliferation by
recruiting EZH2 and regulating thyroid-stimulating hormone receptor
(TSHR). Tumour Biol. 37:3105–3113. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Huang C, Yu W, Wang Q, Cui H, Wang Y,
Zhang L, Han F and Huang T: Increased expression of the lncRNA PVT1
is associated with poor prognosis in pancreatic cancer patients.
Minerva Med. 106:143–149. 2015.PubMed/NCBI
|
|
93
|
Cui D, Yu CH, Liu M, Xia QQ, Zhang YF and
Jiang WL: Long non-coding RNA PVT1 as a novel biomarker for
diagnosis and prognosis of non-small cell lung cancer. Tumour Biol.
37:4127–4134. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Cui M, You L, Ren X, Zhao W, Liao Q and
Zhao Y: Long non-coding RNA PVT1 and cancer. Biochem Biophys Res
Commun. 471:10–14. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Huang C, Liu S, Wang H, Zhang Z, Yang Q
and Gao F: LncRNA PVT1 overexpression is a poor prognostic
biomarker and regulates migration and invasion in small cell lung
cancer. Am J Transl Res. 8:5025–5034. 2016.PubMed/NCBI
|
|
96
|
Sang Y, Zhou F, Wang D, Bi X, Liu X, Hao
Z, Li Q and Zhang W: Up-regulation of long non-coding HOTTIP
functions as an oncogene by regulating HOXA13 in non-small cell
lung cancer. Am J Transl Res. 8:2022–2032. 2016.PubMed/NCBI
|
|
97
|
Zhang GJ, Song W and Song Y:
Overexpression of HOTTIP promotes proliferation and drug resistance
of lung adenocarcinoma by regulating AKT signaling pathway. Eur Rev
Med Pharmacol Sci. 21:5683–5690. 2017.PubMed/NCBI
|
|
98
|
Yang Y, Jiang C, Yang Y, Guo L, Huang J,
Liu X, Wu C and Zou J: Silencing of LncRNA-HOTAIR decreases drug
resistance of non-small cell lung cancer cells by inactivating
autophagy via suppressing the phosphorylation of ULK1. Biochem
Biophys Res Commun. 497:1003–1010. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Liu W, Yin NC, Liu H and Nan KJ: Cav-1
promote lung cancer cell proliferation and invasion through lncRNA
HOTAIR. Gene. 641:335–340. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Jiang C, Yang Y, Yang Y, Guo L, Huang J,
Liu X, Wu C and Zou J: Long noncoding RNA (lncRNA) HOTAIR affects
tumorigenesis and metastasis of non-small cell lung cancer by
up-regulating miR-613. Oncol Res. 26:725–734. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Lin PC, Huang HD, Chang CC, Chang YS, Yen
JC, Lee CC, Chang WH, Liu TC and Chang JG: Long noncoding RNA
TUG1 is downregulated in non-small cell lung cancer and can
regulate CELF1 on binding to PRC2. BMC Cancer. 16:5832016.
View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Zhang EB, Yin DD, Sun M, Kong R, Liu XH,
You LH, Han L, Xia R, Wang KM, Yang JS, et al: P53-regulated long
non-coding RNA TUG1 affects cell proliferation in human non-small
cell lung cancer, partly through epigenetically regulating HOXB7
expression. Cell Death Dis. 5:e12432014. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Liu H, Zhou G, Fu X, Cui H, Pu G, Xiao Y,
Sun W, Dong X, Zhang L, Cao S, et al: Long noncoding RNA TUG1 is a
diagnostic factor in lung adenocarcinoma and suppresses apoptosis
via epigenetic silencing of BAX. Oncotarget. 8:101899–101910.
2017.PubMed/NCBI
|
|
104
|
Zhao Z, Wang J, Wang S, Chang H, Zhang T
and Qu J: LncRNA CCAT2 promotes tumorigenesis by over-expressed
Pokemon in non-small cell lung cancer. Biomed Pharmacother.
87:692–697. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Qiu M, Xu Y, Yang X, Wang J, Hu J, Xu L
and Yin R: CCAT2 is a lung adenocarcinoma-specific long non-coding
RNA and promotes invasion of non-small cell lung cancer. Tumour
Biol. 35:5375–5380. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Wang C, Han C, Zhang Y and Liu F: LncRNA
PVT1 regulate expression of HIF1α via functioning as ceRNA for
miR199a5p in nonsmall cell lung cancer under hypoxia. Mol Med Rep.
17:1105–1110. 2018.PubMed/NCBI
|
|
107
|
Li H, Chen S, Liu J, Guo X, Xiang X, Dong
T, Ran P, Li Q, Zhu B, Zhang X, et al: Long non-coding RNA PVT1-5
promotes cell proliferation by regulating miR-126/SLC7A5 axis in
lung cancer. Biochem Biophys Res Commun. 495:2350–2355. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Guo D, Wang Y, Ren K and Han X: Knockdown
of LncRNA PVT1 inhibits tumorigenesis in non-small-cell lung cancer
by regulating miR-497 expression. Exp Cell Res. 362:172–179. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Wu D, Li Y, Zhang H and Hu X: Knockdown of
lncrna PVT1 enhances radiosensitivity in non-small cell lung cancer
by sponging Mir-195. Cell Physiol Biochem. 42:2453–2466. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Smolle MA, Bauernhofer T, Pummer K, Calin
GA and Pichler M: Current insights into long non-coding RNAs
(lncRNAs) in prostate cancer. Int J Mol Sci. 18:E4732017.
View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Yu X, Zheng H, Chan MT and Wu WK: HULC: An
oncogenic long non-coding RNA in human cancer. J Cell Mol Med.
21:410–417. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Gutschner T, Hämmerle M and Diederichs S:
MALAT1-a paradigm for long noncoding RNA function in cancer. J Mol
Med. 91:791–801. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Huarte M: The emerging role of lncRNAs in
cancer. Nat Med. 21:1253–1261. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
McCleland ML, Mesh K, Lorenzana E, Chopra
VS, Segal E, Watanabe C, Haley B, Mayba O, Yaylaoglu M, Gnad F, et
al: CCAT1 is an enhancer-templated RNA that predicts BET
sensitivity in colorectal cancer. J Clin Invest. 126:639–652. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Zhou Y, Zhang X and Klibanski A:
MEG3 noncoding RNA: A tumor suppressor. J Mol Endocrinol.
48:R45–R53. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Yap TA, Omlin A and de Bono JS:
Development of therapeutic combinations targeting major cancer
signaling pathways. J Clin Oncol. 31:1592–1605. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Liu XH, Sun M, Nie FQ, Ge YB, Zhang EB,
Yin DD, Kong R, Xia R, Lu KH, Li JH, et al: Lnc RNA HOTAIR
functions as a competing endogenous RNA to regulate HER2 expression
by sponging miR-331-3p in gastric cancer. Mol Cancer. 13:922014.
View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Zhang SR, Yang JK, Xie JK and Zhao LC:
Long noncoding RNA HOTTIP contributes to the progression of
prostate cancer by regulating HOXA13. Cell Mol Biology. 62:84–88.
2016.
|
|
119
|
Sørensen KP, Thomassen M, Tan Q, Bak M,
Cold S, Burton M, Larsen MJ and Kruse TA: Long non-coding RNA
HOTAIR is an independent prognostic marker of metastasis in
estrogen receptor-positive primary breast cancer. Breast Cancer Res
Treat. 142:529–536. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Wu ZH, Wang XL, Tang HM, Jiang T, Chen J,
Lu S, Qiu GQ, Peng ZH and Yan DW: Long non-coding RNA HOTAIR is a
powerful predictor of metastasis and poor prognosis and is
associated with epithelial-mesenchymal transition Rep. 32:395–402.
2014.
|
|
121
|
Svoboda M, Slyskova J, Schneiderova M,
Makovicky P, Bielik L, Levy M, Lipska L, Hemmelova B, Kala Z,
Protivankova M, et al: HOTAIR long non-coding RNA is a negative
prognostic factor not only in primary tumors, but also in the blood
of colorectal cancer patients. Carcinogenesis. 35:1510–1515. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Kogo R, Shimamura T, Mimori K, Kawahara K,
Imoto S, Sudo T, Tanaka F, Shibata K, Suzuki A, Komune S, et al:
Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin
modification and is associated with poor prognosis in colorectal
cancers. Cancer Res. 71:6320–6326. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Huang L, Liao LM, Liu AW, Wu JB, Cheng XL,
Lin JX and Zheng M: Overexpression of long noncoding RNA HOTAIR
predicts a poor prognosis in patients with cervical cancer. Arch
Gynecol Obstet. 290:717–723. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Ji F, Wuerkenbieke D, He Y and Ding Y:
Long noncoding RNA HOTAIR: An oncogene in human cervical cancer
interacting with MicroRNA-17-5p. Oncol Res. 2017.Doi:
10.3727/096504017X15002869385155. View Article : Google Scholar :
|
|
125
|
Wu X, Cao X and Chen F: LncRNA-HOTAIR
activates tumor cell proliferation and migration by suppressing
MiR-326 in cervical cancer. Oncol Res. 2017.Doi:
10.3727/096504017X15037515496840. View Article : Google Scholar :
|
|
126
|
Endo H, Shiroki T, Nakagawa T, Yokoyama M,
Tamai K, Yamanami H, Fujiya T, Sato I, Yamaguchi K, Tanaka N, et
al: Enhanced expression of long non-coding RNA HOTAIR is associated
with the development of gastric cancer. PLoS One. 8:e770702013.
View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Hajjari M, Behmanesh M, Sadeghizadeh M and
Zeinoddini M: Up-regulation of HOTAIR long non-coding RNA in human
gastric adenocarcinoma tissues. Med Oncol. 30:6702013. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Chen WM, Chen WD, Jiang XM, Jia XF, Wang
HM, Zhang QJ, Shu YQ and Zhao HB: HOX transcript antisense
intergenic RNA represses E-cadherin expression by binding to EZH2
in gastric cancer. World J Gastroenterol. 23:6100–6110. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Yan J, Dang Y, Liu S, Zhang Y and Zhang G:
LncRNA HOTAIR promotes cisplatin resistance in gastric cancer by
targeting miR-126 to activate the PI3K/AKT/MRP1 genes. Tumour Biol.
Nov 30–2016.(Epub ahead of print). View Article : Google Scholar :
|
|
130
|
Lin YH, Wu MH, Huang YH, Yeh CT, Cheng ML,
Chi HC, Tsai CY, Chung IH, Chen CY and Lin KH: Taurine upregulated
gene 1 functions as a master regulator to coordinate glycolysis and
metastasis in hepatocellular carcinoma. Hepatology. 67:188–203.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Zhao L, Sun H, Kong H, Chen Z, Chen B and
Zhou M: The lncrna-TUG1/EZH2 axis promotes pancreatic cancer cell
proliferation, migration and EMT phenotype formation through
sponging mir-382. Cell Physiol Biochem. 42:2145–2158. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Liu Q, Liu H, Cheng H, Li Y, Li X and Zhu
C: Downregulation of long noncoding RNA TUG1 inhibits proliferation
and induces apoptosis through the TUG1/miR-142/ZEB2 axis in bladder
cancer cells. Onco Targets Ther. 10:2461–2471. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Yu Y, Nangia-Makker P, Farhana L and
Majumdar AP: A novel mechanism of lncRNA and miRNA interaction:
CCAT2 regulates miR-145 expression by suppressing its maturation
process in colon cancer cells. Mol Cancer. 16:1552017. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Hua F, Li CH, Chen XG and Liu XP: Long
noncoding RNA CCAT2 Knockdown suppresses tumorous progression by
sponging miR-424 in epithelial ovarian cancer. Oncol Res.
26:241–247. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Li T, Meng XL and Yang WQ: Long noncoding
RNA PVT1 acts as a ‘Sponge’ to inhibit microRNA-152 in gastric
cancer cells. Dig Dis Sci. 62:3021–3028. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Huang T, Liu HW, Chen JQ, Wang SH, Hao LQ,
Liu M and Wang B: The long noncoding RNA PVT1 functions as a
competing endogenous RNA by sponging miR-186 in gastric cancer.
Biomed Pharmacother. 88:302–308. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Zhao L, Kong H, Sun H, Chen Z, Chen B and
Zhou M: LncRNA-PVT1 promotes pancreatic cancer cells proliferation
and migration through acting as a molecular sponge to regulate
miR-448. J Cell Physiol. 233:4044–4055. 2018. View Article : Google Scholar : PubMed/NCBI
|