Introduction
Gastric cancer is a common malignancy of the
digestive tract that is highly prevalent in Asia, particularly in
China and Japan. Indeed, the incidence of gastric cancer is ranked
4th and 2nd in malignant cancer rankings in the world and in China,
respectively, and worldwide, its mortality rate remains very high
(1). Although the advancement in
endoscopy has significantly improved the diagnosis and prognosis of
early stage gastric cancer patients, at the time of definitive
diagnosis, most patients already present with lymph node and
distant metastases (2). Since tumor
invasion and metastasis are critical factors that affect patient
prognosis, understanding the mechanisms of tumor invasion and
metastasis are of utmost importance.
Previous studies have shown that a key process,
known as epithelial-mesenchymal transition (EMT), occurs during
tumor invasion and metastasis. EMT refers to the loss of cell
polarity, intercellular adhesion and junctions, increased migratory
and invasive capacities in tumor epithelial cells (3). EMT is a complex process that is
characterized by increased fibrous morphology and invasiveness of
cells, reduced cell apoptosis and an increase in extracellular
matrix content (4). In previous
studies, EMT-induced primary tumor metastases have been reported to
cause poor prognosis in various cancer (5–7).
Therefore, EMT is an important mechanism underlying tumor invasion
and metastasis, that in recent years has become an area of
intensive research.
MicroRNAs (miRNAs) are non-coding RNAs that are
approximately 20–25 nucleotides in length. miRNAs are involved in
several biological processes, including development, cell
proliferation, cell differentiation and cell apoptosis. In
addition, miRNAs are associated with the development and
progression of various cancers (8).
The biological effects of miRNAs are mediated through the binding
of miRNAs to the 3′ UTR of target mRNAs via the seed regions (the
2nd to 7th base at the 5′ end) and through Argonaute (Ago)
protein-dependent degradation of mRNAs or blocking of mRNA
translation (9). Zhang et al
demonstrated that miR-181a is highly expressed in gastric cancer,
and was found to promote gastric cancer cell proliferation,
invasion and migration by binding to the 3′ UTR of the
anti-oncogene KLF6, thereby inhibiting KLF6 expression (10). Moreover, Chen et al showed
that miR-379-5p is expressed at a low level in liver cancer tissues
and cells, and inhibited liver cancer cell migration, invasion and
metastasis both in vitro and in vivo. Additional
studies have demonstrated that miR-379-5p binds to the 3′ UTR of
focal adhesion kinase (FAK) to inhibit FAK expression (11). These findings have demonstrated that
miRNAs play a critical role in tumor development, progression and
metastasis. The recent development of miRNA microarray analysis of
gastric cancer has facilitated the identification of gastric cancer
development-, progression- and prognosis-related miRNAs, and
includes members of the miR-200 family, miR-27, miR-373, miR-148,
miR-129 and miR-711 (12–17). However, knowledge concerning these
miRNAs only represent the tip of the iceberg, as the function of
many miRNAs is still unknown.
In our previous study, we used miRNA microarray and
identified miR-711 as an miRNA with low expression in gastric
cancer. In addition, we confirmed that the expression of miR-711 in
gastric cancer tissues was also low. Further investigation
demonstrated that miR-711 inhibited gastric cancer cell invasion
and migration, however the mechanism was unclear (17). In the present study, we demonstrated
that miR-711 regulated EMT of gastric cancer cells by
downregulating CD44 expression both in vivo and in
vitro.
Materials and methods
Cell culture
Human gastric cancer cell lines MGC-803 and SGC-7901
provided by the Cancer Research Institute of the University of
South China (Hengyang, China) were cultured in RPMI-1640 (Gibco;
Thermo Fisher Scientific, Scoresby, VIC, Australia) culture medium,
containing 10% fetal calf serum (FCS) in a 5% CO2
constant-temperature incubator (Thermo Fisher Scientific, Inc.,
Waltham, MA, USA).
Transfection
SGC-7901 and MGC-803 human gastric cancer cells were
seeded and cultured in 6-well plates for ~24 h. Once the cells
reached 40–60% confluency, they were transfected with miR-711
mimics (Shanghai GenePharma, Co., Ltd., Shanghai, China), miR-711
inhibitor (Shanghai GenePharma, Co., Ltd.), or CD44 inhibitor
(Shanghai GenePharma, Co., Ltd.) using Lipofectamine 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's instructions. Empty plasmid-transfected and
non-transfected (blank) cells were used as controls. The 6-well
plates were placed into the incubator, and after 6 h, the medium in
each well was replaced with fresh 10% FCS-containing RPMI-1640
medium. The transfection efficiency was calculated at 48 h
post-transfection, and cells with >60% transfection efficiency
were used for subsequent experiments. The transfection efficiency =
number of cells containing green fluorescence/a randomly observed
visual field consisting of 100 cells using an optical microscope
(Fig. 1).
Transwell invasion assay
After coating of the Transwell chamber with
Matrigel, 500 µl of 10% FCS-containing RPMI-1640 medium was added
into the lower chamber, and 100 µl of serum-free RPMI-1640 medium
was added to the upper chamber. At 48 h post-transfection, the
cells were digested with trypsin, prepared into a single-cell
suspension, seeded at a density of 1×105 cells/100 µl
into the upper chamber and cultured for 24 h. Cells were fixed with
4% formaldehyde, stained with crystal violet and observed under a
light microscope (Olympus Corp., Tokyo, Japan) at a magnification
of ×100. Ten fields were randomly chosen and the number of
transmigrated cells in each field was calculated by summing the
Transwell cells. The assay was performed in triplicate and the mean
± SD of the number of transmigrated cells was calculated.
Scratch wound healing assay
At 48 h post-transfection, cells were digested,
prepared into single-cell suspensions and seeded into 6-well
plates. Once the cells reached 80% confluency, a horizontal line
was scratched across each well using a pipette, and the wells were
washed twice in phosphate-buffered saline (PBS). Cells were
photographed at 0 and 48 h post-scratch using a light microscope
(Olympus Corp.) to evaluate healing at the scratch site.
Dual-Luciferase reporter assay
Wild-type and mutated CD44 3′ UTR vectors (Shanghai
GeneChem, Co., Ltd., Shanghai China) were transfected into gastric
cancer cells SGC-7901 that were transfected with miR-711 mimics or
miR-711 NC (empty-transfected and used as a control) using the
Dual-Glo™ Luciferase Assay System kit protocol (Promega, Madison,
WI, USA). Firefly luminescence (M1) and Renilla luminescence
(M2) were measured using a fluorescence detector of light signal by
fluorescence microscope (Olympus Corp.).
Hematoxylin and eosin staining
(H&E)
Xenograft tumors were harvested and
paraffin-embedded. Sections were cut and stained with hematoxylin
and eosin (H&E). Cell nuclei were stained blue-purple by
hematoxylin and the cytoplasm was stained pink with eosin. Tumor
cell morphology and tumor cell number were determined at a
magnification of ×100 using a light microscope (Olympus Corp.).
Western blot analysis
Proteins were extracted from SGC-7901 and MGC-803
gastric cancer cells and mouse xenografts using methanesulfonyl
fluoride (PMSF) (Beyotime Institute of Biotechnology,
Haimen, China), and the protein concentration was determined using
the bicinchoninic acid (BCA) assay (Beyotime Institute of
Biotechnology). Target protein bands were cut from the 5%
polyacrylamide gels and immersed in transfer buffer. Polyvinylidene
fluoride (PVDF) membranes were soaked in methanol for 3–5 min and
transferred to transfer buffer. Proteins (40 µg/µl) were
transferred to PVDF membranes and membranes were incubated in
blocking buffer for 2 h at room temperature on a rocker at low
speed. Membranes were initially incubated for 1 h at room
temperature with anti-rabbit CD44 (bs-0521R), anti-rabbit
E-cadherin (3195P) and anti-rabbit vimentin antibodies (5741P) at
1:500 dilution (Cell Signaling Technology, Danvers, MA, USA) on a
shaker, and then placed overnight at 4°C. Next, PVDF membranes were
washed and then incubated with goat anti-rabbit IgG secondary
antibody (11040914) at 1:1,000 dilution (Cell Signaling Technology)
for 1–2 h at room temperature on a shaker. Proteins were visualized
using a gel imaging processing and analysis system (Uvitec Ltd.,
Cambridge, UK).
Real-time quantitative polymerase
chain reaction (real-time PCR)
RNA was extracted according to the RNA kit
instructions (Takara Bio, Inc., Otsu, Japan) and synthesized into
cDNA using a Reverse Transcription kit (Takara Bio). Next, cDNA was
amplified using a real-time PCR kit (Takara Bio) in a reaction
mixture containing 10 µl SYBR-Green qPCR Mix, 2 µl miR-711 primers
(Applied Biosystems; Life Technologies, Foster City, CA, USA), 1 µl
cDNA and 7 µl dH2O. Real-time PCR was performed at 95°C
for 3 min, followed by 95°C for 10 sec, and 58°C for 30 sec for a
total of 40 cycles.
Tumor xenografts in nude mice
The study was approved by The First Affiliated
Hospital of the University of South China Institutional Ethics
Committee (no. 201708). The 10 male Balb/c nude mice (4 weeks old)
provided by Beijing Vital River Laboratory Animal Technology Co.,
Ltd. (Beijing, China (weight, 15.5±0.25 g), were bred and housed in
the same temperature and humidity-controlled room on a 12-h
light/dark cycle in laminar air flow room (LAFR). The mice were
provided with autoclaved tap water and autoclaved standard
laboratory chow ad libitum. The gastric cancer SGC-7901
cells were cultured and then the lentivirus carrying miR-711 mimics
or miR-711 NC were transfected into gastric cancer SGC-7901 cells.
SGC-7901 cells with stable miR-711 mimics, or miR-711 NC expression
were cultured for subsequent use. Harvested SGC-7901 cells
(1×107) were subcutaneously implanted into the bilateral
axilla of the mice. After 1 week, tumor volumes were measured every
7 days and the corresponding volumes were calculated by multiplying
the length by the width. The mice were sacrificed after 4 weeks by
decapitation as dictated by the ethical guidelines, and tumor
tissues were used for further research.
Measurement of xenograft tumor
volume
The length (a) and width (b) of the tumors were
measured weekly, and tumor volume (V) (V = ab2/2) was
calculated to create a growth curve of the xenograft tumors in the
nude mice. Four weeks after tumor inoculation, the mice were
euthanized, solid tumors were collected, and stored at −80°C for
subsequent use.
Cell morphology and cell number were determined by
H&E staining. The expression of miR-711 was measured by RT-qPCR
and CD44, E-cadherin and vimentin protein expression were examined
by western blot analysis and immunohistochemistry.
Statistical analysis
All statistical analyses were performed using SPSS
18.0 statistical software (SPSS, Inc., Chicago, IL, USA). In this
study, means and standard deviations were used for continuous
variables. One-way ANOVA followed by Bonferroni was performed to
test multiple variables between experimental groups, normal control
groups and blank groups in Figs.
2–4. Student's t-test was
performed in Figs. 5, and 7D and F. Independent samples
non-parametric test was performed in xenograft tumor volume for
non-normal variables. P<0.05 was considered to indicate a
statistically significant result.
Results
miR-711 inhibits gastric cancer cell
invasion
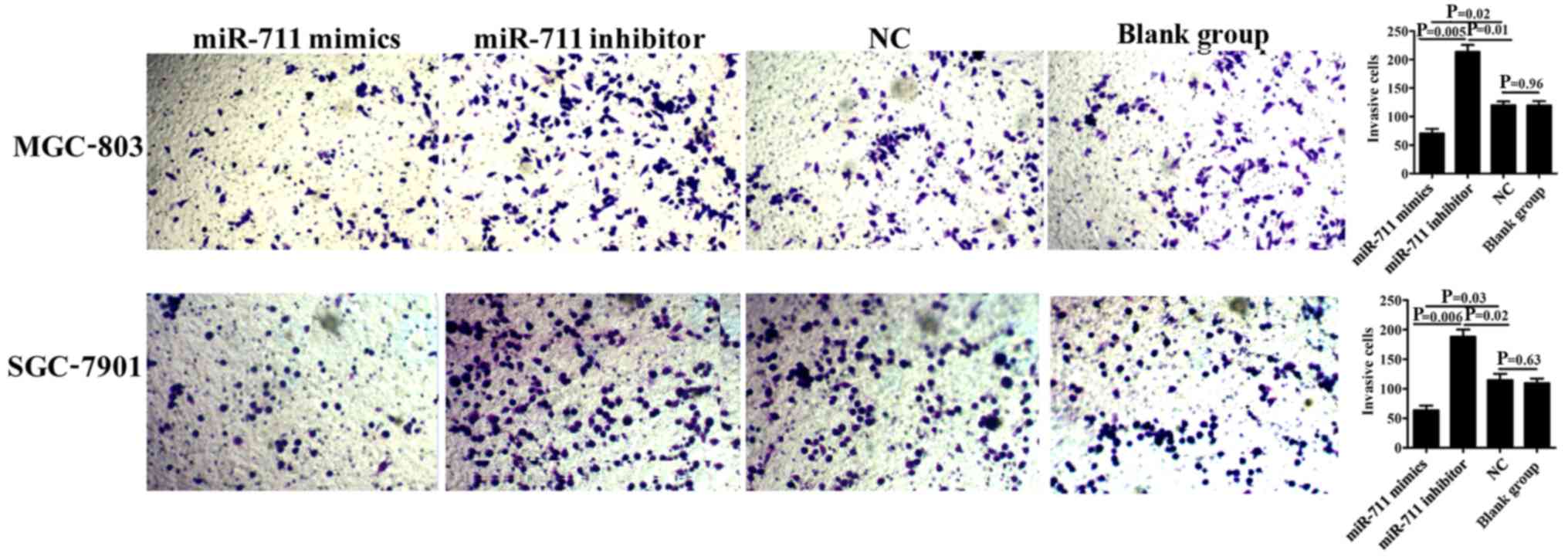
The Transwell invasion assay showed that the number
of transmigrated MGC-803 and SGC-7901 cells was significantly lower
in the miR-711 mimics group compared to that noted in the miR-711
inhibitor (P=0.005 and P=0.006, respectively) and NC groups (P=0.02
and P=0.03, respectively), demonstrating that exogenous
overexpression of miR-711 in SGC-7901 or MGC-803 gastric cancer
cells diminished the invasiveness of these cells. Moreover,
inhibition of miR-711 expression in MGC-803 or SGC-7901 cells
significantly increased the number of transmigrated cells compared
to the NC, indicating that miR-711 inhibition enhanced the
invasiveness of these cells (P=0.01 and P=0.02, respectively).
There was no difference between the NC and blank group (P=0.96 and
P=0.63) (Fig. 2).
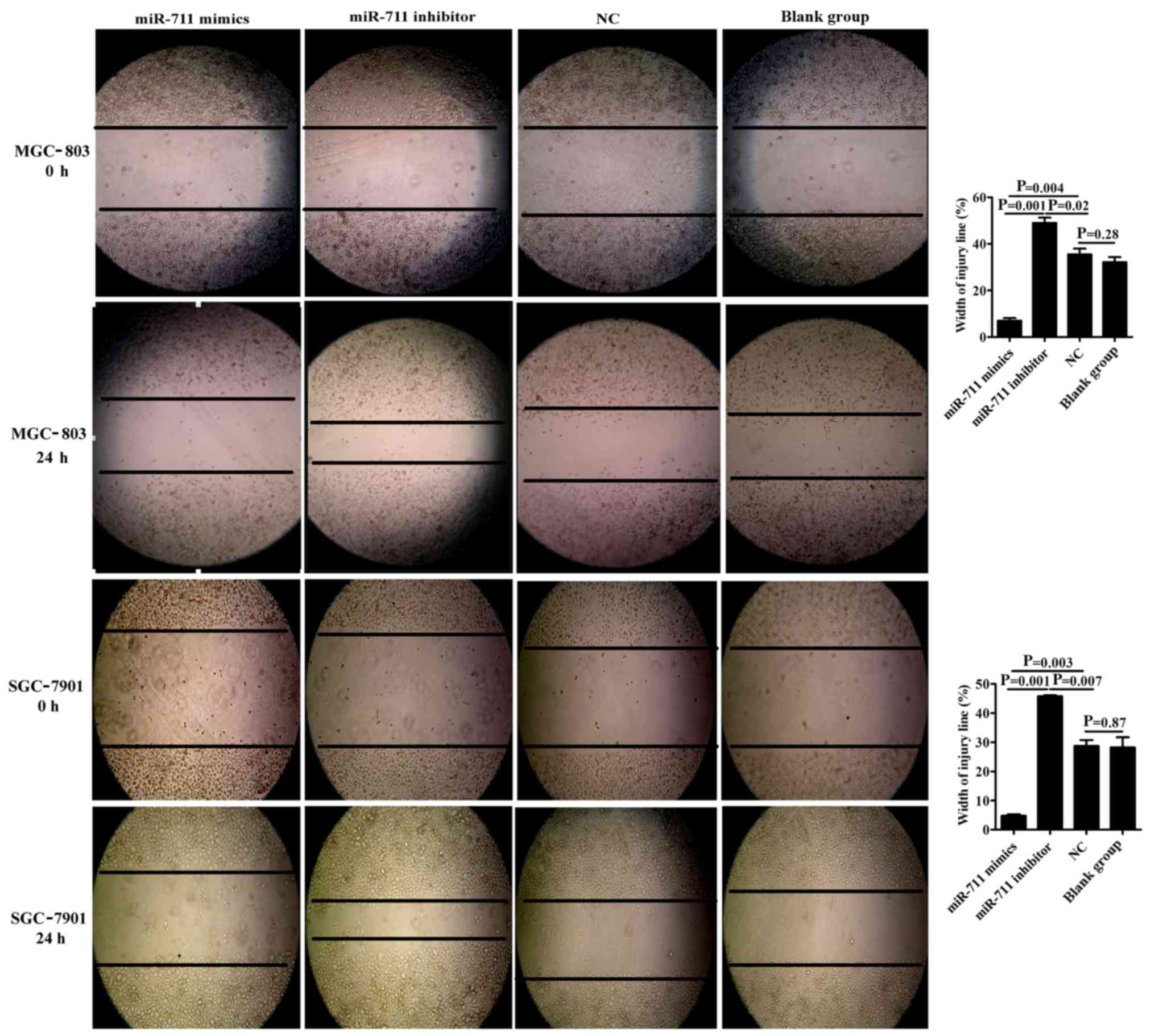
miR-711 inhibits gastric cancer cell
migration
The scratch wound healing assay revealed that cancer
cell migration distance and ‘scratch’ healing abilities were
significantly reduced in the miR-711 mimics group (7.08±0.73) when
compared to the miR-711 NC (32.52±1.73) and blank groups
(32.15±1.55). These findings indicated that exogenous
overexpression of miR-711 significantly inhibited the migratory
ability of MGC-803 or SGC-7901 cells (P=0.004, P=0.001, P=0.003 and
P=0.001, respectively). In addition, inhibition of miR-711
expression enhanced the migration of MGC-803 or SGC-7901 cells, and
the ‘scratch’ healing abilities were significantly higher in the
miR-711 inhibitor group (49.03±1.59) compared to that noted in the
NC group (32.52±1.73) (P=0.02 and P=0.007). The NC group was not
significantly different comparing with the blank group (P=0.28 and
P=0.87) (Fig. 3).
miR-711 regulates CD44, E-cadherin and
vimentin expression
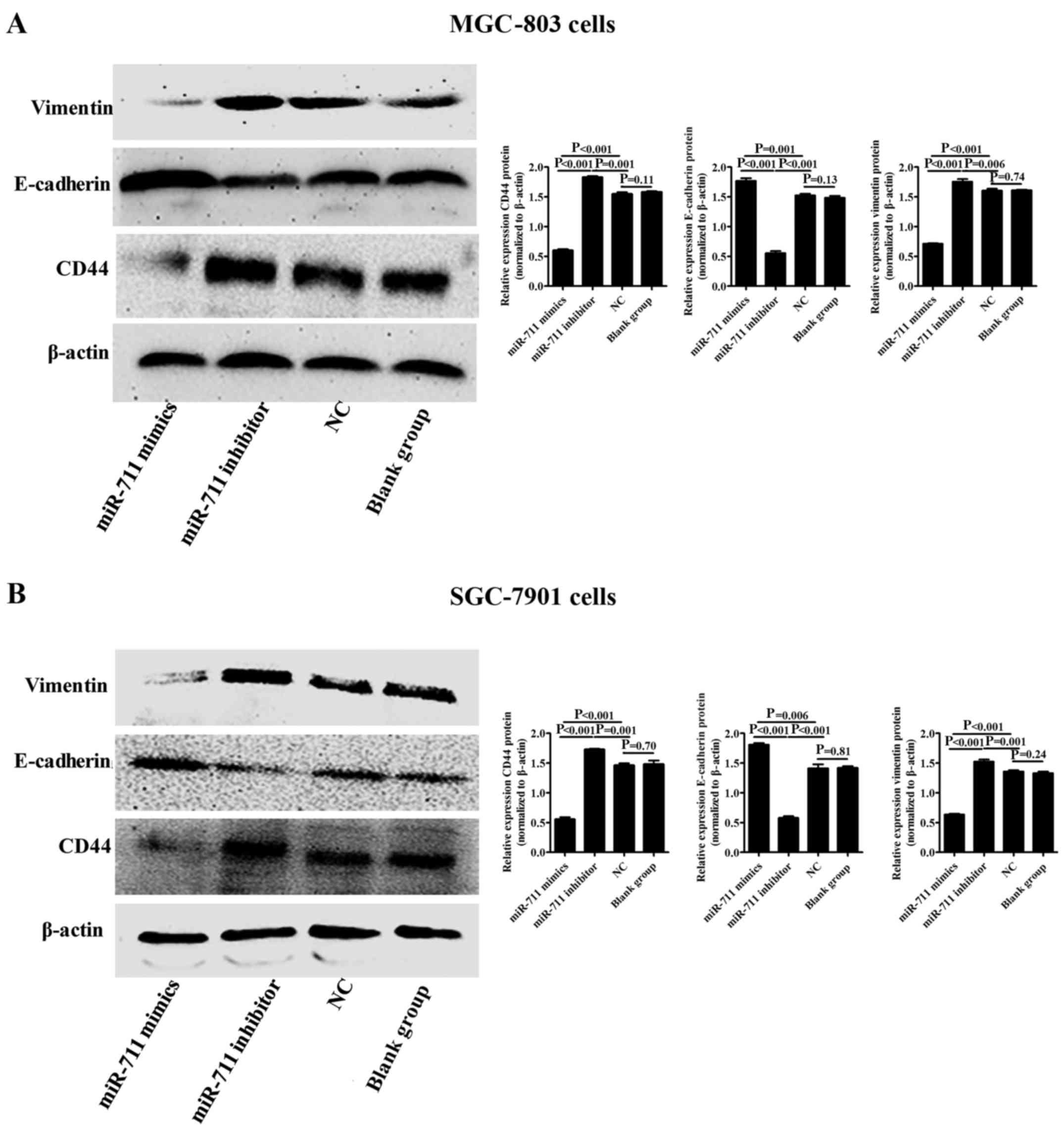
The expression of CD44, E-cadherin and vimentin in
MGC-803 or SGC-7901 cells with exogenous miR-711 overexpression or
inhibition was evaluated using western blot analysis. Compared with
the miR-711 NC and blank groups, CD44 and vimentin protein
expression was downregulated, whereas E-cadherin protein expression
was upregulated in the MGC-803 or SGC-7901 cells with miR-711
overexpression (P<0.001 and P<0.001) (Fig. 4A and B). However, CD44 and vimentin
protein expression were upregulated, and E-cadherin protein
expression was downregulated when miR-711 was inhibited in the
MGC-803 or SGC-7901 cells (P<0.001 and P<0.001) (Fig. 4A and B).
CD44 regulates E-cadherin and vimentin
expression
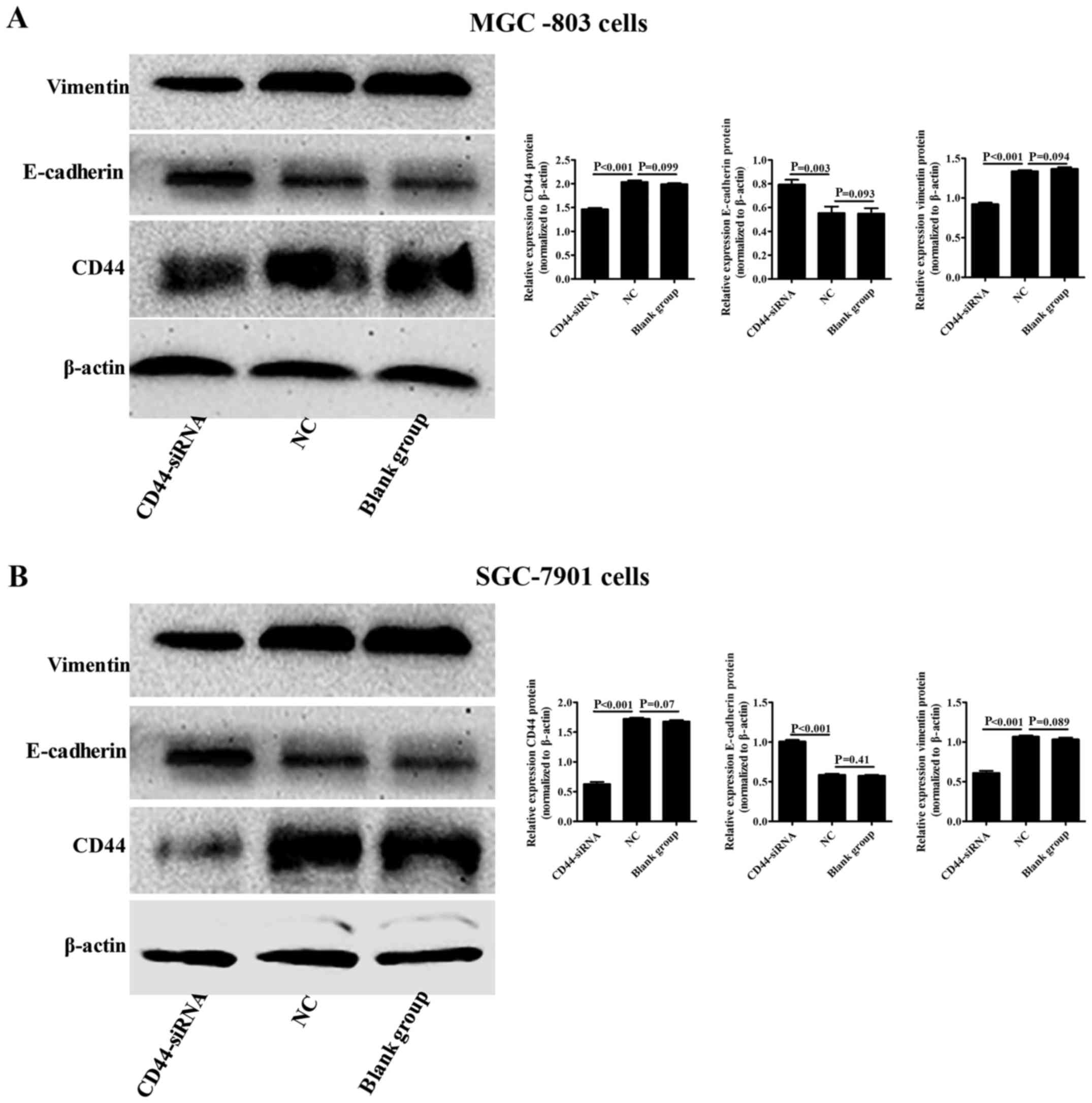
Since CD44 has been shown to be highly expressed in
gastric cancer tissues and cells, we blocked CD44 expression in
MGC-803 or SGC-7901 cells and evaluated E-cadherin and vimentin
expression in these cell types using western blot analysis. We
found that inhibition of CD44 expression in MGC-803 or SGC-7901
cells significantly downregulated vimentin protein expression
(P<0.001 and P<0.001) and upregulated E-cadherin protein
expression (P=0.003 and P<0.001) when compared to the NC group
(Fig. 5A and B).
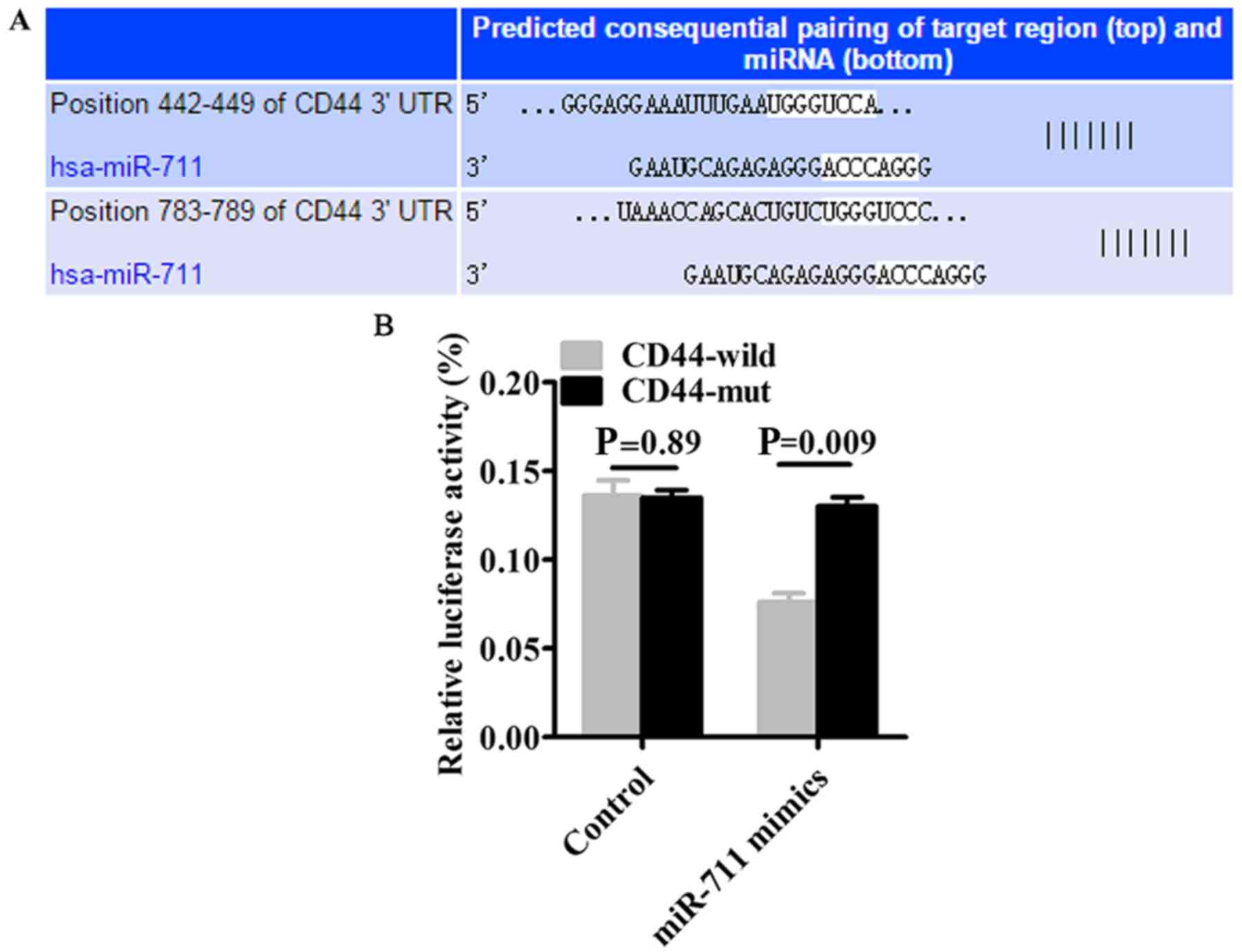
miR-711 specifically binds to
CD44
Based on the TargetScan prediction that miR-711
specifically binds to CD44 3′ UTR (Fig.
6A), we constructed CD44 3′ UTR-wild (wild-type) and CD44
3′-UTR-mut (mutant) vectors and co-transfected SGC-7901 cells with
either vectors and miR-711 mimics. Dual-Luciferase reporter assays
revealed that in the CD44 3′ UTR-wild group, luciferase activity
was significantly reduced compared to the CD44 3′ UTR-mut group
(P=0.009) (Fig. 6B).
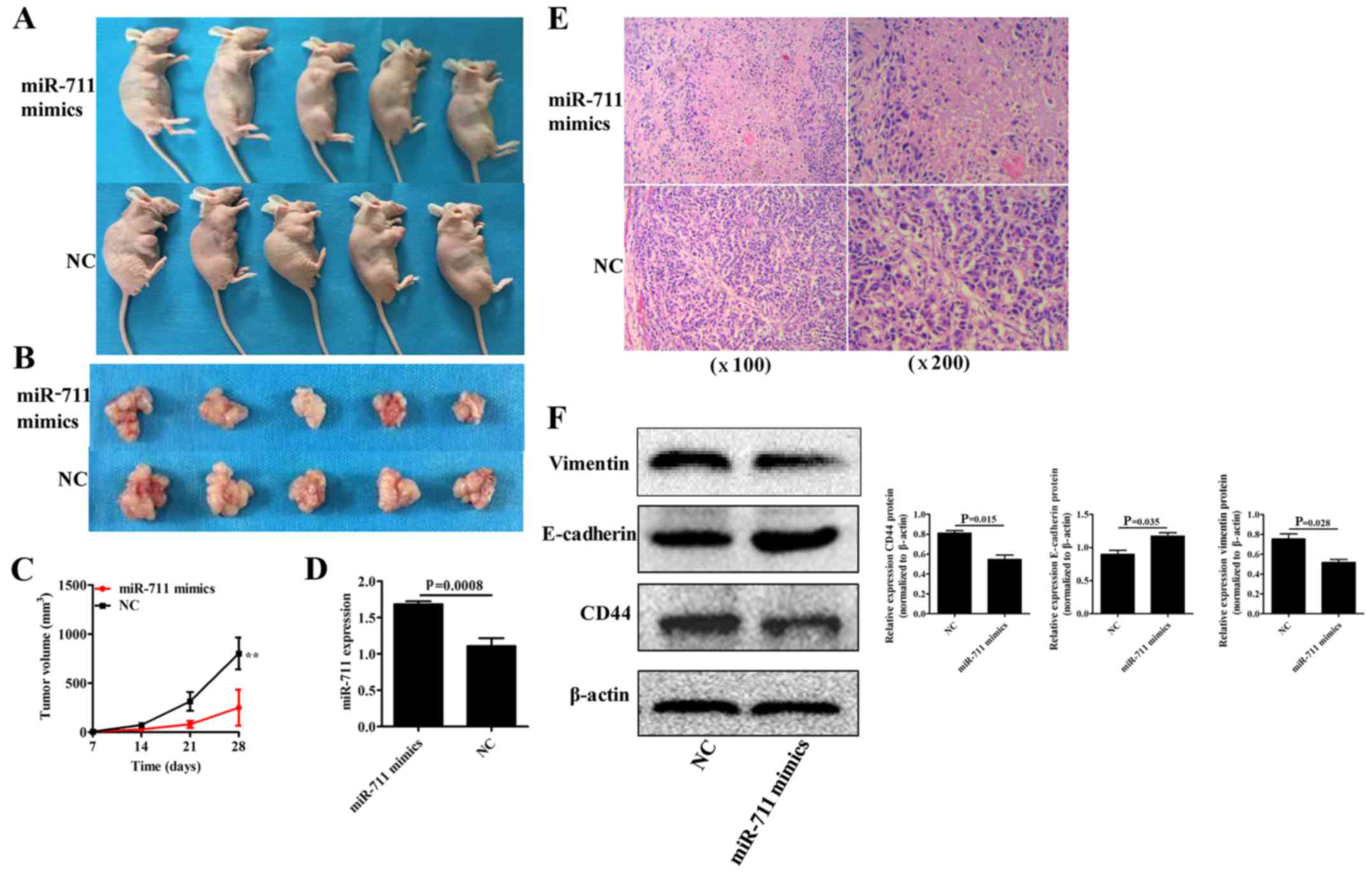
miR-711-mediated upregulation of CD44
inhibits EMT in gastric cancer in vivo
SGC-7901 gastric cancer cells with stable miR-711
mimcs (overexpression) or miR-711 NC were grafted into nude mice
under the armpit, and nude mice were euthanized 4 weeks
post-grafting. Tumor formation assay indicated that the xenograft
tumor volume was significantly smaller in the miR-711 mimics group
compared to that in the miR-711 NC group (P<0.001) at day 28
(Fig. 7A-C). Moreover, qRT-PCR
revealed that miR-711 expression was significantly higher in the
xenograft tumors with miR-711 overexpression when compared to the
NC group (P=0.0008) (Fig. 7D).
Immunohistochemistry using H&E staining showed that, at each
magnification used, the number of tumor cells was significantly
lower in the miR-711 mimics group when compared to the miR-711 NC
group (P<0.05) (Fig. 7E).
Furthermore, western blot analysis demonstrated that CD44 and
vimentin protein expression levels were significantly
downregulated, and E-cadherin protein expression was significantly
upregulated in the xenograft tumors in the miR-711 mimics group
compared to those in the miR-711NC group (P=0.015, P=0.035 and
P=0.028, respectively) (Fig. 7F).
Therefore, it was discovered that miR-711 can inhibit cell invasion
and migration in vitro, and inhibit cell proliferation and
xenograft tumor growth in vivo.
Discussion
Over 90% of cancer-related deaths are associated
with tumor epithelial-mesenchymal transition (EMT). Regulation of
tumor cells by EMT-related proteins during tumor development and
progression results in weakened intercellular adhesion, varying
degrees of mesenchymal phenotypes, and enhanced mobility and
invasiveness. This allows cells to cross the basement membrane with
the help of proteases in the surrounding microenvironment, and
results in local infiltration and distant metastasis. Tumor cells
that arrive at the site of metastatic lesions can reverse EMT and
re-transform into tumor epithelial cells, which then proliferate to
form distant metastatic lesions and promote tumor progression
(18–21). Several factors induce or regulate
the biological processes of EMT, such as the tumor
microenvironment, cytokines, signaling kinases and transcription
factors. Moreover, changes in EMT-related proteins including the
expression of epithelial cell adhesion proteins, cell
morphology-related proteins and mesenchymal cell-related proteins,
such as E-cadherin and vimentin have also been reported (22–25).
In fact, several studies have demonstrated that the biological
processes of EMT play a critical role in the development,
progression, invasion and migration.
EMT is often accompanied by an increase in cancer
stem cells (CSCs), and is therefore believed to be one of the
inducing factors for CSC formation (26). Previous studies have suggested that
prostate CSCs may be the source of prostate cancer and that they
have an important role in the development and progression of
prostate cancer. Prostate CSCs can be identified by several unique
markers, including CD133, CD44, BCRP-1/ABG2 and telomerase
(27,28). Ishimoto et al found that CD44
is an important marker for CSCs (29). In another study, it was demonstrated
that the use of an anti-CD44 monoclonal antibody eliminated acute
myeloid leukemia (AML) CSCs (30).
CD44 is a multifunctional cell surface adhesion receptor that plays
a key role in the invasion and metastasis of multiple cancers.
In vitro and in vivo studies have revealed that
miR-647 may inhibit gastric cancer metastasis via inhibition of
CD44 expression (31). In a study
by Lee et al, it was reported that downregulation of CD44
expression in HCT116 colon cancer cells inhibited cancer cell
proliferation, migration and invasion and promoted cell apoptosis
(32). Moreover, it was found that
exogenous asporin bound to the CD44 receptor on pancreatic cancer
cells to downregulate epithelial phenotype-related proteins and
upregulate mesenchymal phenotype-related proteins, which induced
tumor cell EMT, and thereby promoted tumor cell invasion and
migration (33).
At present, studies on the biological roles of
miR-711 are limited. In a previous study, it was shown that
pioglitazone inhibited collagen-I synthesis and post-infarction
myocardial fibrosis by upregulating miR-711 expression in
cardiomyocytes (34).
miR-711-mediated upregulation of adiponectin (ApN) inhibited
Toll-like receptor 4 (TLR4) signaling and thereby suppressed
inflammation during myositis (35).
In a recent study by Hu et al, a new role of miR-711 in
cancer was revealed. Specifically, the authors showed that high
miR-711 expression in breast cancer tissues was a risk factor for
breast cancer patients, and that high miR-711 expression promoted
breast cancer cell proliferation, invasion and migration in
vitro, suggesting that miR-711 served as an oncogene (36). However, Waseem et al found
that lower miR-711 expression in prostate cancer resulted in a
higher Gleason score, greater malignancy, and higher cancer
metastatic rate, indicating that miR-711 was an anti-oncogene
(37). Because of the inconsistent
findings, the biological functions of miR-711 remain unclear. In
our previous study, we showed that the expression of miR-711 in
gastric cancer tissues and cells was low, and that exogenous
miR-711 expression inhibited gastric cancer cell invasion and
migration by an unidentified mechanism (17). In this study, exogenous miR-711
overexpression in SGC-7901 or MGC-803 gastric cancer cells
inhibited cancer cell invasion and migration. In contrast,
inhibition of miR-711 expression promoted gastric cancer invasion
and migration, indicating that miR-711 inhibited gastric cancer
progression. Moreover, exogenous miR-711 overexpression can inhibit
xenograft tumor growth in vivo and may therefore be an
anti-oncogene. It was discovered that the process of EMT is
dispensable for metastasis, and caused chemo-resistance in lung and
pancreatic cancer (38,39). In this study, EMT contributed to
metastasis in gastric cancer. Since bioinformatics shows that
miR-711 specifically binds to the 3′ UTR of CD44, we speculated
that the biological roles of miR-711 may be mediated through CD44.
To test this hypothesis, we performed western blot analysis and
confirmed that exogenous miR-711 overexpression inhibited CD44 and
vimentin protein expression and promoted E-cadherin protein
expression. In contrast, inhibition of miR-711 expression promoted
CD44 and vimentin protein expression and inhibited E-cadherin
protein expression. These findings indirectly demonstrated that
miR-711 may inhibit EMT in gastric cancer by regulating CD44,
vimentin and E-cadherin expression. In a previous study, CD44 was
shown to be highly expressed in gastric cancer tissues and cells,
and was involved in the regulation of gastric cancer metastasis
(40). We inhibited CD44 expression
in SGC-7901 and MGC-803 gastric cancer cells and demonstrated that
CD44 inhibition downregulated vimentin protein expression and
upregulated E-cadherin protein expression. Therefore, we believed
that miR-711 may inhibit EMT in gastric cancer by downregulating
CD44 expression, which in turn inhibited vimentin expression and
promoted E-cadherin expression. Further investigation using
dual-fluorescence assays revealed that the relative luciferase
activity was significantly lower in miR-711 mimics-transfected CD44
3′ UTR-wt SGC-7901 cells compared to miR-711 mimics-transfected
CD44 3′ UTR-mut SGC-7901 cells, thereby indicating that miR-711
specifically bound to the CD44 3′ UTR. It further demonstrated that
miR-711 inhibited EMT in gastric cancer via inhibition of CD44 3′
UTR. To further confirm our hypothesis, we cultured and grafted
SGC-7901 cells with stable miR-711 overexpression into nude mice,
and showed that exogenous miR-711 overexpression significantly
inhibited xenograft tumor growth in the miR-711 mimics group
compared to the NC group, demonstrating that miR-711 inhibited
xenograft tumor growth in nude mice. In addition, H&E staining
showed that the number of tumor cells was significantly reduced in
the miR-711 mimics group compared to that in the NC group,
indicating that miR-711 indeed inhibited tumor growth. qRT-PCR
confirmed that miR-711 expression was significantly elevated in the
miR-711 mimics group when compared to the NC group. Moreover,
western blot analyses demonstrated that exogenous miR-711
overexpression inhibited EMT in gastric cancer by inhibiting CD44
and vimentin protein expression and by promoting E-cadherin
protein. In summary, this study was the first to show that
miR-711-mediated inhibition of CD44 expression inhibited EMT of
gastric cancer cells in vitro and in vivo via
downregulation of vimentin expression and upregulation of
E-cadherin expression. Thus, our findings provide novel insights
into the development of miR-711-based targeted therapy for EMT in
gastric cancer.
Acknowledgements
We wish to thanks Professor Qi Su the for technical
support.
Funding
The present study was supported by the Natural
Science Foundation of Hunan Province (nos. 2017JJ3270 and
2018JJ2356), the Three Engineering Training Funds in Shenzhen (nos.
SYLY201718 and SYJY201801) and the Hunan Provincial 2018 Annual
Clinical Key Specialist Construction Project (no. 12).
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
WSX, AJL and DFL conceived and designed the study.
YPT, YZC, WBD and JC completed the experiments and collected the
data. WWZ, YPT, YZC, WBD and JC conducted the statistical analysis.
DFL wrote the manuscript. AJL, WWZ and WSX revised the manuscript.
AJL and DFL obtained funding. All authors approved the manuscript
and agree to be accountable for all aspects of the research in
ensuring that the accuracy or integrity of any part of the work are
appropriately investigated and resolved.
Ethics approval and consent to
participate
The study was approved by The First Affiliated
Hospital of the University of South China Institutional Ethics
Committee (no. 201708).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Martin-Richard M, Custodio A, Garcia-Giron
C, Gravalos C, Gomez C, Jimenez-Fonseca P, Manzano JL, Pericay C,
Rivera F and Carrato A: Seom guidelines for the treatment of
gastric cancer 2015. Clin Transl Oncol. 17:996–1004. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Thiery JP, Acloque H, Huang RY and Nieto
MA: Epithelial-mesenchymal transitions in development and disease.
Cell. 139:871–890. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yan W, Cao QJ, Arenas RB, Bentley B and
Shao R: GATA3 inhibits breast cancer metastasis through the
reversal of epithelial-mesenchymal transition. J Biol Chem.
285:14042–14051. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chen Y, Liu J, Wang W, Xiang L, Wang J,
Liu S, Zhou H and Guo Z: High expression of hnRNPA1 promotes cell
invasion by inducing EMT in gastric cancer. Oncol Rep.
39:1993–1701. 2018.
|
|
6
|
Huang M, Wu S, Hu Q, Wu H, Wei S, Xie H,
Sun K, Li X and Fang L: Agkihpin, a novel SVAE may inhibit the
migration and invasion of liver cancer cells associated with the
inversion of EMT induced by Wnt/beta-catenin signaling inhibition.
Biochem Biophys Res Commun. 479:283–289. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang J, Wang X, Liu F and Fu Y:
microRNA-335 inhibits colorectal cancer HCT116 cells growth and
epithelial-mesenchymal transition (EMT) process by targeting
Twist1. Pharmazie. 72:475–481. 2017.PubMed/NCBI
|
|
8
|
Ventura A and Jacks T: MicroRNAs and
cancer: Short RNAs go a long way. Cell. 136:586–591. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang X, Nie Y, Du Y, Cao J, Shen B and Li
Y: MicroRNA-181a promotes gastric cancer by negatively regulating
tumor suppressor KLF6. Tumour Biol. 33:1589–1597. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen JS, Li HS, Huang JQ, Dong SH, Huang
ZJ, Yi W, Zhan GF, Feng JT, Sun JC and Huang XH: MicroRNA-379-5p
inhibits tumor invasion and metastasis by targeting FAK/AKT
signaling in hepatocellular carcinoma. Cancer Lett. 375:73–83.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Song F, Yang D, Liu B, Guo Y, Zheng H, Li
L, Wang T, Yu J, Zhao Y, Niu R, et al: Integrated microRNA network
analyses identify a poor-prognosis subtype of gastric cancer
characterized by the miR-200 family. Clin Cancer Res. 20:878–889.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang Z, Liu S, Shi R and Zhao G: miR-27
promotes human gastric cancer cell metastasis by inducing
epithelial-to-mesenchymal transition. Cancer Genet. 204:486–491.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Shi Y, Shi H, Zhang B, Yan Y, Han X, Jiang
W, Qian H and Xu W: miR-373 suppresses gastric cancer metastasis by
downregulating vimentin. Mol Med Rep. 17:4027–4034. 2018.PubMed/NCBI
|
|
15
|
Chen X, Wang G, Lu X, Gao P, Song Y, Sun
J, Li A, Xu Y, Xu H and Wang Z: Polymorphisms and haplotypes of the
miR-148/152 family are associated with the risk and
clinicopathological features of gastric cancer in a Northern
Chinese population. Mutagenesis. 29:401–407. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yu X, Luo L, Wu Y, Yu X, Liu Y, Yu X, Zhao
X, Zhang X, Cui L, Ye G, et al: Gastric juice miR-129 as a
potential biomarker for screening gastric cancer. Med Oncol.
30:3652013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liao A, Tan G, Chen L, Zhou W and Hu H:
RASSF1A inhibits gastric cancer cell proliferation by
miR-711-mediated downregulation of CDK4 expression. Oncotarget.
7:5842–5851. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ma Z, Xin Z, Hu W, Jiang S, Yang Z, Yan X,
Li X, Yang Y and Chen F: Forkhead box O proteins: Crucial
regulators of cancer EMT. Semin Cancer Biol. 50:21–31. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Boulding T, McCuaig RD, Tan A, Hardy K, Wu
F, Dunn J, Kalimutho M, Sutton CR, Forwood JK, Bert AG, et al: LSD1
activation promotes inducible EMT programs and modulates the tumour
microenvironment in breast cancer. Sci Rep. 8:732018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang Z, Zou Y, Liang M, Chen Y, Luo Y,
Yang B, Liu F, Qin Y, He D, Wang F and Huang O: Suppressor of fused
(Sufu) promotes epithelial-mesenchymal transition (EMT) in cervical
squamous cell carcinoma. Oncotarget. 8:114226–114238. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang J, Tian XJ, Zhang H, Teng Y, Li R,
Bai F, Elankumaran S and Xing J: TGF-beta-induced
epithelial-to-mesenchymal transition proceeds through stepwise
activation of multiple feedback loops. Sci Signal. 7:ra912014.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Asakura T, Yamaguchi N, Ohkawa K and
Yoshida K: Proteasome inhibitor-resistant cells cause EMT-induction
via suppression of E-cadherin by miR-200 and ZEB1. Int J Oncol.
46:2251–2260. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Qin Y, Tang B, Hu CJ, Xiao YF, Xie R, Yong
X, Wu YY, Dong H and Yang SM: An hTERT/ZEB1 complex directly
regulates E-cadherin to promote epithelial-to-mesenchymal
transition (EMT) in colorectal cancer. Oncotarget. 7:351–361. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Satelli A, Batth I, Brownlee Z, Mitra A,
Zhou S, Noh H, Rojas CR, Li H, Meng QH and Li S: EMT circulating
tumor cells detected by cell-surface vimentin are associated with
prostate cancer progression. Oncotarget. 8:49329–49337. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhai X, Zhu H, Wang W, Zhang S, Zhang Y
and Mao G: Abnormal expression of EMT-related proteins, S100A4,
vimentin and E-cadherin, is correlated with clinicopathological
features and prognosis in HCC. Med Oncol. 31:9702014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ikezono Y, Koga H, Akiba J, Abe M, Yoshida
T, Wada F, Nakamura T, Iwamoto H, Masuda A, Sakaue T, et al:
Pancreatic neuroendocrine tumors and EMT behavior are driven by the
CSC marker DCLK1. Mol Cancer Res. 15:744–752. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Jayaraman A, Kumar P, Marin S, de Atauri
P, Mateo F, Thomson M T, Centelles J J, Graham F S and Cascante M:
Untargeted metabolomics reveals distinct metabolic reprogramming in
endothelial cells co-cultured with CSC and non-CSC prostate cancer
cell subpopulations. PLoS One. 13:e01921752018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Mateo F, Meca-Cortes O, Celia-Terrassa T,
Fernandez Y, Abasolo I, Sanchez-Cid L, Bermudo R, Sagasta A,
Rodriguez-Carunchio L, Pons M, et al: SPARC mediates metastatic
cooperation between CSC and non-CSC prostate cancer cell
subpopulations. Mol Cancer. 13:2372014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ishimoto T, Oshima H, Oshima M, Kai K,
Torii R, Masuko T, Baba H, Saya H and Nagano O: CD44+
slow-cycling tumor cell expansion is triggered by cooperative
actions of Wnt and prostaglandin E2 in gastric tumorigenesis.
Cancer Sci. 101:673–678. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Gadhoum SZ, Madhoun NY, Abuelela AF and
Merzaban JS: Anti-CD44 antibodies inhibit both mTORC1 and mTORC2: A
new rationale supporting CD44-induced AML differentiation therapy.
Leukemia. 30:2397–2401. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Cao W, Wei W, Zhan Z, Xie D, Xie Y and
Xiao Q: Role of miR-647 in human gastric cancer suppression. Oncol
Rep. 37:1401–1411. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lee SY, Kim KA, Kim CH, Kim YJ, Lee JH and
Kim HR: CD44-shRNA recombinant adenovirus inhibits cell
proliferation, invasion, and migration, and promotes apoptosis in
HCT116 colon cancer cells. Int J Oncol. 50:329–336. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wang L, Wu H, Wang L, Zhang H, Lu J, Liang
Z and Liu T: Asporin promotes pancreatic cancer cell invasion and
migration by regulating the epithelial-to-mesenchymal transition
(EMT) through both autocrine and paracrine mechanisms. Cancer Lett.
398:24–36. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhao N, Yu H, Yu H, Sun M, Zhang Y, Xu M
and Gao W: MiRNA-711-SP1-collagen-I pathway is involved in the
anti-fibrotic effect of pioglitazone in myocardial infarction. Sci
China Life Sci. 56:431–439. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Boursereau R, Abou-Samra M, Lecompte S,
Noel L and Brichard SM: New targets to alleviate skeletal muscle
inflammation: Role of microRNAs regulated by adiponectin. Sci Rep.
7:434372017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Hu JY, Yi W, Zhang MY, Xu R, Zeng LS, Long
XR, Zhou XM, Zheng XS, Kang Y and Wang HY: MicroRNA-711 is a
prognostic factor for poor overall survival and has an oncogenic
role in breast cancer. Oncol Lett. 11:2155–2163. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Waseem M, Ahmad MK, Srivatava VK, Rastogi
N, Serajuddin M, Kumar S, Mishra DP, Sankhwar SN and Mahdi AA:
Evaluation of miR-711 as novel biomarker in prostate cancer
progression. Asian Pac J Cancer Prev. 18:2185–2191. 2017.PubMed/NCBI
|
|
38
|
Zheng X, Carstens JL, Kim J, Scheible M,
Kaye J, Sugimoto H, Wu CC, LeBleu VS and Kalluri R:
Epithelial-to-mesenchymal transition is dispensable for metastasis
but induces chemoresistance in pancreatic cancer. Nature.
527:525–530. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Fischer KR, Durrans A, Lee S, Sheng J, Li
F, Wong ST, Choi H, El Rayes T, Ryu S, Troeger J, et al:
Epithelial-to-mesenchymal transition is not required for lung
metastasis but contributes to chemoresistance. Nature. 527:472–476.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Hu Y, Wang J, Qian J, Kong X, Tang J, Wang
Y, Chen H, Hong J, Zou W, Chen Y, et al: Long noncoding RNA GAPLINC
regulates CD44-dependent cell invasiveness and associates with poor
prognosis of gastric cancer. Cancer Res. 74:6890–6902. 2014.
View Article : Google Scholar : PubMed/NCBI
|