Introduction
Breast cancer is the most common type of invasive
cancer in women and the second main cause of cancer mortality in
females, following lung cancer. Chemoprevention in combination with
anticancer treatment is therefore important to reduce incidence and
mortality rates. Epidemiological and experimental studies have
demonstrated that natural nutritional compounds are potent
chemopreventive agents against mammary carcinogenesis (1,2). One
of these food constituents is Curcuma, a yellowish-orange
polyphenol from the rhizome of Curcuma longa. Since ancient
times, it has been used as herbal therapy in China and India
against numerous diseases (3).
Curcumin is also an anticancer agent, as recently revealed in a
mouse xenograft breast cancer model, where it significantly
prevented tumor growth and angiogenesis (4). In hormone-dependent and independent
human breast cancer cells, curcumin interferes with apoptosis by
regulating STAT3, NF-κB, AP-1, HER2, CXCR4, EGFR, ERK, αJAK, TNF,
IL and MP activity (5–7). Curcumin attenuates the expression of
matrix metalloproteinases (MMPs) via reduced activity of NF-κB and
transcriptional downregulation of AP-1 (8), thereby decreasing the number of lung
metastases in mice upon intracardiac injection of estrogen
receptor-negative human breast cancer MDA-MB-231 cells (8). Curcumin exhibited a synergistic effect
with paclitaxel against human MCF-7 and MDA-MB-231 cells (9). Furthermore, curcumin demonstrated
better clinical responses also in a phase-I clinical trial with
docetaxel in patients with prolonged and metastatic breast cancer
(10). Notably, despite its
activity against breast cancer cells, curcumin exerts little
toxicity against normal cells, even upon long-term exposure.
Previous studies in mice and rats demonstrated
extensive metabolism of curcumin in the small intestine and liver
mainly to curcumin sulfate, curcumin glucuronide and
tetrahydrocurcumin (11).
Biotransformation of curcumin is also extensive in humans, as shown
in a pilot study of a standardized Curcuma-derived extract
in patients with colorectal cancer (12). In addition to the formation of
curcumin glucuronide and curcumin sulfate, curcumin undergoes
phase-I bioreduction mainly to tetrahydrocurcumin and to a minor
part to hexahydrocurcumin, octahydrocurcumin and
hexahydrocurcuminol (13,14), which are subsequently further
conjugated to glucuronides and sulfates (13). Biotransformation is cell-specific,
as a recent study from our laboratory revealed that in
hormone-dependent ZR-75-1 and hormone-independent MDA-MB-231 breast
cancer cells the main metabolite was curcumin sulfate, while
curcumin glucuronide formation was below the detection limit
(15). Intracellular curcumin
sulfate was subsequently discharged released into the cellular
medium, leading to <12-fold higher concentrations compared with
the cytoplasm indicating an active efflux mechanism. A possible
candidate for this efflux is the breast cancer resistance protein
(BCRP and ABCG2), which is expressed in numerous tissues, including
breast ductal cells, and serves an important role in the efflux of
sulfated conjugates of steroids and drugs (16). An interplay of curcumin with BCRP
has already been described and may also apply to its sulfated
metabolite (17).
Currently, limited information is available about
the activities of curcumin biotransformation products. Previous
in vitro data suggest that the main metabolites of curcumin,
i.e. curcumin sulfate and curcumin glucuronide are less potent than
curcumin against various tumor cell lines. In vivo, however,
curcumin conjugates may markedly contribute to the pharmacological
activity of curcumin, since ubiquitously expressed sulfatases or
β-glucuronidases may rapidly cleave the conjugates back into
curcumin. However, tetrahydrocurcumin, a major metabolite of
curcumin, has demonstrated similar anticancer activities to
curcumin. In H22 ascites hepatocarcinoma tumor-bearing mice and in
an animal carcinogenesis model tetrahydrocurcumin was even more
effective than curcumin (18,19).
Moreover, intragastric treatments of tetrahydrocurcumin (40 mg/kg)
to mice were more effective than curcumin (100 mg/kg) in inhibiting
the expression of cyclooxygenase-2 and suppressing NF-κB (20). Unlike curcumin, tetrahydrocurcumin
is stable in 0.1 M phosphate buffer at pH 7.2 and in plasma
(21), and may therefore
significantly contribute to the anticancer activity of
curcumin.
Active uptake mechanisms into the cytoplasm of tumor
cells may be more significant than metabolism for the efficiency of
curcumin. One of the main membrane transport proteins are
organic-anion-transporting polypeptides (OATPs), which mediate the
uptake of numerous clinically used drugs and natural compounds such
as polyphenols and their sulfates (22–26).
The strongest effect on bioavailability is due to OATP2B1, which is
highly expressed in the intestine, and to OATP1B1 and OATP1B3,
which are expressed in the liver. Various OATPs are also highly
expressed in human hormone receptor-positive (MCF-7) and negative
(MDA-MB-231) breast cancer cell lines (27). Zhou et al recently
demonstrated that curcumin is a substrate of OATP1B1, OATP1B3 and
OATP2B1, and that curcumin glucuronide is a substrate of OATP1B1
and OATP1B3 but not of OATP2B1, using OATP-transfected 293 cells
(28). Additional experiments by
the same authors (28) and by Sun
et al (29) revealed that
curcumin and curcumin glucuronide are also inhibitors of the
OATP1B1 and 1B3 substrates rosuvastatin and docetaxel. However, no
data are available to date on whether curcumin sulfate and
tetrahydrocurcumin are also transported by OATPs. The present study
therefore investigated the time and concentration-dependent
transport of curcumin, curcumin sulfate, curcumin glucuronide and
tetrahydrocurcumin in Chinese hamster ovary (CHO) cells
stably-transfected with OATP1B1, OATP1B3 and OATP2B1. Furthermore,
the kinetics of the uptake of curcumin and its main metabolites was
calculated in order to determine the affinity of each substrate for
the three transporters. Finally, the importance of OATPs in the
anticancer activity of curcumin and tetrahydrocurcumin was
elucidated in wild-type and OATP1B1-knockdown human breast cancer
cell line ZR-75-1, and any differences in cellular uptake
associated with cytotoxicity were evaluated.
Materials and methods
Materials
Curcumin (98% pure) and tetrahydrocurcumin (95%
pure) were purchased from Sigma-Aldrich (Merck KGaA, Darmstadt,
Germany). Curcumin sulfate and curcumin glucuronide were obtained
from TLC Pharmaceutical Standards Ltd. (Aurora, ON, Canada).
Methanol and water were of high-performance liquid chromatography
(HPLC) grade (Merck KGaA). All other chemicals and solvents were
commercially available and of analytical grade, and were used
without further purification.
Cell culture
Chinese hamster ovary (CHO) cells that were stably
transfected with OATP1B1, OATP1B3 and OATP2B1, as well as wild-type
CHO cells, were provided by the Department of Clinical Pharmacology
and Toxicology, University Hospital Zurich (Zurich, Switzerland).
These cells have been extensively characterized previously
(30,31). The CHO cells were grown in
Dulbecco's modified Eagle's medium (DMEM) supplemented with 10%
fetal calf serum (FCS), 50 µg/ml L-proline, 100 U/ml penicillin and
100 µg/ml streptomycin (Life Technologies; Thermo Fisher
Scientific, Inc., Waltham MA, USA). The selection medium for stably
transfected CHO cells additionally contained 500 µg/ml geneticin
sulfate (G418) (32). All of the
media and supplements were obtained from Life Technologies (Thermo
Fisher Scientific, Inc.). The mammalian ZR-75-1 breast cancer cell
line was purchased from the American Type Culture Collection (ATCC;
Manassas, VA, USA) and was maintained in RPMI-1640 medium (Life
Technologies; Thermo Fisher Scientific, Inc.) supplemented with 10%
FCS, 100 U/ml penicillin, 100 µg/ml streptomycin and 1% GlutaMAX
(Life Technologies; Thermo Fisher Scientific, Inc.). The cells were
grown in T-flasks with a 25-cm2 growth area (BD
Biosciences, Franklin Lakes, NJ, USA) and maintained at 37°C under
5% CO2 and 95% relative humidity. The cells were
passaged once per week and were used at passages ≤55 (24).
OATP1B1 knockdown in ZR-75-1
cells
For lentiviral transduction, ZR-75-1 cells were
plated in 24-well tissue culture plates at a density of 40,000
cells/well in 0.5 ml of growth medium. After 24 h, 250 µl of medium
supplemented with 8 µg/ml polybrene (Sigma-Aldrich; Merck KGaA) was
added. Transductions were performed by the addition of 10 µl of
shRNA (Mission® Transduction Particles NM_006446;
Sigma-Aldrich; Merck KGaA), and the TRCN0000043203 coding sequence
was as follows:
5′-CCGGGCCTTCATCTAAGGCTAACATCTCGAGATGTTAGCCTTAGATGAAGGCTTTTTG-3′.
At 24 h post-transduction, the cell culture medium was changed, and
1 ml of growth medium supplemented with 1 or 5 µg/ml puromycin
(Sigma-Aldrich; Merck KGaA) was added to select cells after an
additional 24 h. The obtained silencing efficiency was evaluated
after 3 weeks via reverse transcription-quantitative polymerase
chain reaction (RT-qPCR).
RT-qPCR
Total RNA was extracted from cells using TRIzol
reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to
the manufacturer's instructions as previously described (28). TaqMan® Gene Expression
assays (Applied iosystems; Thermo Fisher Scientific, Inc.) were
applied for human OATP1B1 detection. The 18S gene was used as a
reference gene as previously described (27). Multiplex RT-qPCR was performed in an
amplification mixture (25 µl) containing 10 µl of 2X
TaqMan® Universal PCR Master Mix, 1 µl appropriate Gene
Expression assay, 1 µl TaqMan® endogenous control (human
β-actin or 18S), 10 ng template complementary DNA (cDNA) diluted in
3 µl nuclease-free water. The thermal cycling conditions were as
previously described (33).
Fluorescence generation due to the cleavage of the
TaqMan® probe via the 5′→3′ exonuclease activity of the
DNA polymerase was detected with an ABI PRISM 7700 Sequence
Detection System (Applied Bioystems; Thermo Fisher Scientific,
Inc.). All samples were amplified in triplicate. To cover the range
of expected quantitative cycle (Cq)-values for the target mRNA
(34), a standard curve of 6 serial
dilutions from 50 ng to 500 pg pooled cDNA was analyzed using
Sequence Detection software 1.9.1. (Applied iosystems; Thermo
Fisher Scientific, Inc.). Relative gene expression data are given
as the n-fold change in transcription of target genes normalized to
the endogenous control. Real-time RT-PCR was performed with the
following prefabricated TaqMan® Gene Expression assays
(Applied iosystems; Thermo Fisher Scientific, Inc.) which contained
the intron-spanning primer Hs00272374_m1 for OATP1B1.
Cellular uptake
Transport assays were performed on 12-well plates as
previously described (33).
Briefly, OATP-transfected CHO cells were seeded at a density of
350,000 cells/well on 12-well plates (BD Biosciences). Uptake
assays were generally performed on day 3 after seeding when the
cells had grown to confluence. After 24 h of initiation before
starting the transport experiments, cells were treated with 5 mM
sodium butyrate (Sigma-Aldrich; Merck KGaA) to induce non-specific
gene expression (35). Curcumin and
its metabolites were dissolved in dimethyl sulfoxide (DMSO) and
diluted in uptake buffer (pH 7.4; final DMSO concentration of 0.5%)
to 25–600 µM. Control experiments contained DMSO in the medium
instead of curcumin and its biotransformation products,
respectively. Prior to the transport experiment, cells were rinsed
twice with 2 ml of pre-warmed (37°C) uptake buffer (116.4 mM NaCl,
5.3 mM KCl, 1 mM NaH2PO4, 0.8 mM
MgSO4, 5.5 mM D-glucose and 20 mM HEPES; pH adjusted to
7.4). Uptake at 37°C was initiated by adding 0.25 ml of uptake
buffer containing the substrate. After 1 min, the uptake was
stopped, and the cells were washed 5 times with 2 ml of buffer (pH
7.4) and subsequently trypsinized by the addition of 100 µl of
trypsin. Cell membranes were then disrupted via repeated (5 times)
shock freezing in liquid nitrogen and thawing. Following
centrifugation at 13,500 × g for 5 min, 100 µl supernatant was
diluted with methanol/water (2:1; v/v) and aliquots (80 µl) were
analyzed via HPLC. All experiments were repeated at least 3
times.
Transport of curcumin and
tetrahydrocurcumin in wild-type ZR-75-1 and OATP1B1-knockdown
ZR-75-1 cells
Cells were plated on 6-well plates and allowed to
attach overnight. Cells were then incubated for 1 min at 37°C with
curcumin and tetrahydrocurcumin (25–200 µM; final DMSO
concentration <0.5%). Control experiments contained DMSO in the
medium in place of curcumin and tetrahydrocurcumin. Then uptake was
stopped, and the cells were washed 5 times with 2 ml
phosphate-buffered saline (PBS) and subsequently trypsinized by the
addition of 100 µl trypsin. Cell membranes were lysed by repeated
(5 times) shock freezing in liquid nitrogen and thawing. Following
centrifugation at 13,500 × g for 5 min, 80 µl supernatant
(cytoplasm) was analyzed by HPLC for detection of curcumin and
tetrahydrocurcumin. All experiments were repeated at least 3
times.
Cytotoxicity assay
ZR-75-1 wild-type and OATP-knockdown cells (50,000
cells/ml) were seeded into 96-well plates and incubated for 24 h at
37°C under 5% CO2. Then, cells were incubated with
various concentrations of curcumin (2.5–100 µM) for 72 h, the
incubation stopped and CellTiter-Blue reagent (Promega Corp.,
Madison, WI, USA) (20 µl) was added to the wells. The plates were
then incubated for 2 h at 37°C and the absorbance was recorded for
resazurin (605 nm) and resorufin (573 nm) on a Tecan M200 multimode
plate reader (Tecan Group, Ltd., Männedorf, Switzerland). The
viability of the treated cells was expressed as a percentage of the
viability of the corresponding control cells. All experiments were
repeated at least 3 times.
Determination of protein
concentrations
Total protein was determined using the BCA assay kit
(Pierce; Thermo Fisher Scientific, Inc.) with bovine serum albumin
(BSA) as a standard and quantification at a wavelength of 562 nm on
a spectrophotometer (UV-1800; Shimadzu Corp., Kyoto, Japan). Raw
data were analyzed using UV-Probe version 2.31 software (Shimadzu
Corp.). The protein concentrations were consistent among the plates
(0.150±0.005 mg/well).
HPLC analysis
The concentrations of curcumin and its
biotransformation products were quantified by HPLC using the same
Dionex UltiMate 3000 system (Dionex Corp., Sunnyvale, CA, USA),
column, mobile phase and gradient as previously described (15). Calibration of the chromatogram was
accomplished using the external standard method. Linear calibration
curves were performed by spiking drug-free cell culture medium with
standard solutions of curcumin, curcumin sulfate, curcumin
glucuronide, and tetrahydrocurcumin to produce a concentration
range from 0.01 to 10 µg/ml (average correlation coefficients:
>0.999). Coefficients of accuracy and precision for these
compounds were <11%.
NF-κB-luciferase reporter assay
A total of 100,000 ZR-75-1 wild-type and
OATP1B1-knockdown ZR-75-1 cells were seeded in 24-well plates and
grown to 70% confluency for transfection. Simultaneous transfection
with pTAL-NF-κB (NF-κB response element-Firefly luciferase
reporter; Clontech Laboratories, Inc., Mountainview, CA, USA) and
pRL-TK (Control-Renilla luciferase; Promega Corp.) was
performed with Lipofectamine 2000 (Life Technologies; Thermo Fisher
Scientific, Inc.; cat. no. 11668) according to the manufacturer's
protocol. NF-κB was blocked for 30 min with 10 µM BAY11-7082
(Calbiochem; Merck KGaA) or 100 µM curcumin, respectively. Then, 10
ng/ml interleukin 1β (IL-1β; Sigma-Aldrich; Merck KGaA) was added
and incubated for 90 min at 37°C, immediately followed by
luciferase assay (Promega Corp.; cat. no. E1910). Briefly, cells
were lysed with lysis buffer, the first substrate to detect the
firefly luciferase was added, and the samples were immediately
measured for 6 sec. Upon addition of the second substrate signals
for Renilla luciferase were assessed.
Data and statistical analysis
Kinetic parameters were calculated using the
GraphPad Prism version 6.0 software program (GraphPad Software,
Inc., La Jolla, CA, USA) for a Michaelis-Menten kinetic model: V =
Vmax × S/(Km + S), where V is the rate of the
reaction; Vmax is the maximum velocity; Km is
the Michaelis constant and S is the substrate concentration. The
intrinsic clearance, which is defined as the ratio
Vmax/Km, quantifies the transport capacity.
IC50 values of curcumin and tetrahydrocurcumin
cytotoxicity against wild-type and OATP-knockdown ZR-75-1 cells
were calculated by fitting a non-linear model to cell viability vs.
(log)concentrations data using the GraphPad Prism version 6.0
software (GraphPad Software, Inc.). This software was also used for
all statistical analyses. All values were expressed as the mean ±
standard deviation (SD) of 3 independent biological replicates and
one-way analysis of variance (ANOVA) followed by Tukey's post hoc
test was used to compare differences between the wild-type and
OATP1B1-knockdown ZR-75-1 cells. The statistical significance
threshold was defined as P<0.05 for all calculations.
Results
Accumulation of curcumin and its
metabolites in transfected CHO cells
To investigate whether curcumin and its major
conjugates are substrates of OATPs, uptake analyses were performed
in OATP1B1-, OATP1B3- and OATP2B1-transfected CHO cells. CHO cells
only transfected with the vector were used as controls. The uptake
of curcumin (25–200 µM) by all three OATPs was linear for up to 1
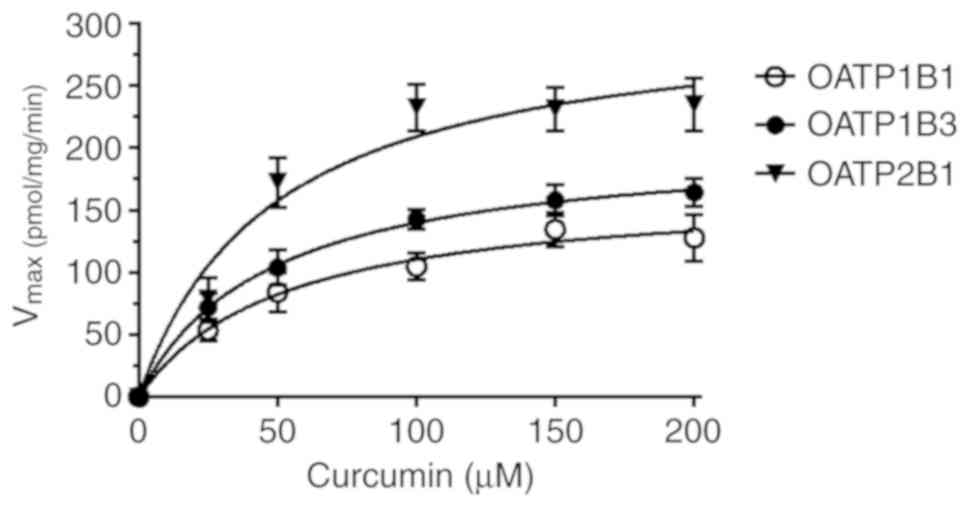
min (data not shown). As revealed in Table I and Fig. 1, the initial OATP1B1-, OATP1B3- and
OATP2B1-mediated accumulation rates (namely, OATP-transfected CHO
cells minus CHO cells only transfected with the vector) for
curcumin followed Michaelis-Menten kinetics, with higher
Vmax values for OATP1B1 than for OATP1B3 and OATP2B1
(Vmax, 310 vs. 205 and 167 pmol/mg protein/min,
respectively). The Km-values were similar for all three
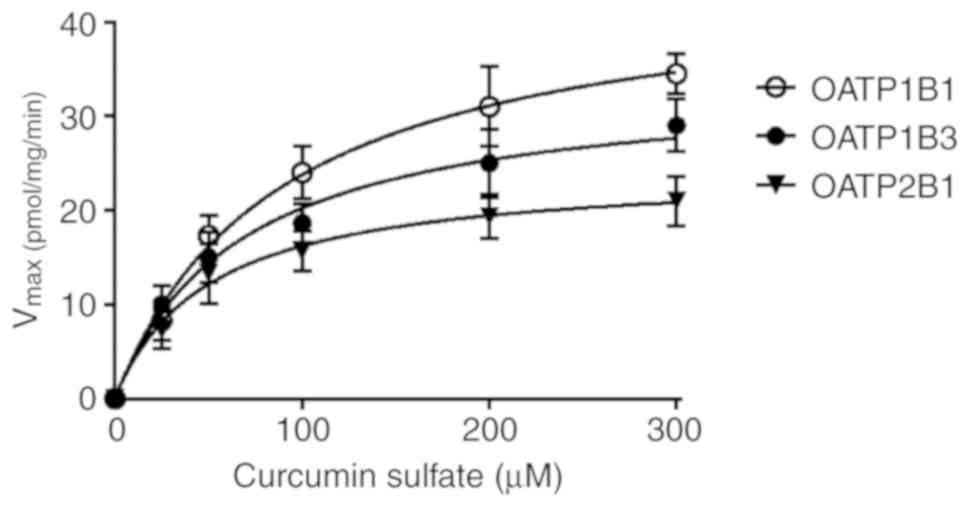
OATPs (range, 46.9–51.9 µM). The uptake of curcumin sulfate (25–300
µM) in OATP1B1-, OATP1B3- and OATP2B1-transfected CHO cells, was
less pronounced, revealing Vmax values of only 45.0,
33.9 and 24.3 pmol/mg protein/min, respectively (Table I and Fig. 2). Its affinity, for OATP1B1 and
OATP1B3, but not for OATP2B1, was 1.9 and 1.4-fold higher with
Km values of 89.1 µM and 67.7 µM compared with curcumin.
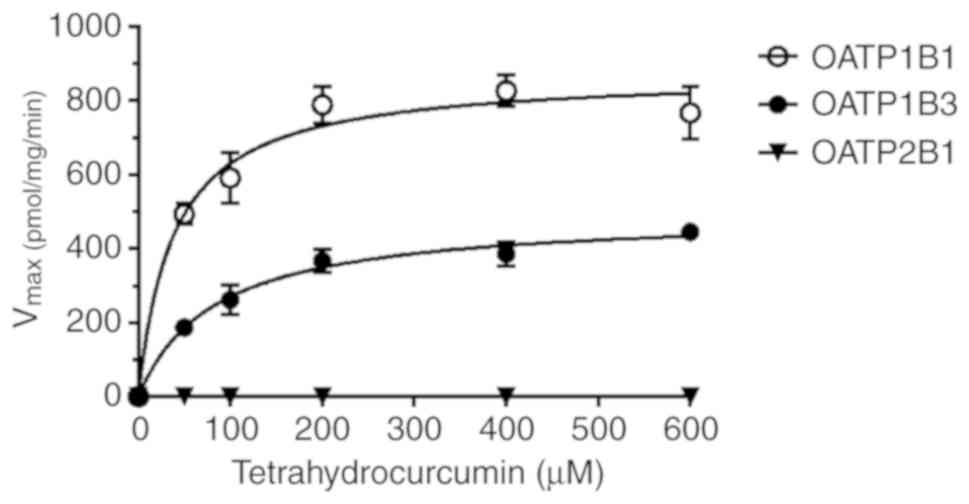
Tetrahydrocurcumin was taken up by OATP1B1 and OATP1B3 with the
highest Vmax values (872 and 493 pmol/mg protein/min,
respectively); Km values were 38.6 and 83.7 µM,
respectively (Table I and Fig. 3). Notably, curcumin glucuronide was
not a substrate for any of the three OATPs as its levels inside the
cytoplasm were below the detection limit.
 | Table I.Michaelis-Menten parameters for the
uptake of curcumin, curcumin sulfate, tetrahydrocurcumin and
curcumin glucuronide in OATP-transfected CHO cells. |
Table I.
Michaelis-Menten parameters for the
uptake of curcumin, curcumin sulfate, tetrahydrocurcumin and
curcumin glucuronide in OATP-transfected CHO cells.
| Substrate | Km
[µM] | Vmax
[pmol/mg/min] |
Vmax/Km [µl/min.
µg] |
|---|
| OATP1B1 |
|
Cur | 51.9±13.6 | 167±14.8 | 3.21±0.64 |
|
Cur-S | 89.1±16.4 | 45.0±3.34 | 0.51±0.06 |
|
TH-cur | 38.6±7.9 | 872±35.6 | 22.6±3.86 |
|
Cur-G | n.d. | n.d. | n.d. |
| OATP1B3 |
|
Cur | 46.9±7.5 | 205±10.7 | 4.37±0.48 |
|
Cur-S | 67.7±15.3 | 33.9±2.76 | 0.50±0.076 |
|
TH-cur | 83.7±13.4 | 493±22.5 | 5.89±0.69 |
|
Cur-G | n.d. | n.d. | n.d. |
| OATP2B1 |
|
Cur | 48.6±12.7 | 310±26.6 | 6.37±1.27 |
|
Cur-S | 50.0±12.6 | 24.3±1.87 | 0.48±0.102 |
|
TH-cur | n.d. | n.d. | n.d. |
|
Cur-G | n.d. | n.d. | n.d. |
OATP1B1-knockdown in ZR-75-1
cells
PCR data from various lentiviral-transfected clones
revealed an up to 10-fold reduction in OATP1B1 expression in
ZR-75-1-cells (relative mRNA expression was reduced from 14.78±0.26
to 1.19±0.02). Cells with the lowest OATP1B1 expression levels were
used for further uptake experiments (27).
Curcumin and tetrahydrocurcumin
accumulation in wild-type and OATP1B1-knockdown ZR-75-1 cells
Based on the markedly higher OATP1B1 mRNA level
noticed in wild-type ZR-75-1 cells compared with the
OATP1B1-knockdown clone and the fact that ZR-75-1 cells do not
express OATP1B3 or OATP2B1, uptake of curcumin and
tetrahydrocurcumin by wild-type cells was expected to be increased.
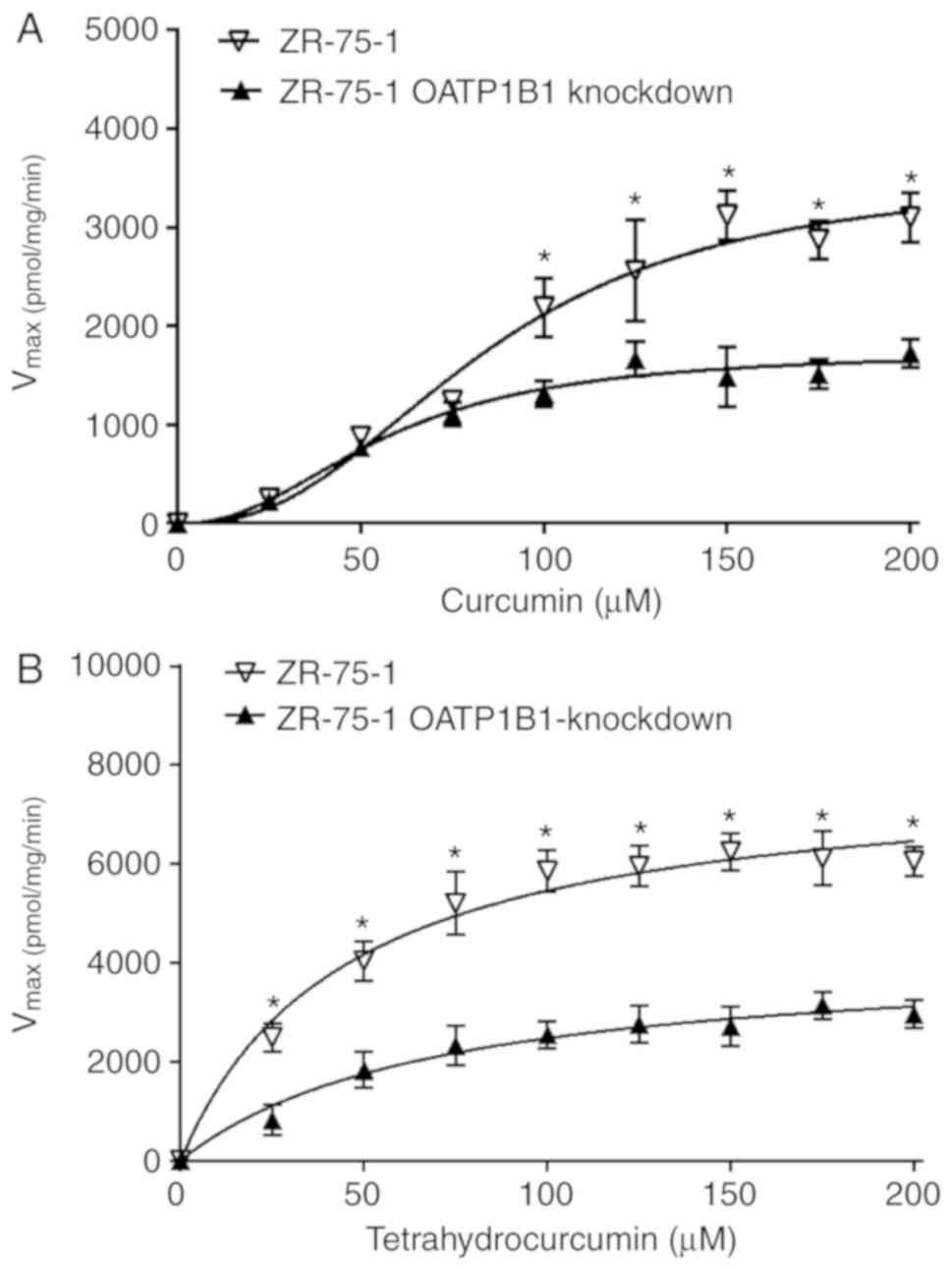
The results revealed that uptake of curcumin by ZR-75-1 cells was
increased as indicated by 2-fold higher Vmax values
compared with OATP1B1-knockdown cells (Vmax, 3535 vs.
1741 pmol/mg protein/min); the Km-values only changed
slightly (Km, 85.1 vs. 56.7 µM) (Fig. 4A and Table II). Differences in the uptake of
tetrahydrocurcumin were also pronounced leading to 2-fold higher
Vmax values in wild-type cells compared with the
OATP1B1-knockdown clone (Vmax, 7904 vs. 4201 pmol/mg
protein/min); the Km-values were significantly reduced
in the wild-type cells (Km, 44.7 vs. 69.6 µM) (Fig. 4B and Table II) supporting the impact of OATP1B1
for curcumin and tetrahydrocurcumin transport. Notably, any
interference in curcumin and tetrahydrocurcumin uptake with efflux
mechanisms (e.g. BCRP) could be excluded as the incubation time was
only 1 min.
 | Table II.Michaelis-Menten parameters of
curcumin and tetrahydrocurcumin determined in ZR-75-1 and
OATP1B1-knockdown ZR-75-1 cells. |
Table II.
Michaelis-Menten parameters of
curcumin and tetrahydrocurcumin determined in ZR-75-1 and
OATP1B1-knockdown ZR-75-1 cells.
|
| ZR-75-1 | OATP1B1-knockdown
ZR-75-1 |
|---|
|
|
|
|
|---|
| Substrate | Km
[µM] | Vmax
[pmol/mg/min] | Km
[µM] | Vmax
[pmol/mg/min] |
|---|
| Curcumin |
85.1±9.1a |
3535±342a |
56.7±5.8a |
1741±122a |
| TH-cur | 44.7±7.9 |
7904±420a | 69.6±14.8 |
4201±337a |
Cytotoxicity of curcumin in ZR-75-1
OATP1B1-knockdown cells
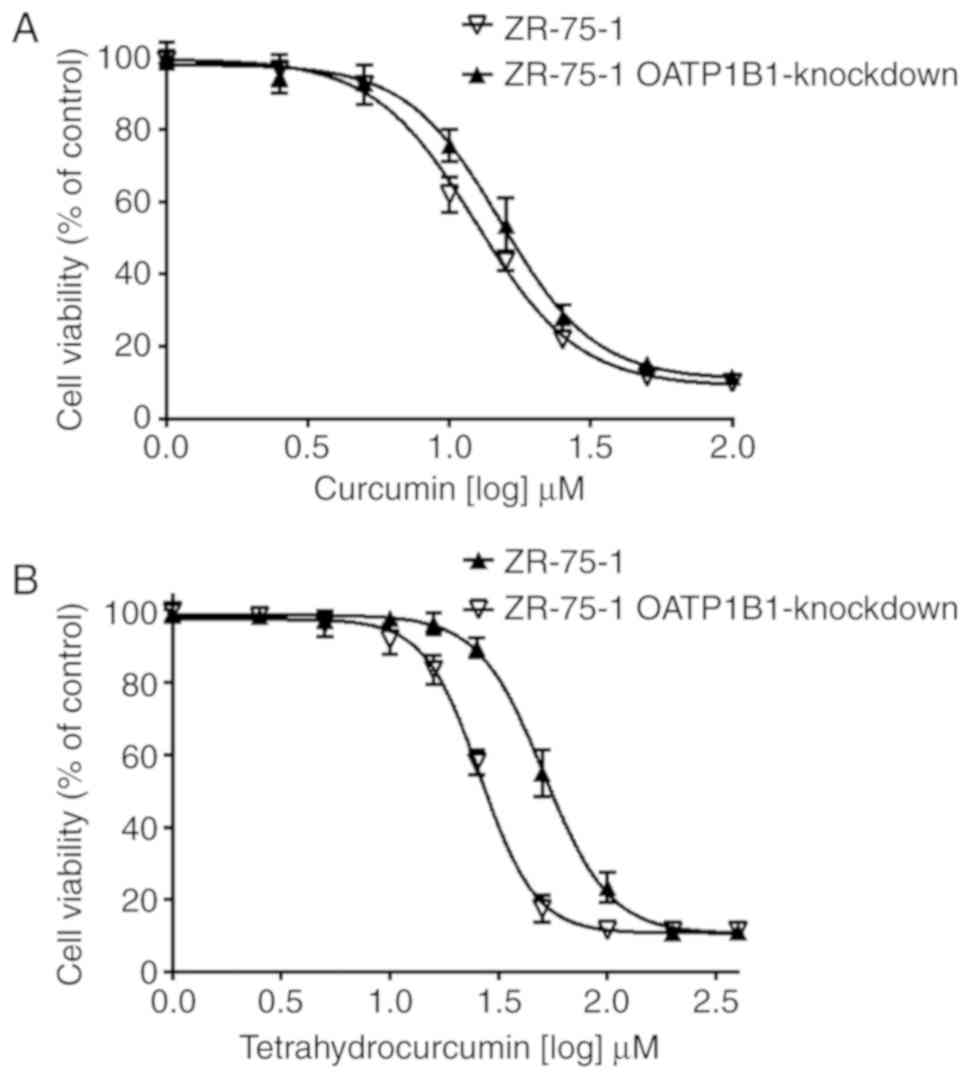
As revealed in Fig.
5A, curcumin exhibited a lower IC50 value in
wild-type ZR-75-1 cells (12.4 µM) compared with the OATP1B1
knockdown clone (15.2 µM), although the differences were not
significant. The IC50 value for tetrahydrocurcumin,
however, was significantly lower in OATP1B1 expressing cells (26.0
vs. 51.3 µM) compared with the OATP1B1-knockdown cells (Fig. 5B) clearly demonstrating that OATP1B1
expression is associated with cytotoxicity.
Inhibition of NF-κB-luciferase by
curcumin in wild-type and OATP1B1-knockdown ZR-75-1 cells
To further evaluate the OATP1B1-dependent
differences in the activity of curcumin, wild-type and
OATP1B1-knockdown ZR-75-1 cells were simultaneously transfected
with an NF-κB promoter sequence connected to a luciferase reporter.
Prior to NF-κB reporter induction by 10 ng/ml IL-1β for 90 min,
cells were treated with curcumin or BAY11-7082 for 30 min, and the
luciferase signals were assessed. As revealed in Fig. 6, curcumin significantly inhibited
IL-1β-induced NF-κB reporter expression by 40.1±7.78% in wild-type
and by 30.9±10.1% in OATP1B1-knockdown cells. Notably, curcumin was
almost as potent as the known NF-κB inhibitor BAY11-7082, used as a
positive control (71.3±12.9 and 62.8±5.3% inhibition in wild-type
and in the OATP-knockdown clone, respectively). As anticipated
inhibition of NF-κB-luciferase by curcumin in OATP1B1-knockdown
cells was less pronounced compared with wild-type cells. However,
differences were not significant.
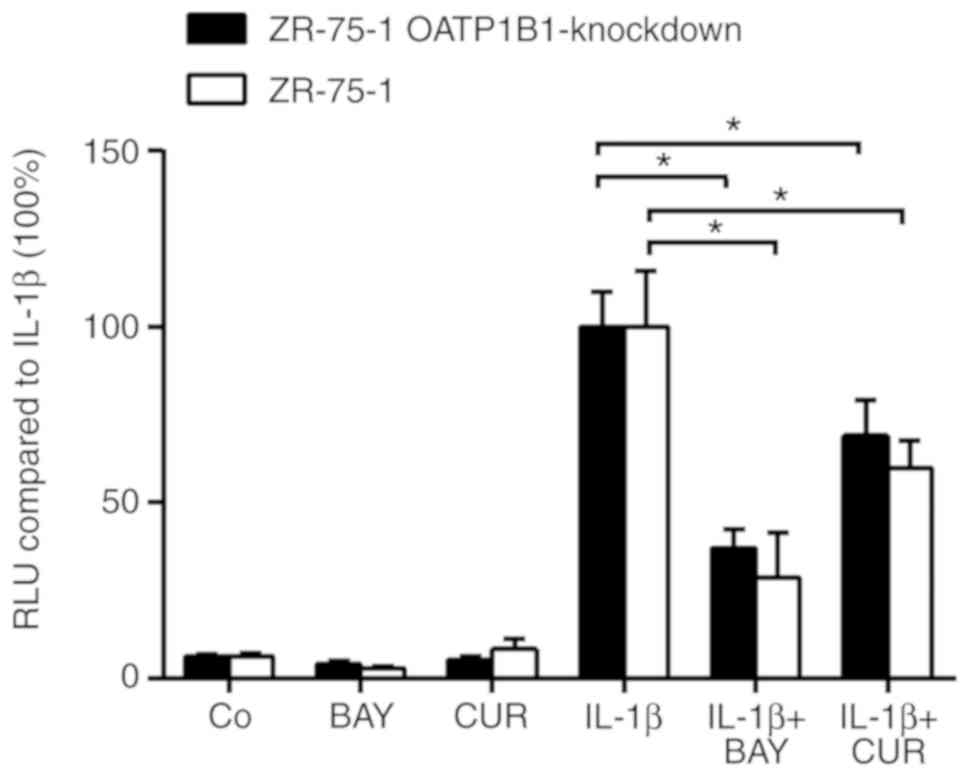 | Figure 6.Inhibition of NF-κB activity by
curcumin. A total of 100,000 ZR-75-1 wild-type and ZR-75-1
OATP1B1-knockdown cells were seeded in 24-well plates and allowed
to grow to 70% confluence. Cells were then pre-treated with 10 µM
BAY or 100 µM CUR for 30 min or with solvent (DMSO). Thereafter,
where indicated, cells were stimulated with IL-1β (10 ng/ml for 90
min), when cells were lysed and firefly luciferase activity was
determined, which was normalized to Renilla luciferase
activity (measured subsequently; RLU, relative light unit).
Experiments were performed in triplicate, error bars indicate ± SD
and asterisks denote significance between the IL-1β-induced
positive controls and the IL-1β-induced BAY and CUR treatment
groups. OATP, organic anion-transporting polypeptide; Co, control;
CUR, curcumin; BAY, Bay11-7082. |
Discussion
The present study aimed to determine the kinetics of
the cellular uptake of curcumin and its major metabolites curcumin
sulfate, curcumin glucuronide and tetrahydrocurcumin and Chinese
hamster ovary (CHO) cells stably transfected with the three-major
organic anion-transporting polypeptides (OATPs) were used. As
indicated in Fig. 1 and Table I, curcumin displayed saturable
uptake kinetics for OATP1B1, OATP1B3 and OATP2B1 with similar
Km values (ranging from 46.9 to 51.9 µM) which indicates
high affinity for the transporter. The affinity of curcumin sulfate
was in a similar range for OATP2B1 but was higher for OATP1B3 and
OATP1B1 (Km values, 50.0, 67.7 and 89.1 µM,
respectively). Notably, tetrahydrocurcumin was only transported by
OATP1B1 and OATP1B3 with reduced affinity in the case of OATP1B1
(Km, 83.7 µM) and increased affinity (Km,
38.6 µM) in the case of OATP1B3. Curcumin glucuronide was not
transported by any of these OATPs. Notably, OATP-dependent uptake
was compound-specific. While the transport capacity
(Vmax/Km) for curcumin sulfate was low for
all three OATPs (0.51, 0.50 and 0.48 µl/min/mg protein,
respectively), the uptake of curcumin by OATP1B1, OATP1B3 and
OATP2B1-transfected cells was 6.3-, 8.7- and 13.7-fold higher,
respectively. The uptake of tetrahydrocurcumin by OATP1B1 was even
more pronounced (i.e. 44.3-fold higher compared with curcumin).
These data revealed that OATP1B1 could be the most important
transporter for tetrahydrocurcumin uptake, whereas the three OATPs
were equally important for the cellular uptake of curcumin and
curcumin sulfate. The involvement of OATP1B1, OAT1B3 and OATP2B1 in
the uptake of curcumin was in line with the data from Zhou et
al (28) which also
demonstrated that this compound was a substrate of all three
transporters. However, contrary to that study, the present study
could not confirm any OATP-dependent uptake of curcumin
glucuronide. Based on our data, it is not possible to predict the
contribution of OATP2B1 in the gut, for that of OATP1B1, OATP1B3
and OATP2B1 in the liver, to the overall uptake of curcumin and its
major metabolites in humans due to large inter individual
variability (up to 10-fold differences) in OATP protein levels
(36–38).
Our data also suggested that unconjugated curcumin
and tetrahydrocurcumin concentrations in human blood were lower
than the Km values calculated for their uptake by
ZR-75-1 cells. A recent phase-I study revealed that administration
of liposomal curcumin at 300 mg/m2 over 8 h to patients
with metastatic cancer resulted in maximal plasma concentrations of
up to 3.48 µg/ml (9.45 µM) (39).
Peak plasma concentrations of up to 22 µM were observed for
tetrahydrocurcumin in rats following an oral dose of 500 mg/kg
tetrahydrocurcumin (40). Notably,
the total tissue concentrations of conjugates (curcumin sulfate and
curcumin glucuronide) were much higher than their blood levels in
mice 1 h after i.p. administration of curcumin (0.1 g/kg) leading
to concentrations of 26.1 µg/g (52.6 µM), 26.9 µg/g (54.3 µM) and
117 µg/g (236 µM) in the spleen, liver and intestine, respectively
(41).
To determine the importance of OATPs in the uptake
of curcumin and tetrahydrocurecumin, hormone-dependent ZR-75-1
breast cancer cells which express high levels of OATP1B1, but do
not express OATP1B3 or OATP2B1 (27), were incubated with both compounds.
The uptake of curcumin by the ZR-75-1 OATP1B1-knockdown cells was
significantly decreased compared with wild-type cells leading to
lower Vmax values (Fig.
4 and Table II). Notably,
differences in curcumin uptake were only observed at >100 µM
curcumin concentrations and not expected based on the uptake
experiments in OATP-transfected CHO cells. This may be explained by
the rapid non-enzymatic hydrolysis of curcumin at pH 7.4,
particularly at low curcumin concentrations, mainly forming ferulic
acid, feruloyl methan and vanillin. These degradation products were
stable under physiological conditions and exhibited antitumor
properties against various cancer cell lines (42–44).
Whether these compounds are substrates for OATPs or other not
identified uptake transporters is not yet known. Compared with
curcumin, the uptake of tetrahydrocurcumin was significantly
different at all the measured concentrations in wild-type and
OATP1B1-knockdown cells, probably due to the higher stability of
tetrahydrocurcumin at physiological pH (21). Differences in the cellular uptake of
tetrahydrocurcumin also resulted in higher Km values in
OATP1B1-knockdown cells indicating a lower affinity for this
transporter, albeit the results were not statistically
significant.
Concomitant with the reduced uptake displayed by
ZR-75-1 OATP1B1 knockdown cells, the present study also observed a
higher IC50 value for curcumin in the cytotoxicity assay
compared with OATP1B1-expressing wild-type cells (15.2 vs. 12.4 µM;
Fig. 5), although this difference
was not statistically significant and may be explained by the
degradation of curcumin in the medium and probably also in the
cytoplasm. For the more stable tetrahydrocurcumin, however, the
difference observed in the IC50 value between wild-type
and OATP1-knockdown ZR-75-1 cells was statistically significant.
Specifically, the IC50 value was reduced in the
OATP1B1-expressing cells (26.0 vs. 51.3 µM) indicating an
association between OATP1B1 expression and cytotoxicity. The
decreased uptake of curcumin by the ZR-75-1 OATP1B1-knockdown cells
also resulted in a reduced inhibition of IL-1β-activated NF-κB
reporter expression (Fig. 6), which
was not significant possibly again due to non-enzymatic hydrolysis
of curcumin to ferulic acid, feruloyl methan and vanillin in the
medium. Formation of these pharmacologically active degradation
products may therefore contribute, in combination with
tetrahydrocurcumin and curcumin sulfate, to the anticancer
properties of curcumin in vitro and in vivo. Since
NF-κB is highly expressed in breast cancer, thereby facilitating
growth and progression (45),
administration of curcumin, either as a single compound or in
combination with anticancer drugs, may lead to cancer
chemoprevention as revealed in human studies (45,46).
Different cellular expression levels of OATPs may
greatly affect the uptake of curcumin sulfate and
tetrahydrocurcumin, and to lesser extent also curcumin, by tumor
cells, thereby altering the efficacy of these compounds. Patients
with little or no detectable expression of OATP1B1, OATP1B3 and
OATP2B1 may, as a result exhibit decreased response rates to
curcumin and its primary metabolites. The same occurs when curcumin
and tetrahydrocurcumin are concomitantly administered with OATP
inhibitors such as clarithromycin, erythromycin and roxithromycin
which inhibit the intake of pravastatin in OATP1B1- and
OATP1B3-transfected 293 cells (37). Cyclosporin A is a potent inhibitor
of both OATPs leading to decreased uptake rates of bosentan
(31) and fexofenadine (47) in 293 and CHO cells. Several natural
occurring flavonoids inhibit OATP-dependent uptake as revealed for
dehydroepiandrosterone sulfate in a cellular model (48). Whether other transporters such
OATP2A1 and OATP4C1, which are expressed in ZR-75-1 wild-type cells
(24), are also involved in the
uptake of curcumin and its main metabolites is not yet known. Other
potential candidates may be the organic anion transporters (OATs)
which are involved in the transport of polyphenol glucuronides and
sulfates (49,50). In fact, Zhou et al reported,
at least for curcumin and curcumin glucuronide uptake, the
involvement of OAT1 and OAT3 (28).
Our data did not demonstrate any passive diffusion mechanism
responsible for the uptake of curcumin, curcumin sulfate, or
tetrahydrocurcumin since the uptake kinetics in wild-type- and
OATP1B1-knockdown ZR-75-1 cells was saturable, indicating
protein-mediated transport. However, the involvement of cellular
efflux mechanism in ZR-75-1 breast cancer cells cannot be excluded,
since uptake and efflux transport works in concert.
In conclusion, our results demonstrated that
curcumin, curcumin sulfate and tetrahydrocurcumin, but not curcumin
glucuronide, are substrates of various OATPs as demonstrated in
OATP-transfected CHO cells. The increased mRNA levels of OATP1B1 in
wild-type human breast cancer ZR-75-1 cells compared with
OATP1B1-knockdown cells were associated with a higher initial
uptake of curcumin and tetrahydrocurcumin leading to decreased
IC50 values. This may occur in patients following the
intravenous application of curcumin and tetrahydrocurcumin as
anticancer agents. Future clinical studies should determine not
only the concentration of curcumin, its main degradation products
and metabolites in blood and tumors but also the expression levels
of OATPs.
Acknowledgements
Not applicable.
Funding
QAJ received a fellowship from OeAD, Austria, in
collaboration with the Higher Education Commission of Pakistan. NJ
was awarded a scholarship financed by the Royal Golden Jubilee
Ph.D. Program of Thailand.
Availability of data and materials
All data generated or analysed during this study are
included in this published article.
Authors' contributions
NJ performed the experiments and analyzed the data
(uptake experiments, cytotoxicity and statistical analysis). QAJ
collected analysed and interpreted the data (uptake experiments and
cytotoxicity). JR designed and supervised the uptake experiments.
DM performed and interpreted the NF-κB experiments. GK wrote the
section concerning NF-κB and interpreted the data. BS was involved
in the study conception, interpretation of data and proofreading of
the manuscript. KJ interpreted the data concerning the uptake
experiments and was involved in the proofreading of the manuscript.
WJ interpreted the data, wrote and edited the manuscript. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Pan MH, Chiou YS, Chen LH and Ho CT:
Breast cancer chemoprevention by dietary natural phenolic
compounds: Specific epigenetic related molecular targets. Mol Nutr
Food Res. 59:21–35. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Aqil F, Jeyabalan J, Munagala R, Ravoori
S, Vadhanam MV, Schultz DJ and Gupta RC: Chemoprevention of rat
mammary carcinogenesis by Apiaceae spices. Int J Mol Sci.
18:E4252017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hatcher H, Planalp R, Cho J, Torti FM and
Torti SV: Curcumin: From ancient medicine to current clinical
trials. Cell Mol Life Sci. 65:1631–1652. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bimonte S, Barbieri A, Palma G, Rea D,
Luciano A, D'Aiuto M, Arra C and Izzo F: Dissecting the role of
curcumin in tumor growth and angiogenesis in a mouse model of human
breast cancer. Biomed Res Int. 2015:8781342015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lv ZD, Liu XP, Zhao WJ, Dong Q, Li FN,
Wang HB and Kong B: Curcumin induces apoptosis in breast cancer
cells and inhibits tumor growth in vitro and in vivo. Int J Clin
Exp Pathol. 7:2818–2824. 2014.PubMed/NCBI
|
|
6
|
Wang Y, Yu J, Cui R, Lin J and Ding X:
Curcumin in treating breast cancer: A review. J Lab Autom.
21:723–731. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kumar G, Mittal S, Sak K and Tuli HS:
Molecular mechanisms underlying chemopreventive potential of
curcumin: Current challenges and future perspectives. Life Sci.
148:313–328. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bachmeier BE, Nerlich AG, Iancu CM, Cilli
M, Schleicher E, Vené R, Dell'Eva R, Jochum M, Albini A and Pfeffer
U: The chemopreventive polyphenol Curcumin prevents hematogenous
breast cancer metastases in immunodeficient mice. Cell Physiol
Biochem. 19:137–152. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Quispe-Soto ET and Calaf GM: Effect of
curcumin and paclitaxel on breast carcinogenesis. Int J Oncol.
49:2569–2577. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bayet-Robert M, Kwiatkowski F, Leheurteur
M, Gachon F, Planchat E, Abrial C, Mouret-Reynier MA, Durando X,
Barthomeuf C and Chollet P: Phase I dose escalation trial of
docetaxel plus curcumin in patients with advanced and metastatic
breast cancer. Cancer Biol Ther. 9:8–14. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ireson C, Orr S, Jones DJ, Verschoyle R,
Lim CK, Luo JL, Howells L, Plummer S, Jukes R, Williams M, et al:
Characterization of metabolites of the chemopreventive agent
curcumin in human and rat hepatocytes and in the rat in vivo, and
evaluation of their ability to inhibit phorbol ester-induced
prostaglandin E2 production. Cancer Res. 61:1058–1064.
2001.PubMed/NCBI
|
|
12
|
Sharma RA, McLelland HR, Hill KA, Ireson
CR, Euden SA, Manson MM, Pirmohamed M, Marnett LJ, Gescher AJ and
Steward WP: Pharmacodynamic and pharmacokinetic study of oral
Curcuma extract in patients with colorectal cancer. Clin Cancer
Res. 7:1894–1900. 2001.PubMed/NCBI
|
|
13
|
Prasad S, Tyagi AK and Aggarwal BB: Recent
developments in delivery, bioavailability, absorption and
metabolism of curcumin: The golden pigment from golden spice.
Cancer Res Treat. 46:2–18. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Stanić Z: Curcumin, a compound from
natural sources, a true scientific challenge-a review. Plant Foods
Hum Nutr. 72:1–12. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jamil QUA, Jaerapong N, Zehl M,
Jarukamjorn K and Jäger W: Metabolism of curcumin in human breast
cancer cells: Impact of sulfation on cytotoxicity. Planta Med.
83:1028–1034. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tian Y, Bian Y, Jiang Y, Qian S, Yu A and
Zeng S: Interplay of breast cancer resistance protein (BCRP) and
metabolizing enzymes. Curr Drug Metab. 16:877–893. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ebert B, Seidel A and Lampen A:
Phytochemicals induce breast cancer resistance protein in Caco-2
cells and enhance the transport of benzo[a]pyrene-3-sulfate.
Toxicol Sci. 96:227–236. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Liu W, Zhang Z, Lin G, Luo D, Chen H, Yang
H, Liang J, Liu Y, Xie J, Su Z, et al: Tetrahydrocurcumin is more
effective than curcumin in inducing the apoptosis of H22 cells via
regulation of a mitochondrial apoptosis pathway in ascites
tumor-bearing mice. Food Funct. 8:3120–3129. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lai CS, Wu JC, Yu SF, Badmaev V,
Nagabhushanam K, Ho CT and Pan MH: Tetrahydrocurcumin is more
effective than curcumin in preventing azoxymethane-induced colon
carcinogenesis. Mol Nutr Food Res. 55:1819–1828. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang ZB, Luo DD, Xie JH, Xian YF, Lai ZQ,
Liu YH, Li WH, Chen JN, Lai XP, Lin ZX, et al: Curcumin's
metabolites, tetrahydrocurcumin and octahydrocurcumin, possess
superior anti-inflammatory effects in vivo through suppression of
TAK1-NF-κB pathway. Front Pharmacol. 9:11812018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Aggarwal BB, Deb L and Prasad S: Curcumin
differs from tetrahydrocurcumin for molecular targets, signaling
pathways and cellular responses. Molecules. 20:185–205. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hagenbuch B and Gui C: Xenobiotic
transporters of the human organic anion transporting polypeptides
(OATP) family. Xenobiotica. 38:778–801. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kim RB: Organic anion-transporting
polypeptide (OATP) transporter family and drug disposition. Eur J
Clin Invest. 33 (Suppl 2):S1–S5. 2003. View Article : Google Scholar
|
|
24
|
Gui C, Obaidat A, Chaguturu R and
Hagenbuch B: Development of a cell-based high-throughput assay to
screen for inhibitors of organic anion transporting polypeptides
1B1 and 1B3. Curr Chem Genomics. 4:1–8. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Obaidat A, Roth M and Hagenbuch B: The
expression and function of organic anion transporting polypeptides
in normal tissues and in cancer. Annu Rev Pharmacol Toxicol.
52:135–151. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Svoboda M, Riha J, Wlcek K, Jaeger W and
Thalhammer T: Organic anion transporting polypeptides (OATPs):
Regulation of expression and function. Curr Drug Metab. 12:139–153.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wlcek K, Svoboda M, Thalhammer T, Sellner
F, Krupitza G and Jaeger W: Altered expression of organic anion
transporter polypeptide (OATP) genes in human breast carcinoma.
Cancer Biol Ther. 7:1450–1455. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhou X, Zhang F, Chen C, Guo Z, Liu J, Yu
J, Xu Y, Zhong D and Jiang H: Impact of curcumin on the
pharmacokinetics of rosuvastatin in rats and dogs based on the
conjugated metabolites. Xenobiotica. 47:267–275. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sun X, Li J, Guo C, Xing H, Xu J, Wen Y,
Qiu Z, Zhang Q, Zheng Y, Chen X, et al: Pharmacokinetic effects of
curcumin on docetaxel mediated by OATP1B1, OATP1B3 and CYP450s.
Drug Metab Pharmacokinet. 31:269–275. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Gui C, Miao Y, Thompson L, Wahlgren B,
Mock M, Stieger B and Hagenbuch B: Effect of pregnane X receptor
ligands on transport mediated by human OATP1B1 and OATP1B3. Eur J
Pharmacol. 584:57–65. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Treiber A, Schneiter R, Häusler S and
Stieger B: Bosentan is a substrate of human OATP1B1 and OATP1B3:
Inhibition of hepatic uptake as the common mechanism of its
interactions with cyclosporin A, rifampicin, and sildenafil. Drug
Metab Dispos. 35:1400–1407. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Leuthold S, Hagenbuch B, Mohebbi N, Wagner
CA, Meier PJ and Stieger B: Mechanisms of pH-gradient driven
transport mediated by organic anion polypeptide transporters. Am J
Physiol Cell Physiol. 296:C570–C582. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Brenner S, Riha J, Giessrigl B, Thalhammer
T, Grusch M, Krupitza G, Stieger B and Jäger W: The effect of
organic anion-transporting polypeptides 1B1, 1B3 and 2B1 on the
antitumor activity of flavopiridol in breast cancer cells. Int J
Oncol. 46:324–332. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2−ΔΔCT method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Palermo DP, DeGraaf ME, Marotti KR,
Rehberg E and Post LE: Production of analytical quantities of
recombinant proteins in Chinese hamster ovary cells using sodium
butyrate to elevate gene expression. J Biotechnol. 19:35–47. 1991.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Otsuka S, Schaefer O, Kawakami H, Inoue T,
Lehner S, Saito A, Ishiguro N, Kishimoto W, Ludwig-Schwellinger E,
Ebner T, et al: Simultaneous absolute protein quantification of
transporters, cytochromes P450, and UDP-glucuronosyltransferases as
a novel approach for the characterization of individual human
liver: Comparison with mRNA levels and activities. Drug Metab
Dispos. 40:83–92. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Prasad B, Evers R, Gupta A, Hop CE,
Salphati L, Shukla S, Ambudkar SV and Unadkat JD: Interindividual
variability in hepatic organic anion-transporting polypeptides and
P-glycoprotein (ABCB1) protein expression: Quantification by liquid
chromatography-tandem mass spectroscopy and influence of genotype,
age, and sex. Drug Metab Dispos. 42:78–88. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Nies AT, Niemi M, Burk O, Winter S, Zanger
UM, Stieger B, Schwab M and Schaeffeler E: Genetics is a major
determinant of expression of the human hepatic uptake transporter
OATP1B1, but not of OATP1B3 and OATP2B1. Genome Med. 5:12013.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Greil R, Greil-Ressler S, Weiss L,
Schönlieb C, Magnes T, Radl B, Bolger GT, Vcelar B and Sordillo PP:
A phase 1 dose-escalation study on the safety, tolerability and
activity of liposomal curcumin (Lipocurc™) in patients with locally
advanced or metastatic cancer. Cancer Chemother Pharmacol.
82:695–706. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Novaes JT, Lillico R, Sayre CL,
Nagabushnam K, Majeed M, Chen Y, Ho EA, Oliveira ALP, Martinez SE,
Alrushaid S, et al: Disposition, metabolism and histone deacetylase
and acetyltransferase inhibition activity of tetrahydrocurcumin and
other curcuminoids. Pharmaceutics. 9:E452017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Pan MH, Huang TM and Lin JK:
Biotransformation of curcumin through reduction and glucuronidation
in mice. Drug Metab Dispos. 27:486–494. 1999.PubMed/NCBI
|
|
42
|
Celińska-Janowicz K, Zaręba I, Lazarek U,
Teul J, Tomczyk M, Pałka J and Miltyk W: Constituents of propolis:
Chrysin, caffeic acid, p-coumaric acid, and ferulic acid induce
PRODH/POX-dependent apoptosis in human tongue squamous cell
carcinoma cell (CAL-27). Front Pharmacol. 9:3362018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Pavelyev RS, Bondar OV, Nguyen TNT,
Ziganshina AA, Al Farroukh M, Karwt R, Alekbaeva GD, Pugachev MV,
Yamaleeva ZR, Kataeva ON, et al: Synthesis and in vitro antitumor
activity of novel alkenyl derivatives of pyridoxine, bioisosteric
analogs of feruloyl methane. Bioorg Med Chem. 26:5824–5837. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Elsherbiny NM, Younis NN, Shaheen MA and
Elseweidy MM: The synergistic effect between vanillin and
doxorubicin in ehrlich ascites carcinoma solid tumor and MCF-7
human breast cancer cell line. Pathol Res Pract. 212:767–777. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Wang W, Nag SA and Zhang R: Targeting the
NFκB signaling pathways for breast cancer prevention and therapy.
Curr Med Chem. 22:264–289. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Sen GS, Mohanty S, Hossain DM,
Bhattacharyya S, Banerjee S, Chakraborty J, Saha S, Ray P,
Bhattacharjee P, Mandal D, et al: Curcumin enhances the efficacy of
chemotherapy by tailoring p65NFκB-p300 cross-talk in favor of
p53-p300 in breast cancer. J Biol Chem. 286:42232–42247. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Matsushima S, Maeda K, Ishiguro N,
Igarashi T and Sugiyama Y: Investigation of the inhibitory effects
of various drugs on the hepatic uptake of fexofenadine in humans.
Drug Metab Dispos. 36:663–669. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Wang X, Wolkoff AW and Morris ME:
Flavonoids as a novel class of human organic anion-transporting
polypeptide OATP1B1 (OATP-C) modulators. Drug Metab Dispos.
33:1666–1672. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Wong CC, Botting NP, Orfila C, Al-Maharik
N and Williamson G: Flavonoid conjugates interact with organic
anion transporters (OATs) and attenuate cytotoxicity of adefovir
mediated by organic anion transporter 1 (OAT1/SLC22A6). Biochem
Pharmacol. 81:942–949. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Wong CC, Akiyama Y, Abe T, Lippiat JD,
Orfila C and Williamson G: Carrier-mediated transport of quercetin
conjugates: Involvement of organic anion transporters and organic
anion transporting polypeptides. Biochem Pharmacol. 84:564–570.
2012. View Article : Google Scholar : PubMed/NCBI
|