Introduction
Lung cancer is a severe disease that causes
extensive death worldwide (1). Once
diagnosed, patients have a poor prognosis and there are no
effective treatments. The overall 5-year survival rate of lung
cancer patients is <14%, less than breast and colon cancer
(2). In the last decade,
pathological molecular mechanisms have been widely established in
lung cancer, such as epidermal growth factor receptor (EGFR)
(3,4) and fibroblast growth factor receptor
(FGFR) (5,6) signaling. Based on these targets,
rational treatments have been developed, among which tyrosine
kinase inhibitors are well-known (7). Although the patients harboring EGFR
mutations are more sensitive to tyrosine kinase inhibitor
treatment, frequent resistance renders its clinical applications
difficult for this disease (8).
Thus, it is urgent to develop novel efficient drug targets for lung
cancer.
BAI1-associated protein 2-like 2 (BAIAP2L2), also
known as Pinkbar, is located on chromosome 22q13.1 (9). Along with BAIAP2L1 (insulin receptor
tyrosine kinase substrate; also known as IRTKS), IRSp53, MIM
(missing in metastasis; also known as MTSS1) and ABBA
(actin-bundling protein with BAIAP2 homology; also known as
MTSS1L), BAIAP2L2 belongs to the Inverse Bin-Amphiphysin-Rvs
(I-BAR) subfamily (10–12). The I-BAR domain interacts with
signaling pathways to induce the formation of actin-based membrane
protrusions (13,14). For example, Tir has been shown to
interact with the I-BAR domain of BAIAP2L1 to regulate actin
pedestal formation (15). Likewise,
it was also revealed that Rif colocalizes with BAIAP2L2 that is
involved in edge ruffling at cell-cell contacts (16).
The I-BAR family has been revealed to participate in
various tumorigenic progression. Interaction of IRSp53 and Eps8 is
essential to cancer cell motility or invasiveness (17). Mice lacking MIM spontaneously
develop tumors characterized by diffusing large B lymphoma at a
late age (18). BAIAP2L1 was
revealed to be upregulated and a potential biomarker in ovarian
cancers (19). Fusion protein of
FGF receptor 3 (FGFR3)-BAIAP2L1 also contributed to bladder cancer
(20). These findings support the
critical role of I-BAR family members in carcinogenesis. However,
little evidence has reported the role of BAIAP2L2 in cancer
development, including lung cancer.
In the present study, BAIAP2L2 was upregulated in
lung adenocarcinoma tissues and lung cancer cells. Based on
knockdown and overexpressing strategies, we revealed that BAIAP2L2
was essential to the proliferation and colony formation of lung
cancer cells. Mechanistically, BAIAP2L2 silencing led to enhanced
apoptosis in A549 and H1299 cells. At the molecular level,
thousands of genes were dysregulated after BAIAP2L2 knockdown.
Pathway enrichment analysis further indicated that
Estrogen-mediated S-phase Entry pathway was significantly
inhibited. In addition, CCNA2, CCND1 and CDK6 were downregulated
and CDKN1B was upregulated in A549 cells with silenced BAIAP2L2.
These findings revealed BAIAP2L2 as a potential biomarker and
therapeutic target for lung adenocarcinoma.
Materials and methods
Cell culture
Lung epithelial cells BEAS-2B and human lung cancer
cells 95D, H1975, H1299 and A549 were purchased from the American
Type Culture Collection (ATCC; Manassas, VA, USA) and cultured in
RPMI-1640 medium (HyClone; GE Healthcare Life Sciences, Logan, UT,
USA). The culture medium was supplemented with 10% fetal bovine
serum and 1% penicillin and streptomycin solution (both from
Corning Inc., Corning, NY, USA). All the cells were maintained at
37°C with 5% CO2.
Microarray of human lung
adenocarcinoma and adjacent normal samples
The HLug-Ade030PG-01 microarrays of lung
adenocarcinoma and adjacent normal samples were obtained from
Shanghai Outdo Biotech Co., Ltd. (Shanghai, China). This array
contained a total of 15 samples with 11 stage II, 4 stage III lung
adenocarcinoma samples and 15 adjacent normal samples. A core
represented a separate case and each sample was fixed in formalin.
Thick 5-µm slices coated with paraffin were subjected to
immunohistochemical staining of BAIAP2L2.
Immunohistochemistry (IHC)
Microarray of human lung adenocarcinoma and adjacent
normal sample were subjected to immunohistochemistry staining of
BAIAP2L2. Briefly, 5 µm sections of tissues were deparaffinized in
xylene and rehydrated in descending alcohol series.
Antigen-retrieval was performed by incubating 0.01 M boiled citrate
buffer in a microwave for 20 min. After cooling to room
temperature, slides were rehydrated in double distilled
H2O for 10 min. The slides were then blocked with 10%
goat serum (cat. no. C0265; Beyotime Institute of Biotechnology,
Beijing, China) at room temperature for 30 min, followed by
incubating with primary anti-BAIAP2L2 antibody (Sigma-Aldrich;
Merck KGaA, Darmstadt, Germany; cat. no. HPA003043, dilution 1:50)
at 4°C overnight. Then, the slides were washed with TBS and
incubating with the secondary antibodies (dilution 1:100; cat. no.
SPN-9001; OriGene Technologies, Inc., Rockville, MD, USA) at room
temperature for 2 h. After washing with TBS, the sections were
stained by Vulcan Fast Red Chromogen kit 2 for 15 min at room
temperature (Biocare, Shanghai, China). The extent and intensity of
BAIAP2L2 immunostaining were taken into consideration. The
intensity of extent of BAIAP2L2 expression was graded as follows:
Negative, 0; weak, 1; moderate, 2; and strong, 3. The extent of
staining was grouped according to the percentage of high-staining
cells in the cancer nest: Negative, 0; 1–25, 1; 26–50, 2; 51–75, 3;
and 76–100%, 4. The final quantitation of each staining was
obtained by multiplying the two scores. Immunoreactivity was
assessed independently by two expert pathologists blinded to all
clinical data.
BAIAP2L2 knockdown in A549 and H1299
cells
A lentiviral system containing pGCSIL-GFP (stably
expressed shRNA fused with a GFP marker), pHelper1.0 (gag/pol
element) and Helper2.0 (VSVG element) were used to silence BAIAP2L2
in A549 and H1299 cells. The vectors were transfected into 293T
cells using Lipofectamine 2000 (Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA). The lentiviral supernatants
were collected 48 h after transfection, filtered through 0.45-µm
filters, and then subjected to infection of A549 and H1299 cells.
Knockdown efficiency was determined by qRT-PCR and western blot
analysis of BAIAP2L2.
BAIAP2L2 overexpression in 95D
cells
The coding sequence of BAIAP2L2 was inserted into
pBABE-puro vector. Targeted vectors or control vectors accompanied
with PIK packaging vectors were co-transfected into 293T cells
using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific,
Inc.). Forty-eight hours later, viral supernatants were collected
and filtered through 0.45-µm filters. Then, they were subjected to
infection of 95D cells. Puromycin was used to select stable cell
lines.
Western blotting
Total proteins were isolated from indicated cells
using lysis buffer [2 g SDS, 1.55 g DTT, 6 ml Tris (1 M, pH 6.8),
10 ml glycerol and ddH2O up to 100 ml]. Cell lysates
were boiled at 98°C for 10 min, and then centrifuged at 16,000 × g
for 5 min. Briefly, the lysates were separated on 10 or 12% sodium
dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and
transferred to polyvinylidene fluoride membranes (PVDF; EMD
Millipore, Billerica, MA, USA). The PVDF membranes were then
blocked in 5% skim milk at room temperature for 60 min and
incubated with primary antibodies at 4°C overnight. Antibodies
against BAIAP2L2 for western blotting were purchased from Abcam
(Cambridge, UK) (dilution 1:1,000; cat. no. ab135897). Antibodies
against CCNA2 (dilution 1:1,000; cat. no. 4656), CDK6 (dilution
1:1,000; cat. no. 3136) and CDKN1B (dilution 1:1,000; cat. no.
3698) were purchased from Cell Signaling Technology (Danvers, MA,
USA). Antibodies against CCND1 (dilution 1:200; cat. no. ab16663)
were purchased from Abcam. Antibodies against GAPDH (dilution
1:2,000; cat. no. sc-32233) and all the secondary antibodies (goat
anti-mouse IgG, dilution 1:5,000; cat. no. sc-2005; goat
anti-rabbit IgG, dilution 1:5,000; cat. no. sc-2004) were obtained
from Santa Cruz Biotechnology, Inc. (Dallas, TX, USA). Following
incubation with the secondary antibodies for 2 h at room
temperature, the membranes were visualized using the ECL Plus kit
(GE Healthcare, Chicago, IL, USA).
RNA isolation and quantitative
real-time PCR
shCtrl and shBAIAP2L2 A549 and H1299 cells were
lysed in TRIzol reagent and subjected to total RNA isolation using
Ultrapure RNA Kit (CWBIO, Beijing, China). Total RNA (0.8 µg) was
subjected to reverse transcription using ReverTra Ace®
qPCR RT Master Mix with gDNA Remover (Toyobo Life Science, Osaka,
Japan). Quantitative real-time PCR was analyzed using TransStart
Top Green qPCR SuperMix (TransGene Biotech Co., Ltd., Beijing,
China) on an iQ5 Real-Time PCR detection system (Bio-Rad
Laboratories, Inc., Hercules, CA, USA). GAPDH served as an internal
control. The primer sequences were as follows: BAIAP2L2 forward,
GTCCCACCATCACCAATGACC and reverse, TCTGCTGCTGTTGCTGCTGTC; GAPDH
forward, 5′-TGACTTCAACAGCGACACCCA-3′ and reverse,
5′-CACCCTGTTGCTGTAGCCAAA-3′.
Colony formation assay
A total of 2,000 Ctrl or BAIAP2L2 overexpressed 95D
cells were seeded in 10-cm plates, and shCtrl or shBAIAP2L2 A549
and H1299 cells (800 cells/well) were seeded in 6-well plates.
After being cultured at 37°C for 12–16 days, the cell colonies were
fixed with methanol for 20 min and stained with 0.5% crystal violet
solution for 15 min. The colony numbers (>50 cells/colony) were
quantified using a fluorescence microscope (Olympus Corp., Tokyo,
Japan).
Cell growth screening
Multiparametric high-content screening (HCS) assay
was used to determine cell viability. In brief, indicated cells
were cultured in 96-well plates for 5 days. The cell density was
assessed using a fluorescence-imaging microscope each day from day
1 to day 5. The density and distribution of the fluorescence
determined the cell viability of shCtrl and shBAIAP2L2 A549 and
H1299 cells. Quantitative results were analyzed using the
ArrayScan™ HCS software (Cellomics, Inc., Pittsburgh, PA, USA).
MTT assay
shCtrl and shBAIAP2L2 A549 and H1299 cells were
seeded at a density of 3,000 cells/well in 96-well plates. After 1,
2, 3, 4 or 5 days, the cells were washed with phosphate-buffered
saline (PBS) and incubated with
3-(4,5-dimethyl-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
solution (5 mg/ml) for 3 h at 37°C. Then, cell viability was
detected at 490 nm of spectrophotometric absorbance.
Caspase-3/7 activity assessment
Caspase-Glo® reagent (Promega
Corporation, Madison, WI, USA) was prepared according to the
manufacturer's instructions. After seeding a total of 10,000
cells/well into 96-well plates, 100 µl Caspase-Glo®
reagent was added into the plates. Then, the plates were rotated at
9.99–25 × g for 0.5 h and maintained at room temperature for 1.5 h.
The activity was detected by a microplate reader.
Apoptosis assay
Apoptosis of shCtrl and shBAIAP2L2 A549 and H1299
cells was examined using Annexin V-APC apoptosis detection kit
(eBioscience; Thermo Fisher Scientific, Inc.), following the
manufacturer's instructions. Briefly, after washing with PBS, the
cells were resuspended with 100 µl staining buffer and incubated
with 5 µl Annexin V-APC at room temperature for 15 min, and
subsequently subjected to flow cytometric detection of
apoptosis.
Microarray assay
Total RNA from A549 cells was extracted using TRIzol
reagent (Invitrogen; Thermo Fisher Scientific, Inc.). NanoDrop 2000
(Thermo Fisher Scientific, Inc., Wilmington, DE, USA) and Agilent
Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA) were
used to detect the RNA quantity and quality. Affymetrix Human
GeneChip PrimeView (Affymetrix; Thermo Fisher Scientific, Inc.) was
used for microarray processing to determine gene expression
profiles according to the manufacturer's instructions.
Significantly different genes between A549 cells treated with
shCtrl and shBAIAP2L2 were identified depending on the follow
criteria: P<0.05 and the absolute fold change >1.5. Pathway
enrichment analysis was performed for all significantly different
genes based on Ingenuity Pathway Analysis v9.0 (IPA; www.ingenuity.com; Ingenuity Systems, Redwood City,
CA, USA).
Statistical analysis
The statistical analysis was performed using
GraphPad Prism 6.0 (GraphPad Software, Inc., La Jolla, CA, USA).
The data were presented as the mean ± SEM of at least 3 independent
repeats. Paired Student's t-tests were used to analyze the
differences of the IHC scores between the adenocarcinoma and
adjacent tissues. Unpaired Student's t-tests were used to analyze
the difference between two other groups. Differences among groups
were determined by one-way analysis of variance (ANOVA) with
repeated measures, followed by the Bonferroni post hoc test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
BAIAP2L2 is upregulated in lung
adenocarcinoma and lung cancer cells
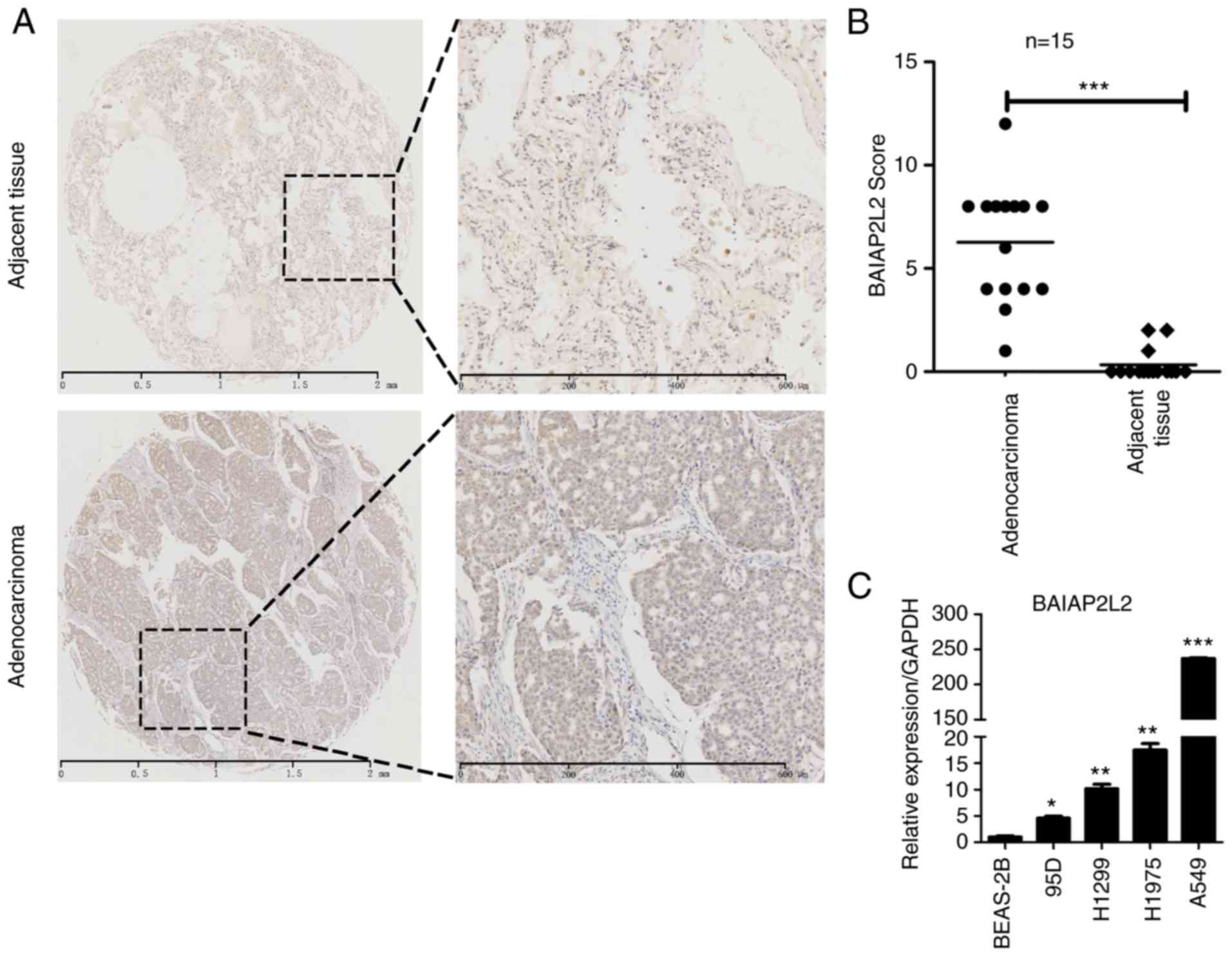
To explore the clinical relevance of BAIAP2L2 in
lung cancer, we collected 15 pairs of lung adenocarcinoma specimens
and detected the expression of BAIAP2L2 using immunohistochemical
assay. We revealed that BAIAP2L2 was overexpressed in the tumor
areas compared to adjacent normal tissues (Fig. 1A and B). In addition, we analyzed
the expression of BAIAP2L2 in normal lung epithelial cells and lung
cancer cells. The qRT-PCR results revealed that the mRNA level of
BAIAP2L2 was higher in lung cancer cells compared to BEAS-2B cells
(Fig. 1C). These results indicated
that BAIAP2L2 was a potential biomarker for lung cancer and that it
may be essential for lung cancer progression.
BAIAP2L2 is critical for lung cancer
cell proliferation and growth
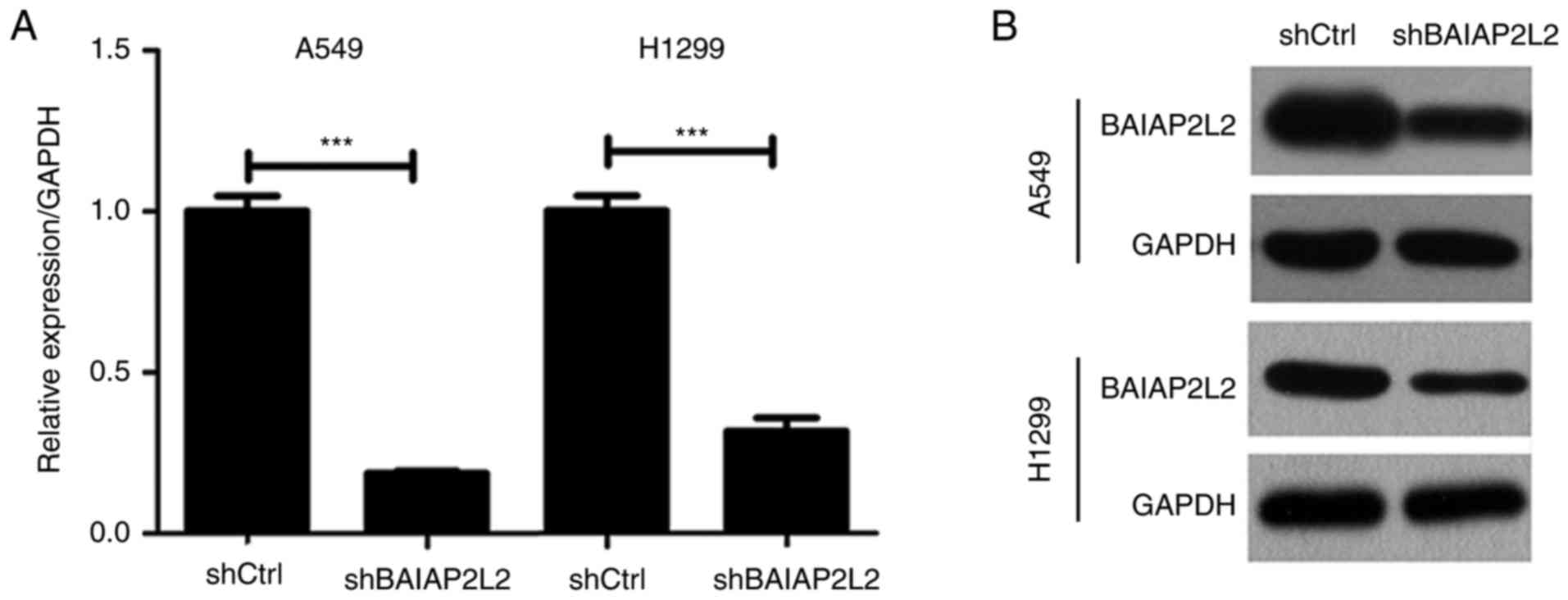
We next determined the role of BAIAP2L2 in lung
cancer. BAIAP2L2 was silenced in A549 cells using
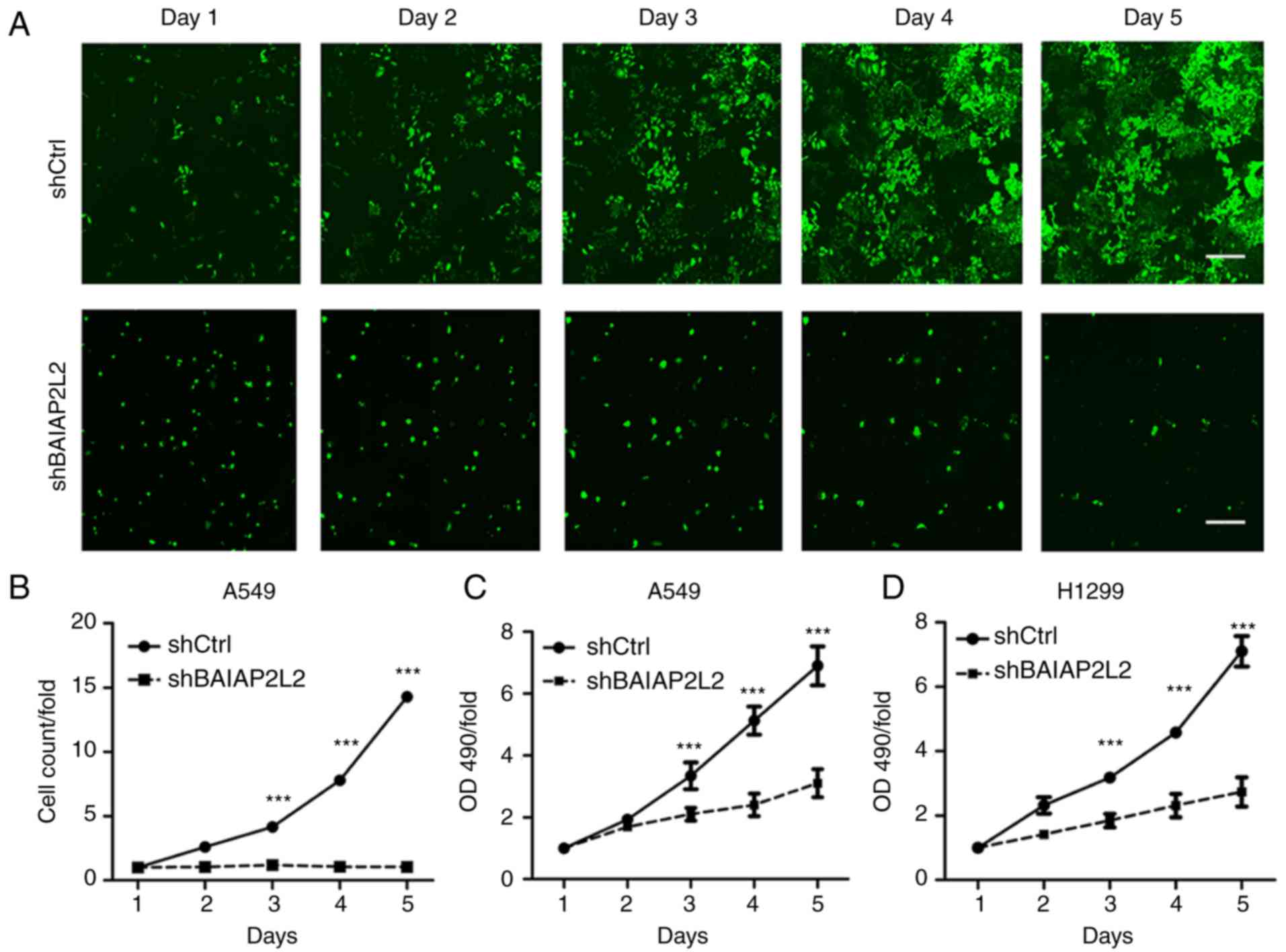
lentivirus-mediated knockdown strategy (Fig. 2). Then, we analyzed the
proliferation of shCtrl and shBAIAP2L2 in A549 cells from day 1 to
day 5 through HCS. We found that A549 cells expressing shCtrl
lentivirus proliferated robustly from day 1 to day 5 (Fig. 3A). However, BAIAP2L2 knockdown
completely suppressed the proliferation of A549 cells (Fig. 3A). Cell count analysis was
consistent with the results (Fig.
3B). Likewise, MTT detection also demonstrated that blockage of
BAIAP2L2 largely inhibited the proliferation of A549 cells
(Fig. 3C). In addition, we silenced
BAIAP2L2 in another lung cancer cell line, H1299. qRT-PCR and
western blot results revealed that BAIAP2L2 was knocked down in
H1299 cells (Fig. 2). Consistently,
blockage of BAIAP2L2 significantly decreased the viability of H1299
cells as indicated by the suppressed cell proliferation rate
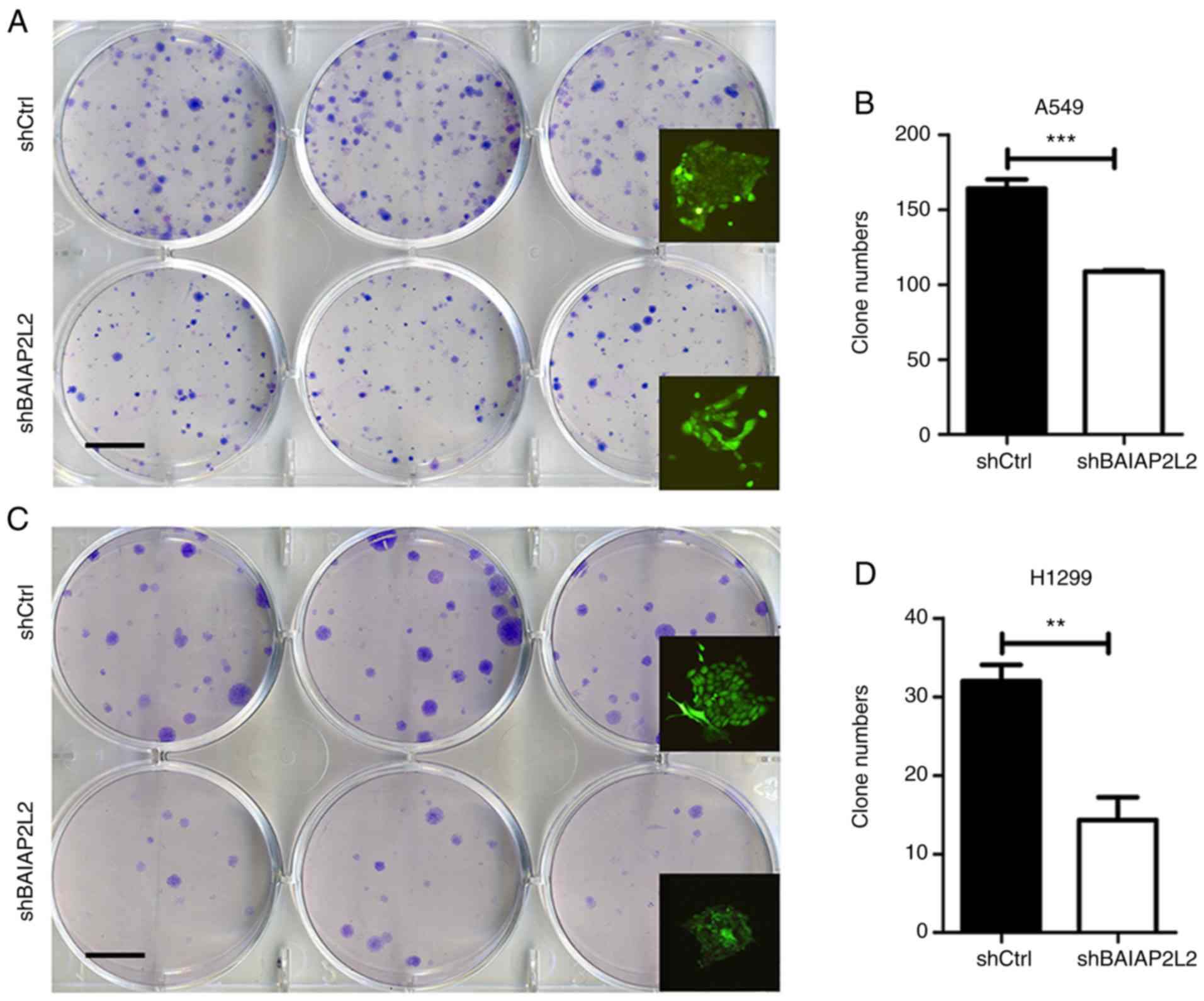
(Fig. 3D). Moreover, we detected a
reduced number of colonies in shBAIAP2L2 A549 or H1299 cells
compared with shCtrl A549 or H1299 cells (Fig. 4).
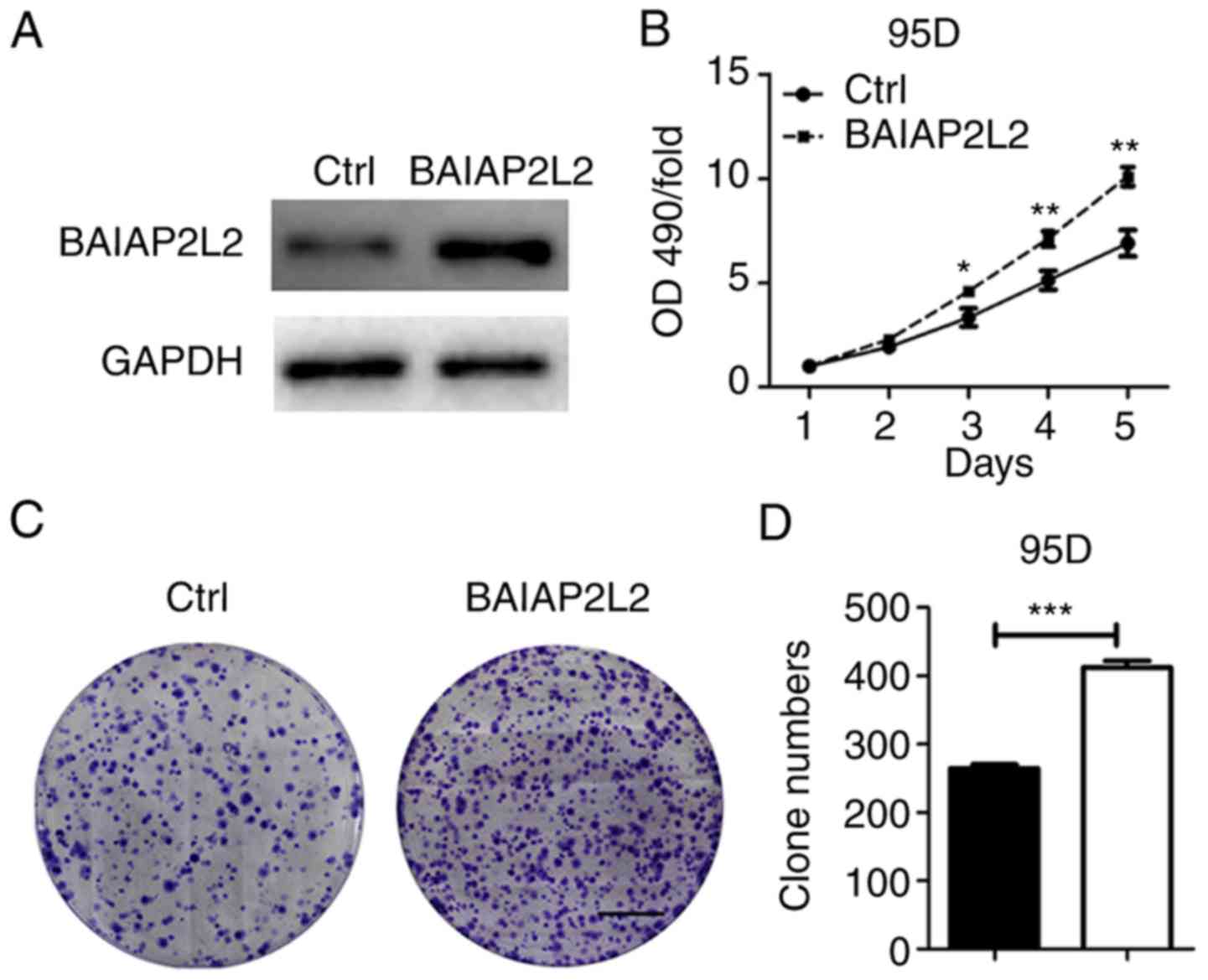
To ascertain our results, we overexpressed BAIAP2L2
in 95D cells. It was revealed that BAIAP2L2 ectopic expression
accelerated the proliferation and growth of 95D cells (Fig. 5). Thus, we concluded that BAIAP2L2
is critical for lung cancer cell viability.
BAIAP2L2 knockdown leads to increased
apoptosis
Suppressed cell proliferation and growth may be a
consequence of increased cell death. Apoptosis is a common cell
death type and plays an important role in cancer development.
Therefore, we investigated whether apoptosis participated in
BAIAP2L2-mediated lung cancer cell proliferation and growth. To
address this question, Annexin V-APC was used to detect the
apoptosis of A549 and H1299 cells expressing shCtrl or shBAIAP2L2
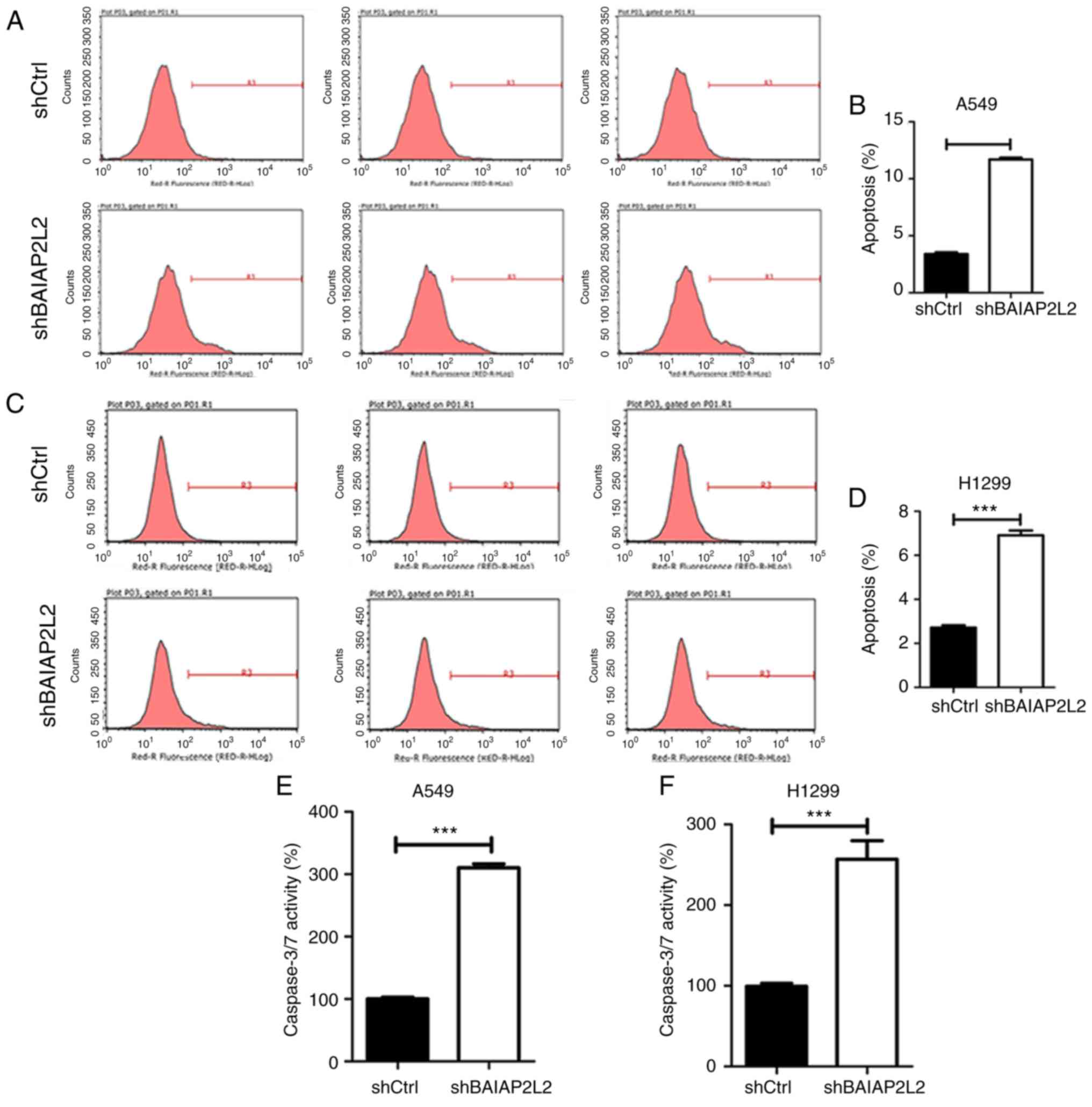
lentivirus. Our results revealed that BAIAP2L2 silencing increased
the apoptosis of A549 and H1299 cells (Fig. 6A and C). Quantitative results
revealed that >2-fold enhanced apoptosis was found in A549 and
H1299 cells after BAIAP2L2 knockdown (Fig. 6B and D). Furthermore, we also
detected increased caspase-3/7 activity in shBAIAP2L2 A549 cells
compared with shCtrl A549 cells (Fig.
6E). Likewise, shBAIAP2L2 H1299 cells exhibited increased
apoptosis and caspase-3/7 activity (Fig. 6F). These results indicated that
BAIAP2L2 knockdown triggered lung cancer cell apoptosis, mainly by
enhancing caspase-3/7 activity. Collectively, our results revealed
that BAIAP2L2 silencing may suppress cell proliferation and growth
at least partly by increasing apoptosis.
Depletion of BAIAP2L2 results in
inhibition of Estrogen-mediated S-phase Entry pathway and
dysregulation of numerous genes
By revealing that BAIAP2L2 is critical for
apoptosis, proliferation and growth of lung cancer cells, we
further explored the potential molecular mechanism. Firstly,
GeneChip was performed to determine dysregulated genes in A549
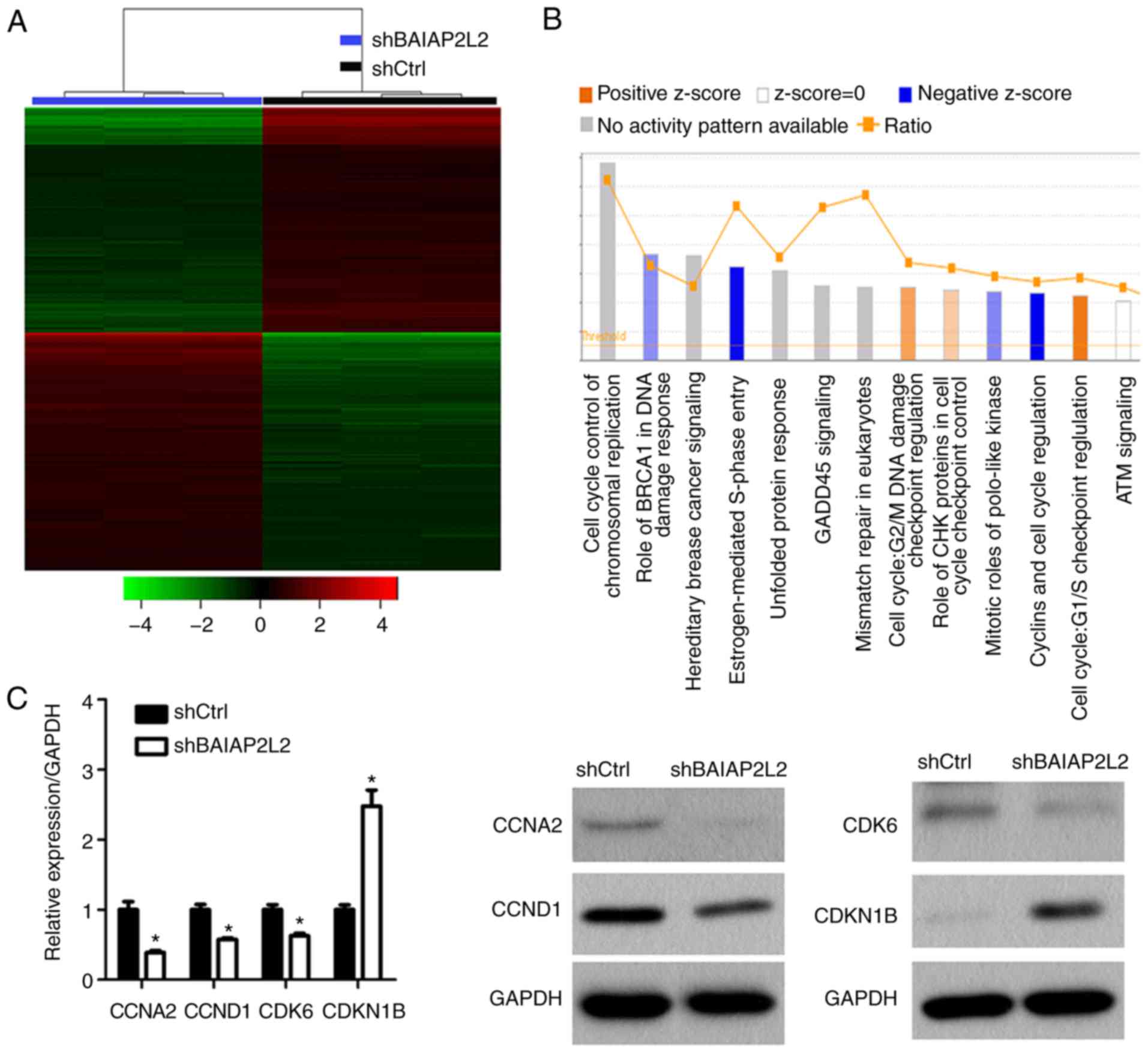
cells after BAIAP2L2 knockdown. We found that 1,240 genes were
upregulated and 1,157 genes were downregulated in BAIAP2L2 silenced
A549 cells (Fig. 7A). By pathway
enrichment analysis, we revealed that the Estrogen-mediated S-phase
Entry pathway was significantly inhibited after BAIAP2L2 knockdown
(Fig. 7B). Cyclins and cell cycle
regulation signaling was suppressed (Fig. 7B). However, the predictive analysis
revealed that the G1/S checkpoint regulation pathway involved in
the cell cycle was upregulated (Fig.
7B). Additionally, BAIAP2L2 knockdown upregulated the G2/M DNA
damage checkpoint regulation pathway to a lesser extent (Fig. 7B). Next, we confirmed our predictive
analysis using qRT-PCR and western blot assays. The results
revealed that CCNA2, CCND1, CDK6 were downregulated and CDKN1B was
upregulated in shBAIAP2L2 A549 cells (Fig. 7C).
Discussion
Lung cancer is continuously a threat to the health
of people due to its high lethality. During the last decade,
crucial signaling pathways triggering this disease have been
identified, including epidermal growth factor receptor (21), vascular endothelial growth factor
(22) and insulin-like growth
factor-1 network (23).
Subsequently, newly targeted drugs against lung cancer have been
developed. However, the outcomes of these targeted agents are less
than satisfactory. Therefore, there is constant need to explore
novel critical molecular mechanisms involved in lung cancer
progression. In the present study, BAIAP2L2 was revealed as a novel
oncogene for lung adenocarcinoma. Upregulation of BAIAP2L2 was
found in lung adenocarcinoma specimens. Knockdown of BAIAP2L2
reduced the proliferation and growth of lung cancer cells.
BAIAP2L2 is identified as an I-BAR family member
that plays an important role in regulating membrane protrusions
(13). Although some studies have
demonstrated that BAIAP2L1, another I-BAR family member, is a
promising biomarker for ovarian cancer (19), the precise function of BAIAP2L2 in
cancer development, particularly in lung cancer, remains largely
unknown. This study provided the first evidence that BAIAP2L2 was a
proto-oncogene for lung cancer. Since it was highly expressed in
lung adenocarcinoma tissues and lung cancer cells, we attempted to
explore the role of BAIAP2L2 through depletion of this gene in lung
cancer cells A549 and H1299. Our results revealed that silencing of
BAIAP2L2 markedly blunted the proliferation of A549 and H1299
cells. In addition, colony formation results also supported the
oncogenic function of BAIAP2L2 since reduction of BAIAP2L2
significantly suppressed the colony formation of A549 and H1299
cells. Collectively, it was concluded that BAIAP2L2 was critical
for lung cancer proliferation and growth.
It has been demonstrated that BAIAP2L1 prevented the
apoptosis of ovarian cancer cells (19). Additionally, BAIAP2L2 has also been
reported to activate EGFR/ERK signaling and promote cell
proliferation of hepatocellular carcinoma (24). Our mechanistic study revealed that
BAIAP2L2 silencing promoted the apoptosis of lung cancer cells.
Caspase-3/7 activity was enhanced in A549 and H1299 cells after
BAIAP2L2 knockdown. These results demonstrated the crucial role of
BAIAP2L2 in apoptosis. Furthermore, our pathway enrichment analysis
indicated that cell cycle regulation pathways were dysregulated
after BAIAP2L2 knockdown. Western blotting results revealed that
CCNA2, CCND1 and CDK6 were downregulated and CDKN1B was upregulated
in shBAIAP2L2 A549 cells. These findings indicated that BAIAP2L2
may be critical for cell cycle progression. It is well-known that
estrogen has different functions in cancer development. Estrogen
prevents hepatocellular carcinoma (25) and is shown to promote lung cancer
(26). In this study, pathway
enrichment analysis revealed that the Estrogen-mediated S-phase
Entry pathway was significantly inhibited after BAIAP2L2 knockdown
in A549 cells. Thus, further study must be performed to elucidate
how this signaling is regulated by BAIAP2L2 and the functional role
of this pathway in lung cancer development.
In summary, the study demonstrated for the first
time, to the best of our knowledge, that BAIAP2L2 was upregulated
in lung adenocarcinoma and functioned as an oncogene in lung
cancer. BAIAP2L2 was essential for lung cancer cell survival since
depletion of BAIAP2L2 led to increased apoptosis, decreased cell
proliferation and growth. Mechanistically, caspase-3/7 activity was
increased in shBAIAP2L2 lung cancer cells. In addition, cell cycle
progression may also be blunted since dysregulation of cell
cycle-related proteins was also found in shBAIAP2L2 lung cancer
cells. Nevertheless, further study should be performed to
investigate the involvement of estrogen in BAIAP2L2-mediated lung
cancer development since the Estrogen-mediated S-phase Entry
pathway was markedly suppressed after BAIAP2L2 knockdown.
Therefore, BAIAP2L2 was revealed to be a potential biomarker and
promising therapeutic target for lung cancer.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China (no. 81460011), the Doctoral
Starting up Foundation of The Affiliated Hospital of Inner Mongolia
Medical University, The project of health and family planning
commission of Inner Mongolia autonomous region (no. 201702084) and
the Natural Science Foundation of Inner Mongolia autonomous region
(no. 2018LH0823).
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
XF conceived the study, carried out the experimental
design and the data interpretation, prepared and revised the
manuscript. LX performed most of the experiments, wrote, reviewed
and edited the manuscript. HD and QZ performed the
immunohistochemical assay and packaging of lentivirus. CW and LY
performed the colony formation and the qPCR assays. GT performed
the western blot assay and the statistical analysis. All authors
read and approved the manuscript and agree to be accountable for
all aspects of the research in ensuring that the accuracy or
integrity of any part of the work are appropriately investigated
and resolved.
Ethics approval and consent to
participate
The present study was approved by the Research
Ethics Committee of the Affiliated Hospital of Inner Mongolia
Medical University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
BAIAP2L1
|
BAI1-associated protein 2-like 1
|
|
EGFR
|
epidermal growth factor receptor
|
|
FGFR
|
fibroblast growth factor receptor
|
|
BAIAP2L2
|
BAI1-associated protein 2-like 2
|
|
MIM
|
missing in metastasis
|
|
I-BAR
|
Inverse Bin-Amphiphysin-Rvs
|
|
HCS
|
high-content screening
|
References
|
1
|
Jemal A, Thomas A, Murray T and Thun M:
Cancer statistics, 2002. CA Cancer J Clin. 52:23–47. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Belinsky SA: Gene-promoter
hypermethylation as a biomarker in lung cancer. Nat Rev Cancer.
4:707–717. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Weir BA, Woo MS, Getz G, Perner S, Ding L,
Beroukhim R, Lin WM, Province MA, Kraja A, Johnson LA, et al:
Characterizing the cancer genome in lung adenocarcinoma. Nature.
450:893–898. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ding L, Getz G, Wheeler DA, Mardis ER,
McLellan MD, Cibulskis K, Sougnez C, Greulich H, Muzny DM, Morgan
MB, et al: Somatic mutations affect key pathways in lung
adenocarcinoma. Nature. 455:1069–1075. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yang W, Yao YW, Zeng JL, Liang WJ, Wang L,
Bai CQ, Liu CH and Song Y: Prognostic value of FGFR1 gene
copy number in patients with non-small cell lung cancer: A
meta-analysis. J Thorac Dis. 6:803–809. 2014.PubMed/NCBI
|
|
6
|
Weiss J, Sos ML, Seidel D, Peifer M,
Zander T, Heuckmann JM, Ullrich RT, Menon R, Maier S, Soltermann A,
et al: Frequent and focal FGFR1 amplification associates
with therapeutically tractable FGFR1 dependency in squamous cell
lung cancer. Sci Transl Med. 2:62ra932010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mok TS, Wu YL, Thongprasert S, Yang CH,
Chu DT, Saijo N, Sunpaweravong P, Han B, Margono B, Ichinose Y, et
al: Gefitinib or carboplatin-paclitaxel in pulmonary
adenocarcinoma. N Engl J Med. 361:947–957. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Pao W and Chmielecki J: Rational,
biologically based treatment of EGFR-mutant non-small-cell lung
cancer. Nat Rev Cancer. 10:760–774. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Pykäläinen A, Boczkowska M, Zhao H,
Saarikangas J, Rebowski G, Jansen M, Hakanen J, Koskela EV, Peränen
J, Vihinen H, et al: Pinkbar is an epithelial-specific BAR domain
protein that generates planar membrane structures. Nat Struct Mol
Biol. 18:902–907. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Saarikangas J, Zhao H, Pykäläinen A,
Laurinmäki P, Mattila PK, Kinnunen PK, Butcher SJ and Lappalainen
P: Molecular mechanisms of membrane deformation by I-BAR domain
proteins. Curr Biol. 19:95–107. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mattila PK, Pykäläinen A, Saarikangas J,
Paavilainen VO, Vihinen H, Jokitalo E and Lappalainen P:
Missing-in-metastasis and IRSp53 deform PI(4,5)P2-rich
membranes by an inverse BAR domain-like mechanism. J Cell Biol.
176:953–964. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhao H, Pykäläinen A and Lappalainen P:
I-BAR domain proteins: Linking actin and plasma membrane dynamics.
Curr Opin Cell Biol. 23:14–21. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chen Z, Shi Z and Baumgart T: Regulation
of membrane-shape transitions induced by I-BAR domains. Biophys J.
109:298–307. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Scita G, Confalonieri S, Lappalainen P and
Suetsugu S: IRSp53: Crossing the road of membrane and actin
dynamics in the formation of membrane protrusions. Trends Cell
Biol. 18:52–60. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Vingadassalom D, Kazlauskas A, Skehan B,
Cheng HC, Magoun L, Robbins D, Rosen MK, Saksela K and Leong JM:
Insulin receptor tyrosine kinase substrate links the E. coli
O157:H7 actin assembly effectors Tir and EspFU during
pedestal formation. Proc Natl Acad Sci USA. 106:6754–6759. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sudhaharan T, Sem KP, Liew HF, Yu YH, Goh
WI, Chou AM and Ahmed S: The Rho GTPase Rif signals through IRTKS,
Eps8 and WAVE2 to generate dorsal membrane ruffles and filopodia. J
Cell Sci. 129:2829–2840. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Funato Y, Terabayashi T, Suenaga N, Seiki
M, Takenawa T and Miki H: IRSp53/Eps8 complex is important for
positive regulation of Rac and cancer cell motility/invasiveness.
Cancer Res. 64:5237–5244. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yu D, Zhan XH, Zhao XF, Williams MS, Carey
GB, Smith E, Scott D, Zhu J, Guo Y, Cherukuri S, et al: Mice
deficient in MIM expression are predisposed to lymphomagenesis.
Oncogene. 31:3561–3568. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chao A, Tsai CL, Jung SM, Chuang WC, Kao
C, Hsu A, Chen SH, Lin CY, Lee YC, Lee YS, et al: BAI1-associated
protein 2-like 1 (BAIAP2L1) is a potential biomarker in ovarian
cancer. PLoS One. 10:e01330812015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Williams SV, Hurst CD and Knowles MA:
Oncogenic FGFR3 gene fusions in bladder cancer. Hum Mol Genet.
22:795–803. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Giaccone G: Epidermal growth factor
receptor inhibitors in the treatment of non-small-cell lung cancer.
J Clin Oncol. 23:3235–3242. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Herbst RS, Onn A and Sandler A:
Angiogenesis and lung cancer: Prognostic and therapeutic
implications. J Clin Oncol. 23:3243–3256. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Karamouzis MV and Papavassiliou AG: The
IGF-1 network in lung carcinoma therapeutics. Trends Mol Med.
12:595–602. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang YP, Huang LY, Sun WM, Zhang ZZ, Fang
JZ, Wei BF, Wu BH and Han ZG: Insulin receptor tyrosine kinase
substrate activates EGFR/ERK signalling pathway and promotes cell
proliferation of hepatocellular carcinoma. Cancer Lett. 337:96–106.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Naugler WE, Sakurai T, Kim S, Maeda S, Kim
K, Elsharkawy AM and Karin M: Gender disparity in liver cancer due
to sex differences in MyD88-dependent IL-6 production. Science.
317:121–124. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hsu LH, Chu NM and Kao SH: Estrogen,
estrogen receptor and lung cancer. Int J Mol Sci. 18:E17132017.
View Article : Google Scholar : PubMed/NCBI
|