Introduction
CD105, also referred to as endoglin, is a type I
membrane glycoprotein that forms part of the TGF-β receptor complex
on cell membranes (1). CD105 is a
180 kDa homodimeric transmembrane protein composed of 633 amino
acids, which consists of a large extracellular domain, a
hydrophobic transmembrane domain and a short intracellular domain
(2). The CD105 external domain
binds to the TGF-β1 and TGF-β3 isoforms with a high affinity,
forming the TGF-β receptor complex (3). It serves an important role in
regulating cellular proliferation, differentiation, migration,
adhesion and angiogenesis, which are vital processes for tumour
growth, survival and metastasis (4,5). CD105
is predominantly expressed in angiogenic endothelial cells, or
blood vessels surrounding tumour tissues and mesenchymal stem cells
(MSCs) (6–8). The CD105 antibody has been used to
label tumour microvessel density, separate MSCs and conjugate drug
for targeting therapy (9–11).
Osteosarcoma is one of the most common primary
malignant bone sarcomas, predominantly targeting the long
cylindrical bones. Approximately 5% of all osteosarcomas are
reported to occur in the maxillofacial region (12). This tumour is more prevalent in
adolescents and children, with an incidence rate of 3–4.5 million
individuals per year globally, and can aggressively metastasise to
the lung in the early stages (13,14).
In a previous study, a subset of bone sarcoma cells that expressed
the MSC marker CD105 was reported to be more aggressive in
comparison with other bone sarcoma cell lineages (15). Therefore, the CD105 antibody may be
a marker for identifying the malignancy of osteosarcoma.
The complexity of production, heat instability, and
poor penetration to the cancer microenvironment has limited the
application of antibodies in vivo. Non-antibody-binding
protein (nABP) is a heat-stable, artificially synthesised peptide
with a potent penetration effect to the tumour microenvironment due
to its small size and positive charge (16). Owning to the advantages of this
peptide, it has attracted previous attention and has been used in
various antitumour studies and tumour-targeting therapies (17,18).
In the present study, 13 novel peptides binding to
CD105 were identified using M13 phage display. Among these 13
peptides, the nABP296 peptide had a higher affinity for the
CD105-positive osteosarcoma MNNG/HOS cell line as compared with
that of other peptides, and a lower affinity for the CD105-negative
Cal27 cell line. In addition, nABP296 was used to visualise the
tumour in a MNNG/HOS tumour-bearing mouse model. It was observed
that nABP296 was able to label osteosarcoma histological sections
derived from an animal MNNG/HOS xenograft tumour model and an
osteosarcoma patient.
Materials and methods
Phage display biopanning
Phage display biopanning, an affinity selection
technique that selects peptides binding to a given target, was
conducted according to standard procedures (19). A peptide phage display library was
commercially constructed (Ph.D.-12 Phage Display Peptide Library;
New England Biolabs, Inc., Beverly, MA, USA) and used in subsequent
experiments. The recombinant human CD105 protein (R&D Systems,
Inc., Minneapolis, MN) at 100 µg/ml in sterile phosphate-buffered
saline (PBS) was added to individual sterilised MaxiSorp plates
(Thermo Fisher Scientific, Inc., Waltham, MA, USA) and incubated
overnight at 4°C. The plate was washed six times with Tris-buffered
saline/0.1% Tween-20 (TBST) and then blocked for 1 h at 4°C using
1% bovine serum albumin (BSA) in PBS. M13 phage display libraries
were allowed to bind for 1 h at room temperature, and the unbound
phages were washed away 10 times with TBST. Subsequently, the
binding phages were eluted by an elution buffer, and the eluted
phages were then amplified for another round of biopanning. For
each round of biopanning, the titering of M13 phage was calculated.
The recovery rate was calculated as follows: Recovery rate =
Titering output/titering input.
Phage ELISA binding assay
Subsequent to three rounds of biopanning, the target
M13 phages were enriched. A total of 16 monoclonal phages were
selected randomly for the ELISA binding essay. Recombinant human
CD105 protein at 100 µg/ml in sterile PBS was added to 16
individual sterilised wells of MaxiSorp plates and incubated
overnight at 4°C. Additionally, 16 individual sterilised wells were
coated using 1% BSA as control. The plate was washed six times with
TBST and blocked for 1 h at 4°C using 1% BSA in PBS. Next, the 16
monoclonal phages were amplified and added to the individual wells
with CD105 protein and 1% BSA and incubated for 1 h at room
temperature. Following repeated washing of the MaxiSorp ELISA plate
with 0.1% TBST, bound phages were detected by incubation with mouse
anti-M13 phage antibody (HRP conjugated; cat. no. ab6188; 1:20;
Abcam, Cambridge, UK). The ABTS/H2O2
substrate (internally prepared) was used to measure the amount of
bound HRP, and the absorbance was monitored at 405 nm. The inserted
DNA sequences of 13 positive affinity monoclonal phages were
determined using the primer 5′-CCCTCATAGTTAGCGTAACG-3′ (New England
Biolabs, Inc.).
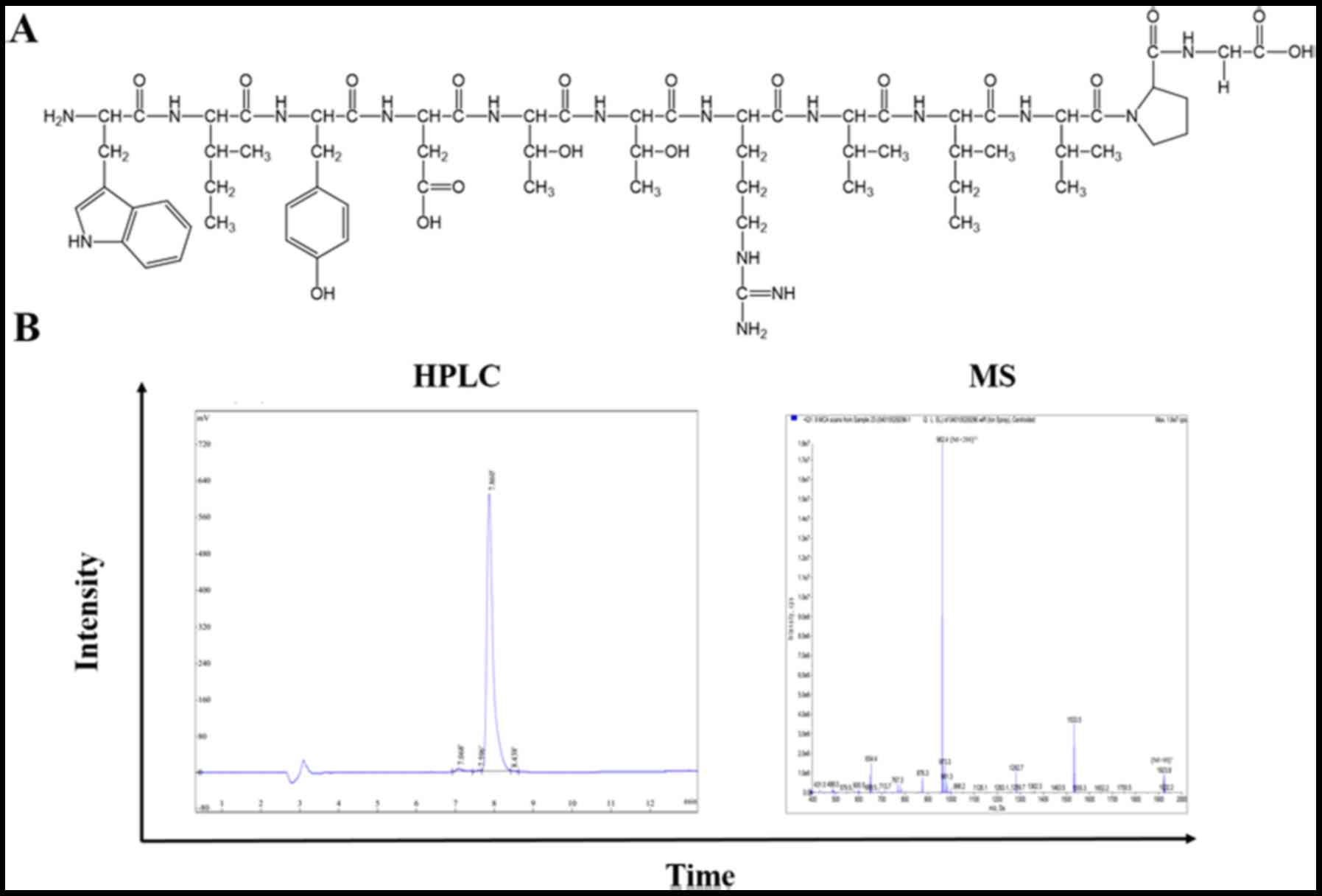
Synthesis of peptides
Based on aforementioned procedures, 13 peptides were
synthesised through solid-phase peptide synthesis using Fmoc
chemistry by Chinese Peptide Company (Jiangsu, China). An extra
fluorescein-5-isothiocyanate (FITC) was linked at the amino (N)
terminus of all peptides for labelling. Prepared peptides were
identified by mass spectrometry (MS), and the purity (>95%) was
assayed by high-performance liquid chromatography (HPLC; Fig. 1). The FITC-labelled peptides were
stored at −20°C. For further usage, peptides were dissolved in
sterilised water at 1 mg/kg and diluted according to the
experimental requirements.
Determination of cell lines for
binding assay
MNNG/HOS and Cal27 cell lines were obtained from the
Type Culture Collection of the Chinese Academy of Sciences
(Shanghai, China). MNNG/HOS cells were cultured in Dulbecco's
modified Eagle's medium (DMEM)/F12 (Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany) supplemented with 5% foetal bovine serum (FBS),
while Cal27 cells were cultured in DMEM (Sigma-Aldrich; Merck KGaA)
supplemented with 10% FBS. The cells were plated onto 10
cm2 flasks and maintained at 37°C in 5% CO2
with 90% relative humidity. In order to investigate the CD105
expression in MNNG/HOS and Cal27 cells, the cell lines were then
examined by reverse transcription-polymerase chain reaction
(RT-PCR), immunofluorescence and flow cytometry.
RT-PCR for CD105 expression
measurement
A NanoDrop 1000 (Thermo Fisher Scientific, Inc.,
Wilmington, DE, USA) was used to measure the amount of RNA by
spectrophotometry. A total of 300 ng total RNA was extracted from
tumour cells, and converted to complementary DNA using the ReverTra
Ace® qPCR RT kit (Toyobo Life Science, Osaka, Japan) for
PCR analysis. The semi-quantitative RT-PCR analysis was performed
using the Takara Ex Taq® kit (Takara Bio, Inc., Otsu,
Japan). The PCR reaction conditions were performed as follows: 95°C
for 5 min, and 30 cycles of 95°C for 30 sec, 60°C for 30 sec and
72°C for 1 min, following by a final elongation at 72°C for 10 min
and held at 16°C for 10 min. The CD105 primers used were
synthesised by Synbio Technologies (Suzhou, China) and were as
follows: CD105 forward, 5′-CACCACAGCGGAAAAAGGTG-3′ and reverse,
5′-GCCGGTTTTGGGTATGGGTA-3′; and GAPDH forward,
5′-AATGGGCAGCCGTTAGGAAA-3′ and reverse,
5′-GCGCCCAATACGACCAAATC-3′). PCR products were then analysed by
electrophoresis on a 1% agarose gel and visualised by ethidium
bromide staining under UV illumination in Iquant Capture RT ECL
(version 1.0.1; GE Healthcare, Chicago, IL, USA).
Immunofluorescence assay
Following collection in a 0.25% (w/v) trypsin
solution with 0.025% (w/v) ethylene diamine tetraacetic acid
(EDTA), MNNG/HOS and Cal27 cells were seeded in a 6-well flask with
slides at a density of 1×105 cells/well and incubated at
37°C in 5% CO2 with 90% relative humidity overnight. On
the second day, the cells were fixed in 4% paraformaldehyde at room
temperature for 15 min, and non-specific binding sites were blocked
by 5% BSA (Guangzhou Xiang Bo Biological Technology Co., Ltd.,
Guangzhou, China) for 1 h at room temperature. The cells were then
incubated with the CD105 antibody (cat. no. MA1-19408; 1:200;
Invitrogen; Thermo Fisher Scientific, Inc.) overnight at 4°C.
Unbound antibodies were eluted by washing three times with PBS.
Subsequently, the cells were further treated with donkey anti-mouse
IgG (H+L) highly cross-adsorbed Alexa Fluor 568 secondary antibody
(cat. no. A10037; 1:1,000; Invitrogen; Thermo Fisher Scientific,
Inc.) in blocking buffer for 1 h at room temperature. Following
further washing with PBS, the nucleus was counterstained by DAPI,
and the samples were visualised under confocal laser scanning
microscopy (CLSM; Leica Microsystems GmbH, Wetzlar, Germany). For
polypeptide labelling, the MNNG/HOS/HOS and Cal27 cells were
incubated in polypeptide at a concentration of 100 µg/ml in 1 ml
PBS for 30 min at 4°C prior to fixation.
Flow cytometry assay
Subsequent to collection in a 0.25% (w/v) trypsin
solution with 0.025% (w/v) EDTA, MNNG/HOS and Cal27 cells were
incubated with the CD105 antibody (cat. no. 130-102-819; 1:10;
Miltenyi Biotec GmbH, Bergisch Gladbach, Germany) at 4°C for 10
min. The cell pellets were washed three times in PBS, suspended in
500 µl PBS and then analysed using a FACSCalibur flow cytometer (BD
Biosciences, Franklin Lakes, NJ, USA). For polypeptides labelling
experiments, the MNNG/HOS/HOS or Cal27 cells were incubated in 100
µg/ml peptides in 100 µl PBS at 4°C for 30 min. Subsequent to
further washing with PBS, cell pellets were suspended in 500 µl PBS
and then analysed using a FACSCalibur flow cytometer.
ELISA binding assay
ELISA binding assay of nABP296 to CD105 protein was
performed, with CD106 protein (R&D Systems, Inc.) used as a
control. Recombinant human CD105 protein and CD106 protein at a
concentration of 100 µg/ml in sterile PBS were added to six
individual sterilised MaxiSorp plates and incubated overnight at
4°C. The plates were washed six times with TBST and blocked for 1 h
at 4°C using 1% BSA in PBS. Subsequently, nABP296 peptide was added
at a concentration of 100 µg/ml and incubated for 1 h at room
temperature. The unbound peptide was removed by PBS washing for 10
times. The absorbance was measured using a VICTOR X5 Multilabel
plate reader (PerkinElmer, Inc., Singapore) at 490 nm, and the
results are reported as the optical density.
Cytotoxicity assay
For the cytotoxicity assay, 1×105
cells/well were seeded in 96-well plates in 100 µl complete culture
medium. Subsequent to culturing overnight at 37°C in a 5%
CO2 atmosphere, the cells were incubated in different
concentrations of nABP296 solution ranging between 0.5 and 5 µMol.
At 24 and 48 h, 10 µl Cell Counting Kit-8 (CCK-8; Dojindo Molecular
Technologies, Inc., Kumamoto, Japan) solution was added to the
cells and incubated for another 1 h. The absorbance was then
determined at 450 nm using the VICTOR X5 Multilabel plate reader,
and the survival rate was calculated.
In vivo assay
Based on the aforementioned experiments, the nABP296
peptide was used in the in vivo experiment. The animal use
protocol was reviewed and approved by the Animal Ethical and
Welfare Committee of Sun Yat-sen University (Guangzhou, Guangdong,
China). Briefly, MNNG/HOS cells were subcutaneously transplanted
into 5-week-old BALB/c Nod mice at 1×107 cells/ml in DF.
When the volume of the xenograft tumours reached ~200
mm3, 1 mg/ml nABP296 in 100 µl sterile Milli-Q water was
intravenously administered to the mice. The mice were observed
using the IVIS Spectrum In Vivo Imaging system (PerkinElmer,
Akron, OH, USA), and then images were captured after 1 h of
administration. Next, the mice were euthanised, tumours were
excised from the MNNG/HOS tumour-bearing mice and images were
captured using the IVIS Spectrum In Vivo Imaging system.
Tumours and hearts were freeze-sectioned into 6 µm and fixed in
100% acetone for 15 min. Following further washing with PBS, the
nuclei of the sections were stained with DAPI. The results were
analysed by CLSM (Leica Microsystems GmbH).
Histological staining
All patients participating in this experiment
provided informed consent prior to tissue collection and agreed to
the use of their samples in scientific research. The described
experiments were approved by the Ethics Committee of The Hospital
of Stomatology at Sun Yat-sen University (approval no.
ERC-2017-11). A total of 2 patients (osteosarcoma patient: Male, 33
years old, hospitalised for 15 days in July 2017; tongue carcinoma
patient: Male, 25 years old, hospitalised for 16 days in October
2017) who were treated in Guanghua School of Stomatology (Hospital
of Stomatoloty, Sun Yat-sen University) and diagnosed with
osteosarcoma and tongue carcinoma by the Department of Pathology
were included in these experiments. Osteosarcoma and tongue
carcinoma tissues were obtained from the patients during
surgery.
Tissue slices derived from a xenograft tumour model,
an osteosarcoma patient and a tongue carcinoma patient were used
for histological staining. The tumour samples were sectioned (6-µm
thick) using a cryostat microtome (Microm HM560; Thermo Fisher
Scientific, Inc.). Non-specific binding sites were blocked by
incubation with 5% BSA for 1 h at room temperature. Next,
histological sections were incubated using a combination of 50 µMol
nABP296 peptide and mouse monoclonal CD105 antibody (cat. no.
MA1-19408; 1:200; Invitrogen; Thermo Fisher Scientific, Inc.) in
blocking buffer at 4°C overnight. Unbound peptide and antibody were
eluted by washing three times with PBS, following which cells were
further treated with donkey anti-mouse IgG (H+L) highly
cross-adsorbed Alexa Fluor 568 secondary antibody (cat. no. A10037;
1:1,000; Invitrogen; Thermo Fisher Scientific, Inc.) for 1 h at
room temperature. Following further washing with PBS, the nucleus
was stained with DAPI, and the results were analysed under CLSM
(Leica Microsystems GmbH).
Statistical analysis
Statistical analysis was performed using the
GraphPad Prism software (version 5; GraphPad Software, Inc., La
Jolla, CA, USA). One-way analysis of variance was used to evaluate
the survival rate of MNNG/HOS cells at 24 and 48 h. P-values of
<0.05 were considered to denote statistically significant
differences.
Results
Determining peptide binding to
CD105
The recovery rate of three rounds of biopanning is
shown in Table I. Subsequent to
three rounds of biopanning, the recovery rate of M13 phage was
raised from 4.9×10−6 to 4.5×10−5. M13 phage
binding to recombinant human CD105 protein was enriched. To
identify a monoclone phage that can bind to CD105, ELISA was
performed to M13 phage single clones (A-P). A total of 13
monoclonal phages were confirmed to have high affinity for CD105,
as shown in Table II. The
sequences and characteristics of the 13 positive monoclonal phages
are listed in Table III in the
range between nABP295 and nABP307.
 | Table I.Recovery rate of each round of
biopanning. |
Table I.
Recovery rate of each round of
biopanning.
| Round | Input, pfu | Output, pfu | Ratio
(output/input) |
|---|
| 1st |
1.8×1011 |
8.9×105 |
4.9×10−6 |
| 2nd |
1.56×1011 |
2.4×106 |
1.54×10−5 |
| 3rd |
1.0×1011 |
4.5×106 |
4.5×10−5 |
 | Table II.Binding affinity of M13 phage for
CD105 protein. |
Table II.
Binding affinity of M13 phage for
CD105 protein.
| M13 monoclonal
phages | Human recombinant
CD105 protein | NC |
|---|
| A | 0.481 | 0.371 |
| B | 0.642 | 0.574 |
| C | 0.651 | 0.593 |
| D | 0.603 | 0.507 |
| E | 0.572 | 0.611 |
| F | 0.541 | 0.512 |
| G | 0.388 | 0.326 |
| H | 0.355 | 0.330 |
| I | 0.632 | 0.521 |
| J | 0.395 | 0.293 |
| K | 0.429 | 0.336 |
| L | 0.517 | 0.461 |
| M | 0.427 | 0.376 |
| N | 0.521 | 0.456 |
| O | 0.569 | 0.460 |
| P | 0.436 | 0.335 |
 | Table III.Sequences of 13 clones obtained from
screening that exhibited a positive ELISA result, and their
characteristics. |
Table III.
Sequences of 13 clones obtained from
screening that exhibited a positive ELISA result, and their
characteristics.
| No. | Sequence | Molecular
weight | Net charge | Protein-binding
potential, kcal/mol |
|---|
| nABP295 | NWTTLSRSVNWP | 1,460.6 | +1 | 2.21 |
| nABP296 | WIYDTTRVIVPG | 1,419.6 | 0 | 0.64 |
| nABP297 | HAMSPVFLSKYA | 1,350.6 | +1 | −0.06 |
| nABP298 | LVPSILGATFIH | 1,267.5 | 0 | −1.56 |
| nABP299 | DISASLQSNRWH | 1,413.5 | 0 | 3.05 |
| nABP300 | AAGTFLMSMMSR | 1,302.6 | +1 | 0.39 |
| nABP301 | NNLPTSRTLAGN | 1,257.4 | +1 | 2.56 |
| nABP302 | GNNPLHVHHDKR | 1,423.6 | +1 | 3.87 |
| nABP303 | HHLRIPYALDQT | 1,463.7 | 0 | 2.05 |
| nABP304 | GTGAALAKVSEA | 1,074.2 | 0 | 0.02 |
| nABP305 | IKPVRALYTLAD | 1,359.6 | +1 | 0.78 |
| nABP306 | GTIRTSFWHTNT | 1,420.5 | +1 | 2.39 |
| nABP307 | GVHSVFAPLTPN | 1,238.4 | 0 | −0.12 |
Peptide labelling of the
CD105-positive MNNG/HOS cell line
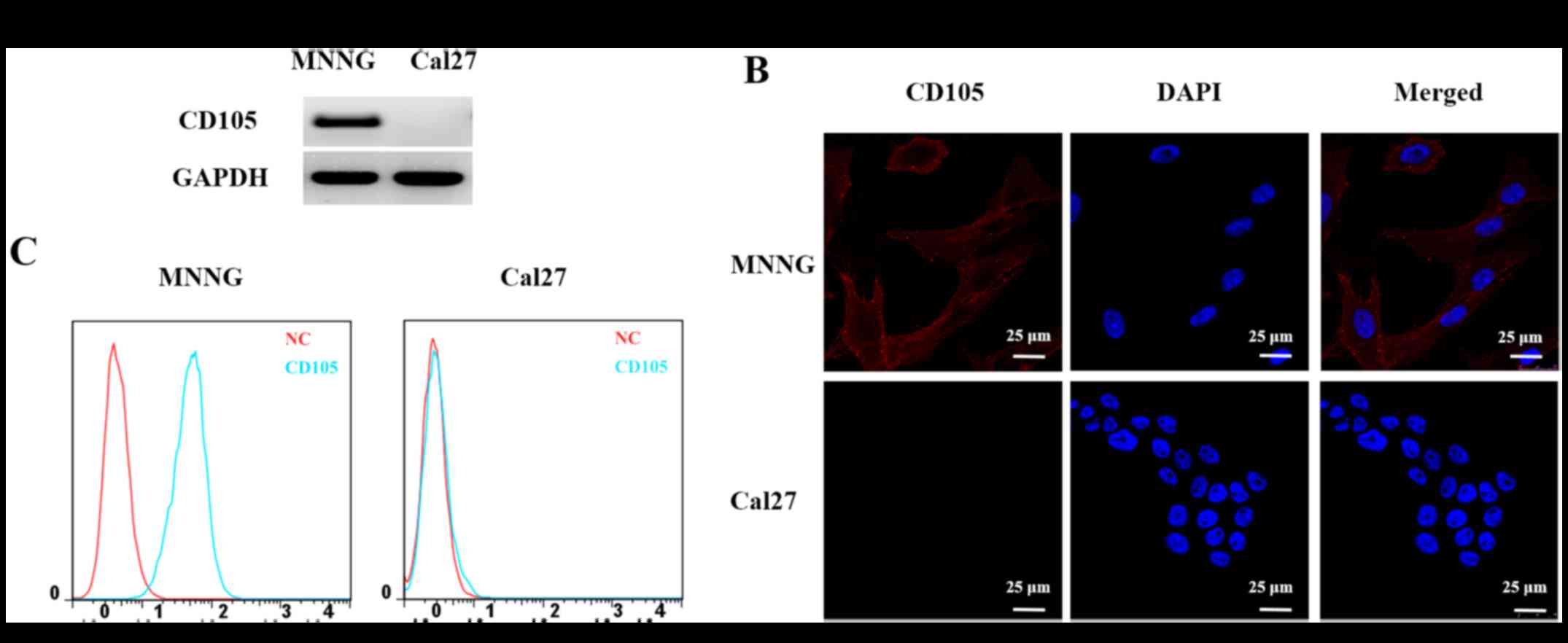
Based on the results of RT-PCR, fluorescence and
flow cytometry (Fig. 1), the
MNNG/HOS cell line was found to be CD105-positive and the Cal27
cell line was observed to be CD105-negative. These two cell lines
were then used in subsequent experiments to identify the peptide
with high binding affinity for CD105 expressed in the cell
membranes.
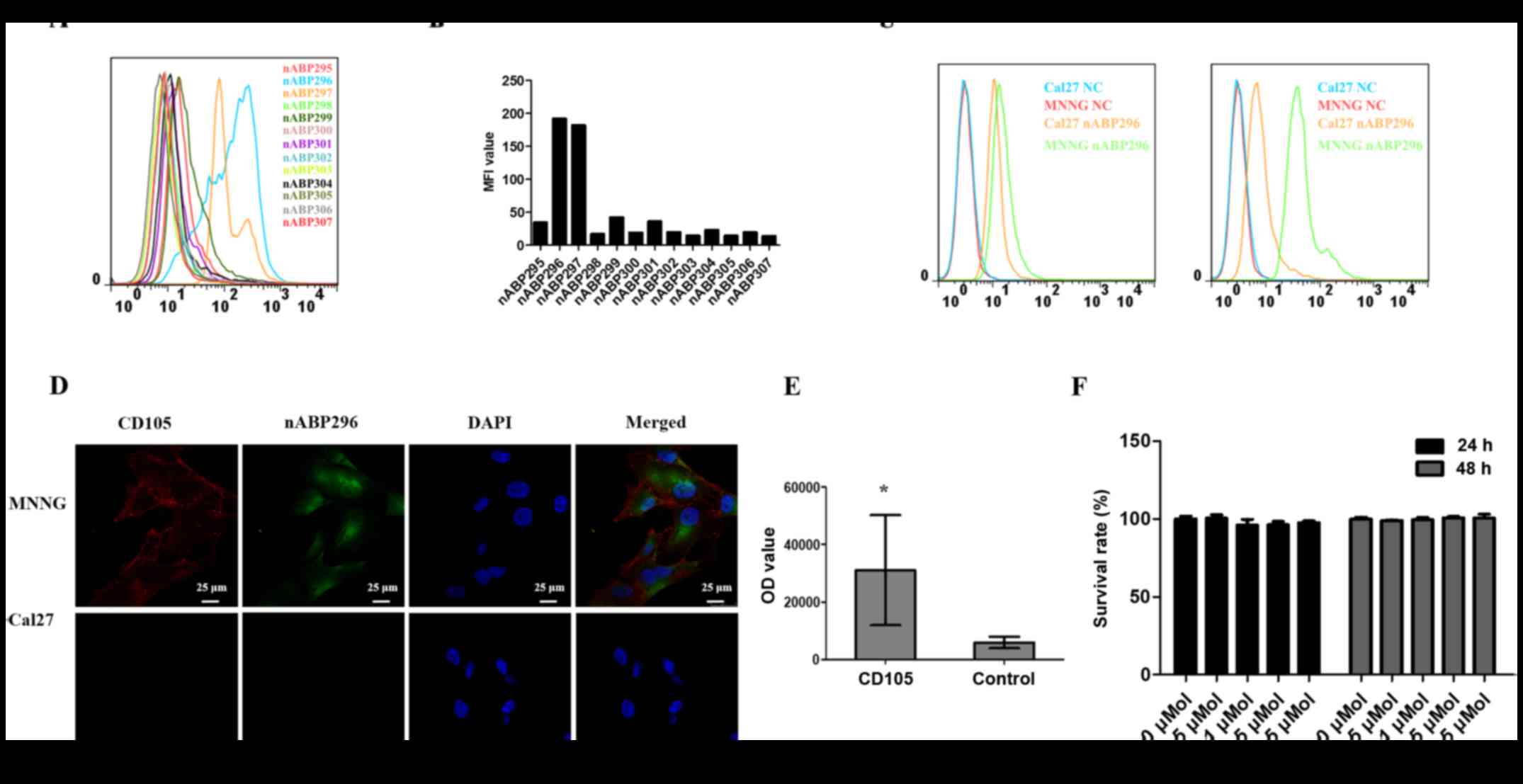
By flow cytometry, two peptides, namely nABP296 and
nABP297, with high affinity for the CD105-positive MNNG/HOS cell
line were identified (Fig. 2A and
B). To determine whether these two peptides had a specific
affinity for CD105, the binding of nABP296 and nABP297 to the
CD105-positive MNNG/HOS and CD105-negative Cal27 cell lines was
analysed by a flow cytometry assay. As shown in Fig. 2C, nABP296 exhibited higher binding
efficiency to MNNG/HOS than Cal27 cells, while nABP297 did not
exhibit different binding efficiencies between MNNG/HOS or Cal27
cells. According to the results of the immunofluorescence assay,
nABP296 co-localised with the CD105 antibody in the MNNG/HOS cell
line and had no binding affinity for Cal27 cells (Fig. 2D). Furthermore, the ELISA experiment
of nABP296 binding to CD105 and control proteins revealed that
nABP296 had a higher affinity for CD105 protein in comparison with
that for the control protein (Fig.
2E). Finally, nABP296 exhibited no cytotoxicity on the MNNG/HOS
cell line after 24 and 48 h (Fig.
2F). Taken together, nABP296 can selectively bind to the
CD105-positive MNNG/HOS cell line and is biocompatible with this
cell line, suggesting that nABP296 may be able to target human
osteosarcoma in vivo. The chemical structures of nABP296 and
the HPLC/mass spectrometry results are shown in Fig. 3A and B, respectively.
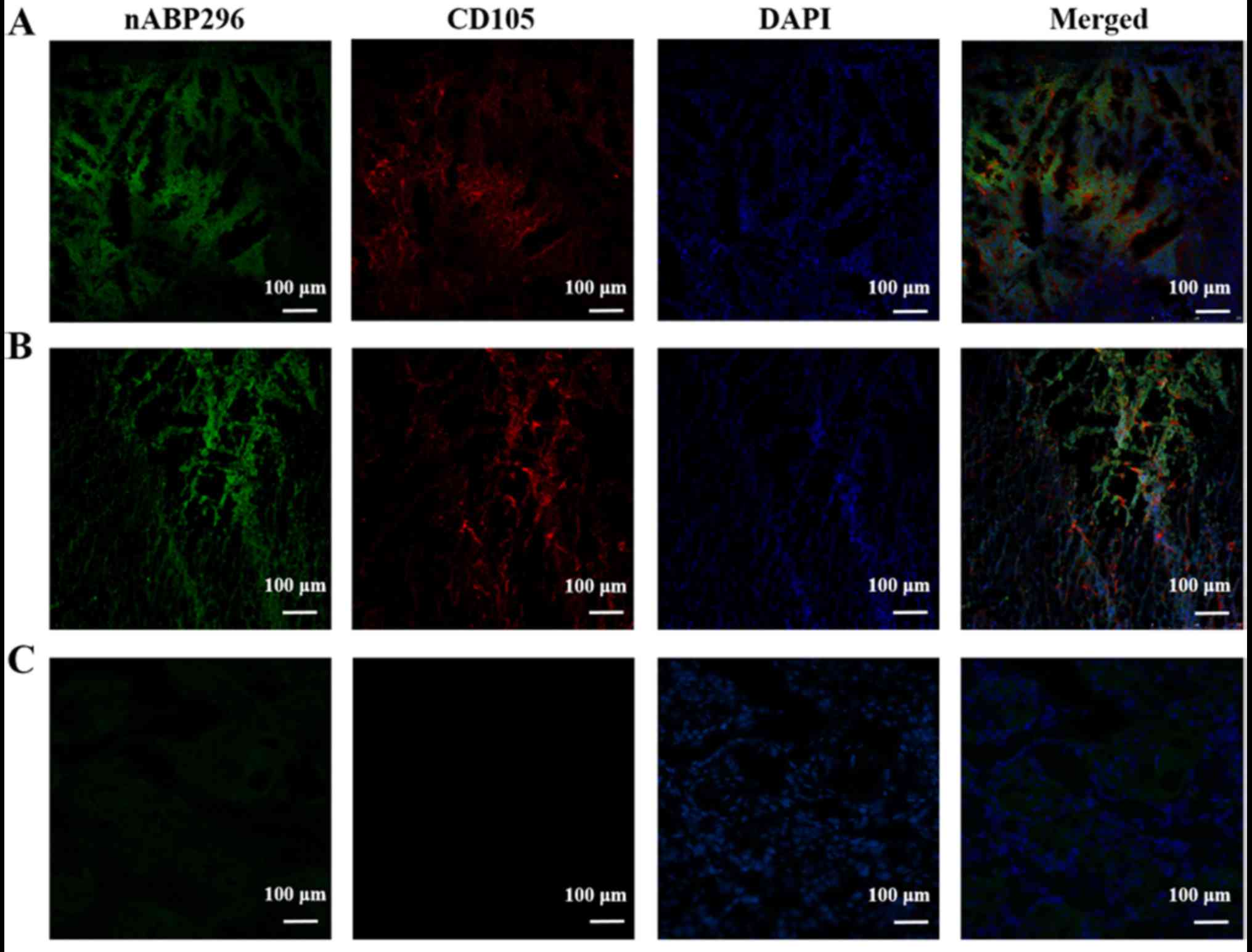
In vitro labelling of
osteosarcoma
Following incubation of osteosarcoma histological
sections derived from an MNNG/HOS xenograft tumour model and an
osteosarcoma patient with nABP296, green fluorescence indicated
nABP296 peptide and red flourescence indicated CD105 antibody.
Results showed that the MNNG/HOS xenograft tumour model and
osteosarcoma derived from the patient were successfully co-labelled
with the nABP296 peptide and CD105 antibody (Fig. 4A and B). By contrast, absence of
CD105 antibody and only weak nABP296 fluorescence were observed in
the tongue carcinoma section (Fig.
4C). The results indicated that nABP296 was able to bind to
osteosarcoma derived from a xenograft tumour model and a patient
in vitro. Thus, nABP296 may be used in the diagnosis of
osteosarcoma in place of CD105 in certain situations.
In vivo imaging of MNNG/HOS
tumour
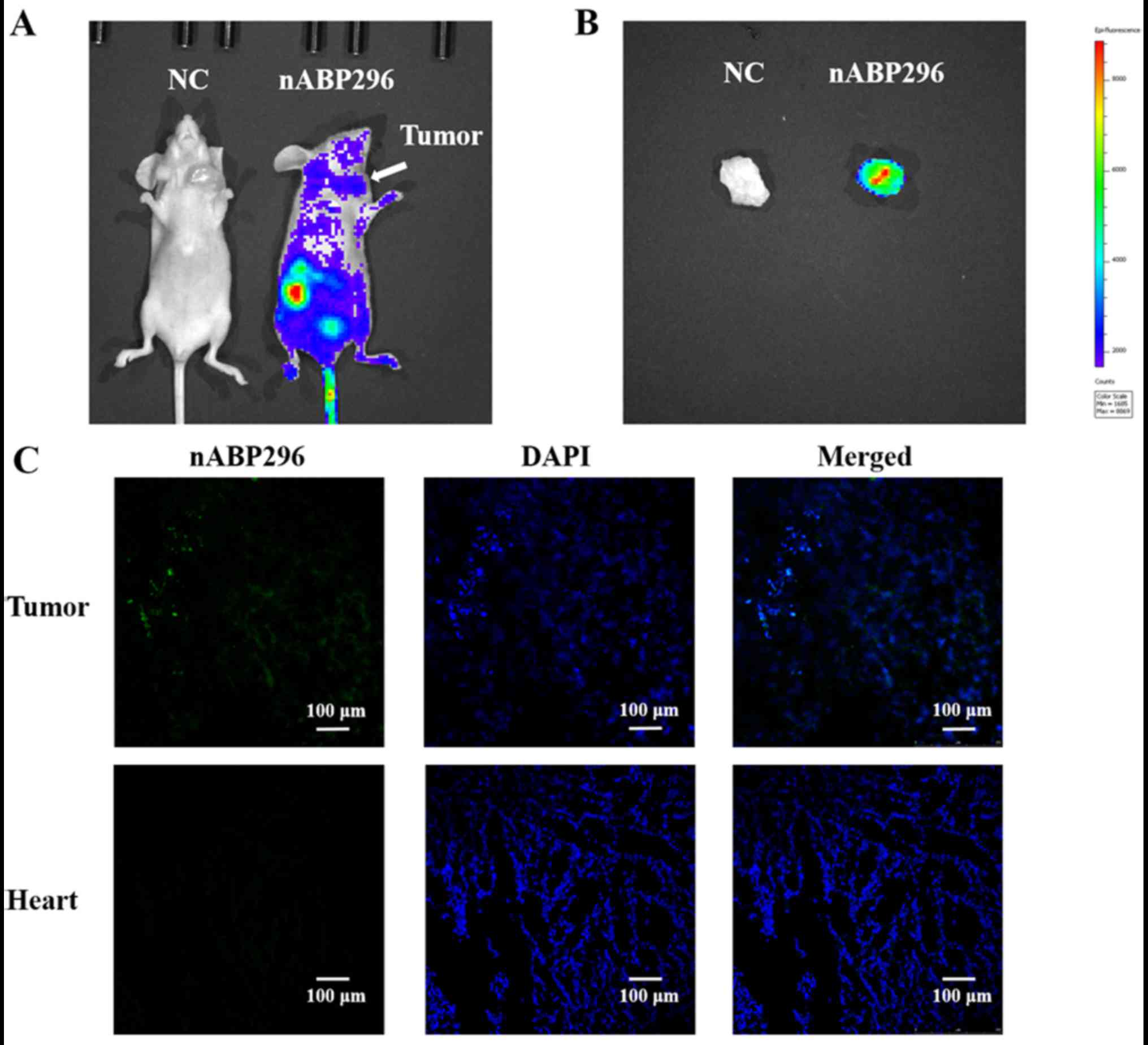
As shown in Fig. 5A,
at 1 h after intravascular administration of nABP296 to MNNG/HOS
tumour-bearing mice, fluorescence was detected in the MNNG/HOS
xenograft tumour. Fluorescence detected in the tumour was higher
compared with that in the surrounding tissues, whereas there was no
fluorescence detected in the NC group. Furthermore, ex vivo
imaging of tumours that were excised from the MNNG/HOS
tumour-bearing mice was performed at 1.5 h. It was observed that
FITC-labelled nABP296 was accumulated in the xenograft tumour
(Fig. 5B). Fluorescence was
detected in frozen tumour histological sections, while there was no
FITC fluorescence in the heart sections (Fig. 5C). These results supported the high
affinity and specificity for nABP296 peptide for the identified
MNNG/HOS xenograft tumour in vivo.
Discussion
In the present study, it was demonstrated that
nABP296, a 12 amino acid peptide, can specifically bind to
recombinant human CD105 protein (Table
II, and Fig. 2C-E). CD105 is a
biomarker of MSCs that is used for the isolation and enrichment of
these cells. The CD105 antibody has previously been used to isolate
CD105+ phenotype MSCs from human cardiac mesenchymal
stromal cells to enhance the function of the post-infarction heart
in mice (20). In addition, the
CD105 antibody is a reliable marker to distinguish the
polydirectional differentiation potential of MSCs (21,22).
The nABP296 peptide discovered in the present study is relatively
short and can specifically bind to the CD105-positive MNNG/HOS cell
line. nABP296 was not found to have a cytotoxic effect on MNNG/HOS
and Cal27 cells (Fig. 2E). Thus,
nABP296 has the characteristics of specific affinity and good
biocompatibility that can solve the problems of the usage of the
CD105 antibody in vivo, and has the potential to serve as a
novel probe to isolate MSCs from tissues.
Angiogenesis, the process of new blood vessel
formation in tissues, is necessary for tumour growth and
metastasis. CD105 serves a crucial role in angiogenesis (23) and is thus overexpressed in actively
proliferating tumours (24,25). In vivo targeting by CD105 can
be used for tumour positron-emission tomography imaging and
visualization. In a previous study, CD105 labelled with
64Cu was used in BXPC-3 ×enograft tumour imaging
(26). CD105 conjugated with IRDye
800CW was used to label 4T1 tumour-bearing mice, which were then
visualised by near-infrared fluorescence imaging (27). Peptides have been used for
osteosarcoma imaging in numerous studies. Ma et al (28) reported that several peptides derived
from a T7 phage display were used for in vivo specific
photoacoustic imaging of osteosarcomas. The contrast of
osteosarcoma images was enhanced by 170–230%. Other peptides,
including the hydroxyapatite binding peptide, were used to
characterise hydroxyapatite on human osteotropic tumour tissue
(29). OSP-1 derived from the
Ph.D.-12 Phage Display Peptide Library was able to specifically
bind to 143B osteosarcomas, other than 293T human embryonic kidney
cells. The binding site of OSP-1 may be associated with heparin
sulphate proteoglycans (30). In
the present study, it was reported that nABP296 was able to locate
osteosarcomas in MNNG/HOS tumour-bearing mice (Fig. 5). The nABP296 peptide is a
relatively short, human-made peptide that is synthesised in
vitro, and has no animal heterologous form or immune response.
nABP296 is nontoxic to cells, thus, it can be safely used in
humans. This peptide can be activated by fluorescence, and it is
therefore useful for the detection of tumour cells. Owning to these
advantages, the nABP296 peptide may be a new strategy for the early
diagnosis and detection of metastasis of osteosarcoma in
patients.
nABPs usually consist of <50 amino acids, and
their penetrating ability to the tumour microenvironment is
superior to that of antibodies. Owing to the potent penetrating
ability and targeting effect, peptides have been used as cargo in
antitumour therapy. Peptides favoured the cellular absorption of
Ara-C in Caco-2 cells to increase their internalization rate
(31). In a study by Ma et
al (32), an αvβ3-targeting
peptide conjugated with methotrexate was demonstrated to increase
the efficacy of methotrexate and reduce its side effects. In the
immunofluorescence assay conducted in the present study, nABP296
was detected in the cytoplasm, suggesting that nABP296 was able to
penetrate the cell membrane. Thus, nABP296 may serve as a CD105
protein target cargo to transfer antitumour drugs to CD105-positive
tumour cells or therapeutic agents to target cells.
Future research must focus on the targeting
properties of nABP296. In addition, the mechanism underlying the
penetrating effect of nABP296 remains unclear and needs to be
determined. Based on the aforementioned findings, nABP296 may serve
as a targeted therapeutic cargo conjugate with other therapeutic
agents.
In conclusion, in the present study, a novel
peptide, namely nABP296, was discovered by M13 phage biopanning,
and this peptide specifically binds to the recombinant human CD105
protein. nABP296 labels frozen osteosarcoma sections derived from
patients and an MNNG/HOS xenograft tumour model in vitro. In
addition, the MNNG/HOS xenograft tumour model was labelled by
nABP296 in vivo. Therefore, this peptide may be used for
diagnosis and osteosarcoma imaging in vivo, as well as for
MSC isolation and CD105 targeted therapy in the future.
Acknowledgements
Not applicable.
Funding
The authors acknowledge the financial support from
the National Natural Science Foundation of China (grant no.
31371390), the Doctoral Foundation of Ministry of Education of
China (grant no. 20130171110010), the Program of the State
High-Tech Development Project (grant no. 2014AA020702) and the
Program of Guangdong Science and Technology (grant nos.
2016B030231001 and 2017B020230002).
Availability of data and materials
The datasets used and/or analysed in the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
XL and HW conceived and designed the experiments. XL
and XH performed the experiments. JZ, HH and LZ analysed the data.
XL and HW wrote the manuscript. MY and YZ reviewed and polished the
paper.
Ethics approval and consent to
participate
The animal use protocol was reviewed and approved by
the Animal Ethical and Welfare Committee of Sun Yat-sen University.
The described patient experiments were approved by the Ethics
Committee of The Hospital of Stomatology at Sun Yat-sen University
(approval no. ERC-2017-11). All patients participating in this
study provided writtem informed consent prior to tissue collection
and agreed to the use of their samples in scientific research.
Patient consent for publication
All patients participating in this study provided
written informed consent.
Competing interests
The authors declare no conflict of interest. The
founding sponsors had no role in the design of the study; in the
collection, analyses, or interpretation of data; in the writing of
the manuscript, and in the decision to publish the results.
Glossary
Abbreviations
Abbreviations:
|
nABP
|
non-antibody-binding peptide
|
|
CCK-8
|
Cell Counting Kit-8
|
|
CLSM
|
confocal laser scanning microscopy
|
References
|
1
|
Li C, Hampson IN, Hampson L, Kumar P,
Bernabeu C and Kumar S: CD105 antagonizes the inhibitory signaling
of transforming growth factor beta1 on human vascular endothelial
cells. FASEB J. 14:55–64. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gougos A and Letarte M: Primary structure
of endoglin, an RGD-containing glycoprotein of human endothelial
cells. J Biol Chem. 265:8361–8364. 1990.PubMed/NCBI
|
|
3
|
Barbara NP, Wrana JL and Letarte M:
Endoglin is an accessory protein that interacts with the signaling
receptor complex of multiple members of the transforming growth
factor-beta superfamily. J Biol Chem. 274:584–594. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Li C, Guo B, Wilson PB, Stewart A, Byrne
G, Bundred N and Kumar S: Plasma levels of soluble CD105 correlate
with metastasis in patients with breast cancer. Int J Cancer.
89:122–126. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Duff SE, Li C, Garland JM and Kumar S:
CD105 is important for angiogenesis: Evidence and potential
applications. FASEB J. 17:984–992. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Marioni G, D'Alessandro E, Giacomelli L
and Staffieri A: CD105 is a marker of tumour vasculature and a
potential target for the treatment of head and neck squamous cell
carcinoma. J Oral Pathol Med. 39:361–367. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Minhajat R, Mori D, Yamasaki F, Sugita Y,
Satoh T and Tokunaga O: Organ-specific endoglin (CD105) expression
in the angiogenesis of human cancers. Pathol Int. 56:717–723. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Muniz C, Teodosio C, Mayado A, Amaral AT,
Matarraz S, Barcena P, Sanchez ML, Alvarez-Twose I, Diez-Campelo M,
Garcia-Montero AC, et al: Ex vivo identification and
characterization of a population of CD13high
CD105+ CD45− mesenchymal stem cells in human
bone marrow. Stem Cell Res Ther. 6:1692015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Odabas S, Sayar F, Guven G,
Yanikkaya-Demirel G and Piskin E: Separation of mesenchymal stem
cells with magnetic nanosorbents carrying CD105 and CD73 antibodies
in flow-through and batch systems. J Chromatogr B Analyt Technol
Biomed Life Sci. 861:74–80. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cavar S, Jelasic D, Seiwerth S, Milosevic
M, Hutinec Z and Misic M: Endoglin (CD 105) as a potential
prognostic factor in neuroblastoma. Pediatr Blood Cancer.
62:770–775. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhou Y, Gu H, Xu Y, Li F, Kuang S, Wang Z,
Zhou X, Ma H, Li P, Zheng Y, et al: Targeted antiangiogenesis gene
therapy using targeted cationic microbubbles conjugated with CD105
antibody compared with untargeted cationic and neutral
microbubbles. Theranostics. 5:399–417. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Damron TA, Ward WG and Stewart A:
Osteosarcoma, chondrosarcoma, and Ewing's sarcoma: National cancer
data base report. Clin Orthop Relat Res. 459:40–47. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Savage SA and Mirabello L: Using
epidemiology and genomics to understand osteosarcoma etiology.
Sarcoma. 2011:5481512011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Allison DC, Carney SC, Ahlmann ER,
Hendifar A, Chawla S, Fedenko A, Angeles C and Menendez LR: A
meta-analysis of osteosarcoma outcomes in the modern medical era.
Sarcoma. 2012:7048722012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gibbs CP, Kukekov VG, Reith JD,
Tchigrinova O, Suslov ON, Scott EW, Ghivizzani SC, Ignatova TN and
Steindler DA: Stem-like cells in bone sarcomas: Implications for
tumorigenesis. Neoplasia. 7:967–976. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Reubi JC: Peptide receptors as molecular
targets for cancer diagnosis and therapy. Endocr Rev. 24:389–427.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Xiang Z, Yang X, Xu J, Lai W, Wang Z, Hu
Z, Tian J, Geng L and Fang Q: Tumor detection using magnetosome
nanoparticles functionalized with a newly screened EGFR/HER2
targeting peptide. Biomaterials. 115:53–64. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Alexander-Bryant AA, Zhang H, Attaway CC,
Pugh W, Eggart L, Sansevere RM, Andino LM, Dinh L, Cantini LP and
Jakymiw A: Dual peptide-mediated targeted delivery of bioactive
siRNAs to oral cancer cells in vivo. Oral Oncol. 72:123–131. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ma Y, Zhao S, Shen S, Fang S, Ye Z, Shi Z
and Hong A: A novel recombinant slow-release TNF α-derived peptide
effectively inhibits tumor growth and angiogensis. Sci Rep.
5:135952015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Czapla J, Matuszczak S, Wisniewska E,
Jarosz-Biej M, Smolarczyk R, Cichon T, Glowala-Kosinska M, Sliwka
J, Garbacz M, Szczypior M, et al: Human cardiac mesenchymal stromal
cells with CD105+CD34− phenotype enhance the
function of post-infarction heart in mice. PLoS One.
11:e1587452016. View Article : Google Scholar
|
|
21
|
Yamamoto M, Nakata H, Hao J, Chou J,
Kasugai S and Kuroda S: Osteogenic potential of mouse
adipose-derived stem cells sorted for CD90 and CD105 in vitro. Stem
Cells Int. 2014:5763582014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Fan W, Li J, Wang Y, Pan J, Li S, Zhu L,
Guo C and Yan Z: CD105 promotes chondrogenesis of synovium-derived
mesenchymal stem cells through Smad2 signaling. Biochem Biophys Res
Commun. 474:338–344. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li DY, Sorensen LK, Brooke BS, Urness LD,
Davis EC, Taylor DG, Boak BB and Wendel DP: Defective angiogenesis
in mice lacking endoglin. Science. 284:1534–1537. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Takase Y, Kai K, Masuda M, Akashi M and
Tokunaga O: Endoglin (CD105) expression and angiogenesis status in
small cell lung cancer. Pathol Res Pract. 206:725–730. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Miyata Y, Mitsunari K, Asai A, Takehara K,
Mochizuki Y and Sakai H: Pathological significance and prognostic
role of microvessel density, evaluated using CD31, CD34, and CD105
in prostate cancer patients after radical prostatectomy with
neoadjuvant therapy. Prostate. 75:84–91. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Luo H, England CG, Shi S, Graves SA,
Hernandez R, Liu B, Theuer CP, Wong HC, Nickles RJ and Cai W: Dual
targeting of tissue factor and CD105 for preclinical PET imaging of
pancreatic cancer. Clin Cancer Res. 22:3821–3830. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang Y, Hong H, Severin GW, Engle JW,
Yang Y, Goel S, Nathanson AJ, Liu G, Nickles RJ, Leigh BR, et al:
ImmunoPET and near-infrared fluorescence imaging of CD105
expression using a monoclonal antibody dual-labeled with
89Zr and IRDye 800CW. Am J Transl Res. 4:333–346.
2012.PubMed/NCBI
|
|
28
|
Ma Z, Qin H, Chen H, Yang H, Xu J, Yang S,
Hu J and Xing D: Phage display-derived oligopeptide-functionalized
probes for in vivo specific photoacoustic imaging of osteosarcoma.
Nanomedicine. 13:111–121. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lee JS and Tung CH: Osteotropic cancer
diagnosis by an osteocalcin inspired molecular imaging mimetic.
Biochim Biophys Acta. 1830:4621–4627. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sun X, Niu G, Yan Y, Yang M, Chen K, Ma Y,
Chan N, Shen B and Chen X: Phage display-derived peptides for
osteosarcoma imaging. Clin Cancer Res. 16:4268–4277. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Cheon EP, Hong JH and Han HK: Enhanced
cellular uptake of Ara-C via a peptidomimetic prodrug,
L-valyl-ara-C in Caco-2 cells. J Pharm Pharmacol. 58:927–932. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ma P, Yu H, Zhang X, Mu H, Chu Y, Ni L,
Xing P, Wang Y and Sun K: Increased active tumor targeting by an
αvβ3-targeting and cell-penetrating bifunctional peptide-mediated
dendrimer-based conjugate. Pharm Res. 34:121–135. 2017. View Article : Google Scholar : PubMed/NCBI
|