Introduction
Cancer stem cells (CSCs) are considered the source
of cancer recurrence, treatment resistance and distant metastasis
(1). Al-Hajj et al (2) reported that the cluster of
differentiation
(CD)44+CD24−/lowLineage− breast
cancer cells are consistently considered breast CSCs (BCSCs). As
research has progressed, further BCSC markers, such as aldehyde
dehydrogenase 1 (3) and CD133
(4), have been identified. In
clinical analysis, stemness and phenotypic markers exhibit more
heterogeneity in the intra-tumor heterogeneity as partially
attributing to the different CSCs and subclones of cancer cells
(5,6). In addition, researchers have reported
that collective cancer movement promotes tumor progression through
differently labeled cell populations (7,8). Since
asymmetrical division and multi-differentiation potency are the
main features of CSCs (9,10), there is reason to believe that cells
have differentiated and evolved to specialize for different
functions. For example, CSCs have been revealed to differentiate
into endothelial cells and participate in tumor angiogenesis
(11). Notably, already
asymmetrically divided or differentiated cells can, in turn,
maintain CSC stemness; however, this mechanism remains to be
explored. A recent study confirmed that the stemness characteristic
is maintained through the asymmetrical division of aged
mitochondria (12).
Collective invasion has been described as a novel
behavior of tumor cells in cancer metastasis (7,8).
However, the reasons for collective invasion remain unclear. It has
been reported that collective invasion may be associated with the
heterogeneity of cell populations and differences between cell
markers (7). Other studies have
confirmed that vascular and fibronectin-focal adhesion kinase
signaling (8,13), and cytokine networks (14) have evolved from the tumor
microenvironment, and may participate in the collective invasion
process. In the process of collective invasion, it appears that
information is being exchanged and communicated among cells
(8). However, to the best of our
knowledge, there are no reports of intercellular structural
involvement. The association between collective movement, and CSCs
and vascular niches also remains poorly understood (15).
In a recent study, Baker discussed and summarized
the concept of the cell network as well as the role of networks of
nanotubes and microtubules within it (16). Networks of nanotubes are considered
to participate in cellular communication, allowing for the sharing
and exchange of various content and information (16–18). A
previous study demonstrated that the stem cell marker CD133 may be
transferred between hematopoietic cells via tunneling nanotubes
(19). Similar membrane
microtubules have been detected in vitro and are considered
to be, in part, a result of brain CSC differentiation (20). Networks of microtubules have been
reported to markedly promote the malignant progression of brain
tumors (20,21); however, despite reports of nanotubes
in vitro (22,23), reports of structural networks
participating in cellular communication in mammosphere growth and
invasion are rare.
In the present study, cellular communication was
revealed to be widely present in mammosphere growth and collective
invasion, through networks of microtubule-like structures and
angiogenesis in vitro and in vivo, and the mechanism
was explored. The present study aimed to demonstrate a novel
behavior of cellular communication in the progression of mammary
tumors, and to identify the stemness and differentiation
characteristics of mammospheres. In addition, the effects of
anti-vascular endothelial growth factor (VEGF) treatment on the
prevention of mammary tumor progression were analyzed.
Materials and methods
Animals
A total of 26 Female athymic nude mice (age, 3–5
weeks; weight, 16.1±0.4 g) were purchased from Shanghai Silaike
Laboratory Animal Co., Ltd. (Shanghai, China). The experimental
protocol was conducted according to the Regulations of Experimental
Animal Administration issued by the Ministry of Science and
Technology of the People's Republic of China. Mouse care and usage
were approved by the Ethics Committee of the School of Medicine of
Xi'an Jiaotong University (approval no. 0108; Xi'an, China). The
mice were maintained in air-conditioned pathogen-free rooms with
ad libitum access to food and water, at 25±2°C and 55%
humidity under a controlled light-dark cycle (12–12 h).
Cells and culture
MDA-MB-231 and MCF-7 human breast cancer cell lines,
and the MCF-10A human normal breast cell line, at passages 3–15
were obtained from the Cell Bank of the Chinese Academy of Sciences
(Shanghai, China). MCF-10A cells were cultured in Dulbecco's
modified Eagle's medium (DMEM)/F12 (Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) supplemented with 5% horse serum (Thermo Fisher
Scientific, Inc.), 10 µg/ml insulin (Thermo Fisher Scientific,
Inc.), 100 ng/ml cholera toxin (Biomol GmbH, Hamburg, Germany) and
0.5 µg/ml hydrocortisone (Merck KGaA, Darmstadt, Germany).
MDA-MB-231 cells were cultured in RPMI 1640 medium (Thermo Fisher
Scientific, Inc.) supplemented with 10% fetal bovine serum (FBS;
Thermo Fisher Scientific, Inc.), 100 U/ml penicillin and 100 µg/ml
streptomycin (Thermo Fisher Scientific, Inc.). MCF-7 cells were
cultured in DMEM (Thermo Fisher Scientific, Inc.) supplemented with
10% FBS. All cell lines were cultured at 37°C in an atmosphere
containing 5% CO2. Primary MDA-MB-231 or MCF-7 cells
were obtained from xenograft tissues; xenografts were generated by
subcutaneously implanting 1×106 MDA-MB-231 or MCF-7
cells into six athymic nude mice (n=3/group; approval no. 0108),
according to the method described by Al-Hajj et al (2). When the MDA-MB-231 or MCF-7 ×enografts
reached 1 cm3, the fresh tumor tissues were harvested
and digested into a single cell (2)
suspension in DMEM/F12 supplemented with 10% FBS; these cells were
referred to as the primary MDA-MB-231 or MCF-7 cells, respectively.
Subsequently, MDA-MB-231 or MCF-7 mammospheres, and primary
MDA-MB-231 or MCF-7 mammospheres, were generated from parental
MDA-MB-231/MCF-7 and primary MDA-MB-231/MCF-7 cells; for
mammosphere generation, these cells were harvested and were
maintained in mammosphere culture conditions (24). Specifically, all of the mammospheres
were cultured in serum-free DMEM/F12 medium solution mix,
supplemented with B27 (1:50, Thermo Fisher Scientific, Inc.), 20
ng/ml human epidermal growth factor (Merck KGaA) and 20 ng/ml human
basic fibroblast growth factor (Merck KGaA) (24,25).
Mammospheres were collected by gentle centrifugation (1,000 × g)
after 7–10 days and were dissociated enzymatically [10 min in 0.05%
trypsin (Thermo Fisher Scientific, Inc.)] (24). For differentiation assays,
mammospheres were collected and then cultured in DMEM/F12
supplemented with 10, 5 and 1% FBS.
Cell surface markers and cell cycle
analysis
MDA-MB-231 and MCF-7 cells were washed three times
with PBS and collected following trypsinization. A suspension of
mammosphere cells was collected and centrifuged at 1,000 × g for 5
min after trypsinization into single cells. Subsequently, collected
cells (1×105−1×106) were resuspended in
50–100 µl PBS, after which, 5 µl anti-CD44-fluorescein
isothiocyanate (FITC) antibodies (cat. no. 560977; BD Biosciences,
Franklin Lakes, NJ, USA) and 5 µl anti-CD24-phycoerythrin (PE)
antibodies (cat. no. 560991; BD Biosciences) were added in
succession. Subsequently, the labeled cells were incubated at 37°C
for 1 h in the dark; the same volume of isotype control antibodies
[FITC-labeled/PE-labeled immunoglobulin G (IgG), cat. nos. 555742
and 555574; BD Biosciences] was added to the control group. The
labeled cells were then examined by flow cytometry on a FACSCalibur
flow cytometer (BD Biosciences), following two rounds of washing in
cold PBS. For cell cycle analysis, all cells were harvested and
fixed with 75% ethanol on ice for 24 h. Subsequently, the cells
were treated with 0.1% RNase A (Nanjing KeyGen Biotech Co., Ltd.,
Nanjing, China) in PBS at 37°C for 30 min and stained with 50 µg/ml
propidium iodide (Nanjing KeyGen Biotech Co., Ltd.) at room
temperature for 30 min, after which, cell cycle analysis was
conducted using a flow cytometer (FACSCalibur; BD Biosciences) and
FlowJo 10.0 software (FlowJo, LLC, Ashland, OR, USA) was used for
analysis. Experiments were performed in triplicate.
Statistical assessment of polymerase
chain reaction (PCR) datasets
The PCR data of all cell types were analyzed using
the Real-Time PCR analysis software and platform (Bio-Rad
Laboratories, Inc., Hercules, CA, USA), in order to generate
quantification cycle (Cq) values for each gene. Briefly, cells were
collected and total RNA was extracted using TRIzol®
reagent (Invitrogen; Thermo Fisher Scientific, Inc.), according to
the manufacturer's protocol. First-strand cDNA was synthesized from
2 µg total RNA using a Reverse Transcription kit (Takara Bio, Inc.,
Otsu, Japan), according to the manufacturer's protocol. All primer
sequences were obtained from the National Center for Biotechnology
Information (https://www.ncbi.nlm.nih.gov/) and are presented in
Table I. The PCR reaction mixture
(Takara Bio, Inc.) contained 10 µl SYBR premix EX Taq (2X), 0.8 µl
forward and reverse primers (2.5 µM), 5 µl cDNA (2 ng) and 4.2 µl
ddH2O. Cycling conditions were as follows: Denaturation
at 95°C for 1 min, followed by 40 cycles of denaturation at 95°C
for 20 sec, annealing at 60°C for 5 sec and extension at 72°C for
30 sec, with a final 10 min extension at 72°C. β-actin was used as
the internal control. Normalized Cq values were converted to
relative log10 or log2 expression values. All
gene expression levels were calculated using the 2−ΔΔCq
method (26).
 | Table I.Primers for quantitative polymerase
chain reaction. |
Table I.
Primers for quantitative polymerase
chain reaction.
| Gene name | Primer
sequences |
|---|
| Human CD44 | F:
5′-TTACAGCCTCATCAGCAGAGCAC-3′ |
|
| R:
5′-CAATGGTGTGACGCAGGGAT-3′ |
| Human c-Myc | F:
5′-CAAGAGGCGAACACACAACG-3′ |
|
| R:
5′-GTCGTTTCCGCAACAAGTCC-3′ |
| Human SOX2 | F:
5′-AACCAGCGCATGGACAGTTA-3′ |
|
| R:
5′-CGAGCTGGTCATGGAGTTGT-3′ |
| Human NANOG | F:
5′-CAATGGTGTGACGCAGGGAT-3′ |
|
| R:
5′-TGCACCAGGTCTGAGTGTTC-3′ |
| Human POU5F1 | F:
5′-GCCGCTGGCTTATAGAAGGT-3′ |
|
| R:
5′-CTCTCCCCAGCTTGCTTTGA-3′ |
| Human VEGF | F:
5′-AACTTTCTGCTGTCTTGG-3′ |
|
| R:
5′-ACTTCGTGATGATTCTGC-3′ |
| Human MMP1 | F:
5′-AGAAAGAAGACAAAGGCAAGTTGA-3′ |
|
| R:
5′-CCACATCTGGGCTGCTTCAT-3′ |
| Human β-actin | F:
5′-AGCGAGCATCCCCCAAAGTT-3′ |
|
| R:
5′-GGGCACGAAGGCTCATCATT-3′ |
Cell migration and invasion
assays
For the migration assays, 4×104
non-trypsin-treated primary MDA-MB-231 mammosphere cells/well were
seeded into the top compartment of a 24-well plate on 8-µm
transwell filters (EMD Millipore, Billerica, MA, USA) in 200 µl
pure DMEM/F12 medium. In the lower chamber, 600 µl DMEM/F12 medium
supplemented with 20% FBS was added. Subsequently, cells were
incubated for 24 h at 37°C in 5% CO2, and non-migratory
cells were removed using a cotton swab. Cells that adhered to the
underside of the membrane were fixed with 4% paraformaldehyde for
30 min at room temperature and stained with 0.1% crystal violet
solution for 20 min at room temperature in the dark. For the
invasion assay, the bottom of the 25 cm2 suspension
culture bottle (Corning Inc., Corning, NY, USA), which contained
the primary MDA-MB-231 mammospheres in serum-free DMEM/F12 medium
solution supplemented with B27 (1:50, Thermo Fisher Scientific,
Inc.), 20 ng/ml human epidermal growth factor (Merck KGaA) and 20
ng/ml human basic fibroblast growth factor (Merck KGaA), was coated
with 0.75 mg/ml Matrigel (BD Biosciences) and cells were cultured
at 37°C in 5% CO2.
Western blot analysis
Primary MDA-MB-231 mammosphere cells were treated
with 5 µg/ml bevacizumab (Shanghai Roche Pharmaceuticals Co., Ltd.,
Shanghai, China) (27) for 24 h at
37°C in 5% CO2. Harvested mammospheres were lysed using
radioimmunoprecipitation assay lysis buffer (Merck KGaA) containing
2 mg/ml protease inhibitors for 30 min. The lysates were then
centrifuged at 12,000 × g for 20 min at 4°C, followed by boiling in
loading buffer for 5 min. Total protein concentrations were
determined using a Bio-Rad protein assay kit (Bio-Rad Laboratories,
Inc.). Following protein extraction, equal amounts of total
cellular protein (150 µg) were separated by 10% SDS-PAGE and were
transferred onto polyvinylidene difluoride membranes (EMD
Millipore). After blocking with 5% non-fat dry milk dissolved in
Tris-buffered saline containing 0.1% Tween-20 (Bio-Rad
Laboratories, Inc.) for 1 h at room temperature, membranes were
incubated overnight at 4°C with the indicated antibodies.
Subsequently, the immunoblots were probed with horseradish
peroxidase (HRP)-conjugated secondary antibodies (anti-mouse m-IgGκ
BP-HRP: Dilution 1:10,000, cat. no. sc-516102; mouse anti-rabbit
IgG-HRP: Dilution 1:5,000, cat. no. sc-2357; both Santa Cruz
Biotechnology, Inc., Dallas, TX, USA) for 2 h at room temperature
and were visualized using enhanced chemiluminescence (GE
Healthcare, Chicago, IL, USA). Images were captured using a
ChemiDoc™ MP system (Bio-Rad Laboratories, Inc.). β-actin was used
as a control. Signals were semi-quantified using Image-Pro Plus 6.0
software (Media Cybernetics, Inc. Rockville, MD, USA). The primary
antibodies used in the present study were as follows: Mouse
monoclonal anti-VEGF (dilution 1:500; cat. no. ab1316; Abcam,
Cambridge, UK), mouse monoclonal anti-CD31 (dilution 1:1,000; cat.
no. ab24590; Abcam), rabbit polyclonal anti-CD133 (dilution
1:1,000; cat. no. ab19898; Abcam), rabbit polyclonal anti-matrix
metalloproteinase 1 (MMP1; dilution 1:1,000; cat. no. 10371-2-AP;
Wuhan Sanying Biotechnology, Wuhan, China) and mouse monoclonal
anti-β-actin (dilution 1:1,000; cat. no. sc-47778; Santa Cruz
Biotechnology, Inc.).
CSC formation and blocking assay
Anti-CD44-FITC and anti-CD24-PE (cat. nos. 560977
and 560991; BD Biosciences) were used to sort
CD44+CD24− cells (>90% purity) from the
trypsinized primary MDA-MB-231 mammospheres using BD FACSMelody™
(BD Biosciences). Subsequently, equal amounts of cells
(~1×103) were allocated to each group and were
introduced to the in vitro mammosphere culture system, in
which cells were cultured in suspension in 24-well plates (Corning,
Inc.) with serum-free DMEM/F12 medium, supplemented with B27 (1:50;
Thermo Fisher Scientific, Inc.), 20 ng/ml human epidermal growth
factor (Merck KGaA) and 20 ng/ml human basic fibroblast growth
factor (Merck KGaA). After 8 days, the number of mammospheres was
counted and bevacizumab (5 µg/ml) (27) was added, and after 2 weeks, the
number of mammospheres was recounted, and 5 µg/ml bevacizumab was
added at 22 days. No interventions were made to the control group.
Each experimental group had three replicates.
Orthotopic xenograft tumor model
A total of 20 athymic nude mice were randomly
divided into two groups (n=10/group); approximately
1×106 MDA-MB-231 cells were subcutaneously implanted
into the mice of the first group, and ~1×104 MDA-MB-231
mammospheres were subcutaneously implanted into the mice of the
second group. The growth of implanted xenograft tumors was
monitored every 3–4 days, and the tumor volumes were calculated
according to the following formula: Volume=(length ×
width2)/2. After 1 month, mice were sacrificed by
cervical dislocation under anesthesia. For immunohistochemistry and
hematoxylin and eosin staining (H&E), the tumor tissues were
harvested, fixed in 4% paraformaldehyde solution for 12 h at 4°C
and embedded in paraffin. Sections (4 µm) were continuously sliced
and stained with H&E (AR1180; Wuhan Boster Biological
Technology, Ltd., Wuhan, China), according to the manufacturer's
protocol.
Immunohistochemical staining and
evaluation
The protein expression in the xenograft tissues was
characterized by immunohistochemistry. Briefly, the tissue sections
(4 µm) were dewaxed, rehydrated and treated with 3%
H2O2 in methanol for 10 min at room
temperature to inactivate endogenous peroxidase activity, followed
by antigen retrieval in 0.01 mol/l citrate buffer (pH 6.0) in a
microwave for 15 min. After blocking with 10% normal goat serum
(Thermo Fisher Scientific, Inc.) in PBS at room temperature for 30
min, the sections were incubated with mouse monoclonal anti-VEGF
(dilution 1:100; cat. no. ab1316; Abcam), mouse monoclonal
anti-proliferating cell nuclear antigen (PCNA; dilution 1:100; cat.
no. 60097-1-Ig; Wuhan Sanying Biotechnology), rabbit polyclonal
anti-MMP1 (dilution 1:100; cat. no. 10371-2-AP; Wuhan Sanying
Biotechnology) and mouse monoclonal anti-CD31 (dilution 1:50; cat.
no. ab24590; Abcam) antibodies overnight at 4°C. Rabbit IgG
(dilution 1:100; cat. no. ab172730; Abcam) served as the negative
control. After washing with PBS, the bound antibodies were detected
using HRP-anti-rabbit/anti-mouse IgG (mouse anti-rabbit IgG-HRP:
Dilution 1:1,000, cat. no. sc-2357; anti-mouse m-IgGκ BP-HRP;
Dilution 1:1,000, cat. no. sc-516102; both Santa Cruz
Biotechnology, Inc.) for 15 min at 37°C. The sections were treated
using the SABC kit (Wuhan Boster Biological Technology, Ltd.),
according to the manufacturer's protocol. Sections were visualized
using freshly prepared 0.1% DAB (Agilent Technologies, Inc., Santa
Clara, CA, USA) for 5 min at room temperature, and were then
counterstained with 5% hematoxylin for 30 sec at room temperature
to stain the nuclei, after which, they were dehydrated in a graded
series of alcohol solutions. Two investigators independently
evaluated expression levels in a blinded manner. Semi-quantitative
analysis of staining distribution was scored as -, +, ++ and +++
according to the percentage of immunoreactive cells. Specifically,
‘-’ indicated complete absence of staining or weak staining in
<1% of the tumor cells, ‘+’ indicated focal staining in 1–10% of
tumor cells, ‘++’ indicated positive staining in 11–50% of tumor
cells, and ‘+++’ indicated positive staining in >50% of tumor
cells. When >10% of tumor cells exhibited immunoreactivity, the
sample was defined as immunopositive. The membranous or cytoplasmic
expression of VEGF and MMP1, and the nuclear expression of PCNA
were considered positive events using a light microscope (Olympus
BX51; Olympus Corporation, Tokyo, Japan).
Microvessel density (MVD) assay
The MVD of mouse sections was determined by staining
with anti-CD31 (dilution 1:50; cat. no. ab24590; Abcam); staining
was evaluated using a light microscope (Olympus BX51; Olympus
Corporation), according to the method recommended by Weidner et
al (28). Initially,
microvascular staining was observed at a magnification of ×100.
Subsequently, three ‘hot spots’ were selected in the regions of
highest vascular density. Each brown immunostained endothelial cell
or endothelial cell cluster, which was clearly separate from the
adjacent micovessels and stromal structures, was counted as a
single microvessel. To define MVD, the mean number of microvessels
in each field was counted in each of the paraffin-embedded mouse
sections (n=10/group).
Immunohistochemical fluorescence
staining
Frozen sections of xenograft tumors were washed
three times with PBS and were then dried on ice. The cell membranes
were penetrated using 1% Triton X-100 for 10 min and the cells were
then washed a further three times with PBS for 5 min. Following
incubation with 5% bovine serum albumin [Serana (WA) Pty Ltd.,
Bunbury, WA, Australia] for 30 min at room temperature, rabbit
polyclonal anti-CD133 (dilution 1:100; cat. no. ab19898; Abcam) and
mouse monoclonal anti-CD31 antibodies (dilution 1:50; cat. no.
ab24590; Abcam) were added at 4°C overnight. Subsequently, sections
were incubated with the relative FITC-labeled
anti-rabbit/PE-labeled anti-mouse IgG antibodies (dilution 1:1,000;
cat. no. ab6717/dilution 1:200; cat. no. ab5881; Abcam) at 37°C for
1 h in the dark. After washing with PBS, nuclear counterstaining of
the sections was performed using 1 µg/ml DAPI (Santa Cruz
Biotechnology, Inc.) for 10 min at room temperature in the dark.
After washing twice with PBS, the sections were treated with
anti-quenchable sealing oil, and were observed and analyzed using a
fluorescence microscope (ECLIPSE Ti; Nikon Corporation, Tokyo,
Japan).
Statistical analysis
All statistical analyses were performed using SPSS
18.0 software (SPSS, Inc., Chicago, IL, USA). Representative images
are presented or data are expressed as the means ± standard
deviation/standard error of the mean (n≥3). Differences among
groups were determined using one-way analysis of variance (ANOVA)
or two-tailed Student's t-test. Multiple group comparisons were
performed by one-way ANOVA, followed by a Tukey's multiple
comparison test. Qualitative data were compared using the
χ2 test or Fisher's exact test. All statistical
comparisons were two-sided. P<0.05 was considered to indicate a
statistically significant difference.
Results
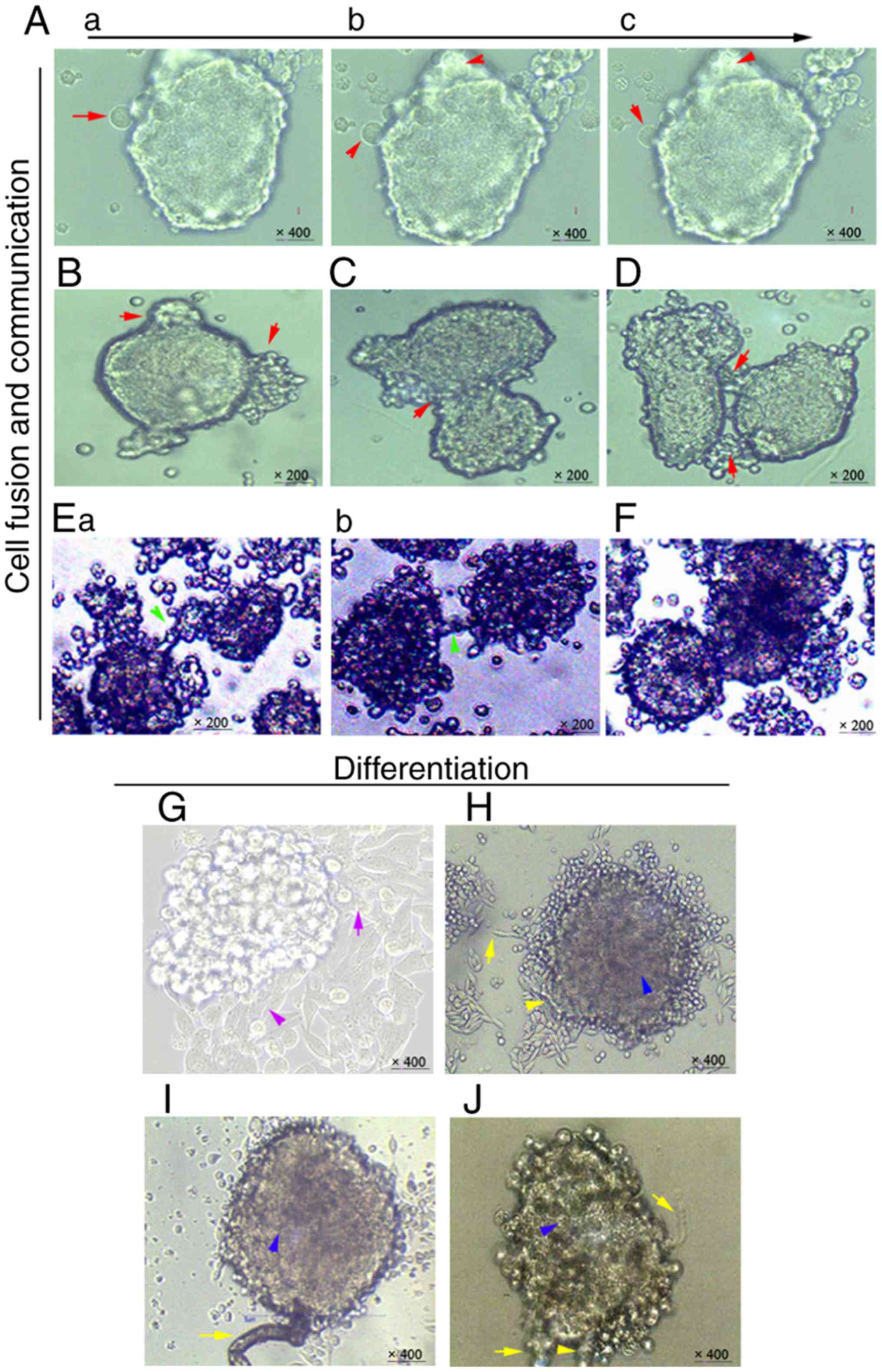
Cellular communication exists widely
in mammosphere fusion and differentiation
The primary MDA-MB-231 cells were harvested and
cultured in the in vitro mammosphere culture systems.
Initially, the primary mammospheres continued to attract and
swallow smaller cell spheres (Fig.
1Aa-c). Subsequently, cellular-like protrusions appeared and
the irregular mammosphere morphology was maintained (Fig. 1B). In addition, cell fusion occurred
when two medium-sized mammospheres came into contact (Fig. 1C). When two larger mammospheres
moved alongside each other, they communicated with each other
through cellular-like protrusions (Fig.
1D) (20,29). Some microtubule-like contacts were
revealed to participate in this mammosphere-to- mammosphere
communication (Fig. 1Ea and b).
Notably, cell fusion also occurred, albeit >1 week after these
connections were formed between the large mammospheres (Fig. 1F).
By adding different concentrations of
serum-containing medium (10, 5 and 1%), it was demonstrated that
the higher the concentration, the faster the cell fusion was
promoted (data not shown). Notably, 1% serum-containing medium made
it easier to observe the slow alterations in cellular
communication. The results demonstrated that the mammospheres
differentiated into linear leaf-shaped cells with cord-like
arrangements from their periphery (Fig.
1G and H). Microtubule-like structures were differentiated and
grew in or around the mammospheres (Fig. 1H-J). Combined, these results
indicated that cellular communication may be present in
mammospheres during the entire process of dynamic formation and
differentiation in vitro.
Networks of cells and microtubules are
differentiated among mammospheres
In order to further clarify the footprints of
cellular communication, mammospheres were continuously cultured in
1% serum-containing medium. The results demonstrated that
microtubule-like structures and fibrous cellular-like cables could
further divide into various branches and were widely distributed in
the cellular space (Fig. 2Aa-c).
Varieties of sphere cells were connected and surrounded with these
network links (Fig. 2B). Networks
of cells were even re-differentiated and transported along these
links (Fig. 2Ca and b). As
expected, nanotube-like structures on membranes were also observed
in cell-to-cell and mammosphere-to-mammosphere communications
(Fig. 2Da and b). Some cord-like
connective structures were differentiated and made up by deformed
cell clusters (Fig. 2E).
Furthermore, a large number of structures, including vascular
epithelium and vasculogenic mimics, were found to emerge from a
change in cell morphology, forming another communication channel
network (Fig. 2F and G). Similar
networks of cells were also observed in these cell channels
(Fig. 2Gc). Combined, these results
indicated that various communication networks may be differentiated
among mammospheres and cells in vitro.
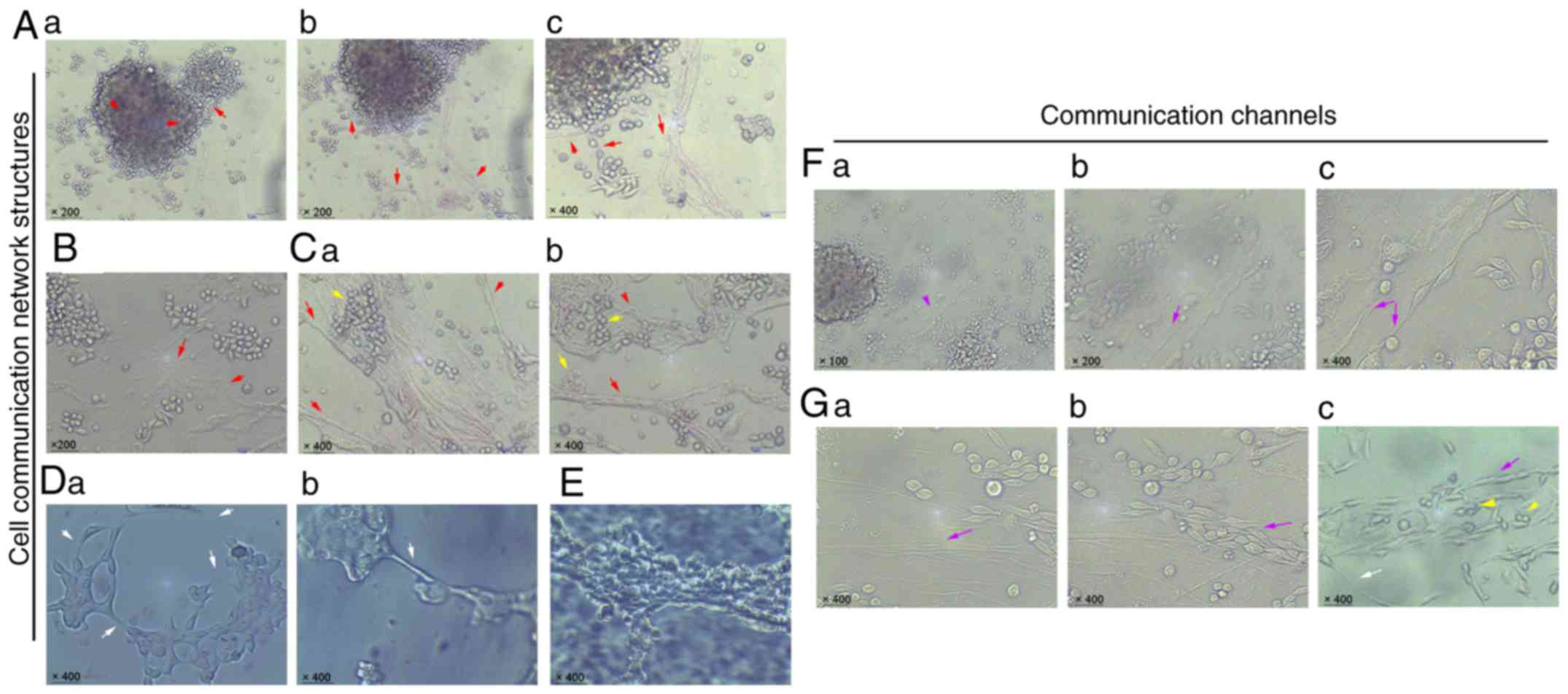 | Figure 2.Networks of cellular communication
structures are distributed in vitro. (Aa-c) Microtubule-like
structures were distributed between mammospheres and mammospheres,
mammospheres and cells (red arrows indicate microtubules;
magnification: Aa and b, ×200; Ac, ×400). (B and C) Networks of
microtubules differentiating and evolving (red arrows indicate
microtubules; yellow arrows indicate connection between cancer
cells and cell cluster; magnification: B, ×200; Ca and b, ×400).
(Da and b) Nanotube-like structures among cell-to-cell and
mammosphere-to-mammosphere links (white arrows; magnification,
×400). (E) Cord-like connective structure of cancer cells
(magnification, ×400). (Fa-c) Similar channel network formation
between mammospheres and cells (purple arrows indicate the deformed
connective cells; magnification: Fa, ×100; Fb, ×200; Fc, ×400).
(Ga-c) Similar channel network formation with pipe-like
vasculogenic mimicry (purple arrows indicate the deformed elongated
cells, yellow arrows indicate the free round cells and the white
arrow indicates the nanotube-like structure; magnification,
×400). |
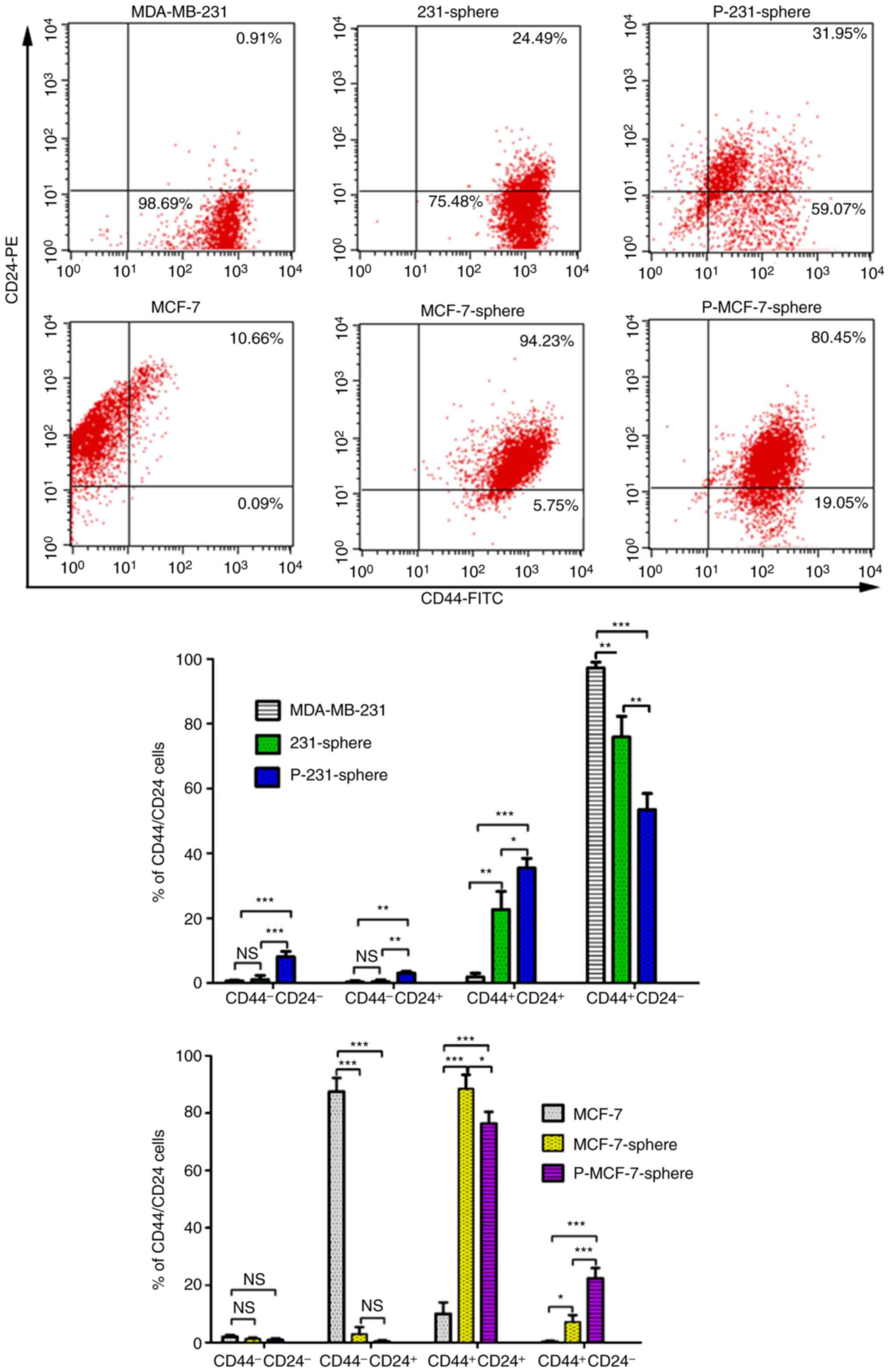
High percentage of
CD44+/CD24+ or
CD44+/CD24− cells is present in
mammospheres
CSC markers were compared between mammospheres and
their parental cells. The results indicated that the percentage of
CD44+/CD24+ cells was significantly increased
in MDA-MB-231 (22.65±5.59%) and primary MDA-MB-231 mammospheres
(35.43±3.03%) compared with the parental MDA-MB-231 cells
(1.81±1.16%; Fig. 3; P<0.01 and
P<0.001) (30,31). However, the percentage of
CD44+/CD24− cells was decreased in both
MDA-MB-231 (75.89±6.34%) and primary MDA-MB-231 mammospheres
(53.48±4.99%) compared with the parental cells (97.25±1.74%;
Fig. 3; P<0.01 and P<0.001).
Conversely, the percentage of CD44+/CD24−
cells was significantly increased in MCF-7 mammospheres
(7.26±2.36%) and primary MCF-7 mammospheres (22.42±3.52%) compared
with the parental MCF-7 cells (0.31±0.32%; P<0.05 and
P<0.001). Similarly, the percentage of
CD44+/CD24+ cells was significantly increased
in both MCF-7 mammospheres (88.49±4.92%) and primary MCF-7
mammospheres (76.33±4.12%) compared with the parental MCF-7 cells
(0.31±0.32%; Fig. 3; P<0.001 and
P<0.001). In conclusion, the percentage of
CD44+/CD24+ cells was significantly increased
in the MDA-MB-231/MCF-7 mammospheres and primary mammospheres,
compared with in their parental cells. The MDA-MB-231 group had a
higher proportion of CD44+/CD24− cells than
the MCF-7 group.
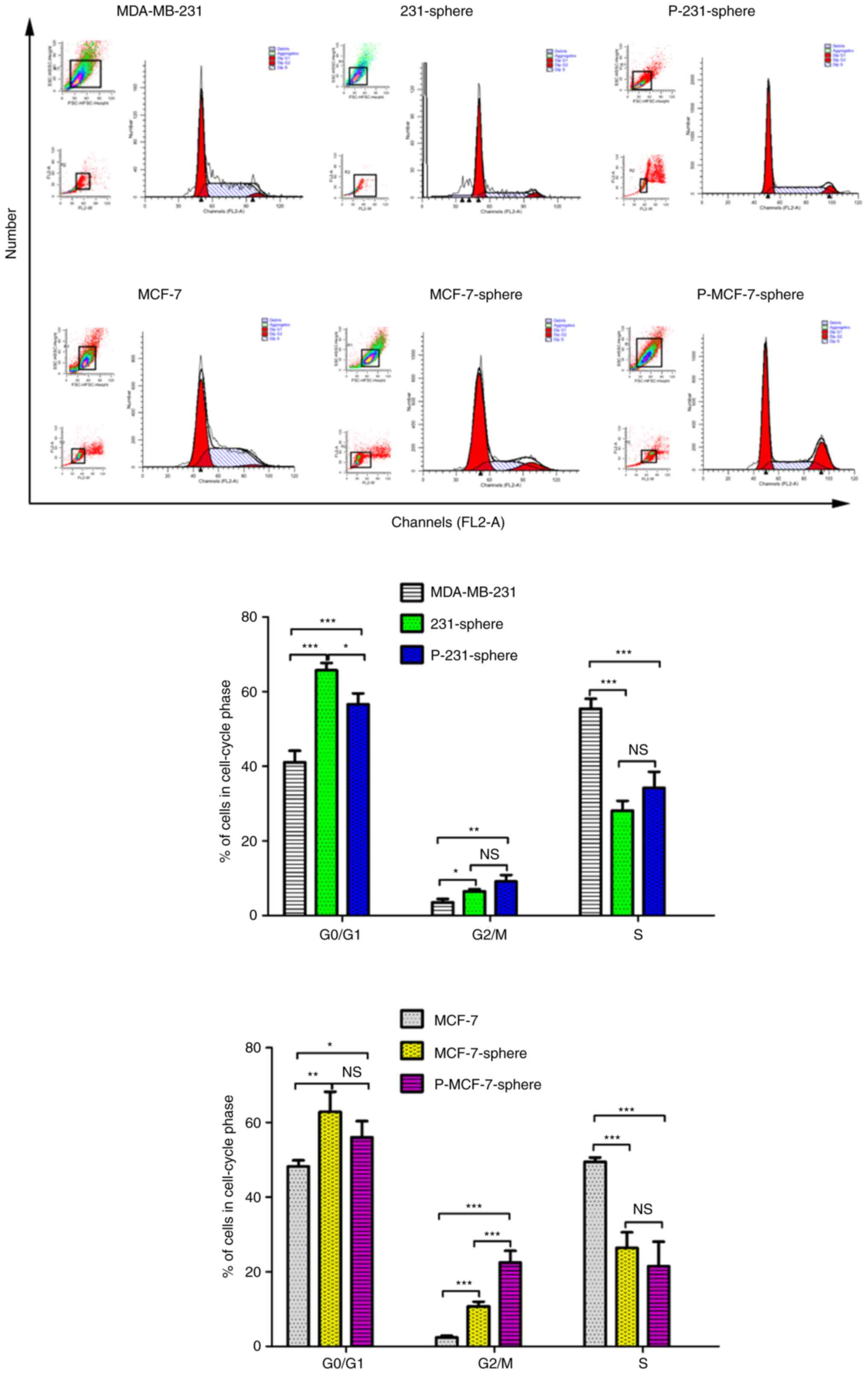
Cell cycle arrest in mammospheres
Cell cycle progression and the distribution of cells
at different phases of the cell cycle are shown in Fig. 4. When compared with the parental
MDA-MB-231 cells, a higher percentage of
G0/G1 and G2/M phase cells was
distributed in MDA-MB-231 and primary mammospheres (Fig. 4; P<0.001 and P<0.001;
P<0.05 and P<0.01). The percentage of S phase cells in the
parental MDA-MB-231 group was higher than that in the MDA-MB-231
and primary mammospheres (Fig. 4;
P<0.001 and P<0.001). The cell cycle distribution trend of
the MCF-7 group was similar to that of the MDA-MB-231 group.
Analysis of cell cycle progression indicated that mammospheres may
carry more dormant cells compared with the parental cells, and thus
may exhibit a more flexible space for proliferation and
differentiation.
High expression of stemness-,
invasiveness- and angiogenesis-associated genes in
mammospheres
Gene expression levels in mammospheres were compared
with those in parental cells using quantitative PCR. The results
demonstrated that CD44, c-Myc and SRY-box 2 genes, which are
associated with stemness, were all significantly increased in
primary MDA-MB-231 and primary MCF-7 mammospheres compared with the
parental cells (Fig. 5A-C). Nanog
homeobox and POU class 5 homeobox 1, which are genes associated
with reproduction and stemness (32), were also significantly increased in
primary MDA-MB-231 and primary MCF-7 mammospheres (Fig. 5D and E, P<0.01). MMP1, which is a
gene associated with invasiveness, and VEGF, which is associated
with angiogenesis (33), were also
significantly increased in primary MDA-MB-231 and primary MCF-7
mammospheres (Fig. 5F and G).
Combined, these results indicated that the aforementioned genes
have been significantly enriched and expressed in primary
mammospheres compared with in the parental cells.
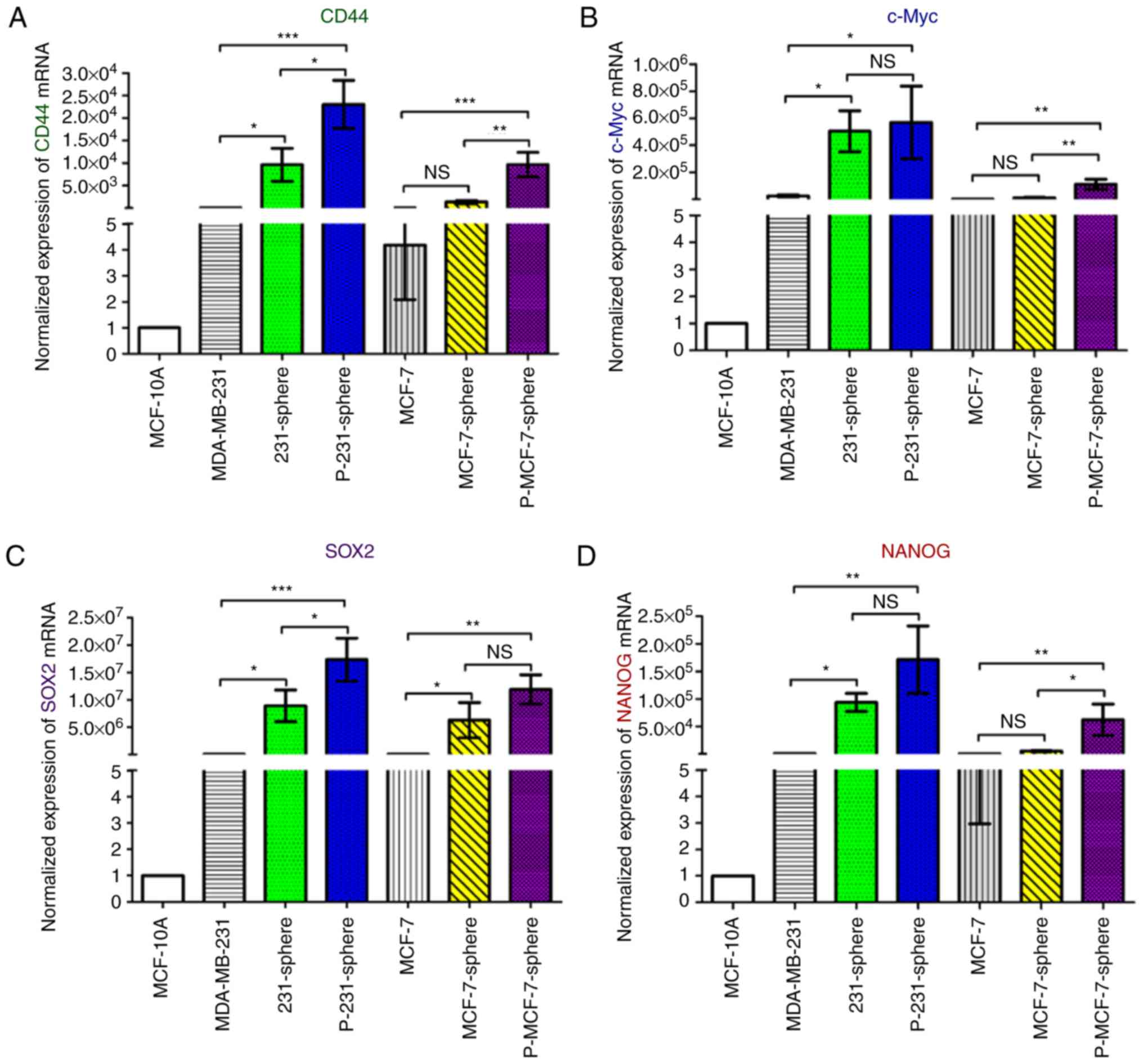 | Figure 5.Expression levels of stemness-,
invasiveness- and angiogenesis-associated genes are increased in
mammospheres. (A-G) Normalized gene expression was compared to
MCF-10A for standardization. All PCR data were detected using the
Bio-Rad Connect Real-Time PCR platform. Normalized Cq values were
converted to relative log10 or log2
expression values, and the gene expression levels were calculated
using the 2−ΔΔCq method. Relative gene expression data
were compared using one-way analysis of variance, followed by
Tukey's test, and are expressed as the means ± standard deviation.
*P<0.05, **P<0.01 and ***P<0.001. CD44, cluster of
differentiation 44; Cq, quantification cycle; MMP1, matrix
metalloproteinase 1; NANOG, Nanog homeobox; PCR, polymerase chain
reaction; POU5F1, POU class 5 homeobox 1; SOX2, SRY-box 2; VEGF,
vascular endothelial growth factor. |
Collective invasion in mammospheres
through cellular communication
To investigate whether cellular communication was
directly involved in collective invasion, a series of invasion
assays were performed in vitro. The results demonstrated
that primary MDA-MB-231 mammospheres started collective invasion
once the mammospheres gathered and joined into larger mammospheres
(Fig. 6A). Subsequently, the front
cells of the mammospheres broke the matrix gel and invaded into the
bottom of the culture bottles (Fig. 6B
and C). Along with the traction of cell cords and
microtubule-like structures, mammospheres and cells gradually
invaded in an almost completely collective manner (Fig. 6D and E). Cellular cable-like
connections between cell clusters were also revealed to participate
in the collective invasion process (Fig. 6E). Subsequently, non-trypsin-treated
mammospheres were analyzed using the transwell migration assay. The
results revealed that mammospheres migrated collectively by
following and surrounding long, strip-shaped connection cells
(Fig. 6F).
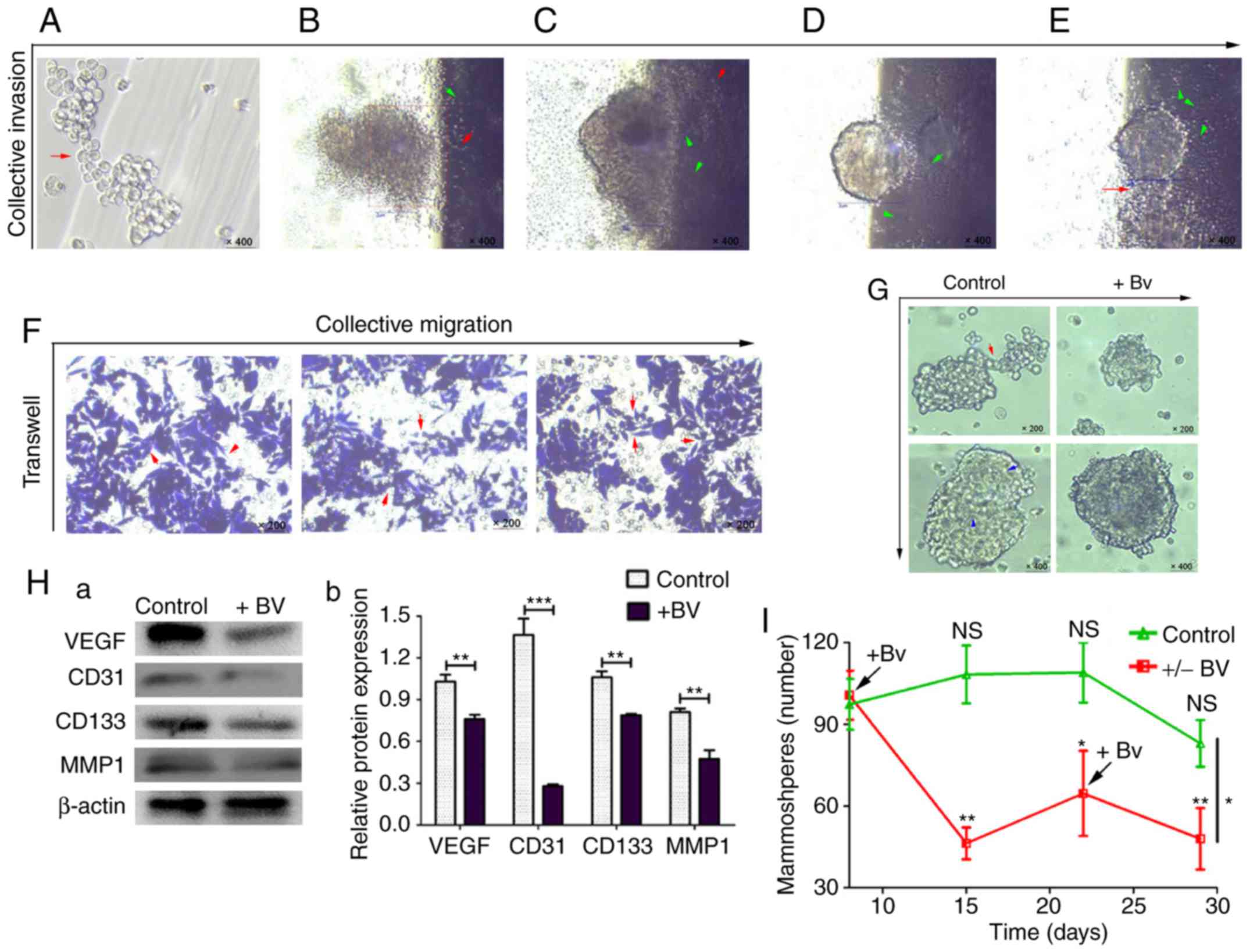 | Figure 6.Mammospheres collectively migrate,
invade and grow through cellular communication and angiogenesis.
(A) Invasive adherent growth (red arrow indicates the connected
cell cluster; magnification, ×400). (B-E) Collective invasion of
mammospheres (green arrows indicate the microtubule-like
structures; red arrows indicate the long strip cell transfer
connections; magnification, ×400). (F) Collective migration, as
observed using the transwell assay on non-trypsin-treated cells
(red arrows indicate the connecting cell bundles in migration;
magnification, ×200). (G) Bevacizumab (5 µg/ml) reduced mammosphere
communication links and weakened mammosphere light transmission
(red arrow indicates the connecting cell bundle; magnification,
×200; blue arrows indicate the microtubule-like structures;
magnification, ×400). (Ha and b) Western blot analysis of the
relative protein expression levels of VEGF, CD31, CD133 and MMP1 in
each group. Data were compared using the Student's t-test and are
expressed as the means ± standard error of the mean. **P<0.01
and ***P<0.001. (I) Formation and inhibition line of stem-like
mammospheres (+Bv represents the addition of 5 µg/ml bevacizumab).
Data between the control and +/− Bv groups were compared using the
Student's t-test at day 29, *P<0.05; and data within each group
were compared using one-way analysis of variance followed by
Tukey's test. Day 22 vs. day 8, *P<0.05; day 15 and day 29 vs.
day 8, **P<0.01. CD, cluster of differentiation; MMP1, matrix
metalloproteinase-1; NS, not significant; VEGF, vascular
endothelial growth factor. |
Since VEGF was considered to serve an important role
in collective cancer cell migration (8), and low dose of bevacizumab could
target the VEGF-dependent transendothelial migration of cancer
cells (34), it was hypothesized
that low dose of anti-VEGF intervention could potentially inhibit
cellular communication. The results demonstrated that following the
addition of bevacizumab (5 µg/ml) (27), after 24 h, the number of peripheral
connective cells and microtubule-like structures in the primary
MDA-MB-231 mammospheres were clearly decreased (Fig. 6G). In addition, anti-VEGF
intervention resulted in reduced activity of mammospheres, with
much lower light transmission compared with the control group
(Fig. 6G). Western blot analysis
revealed that angiogenesis-associated VEGF and CD31,
stemness-associated CD133 (4) and
invasion-associated MMP1 proteins were significantly decreased
following bevacizumab intervention (Fig. 6Ha and b; P<0.01).
In order to further study the effects of
anti-angiogenic therapy on the formation of mammospheres, stem-like
cells were sorted and ~1×103 cells were allocated to
each group. The results demonstrated that following the addition of
bevacizumab, the formation of mammospheres was significantly
inhibited (Fig. 6I, Table II; P<0.01). However, during the
period (~2 weeks) when no bevacizumab was added, the number of
mammospheres formed increased once again. Notably, the number of
mammospheres decreased following the second addition of bevacizumab
(Fig. 6I). Compared with the
control group, mammosphere growth was significantly reduced in the
intervention group during the same period (Table II; P<0.05). These results
indicated that by inhibiting angiogenesis, the formation ability of
stem-like mammospheres may be effectively inhibited.
 | Table II.Mammosphere formation and inhibition
rate. |
Table II.
Mammosphere formation and inhibition
rate.
| Sample group | Initial number of
CSCs (n) | Time (days) | Number of
mammospheres (n) |
P-valuea |
|---|
| Control 1 |
1×103 | 8 | 97.3±9.3 | 0.6779 |
| (−) Bv |
1×103 |
| 100.7±9.0 |
|
| Control 2 |
1×103 | 15 | 108.3±10.6 | 0.0009b |
| (+) Bv |
1×103 |
| 46.3±5.9 |
|
| Control 3 |
1×103 | 22 | 109.0±11.0 | 0.0159c |
| (+/−) Bv |
1×103 |
| 64.7±15.6 |
|
| Control 4 |
1×103 | 29 | 83.0±8.5 | 0.0128c |
| (+/−/+) Bv |
1×103 |
| 48.0±11.3 |
|
CSC and vascular niche formation
promote cancer progression by cellular communication in vivo
In order to further clarify whether cellular
communication exists in vivo, tumorigenesis was compared
between the MDA-MB-231 mammospheres and parental MDA-MB-231 cells.
A minimum of 1×106 MDA-MB-231 cells was required to
induce tumor formation, whereas <1×104 mammospheres
were able to induce tumorigenesis (Fig.
7Aa and b). The results revealed that more tumor nodules, of a
larger size, were formed in the mammosphere group, which had more
irregular nodular-like protrusions (Fig. 7Aa and b, and Table III). In addition, faster growth
and larger tumor volume were observed in the mammosphere group
(Fig. 7Aa and b). Notably, tumor
angiogenesis was often observed around these nodules in the
mammosphere group (Fig. 7Ba and b).
Metastatic lung or liver lesions were also observed in the
mammosphere group (Fig. 7C).
Together, these results indicated that mammospheres may promote
xenograft growth and metastasis through irregular nodules and
angiogenesis.
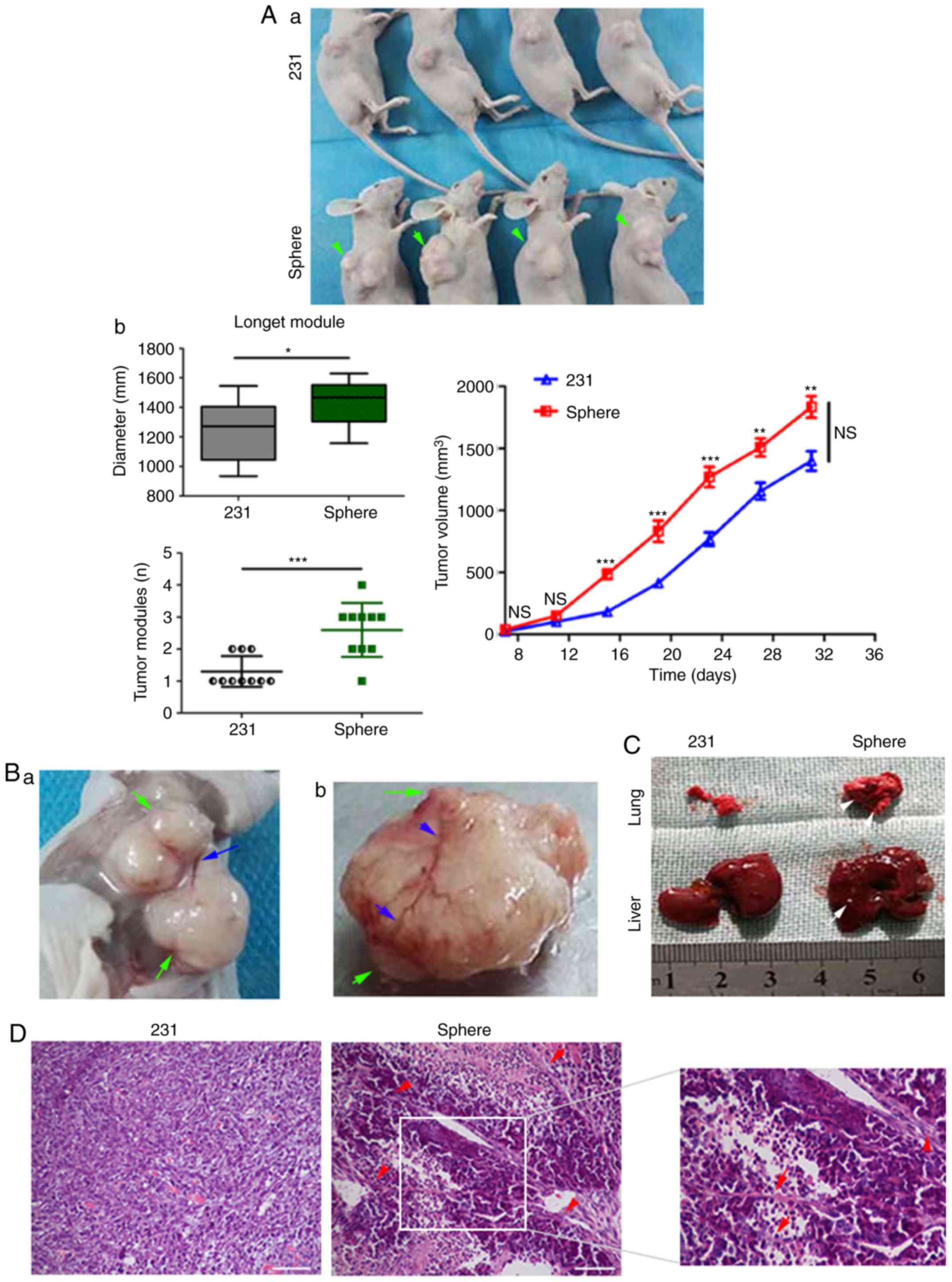 | Figure 7.Cellular communication in vivo
promotes tumor progression. (Aa and b) Tumor size, volume and
growth curve of mice (green arrows indicate multiple fusion or
subcutaneous metastatic nodules). Data were compared using
Student's t-test and are expressed as the means ± standard error of
the mean (n=10/group). *P<0.05, **P<0.01 and ***P<0.001.
(Ba and b) Tumor nodules and protrusions (green arrows) and tumor
angiogenesis vessels (blue arrows). (C) Lung and liver metastases
(white arrows). (D) Large microtubules and cable-like structures
(hematoxylin and eosin staining, red arrows; scale bars, 100 µm).
Cellular communication in vivo promotes tumor progression.
(E) Immunohistochemistry of tumor cells in both groups using
anti-VEGF, -PCNA and -MMP1 (n=10/group; scale bars, 50 µm). (F)
Tumor MVD was calculated using CD31 to mark vascular endothelial
cells (yellow arrows, immunohistochemistry; scale bars, 100 µm).
Data were compared using Student's t-test and are expressed as the
means ± standard error of the mean. ***P<0.001. (G)
Immunofluorescence staining of tumor sections showing the quantity
of CSCs (CD133+, green fluorescence) and tumor
angiogenic cells (CD31+, red fluorescence; scale bars,
100 µm). Nuclei were stained with DAPI. White arrows indicate
CD133+ cells. CD, cluster of differentiation; DAPI,
4′,6-diamidino-2-phenylindole; MMP1, matrix metalloproteinase-1;
PCNA, proliferating cell nuclear antigen; VEGF, vascular
endothelial growth factor. |
 | Table III.Number of subcutaneous tumors and
their diameters. |
Table III.
Number of subcutaneous tumors and
their diameters.
|
| Number of
subcutaneous nodules (n) | Diameter of each
subcutaneous nodule (cm) |
|---|
|
|
|
|
|---|
| Mouse number | MDA-MB-231 | Mammosphere | MDA-MB-231 | Mammosphere |
|---|
| 1 | 1 | 1 | 1.55 | 1.63 |
| 2 | 1 | 2 | 1.43 | 1.60, 0.15 |
| 3 | 1 | 2 | 1.40 | 1.54, 0.25 |
| 4 | 1 | 2 | 1.32 | 1.53, 0.31 |
| 5 | 1 | 3 | 1.22 | 1.50, 0.22,
0.14 |
| 6 | 1 | 3 | 1.38 | 1.43, 0.24,
0.15 |
| 7 | 1 | 3 | 1.11 | 1.38, 0.31,
0.17 |
| 8 | 2 | 3 | 1.06 | 1.32, 0.22,
0.21 |
|
|
|
| 0.48 |
|
| 9 | 2 | 3 | 1.01 | 1.27, 0.30,
0.12 |
|
|
|
| 0.66 |
|
| 10 | 2 | 4 | 0.93 | 1.16, 0.51, 0.15,
0.11 |
|
|
|
| 0.72 |
|
|
P-valuea | 0.0005b |
| 0.0275c |
|
H&E staining indicated that more
microtubule-like structures and similar cellular strand connections
appeared in the tissues of the mammosphere group compared with
those of the parental MDA-MB-231 cell group (Fig. 7D). In addition, immunohistochemistry
detected a higher positive expression of VEGF, PCNA and MMP1 in the
mammosphere group compared with in the parental MDA-MB-231 cell
group (Fig. 7E and Table IV). Analysis of the stained area
revealed that all three proteins were expressed near cancer nests
(Fig. 7E). Subsequently, CD31
staining was used to calculate the MVD; the MVD in the mammosphere
group was clearly higher than that in the parental MDA-MB-231 group
(Fig. 7F; P<0.001).
Immunofluorescence detection demonstrated that a large number of
tumor vascular cells gathered around the CSC niches and tubular
channels in the mammosphere group (Fig.
7G). CD133+ cells were concentrically distributed
along the tumor blood vessels and channel structures. In
particular, CD133 could be delivered or distributed further along
the tissue tubular channels in the mammosphere group (Fig. 7G) (19). These results indicated that CSCs and
vascular niches may be closely associated with angiogenesis and
tubular channels, which are widely distributed in these highly
metastatic tissues. All these may enhance the communication between
CSC niches and cancer cells in vivo.
 | Table IV.Immunohistochemical analysis of
positive expression levels of VEGF, PCNA and MMP1 in two mouse
group samples. |
Table IV.
Immunohistochemical analysis of
positive expression levels of VEGF, PCNA and MMP1 in two mouse
group samples.
|
|
| Expression |
|
|
|---|
|
|
|
|
|
|
|---|
| Protein | n | − | + | ++ | +++ | Proportion of +++
(%) | P-value |
|---|
| VEGF |
|
|
|
|
|
| 0.020a,b |
|
MDA-MB-231 | 10 | 2 | 3 | 4 | 1 | 10.0 |
|
|
Mammosphere | 10 | 1 | 1 | 1 | 7 | 70.0 |
|
| PCNA |
|
|
|
|
|
| 0.007c,d |
|
MDA-MB-231 | 10 | 1 | 3 | 4 | 2 | 20.0 |
|
|
Mammosphere | 10 | 0 | 2 | 0 | 8 | 80.0 |
|
| MMP1 |
|
|
|
|
|
| 0.033a,b |
|
MDA-MB-231 | 10 | 2 | 4 | 4 | 0 | 0.0 |
|
|
Mammosphere | 10 | 1 | 3 | 1 | 5 | 50.0 |
|
Discussion
Cellular communication through synapses and tube
structures has recently attracted the attention of researchers
(16). However, reports of such
structures during conventional cancer cell culture in vitro
are rare (16). Cellular
communication structures, such as nanotubes or microtubules, have
mainly been reported in developmental biology (16–18).
In the present study, such cellular communication structures were
observed during mammosphere formation and differentiation in
vitro, proving that microtubule-like structural networks may be
present in vitro. Notably, although cell protrusion has been
reported in tumor growth of human gliomas (20), to the best of our knowledge, the
present study is the first to detect similar cell protrusions
dynamically forming on mammospheres. An increasing number of
studies have reported that antenna-like cell protrusions
participate in cellular information exchange (20,29,35).
Herein, it was determined that these cell protrusions promoted not
only cell fusion, but also cellular communication. Notably, it was
further observed that some microtubule-like structures and contacts
joined in these mammosphere-to-mammosphere communications. Since
low doses of serum-containing medium can promote CSC
differentiation and facilitate its observation (36,37),
large microtubule-like structures outside or inside mammospheres
were observed when 1% serum-containing medium was added. Further
culture revealed that various networks of microtubules were
differentiated, with some presenting as a net of vascular and
fibrous morphology. In addition, nanotube-like structures on
membranes formed part of cellular networks. Based on these networks
of microtubule-like structures, cell-to-cell,
mammosphere-to-mammosphere and mammosphere-to-cell connection may
occur, potentially allowing information to be exchanged.
The present study aimed to determine how cellular
communication is reflected in mammospheres. Previous studies have
revealed that mammospheres are groups of multi-differentiated cells
(1,10); therefore, it was hypothesized that
there may be a common characteristic between stemness and
differentiation that leads to the continuous exchange of
information in mammospheres. In the present study,
CD44+/CD24+ cells were significantly
increased in all mammosphere groups; however, the proportion of
CD44+/CD24− cells varied according to
different parental cell lines. Since the early stages of high
proliferation, invasion and heterogeneous differentiation are
partly attributed to CD24+ cells in mammary tumors
(30,38,39),
and stemness characteristics are mainly attributed to
CD44+ cells (2,31,40),
the co-expression of CD44 and CD24 may result in high levels of
stemness and differentiation in mammospheres. The cell cycle
results indicated that mammospheres exhibited a more flexible space
for division. Genes associated with stemness, invasiveness and
angiogenesis were all highly expressed in mammospheres. The results
from the breast cancer cell line analysis revealed that
mammospheres and primary mammospheres exhibited a highly invasive
collective cell state, in which stemness and multi-differentiation
factors were detected (41). It may
be hypothesized that the phenotypic alteration of cells gives rise
to an equilibrium state in mammospheres, requiring continuous
information exchange between cells.
Studies regarding CSCs and cell clusters have made
great progress in determining the mechanism of collective tumor
metastasis (7,8,42). In
the present study, it was confirmed that cellular communication may
participate in this process. During mammosphere migration and
invasion, the leader cells, follower cells and mammospheres moved
collectively (8). The results of an
invasion analysis demonstrated that microtubule-like structures and
cellular cord-like connections were constantly accompanying each
other and induced interconnection of cell clusters. Cellular
information appeared to constantly be exchanged through this
collective movement (8,18). Konen et al predicted that
there is some cooperation between leader and follower cells in cell
cultures during their movement (8).
Since it has been reported that collective cancer
movement is mainly associated with VEGF and fibroblast growth
factor (8,43), it was suggested that anti-VEGF or
anti-angiogenic therapy could inhibit communication.
Morphologically, mammosphere growth was significantly inhibited,
and the associated cell connections and microtubule-like structures
were markedly reduced in response to anti-VEGF. Protein detection
revealed that anti-VEGF intervention not only inhibited VEGF and
CD31, but also significantly reduced the expression of proteins
associated with stemness and invasiveness. The results of the
growth and inhibition line analysis demonstrated that anti-VEGF
intervention could effectively inhibit the formation and growth of
stem-like mammospheres. Through comprehensive analysis, it was
hypothesized that the cascade response to anti-VEGF therapy might
effectively inhibit the multilineage differentiation of CSCs. VEGF
has been reported to serve an important role in information
exchange during cell differentiation, including the differentiation
of functional endothelium (44),
stem cell remodeling (45), and to
have extensive effects on tumor microvasculature (46). However, the therapeutic effects of
anti-VEGF treatment can be reduced when the addition of the drug is
interrupted (47,48). It was hypothesized that, due to the
persistence of cellular communication, the inhibition of stem-like
mammosphere growth and invasion may be one of the reasons for the
need for continuous intervention.
In vivo, xenografts from mammospheres
promoted tumor growth and metastasis through the development of
fusion nodules and angiogenesis. Staining confirmed that the number
of microtubule-like channels in mammosphere tissues was increased,
and the channels were much longer and messier than those in the
parental cell tissues. Immunohistochemical analysis indicated that
VEGF, PCNA and MMP1 not only exhibited higher expression in the
mammosphere group tissues, but also around cancer nests. MVD was
significantly increased and was widely distributed in the
mammosphere tissue group, which indicated a large number of
microvascular channels within these tissues. This was more apparent
in CSCs and vascular niches. As previous studies have reported that
CSCs or vascular niches may be the base for metastasis and
network-like information export and exchange (15,49,50),
it was hypothesized that cellular communication would be more
strongly observed near these niches in vivo. However, the
transmission of information to the distant areas of the tissues may
still need to occur through microtubule-like channels,
cell-associated cords or tumor angiogenesis. As previously shown,
CD133+ was not only distributed along the
CD31+ cell strip, but could also be delivered further
along the tissue tubular channels (19).
In conclusion, the results of the present study
indicated that cellular communication was not only widely present
in the growth and differentiation processes of mammospheres in
vitro, but was also reflected in vivo. The collective
characteristics of stemness and differentiation in mammospheres
contributed to the continuous exchange of information. Furthermore,
anti-angiogenic treatment may be an efficient method of blocking
cellular communication; however, more specific mechanisms need to
be explored.
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from the
National Natural Science Foundation of China (grant no.
81272898).
Availability of data and materials
The datasets used and/or analyzed during the
current study are available from the corresponding author on
reasonable request.
Authors' contributions
SH, XZ and LZ conceived and designed the study. SH,
NY, GW, FW and LF performed the experiments. SH wrote the
manuscript. MLu, MLi and AL edited the manuscript and were also
involved in the conception of the study. All authors read and
approved the manuscript, and agree to be accountable for all
aspects of the research, ensuring that the accuracy or integrity of
any part of the work are appropriately investigated and
resolved.
Ethics approval and consent to
participate
Mouse care and usage were approved by the Ethics
Committee of the School of Medicine of Xi'an Jiaotong University
(approval no. 0108; Xi'an, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Adorno-Cruz V, Kibria G, Liu X, Doherty M,
Junk DJ, Guan D, Hubert C, Venere M, Mulkearns-Hubert E, Sinyuk M,
et al: Cancer stem cells: Targeting the roots of cancer, seeds of
metastasis, and sources of therapy resistance. Cancer Res.
75:924–929. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Al-Hajj M, Wicha MS, Benito-Hernandez A,
Morrison SJ and Clarke MF: Prospective identification of
tumorigenic breast cancer cells. Proc Natl Acad Sci USA.
100:3983–3988. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ginestier C, Hur MH, Charafe-Jauffret E,
Monville F, Dutcher J, Brown M, Jacquemier J, Viens P, Kleer CG,
Liu S, et al: ALDH1 is a marker of normal and malignant human
mammary stem cells and a predictor of poor clinical outcome. Cell
Stem Cell. 1:555–567. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Liu TJ, Sun BC, Zhao XL, Zhao XM, Sun T,
Gu Q, Yao Z, Dong XY, Zhao N and Liu N: CD133+ cells
with cancer stem cell characteristics associates with vasculogenic
mimicry in triple-negative breast cancer. Oncogene. 32:544–553.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Martelotto LG, Ng CK, Piscuoglio S,
Weigelt B and Reis-Filho JS: Breast cancer intra-tumor
heterogeneity. Breast Cancer Res. 16:2102014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Prasetyanti PR and Medema JP: Intra-tumor
heterogeneity from a cancer stem cell perspective. Mol Cancer.
16:412017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Cheung KJ, Padmanaban V, Silvestri V,
Schipper K, Cohen JD, Fairchild AN, Gorin MA, Verdone JE, Pienta
KJ, Bader JS and Ewald AJ: Polyclonal breast cancer metastases
arise from collective dissemination of keratin 14-expressing tumor
cell clusters. Proc Natl Acad Sci USA. 113:E854–E863. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Konen J, Summerbell E, Dwivedi B, Galior
K, Hou Y, Rusnak L, Chen A, Saltz J, Zhou W, Boise LH, et al:
Image-guided genomics of phenotypically heterogeneous populations
reveals vascular signalling during symbiotic collective cancer
invasion. Nat Commun. 8:150782017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Santoro A, Vlachou T, Carminati M, Pelicci
PG and Mapelli M: Molecular mechanisms of asymmetric divisions in
mammary stem cells. EMBO Rep. 17:1700–1720. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
van Niekerk G, Davids LM, Hattingh SM and
Engelbrecht AM: Cancer stem cells: A product of clonal evolution?
Int J Cancer. 140:993–999. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tang S, Xiang T, Huang S, Zhou J, Wang Z,
Xie R, Long H and Zhu B: Ovarian cancer stem-like cells
differentiate into endothelial cells and participate in tumor
angiogenesis through autocrine CCL5 signaling. Cancer Lett.
376:137–147. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Katajisto P, Döhla J, Chaffer CL,
Pentinmikko N, Marjanovic N, Iqbal S, Zoncu R, Chen W, Weinberg RA
and Sabatini DM: Stem cells. Asymmetric apportioning of aged
mitochondria between daughter cells is required for stemness.
Science. 348:340–343. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Serres E, Debarbieux F, Stanchi F,
Maggiorella L, Grall D, Turchi L, Burel-Vandenbos F,
Figarella-Branger D, Virolle T, Rougon G and Van
Obberghen-Schilling E: Fibronectin expression in glioblastomas
promotes cell cohesion, collective invasion of basement membrane in
vitro and orthotopic tumor growth in mice. Oncogene. 33:3451–3462.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Korkaya H, Liu S and Wicha MS: Breast
cancer stem cells, cytokine networks, and the tumor
microenvironment. J Clin Invest. 121:3804–3809. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ping YF, Zhang X and Bian XW: Cancer stem
cells and their vascular niche: Do they benefit from each other?
Cancer Lett. 380:561–567. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Baker M: How the Internet of cells has
biologists buzzing. Nature. 549:322–324. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kornberg TB and Gilboa L: Developmental
biology: Nanotubes in the niche. Nature. 523:292–293. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gerdes HH and Carvalho RN: Intercellular
transfer mediated by tunneling nanotubes. Curr Opin Cell Biol.
20:470–475. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Reichert D, Scheinpflug J, Karbanová J,
Freund D, Bornhäuser M and Corbeil D: Tunneling nanotubes mediate
the transfer of stem cell marker CD133 between hematopoietic
progenitor cells. Exp Hematol. 44(1092–1112): e10922016. View Article : Google Scholar
|
|
20
|
Osswald M, Jung E, Sahm F, Solecki G,
Venkataramani V, Blaes J, Weil S, Horstmann H, Wiestler B, Syed M,
et al: Brain tumour cells interconnect to a functional and
resistant network. Nature. 528:93–98. 2015.PubMed/NCBI
|
|
21
|
Osswald M, Solecki G, Wick W and Winkler
F: A malignant cellular network in gliomas: Potential clinical
implications. Neuro Oncol. 18:479–485. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Patheja P and Sahu K: Macrophage
conditioned medium induced cellular network formation in MCF-7
cells through enhanced tunneling nanotube formation and tunneling
nanotube mediated release of viable cytoplasmic fragments. Exp Cell
Res. 355:182–193. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Thayanithy V, Dickson EL, Steer C,
Subramanian S and Lou E: Tumor-stromal cross talk: Direct
cell-to-cell transfer of oncogenic microRNAs via tunneling
nanotubes. Transl Res. 164:359–365. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Dontu G, Abdallah WM, Foley JM, Jackson
KW, Clarke MF, Kawamura MJ and Wicha MS: In vitro propagation and
transcriptional profiling of human mammary stem/progenitor cells.
Genes Dev. 17:1253–1270. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Peleg R, Romzova M, Kogan-Zviagin, Apte RN
and Priel E: Modification of topoisomerases in mammospheres derived
from breast cancer cell line: Clinical implications for combined
treatments with tyrosine kinase inhibitors. BMC Cancer. 14:9102014.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Takahashi H, Nomura Y, Nishida J, Fujino
Y, Yanagi Y and Kawashima H: Vascular endothelial growth factor
(VEGF) concentration is underestimated by enzyme-linked
immunosorbent assay in the presence of anti-VEGF drugs. Invest
Ophthalmol Vis Sci. 57:462–466. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Weidner N, Semple JP, Welch WR and Folkman
J: Tumor angiogenesis and metastasis-correlation in invasive breast
carcinoma. N Engl J Med. 324:1–8. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ramirez-Weber FA and Kornberg TB:
Cytonemes: Cellular processes that project to the principal
signaling center in Drosophila imaginal discs. Cell. 97:599–607.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Rostoker R, Abelson S, Genkin I,
Ben-Shmuel S, Sachidanandam R, Scheinman EJ, Bitton-Worms K, Orr
ZS, Caspi A, Tzukerman M and LeRoith D: CD24(+) cells fuel rapid
tumor growth and display high metastatic capacity. Breast Cancer
Res. 17:782015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li W, Ma H, Zhang J, Zhu L, Wang C and
Yang Y: Unraveling the roles of CD44/CD24 and ALDH1 as cancer stem
cell markers in tumorigenesis and metastasis. Sci Rep. 7:138562017.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang Z, Oron E, Nelson B, Razis S and
Ivanova N: Distinct lineage specification roles for NANOG, OCT4,
and SOX2 in human embryonic stem cells. Cell Stem Cell. 10:440–454.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ebrahem Q, Chaurasia SS, Vasanji A, Qi JH,
Klenotic PA, Cutler A, Asosingh K, Erzurum S and Anand-Apte B:
Cross-talk between vascular endothelial growth factor and matrix
metalloproteinases in the induction of neovascularization in vivo.
Am J Pathol. 176:496–503. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Prager GW, Lackner EM, Krauth MT, Unseld
M, Poettler M, Laffer S, Cerny-Reiterer S, Lamm W, Kornek GV,
Binder BR, et al: Targeting of VEGF-dependent transendothelial
migration of cancer cells by bevacizumab. Mol Oncol. 4:150–160.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Rustom A, Saffrich R, Markovic I, Walther
P and Gerdes HH: Nanotubular highways for intercellular organelle
transport. Science. 303:1007–1010. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Hong X, Chedid K and Kalkanis SN:
Glioblastoma cell line-derived spheres in serumcontaining medium
versus serum-free medium: A comparison of cancer stem cell
properties. Int J Oncol. 41:1693–1700. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
deCarvalho AC, Nelson K, Lemke N, Lehman
NL, Arbab AS, Kalkanis S and Mikkelsen T: Gliosarcoma stem cells
undergo glial and mesenchymal differentiation in vivo. Stem Cells.
28:181–190. 2010.PubMed/NCBI
|
|
38
|
Hosonaga M, Arima Y, Sugihara E, Kohno N
and Saya H: Expression of CD24 is associated with HER2 expression
and supports HER2-Akt signaling in HER2-positive breast cancer
cells. Cancer Sci. 105:779–787. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Jaggupilli A and Elkord E: Significance of
CD44 and CD24 as cancer stem cell markers: An enduring ambiguity.
Clin Dev Immunol. 2012:7080362012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Camerlingo R, Ferraro GA, De Francesco F,
Romano M, Nicoletti G, Di Bonito M, Rinaldo M, D'Andrea F and
Pirozzi G: The role of CD44+/CD24-/low biomarker for screening,
diagnosis and monitoring of breast cancer. Oncol Rep. 31:1127–1132.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Smart CE, Morrison BJ, Saunus JM, Vargas
AC, Keith P, Reid L, Wockner L, Askarian-Amiri M, Sarkar D, Simpson
PT, et al: In vitro analysis of breast cancer cell line
tumourspheres and primary human breast epithelia mammospheres
demonstrates inter- and intrasphere heterogeneity. PLoS One.
8:e643882013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Aceto N, Bardia A, Miyamoto DT, Donaldson
MC, Wittner BS, Spencer JA, Yu M, Pely A, Engstrom A, Zhu H, et al:
Circulating tumor cell clusters are oligoclonal precursors of
breast cancer metastasis. Cell. 158:1110–1122. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Karagiannis GS, Poutahidis T, Erdman SE,
Kirsch R, Riddell RH and Diamandis EP: Cancer-associated
fibroblasts drive the progression of metastasis through both
paracrine and mechanical pressure on cancer tissue. Mol Cancer Res.
10:1403–1418. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Nourse MB, Halpin DE, Scatena M, Mortisen
DJ, Tulloch NL, Hauch KD, Torok-Storb B, Ratner BD, Pabon L and
Murry CE: VEGF induces differentiation of functional endothelium
from human embryonic stem cells: Implications for tissue
engineering. Arterioscler Thromb Vasc Biol. 30:80–89. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Licht T, Rothe G, Kreisel T, Wolf B, Benny
O, Rooney AG, Ffrench-Constant C, Enikolopov G and Keshet E: VEGF
preconditioning leads to stem cell remodeling and attenuates
age-related decay of adult hippocampal neurogenesis. Proc Natl Acad
Sci USA. 113:E7828–E7836. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Beck B, Driessens G, Goossens S, Youssef
KK, Kuchnio A, Caauwe A, Sotiropoulou PA, Loges S, Lapouge G, Candi
A, et al: A vascular niche and a VEGF-Nrp1 loop regulate the
initiation and stemness of skin tumours. Nature. 478:399–403. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Xu Y, Li Q, Li XY, Yang QY, Xu WW and Liu
GL: Short-term anti-vascular endothelial growth factor treatment
elicits vasculogenic mimicry formation of tumors to accelerate
metastasis. J Exp Clin Cancer Res. 31:162012. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Markowska A, Sajdak S, Markowska J and
Huczyński A: Angiogenesis and cancer stem cells: New perspectives
on therapy of ovarian cancer. Eur J Med Chem. 142:87–94. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Plaks V, Kong N and Werb Z: The cancer
stem cell niche: How essential is the niche in regulating stemness
of tumor cells? Cell Stem Cell. 16:225–238. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Hale JS, Li M and Lathia JD: The malignant
social network: Cell-cell adhesion and communication in cancer stem
cells. Cell Adh Migr. 6:346–355. 2012. View Article : Google Scholar : PubMed/NCBI
|