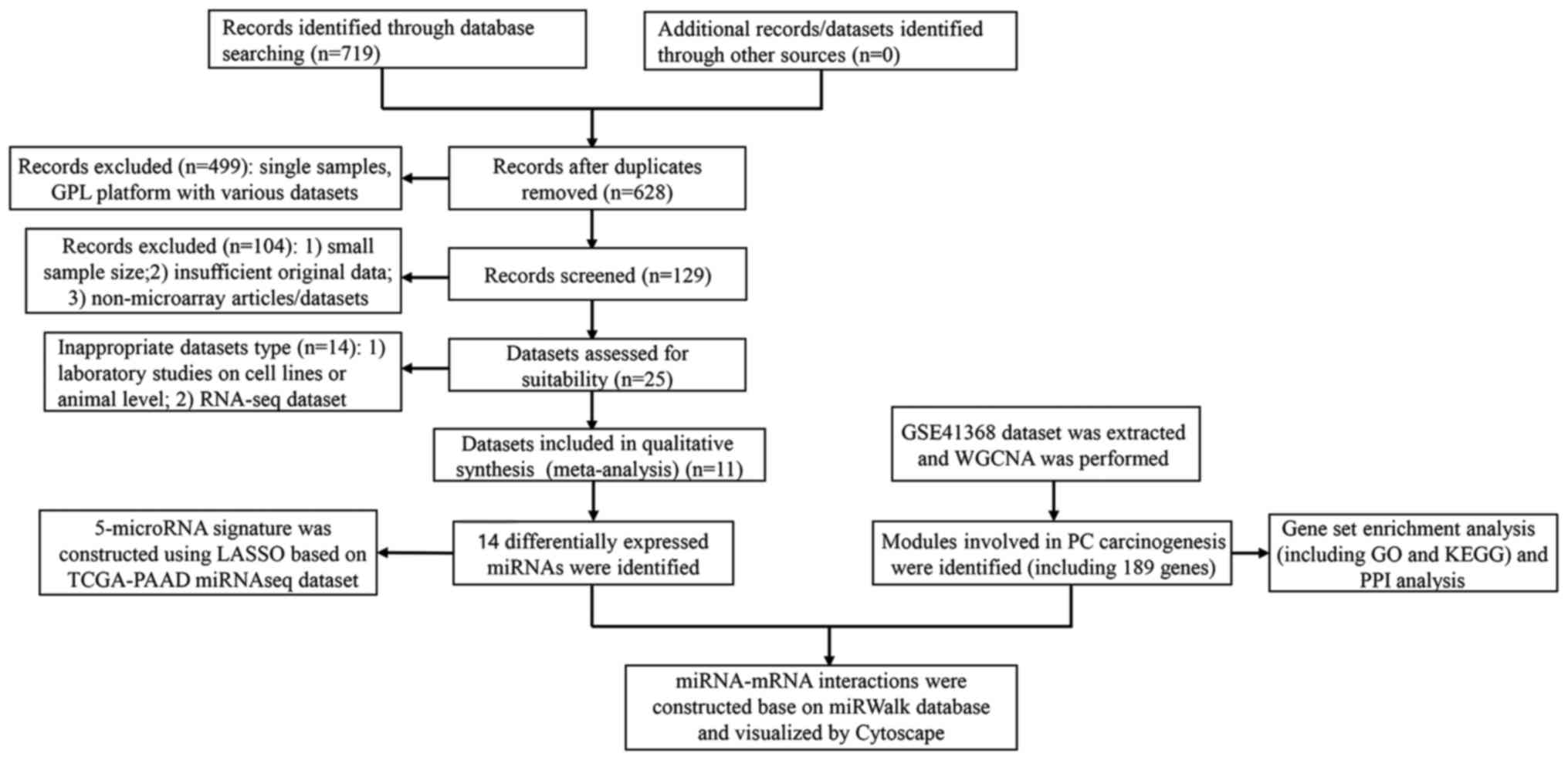
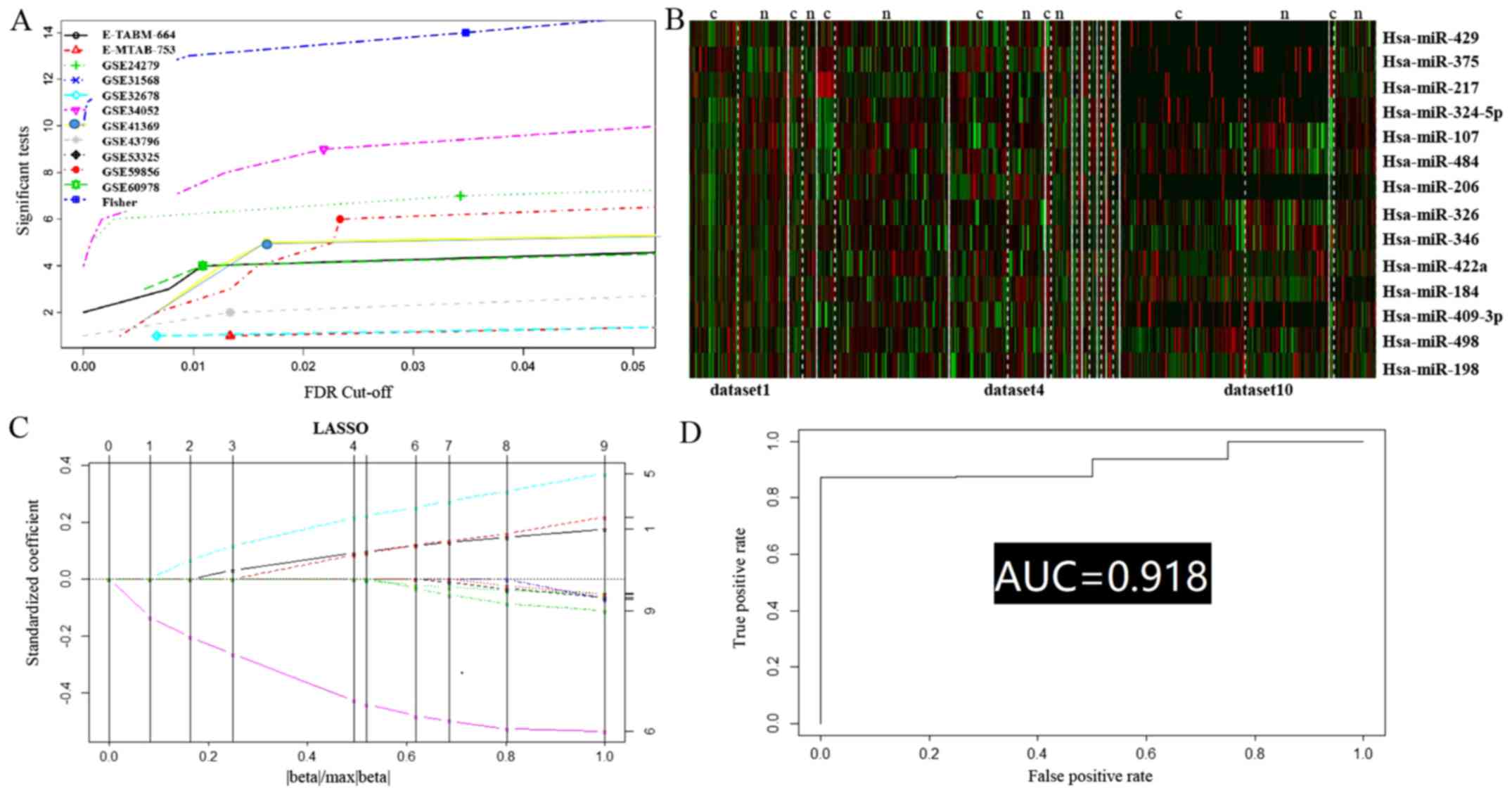
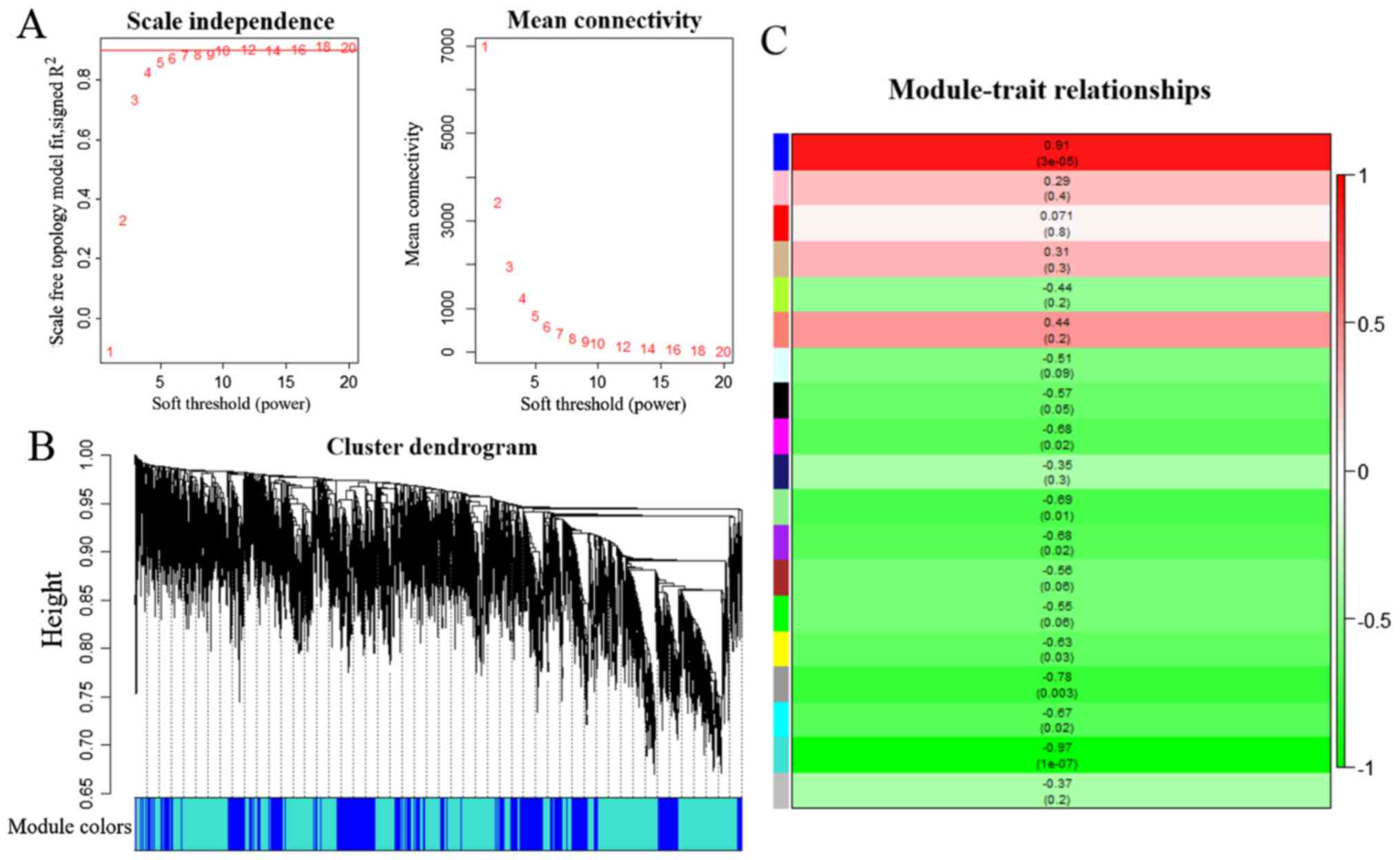
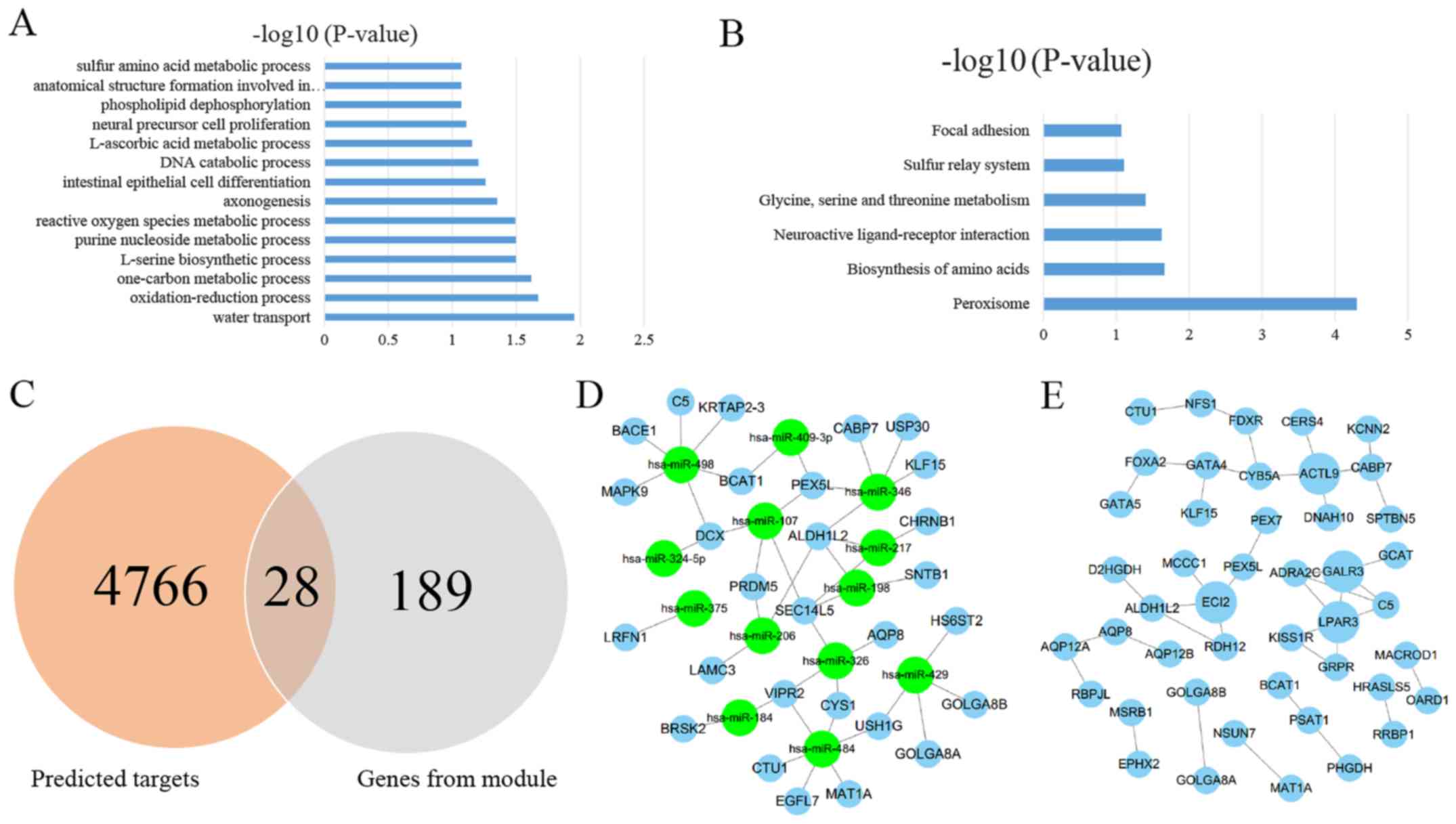
|
1
|
Jemal A, Siegel R, Ward E, Murray T, Xu J
and Thun MJ: Cancer statistics, 2007. CA Cancer J Clin. 57:43–66.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Simianu VV, Zyromski NJ, Nakeeb A and
Lillemoe KD: Pancreatic cancer: Progress made. Acta Oncol.
49:407–417. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hartwig W, Werner J, Jäger D, Debus J and
Büchler MW: Improvement of surgical results for pancreatic cancer.
Lancet Oncol. 14:e476–e485. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Costello E, Greenhalf W and Neoptolemos
JP: New biomarkers and targets in pancreatic cancer and their
application to treatment. Nat Rev Gastroenterol Hepatol. 9:435–444.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Du Y, Liu M, Gao J and Li Z: Aberrant
microRNAs expression patterns in pancreatic cancer and their
clinical translation. Cancer Biother Radiopharm. 28:361–369. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gregory RI, Chendrimada TP, Cooch N and
Shiekhattar R: Human RISC couples microRNA biogenesis and
posttranscriptional gene silencing. Cell. 123:631–640. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yates LA, Norbury CJ and Gilbert RJ: The
long and short of microRNA. Cell. 153:516–519. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Baer C, Claus R and Plass C: Genome-wide
epigenetic regulation of miRNAs in cancer. Cancer Res. 73:473–477.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Singh R and Mo YY: Role of microRNAs in
breast cancer. Cancer Biol Ther. 14:201–212. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Song S and Ajani JA: The role of microRNAs
in cancers of the upper gastrointestinal tract. Nat Rev
Gastroenterol Hepatol. 10:109–118. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bustin SA and Dorudi S: Gene expression
profiling for molecular staging and prognosis prediction in
colorectal cancer. Expert Rev Mol Diagn. 4:599–607. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kulasingam V and Diamandis EP: Strategies
for discovering novel cancer biomarkers through utilization of
emerging technologies. Nat Clin Pract Oncol. 5:588–599. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Nannini M, Pantaleo MA, Maleddu A, Astolfi
A, Formica S and Biasco G: Gene expression profiling in colorectal
cancer using microarray technologies: Results and perspectives.
Cancer Treat Rev. 35:201–209. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lascorz J, Hemminki K and Försti A:
Systematic enrichment analysis of gene expression profiling studies
identifies consensus pathways implicated in colorectal cancer
development. J Carcinog. 10:72011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lu Y, Zhou Y, Qu W, Deng M and Zhang C: A
Lasso regression model for the construction of microRNA-target
regulatory networks. Bioinformatics. 27:2406–2413. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Langfelder P and Horvath S: WGC NA An R
package for weighted correlation network analysis. BMC
Bioinformatics. 9:5592008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Franceschini A, Szklarczyk D, Frankild S,
Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C
and Jensen LJ: STRING v9.1: Protein-protein interaction networks,
with increased coverage and integration. Nucleic Acids Res.
41:D808–D815. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Smoot ME, Ono K, Ruscheinski J, Wang PL
and Ideker T: Cytoscape 2.8: New features for data integration and
network visualization. Bioinformatics. 27:431–432. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Davis S and Meltzer PS: GE Oquery A bridge
between the Gene Expression Omnibus (GEO) and BioConductor.
Bioinformatics. 23:1846–1847. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Diboun I, Wernisch L, Orengo CA and
Koltzenburg M: Microarray analysis after RNA amplification can
detect pronounced differences in gene expression using limma. BMC
Genomics. 7:2522006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Haunsberger SJ, Connolly NM and Prehn JH:
miRNAmeConverter: An R/bioconductor package for translating mature
miRNA names to different miRBase versions. Bioinformatics.
33:592–593. 2017.PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Colaprico A, Silva TC, Olsen C, Garofano
L, Cava C, Garolini D, Sabedot TS, Malta TM, Pagnotta SM,
Castiglioni I, et al: TCGAbiolinks: An R/Bioconductor package for
integrative analysis of TCGA data. Nucleic Acids Res. 44:e712016.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Srihari S and Leong HW: Temporal dynamics
of protein complexes in PPI networks: A case study using yeast cell
cycle dynamics. BMC Bioinformatics. 13((Suppl 17)):
S162012.PubMed/NCBI
|
|
25
|
Dweep H, Gretz N and Sticht C: miRWalk
database for miRNA-target interactions. Methods Mol Biol.
1182:289–305. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Friedman RC, Farh KK, Burge CB and Bartel
DP: Most mammalian mRNAs are conserved targets of microRNAs. Genome
Res. 19:92–105. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Betel D, Koppal A, Agius P, Sander C and
Leslie C: Comprehensive modeling of microRNA targets predicts
functional non-conserved and non-canonical sites. Genome Biol.
11:R902010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kertesz M, Iovino N, Unnerstall U, Gaul U
and Segal E: The role of site accessibility in microRNA target
recognition. Nat Genet. 39:1278–1284. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ramasamy A, Mondry A, Holmes CC and Altman
DG: Key issues in conducting a meta-analysis of gene expression
microarray datasets. PLoS Med. 5:e1842008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yuan C, Morales-Oyarvide V, Babic A, Clish
CB, Kraft P, Bao Y, Qian ZR, Rubinson DA, Ng K, Giovannucci EL, et
al: Cigarette smoking and pancreatic cancer survival. J Clin Oncol.
35:1822–1828. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Carreras-Torres R, Johansson M, Gaborieau
V, Haycock PC, Wade KH, Relton CL, Martin RM, Davey Smith G and
Brennan P: The role of obesity, type 2 diabetes, and metabolic
factors in pancreatic cancer: A mendelian randomization study. J
Natl Cancer Inst. 109:92017. View Article : Google Scholar
|
|
32
|
Guerra C, Collado M, Navas C, Schuhmacher
AJ, Hernández-Porras I, Cañamero M, Rodriguez-Justo M, Serrano M
and Barbacid M: Pancreatitis-induced inflammation contributes to
pancreatic cancer by inhibiting oncogene-induced senescence. Cancer
Cell. 19:728–739. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Molina-Montes E, Gomez-Rubio P, Márquez M,
Rava M, Löhr M, Michalski CW, Molero X, Farré A, Perea J, Greenhalf
W, et al: Risk of pancreatic cancer associated with family history
of cancer and other medical conditions by accounting for smoking
among relatives. Int J Epidemiol. 47:473–483. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Tanday S: Biomarkers in blood could help
to detect pancreatic cancer. Lancet Oncol. 15:e1082014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Li L, Li Z, Kong X, Xie D, Jia Z, Jiang W,
Cui J, Du Y, Wei D, Huang S and Xie K: Down-regulation of
microRNA-494 via loss of SMAD4 increases FOXM1 and β-catenin
signaling in pancreatic ductal adenocarcinoma cells.
Gastroenterology. 147:485–497.e18. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Liu R, Chen X, Du Y, Yao W, Shen L, Wang
C, Hu Z, Zhuang R, Ning G, Zhang C, et al: Serum microRNA
expression profile as a biomarker in the diagnosis and prognosis of
pancreatic cancer. Clin Chem. 58:610–618. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Schultz NA, Werner J, Willenbrock H,
Roslind A, Giese N, Horn T, Wøjdemann M and Johansen JS: MicroRNA
expression profiles associated with pancreatic adenocarcinoma and
ampullary adenocarcinoma. Mod Pathol. 25:1609–1622. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Avalos M, Pouyes H, Grandvalet Y, Orriols
L and Lagarde E: Sparse conditional logistic regression for
analyzing large-scale matched data from epidemiological studies: A
simple algorithm. BMC Bioinformatics. 16((Suppl 6)): S12015.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Song B, Zheng K, Ma H, Liu A, Jing W, Shao
C, Li G and Jin G: miR-429 determines poor outcome and inhibits
pancreatic ductal adenocarcinoma growth by targeting TBK1. Cell
Physiol Biochem. 35:1846–1856. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yang D, Yan R and Zhang X, Zhu Z, Wang C,
Liang C and Zhang X: Deregulation of MicroRNA-375 inhibits cancer
proliferation migration and chemosensitivity in pancreatic cancer
through the association of HOXB3. Am J Transl Res. 8:1551–1559.
2016.PubMed/NCBI
|
|
41
|
Deng S, Zhu S, Wang B, Li X, Liu Y, Qin Q,
Gong Q, Niu Y, Xiang C, Chen J, et al: Chronic pancreatitis and
pancreatic cancer demonstrate active epithelial-mesenchymal
transition profile, regulated by miR-217-SIRT1 pathway. Cancer
Lett. 355:184–191. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Lee KH, Lotterman C, Karikari C, Omura N,
Feldmann G, Habbe N, Goggins MG, Mendell JT and Maitra A:
Epigenetic silencing of MicroRNA miR-107 regulates cyclin-dependent
kinase 6 expression in pancreatic cancer. Pancreatology. 9:293–301.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Mao Y, Shen J, Lu Y, Lin K, Wang H, Li Y,
Chang P, Walker MG and Li D: RNA sequencing analyses reveal novel
differentially expressed genes and pathways in pancreatic cancer.
Oncotarget. 8:42537–42547. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Bloomston M, Frankel WL, Petrocca F,
Volinia S, Alder H, Hagan JP, Liu CG, Bhatt D, Taccioli C and Croce
CM: MicroRNA expression patterns to differentiate pancreatic
adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA.
297:1901–1908. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Lin D, Shkedy Z, Burzykowski T, Ion R,
Göhlmann HW, Bondt AD, Perer T, Geerts T, Van den Wyngaert I and
Bijnens L: An investigation on performance of Significance Analysis
of Microarray (SAM) for the comparisons of several treatments with
one control in the presence of small-variance genes. Biom J.
50:801–823. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Yang S, Zhang L, Purohit V, Shukla SK,
Chen X, Yu F, Fu K, Chen Y, Solheim J, Singh PK, et al: Active YAP
promotes pancreatic cancer cell motility, invasion and
tumorigenesis in a mitotic phosphorylation-dependent manner through
LPAR3. Oncotarget. 6:36019–36031. 2015. View Article : Google Scholar : PubMed/NCBI
|