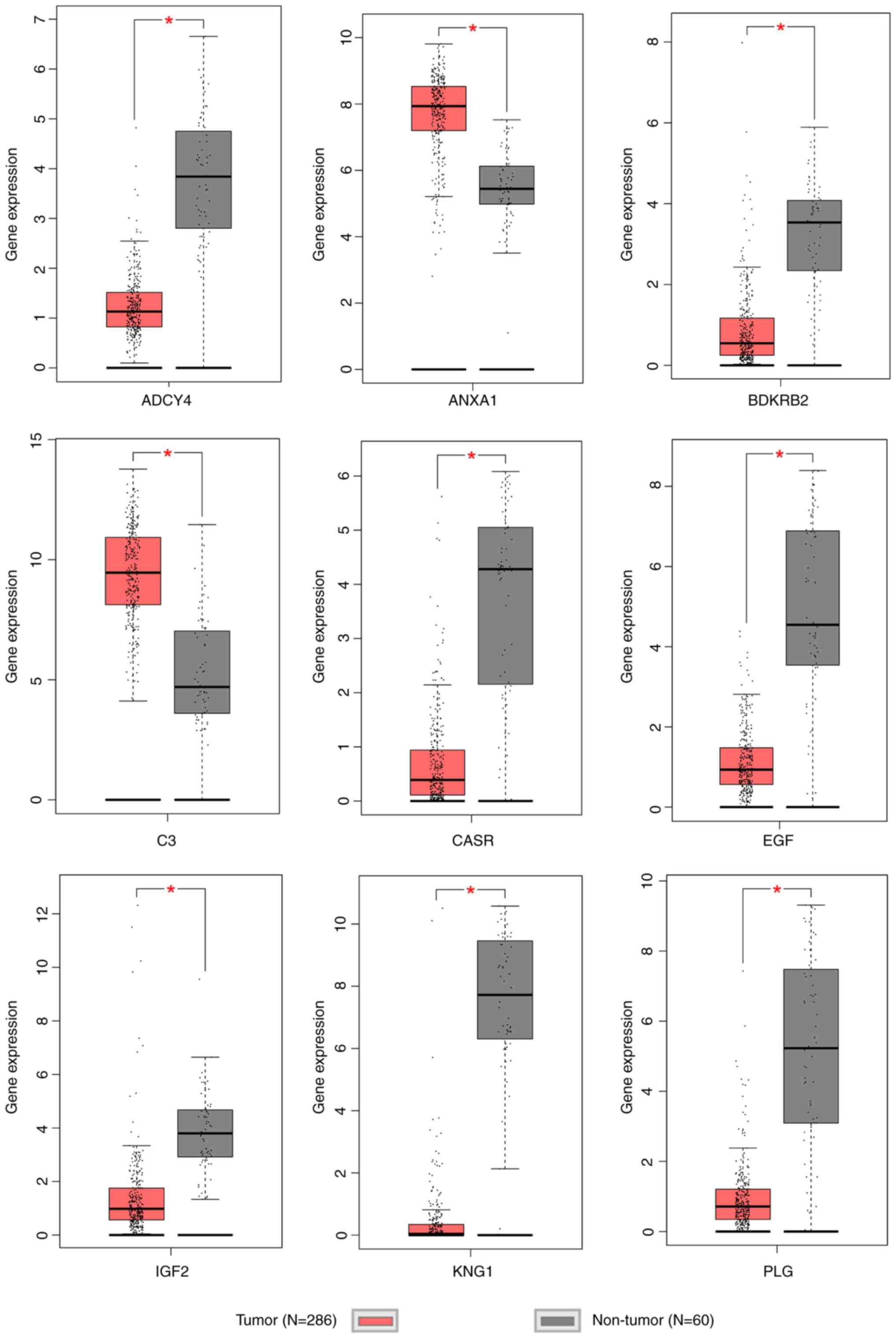
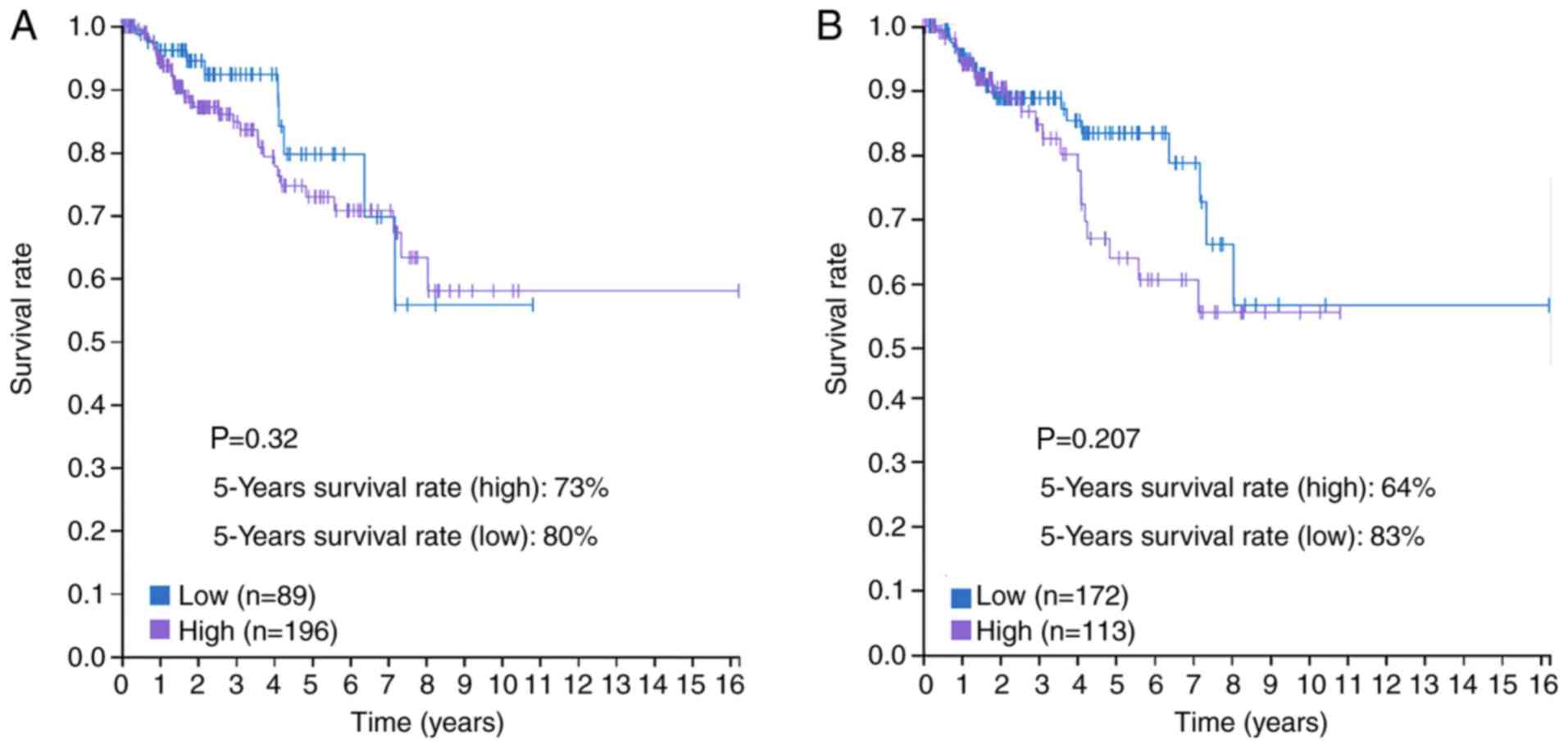
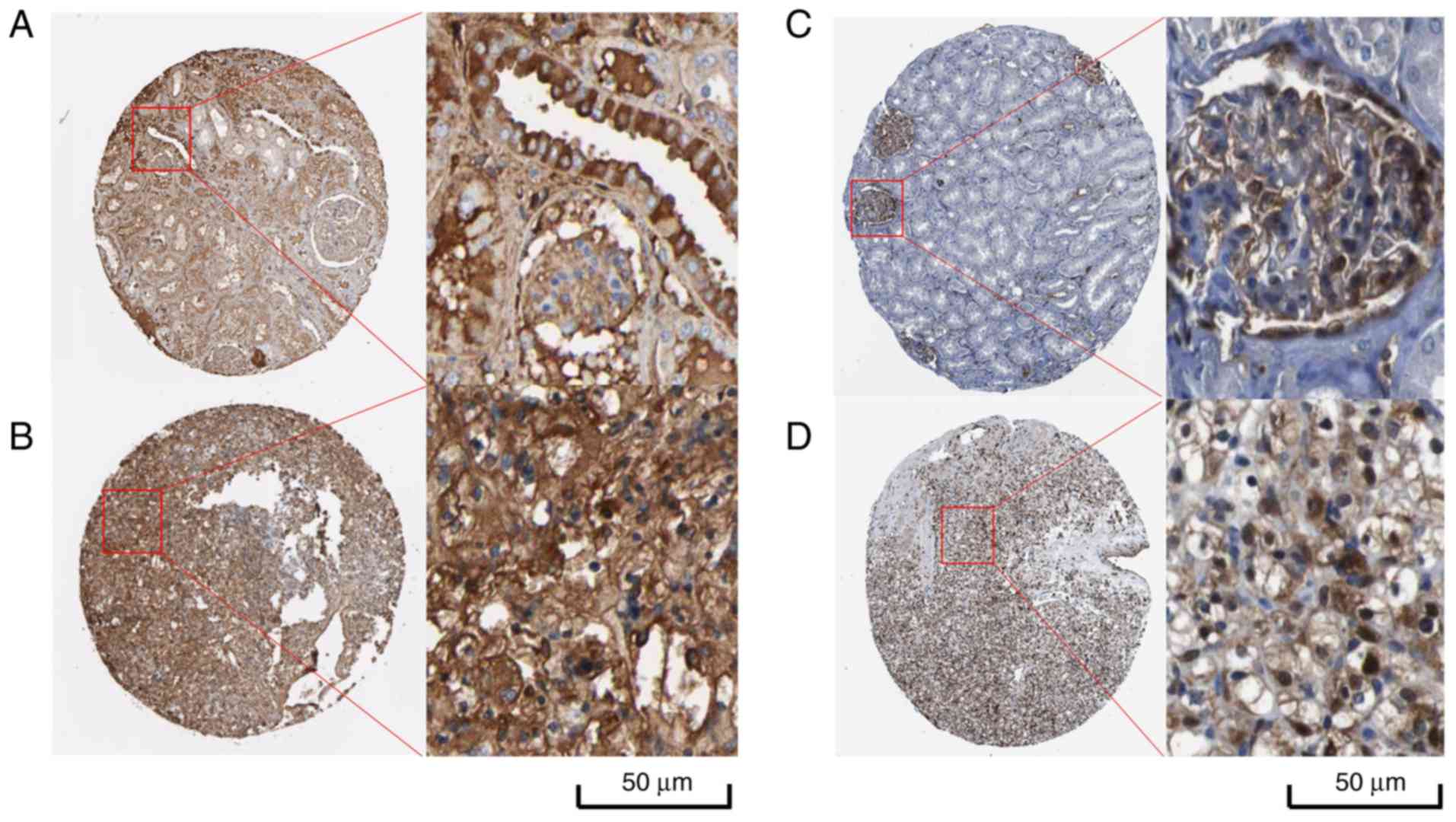
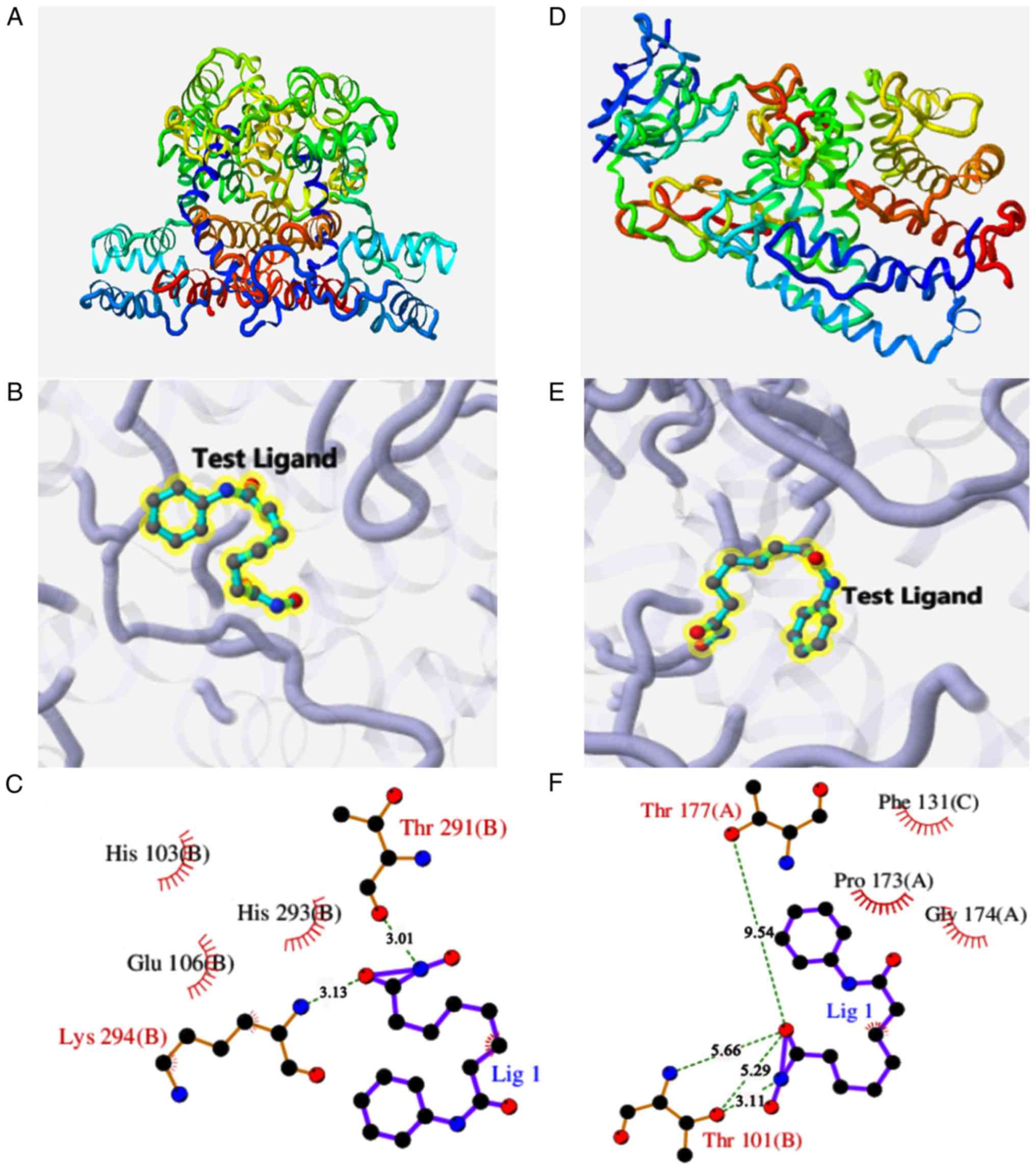
|
1
|
Wu X, Liu D, Gao X, Xie F, Tao D, Xiao X,
Wang L, Jiang G and Zeng F: Inhibition of BRD4 suppresses cell
proliferation and induces apoptosis in renal cell carcinoma. Cell
Physiol Biochem. 41:1947–1956. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wang C, Cai L, Liu J, Wang G, Li H, Wang
X, Xu W, Ren M, Feng L, Liu P, et al: MicroRNA-30a-5p inhibits the
growth of renal cell carcinoma by modulating GRP78 expression. Cell
Physiol Biochem. 43:2405–2419. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wang J, Li M, Wang Y and Liu X:
Integrating subpathway analysis to identify candidate agents for
hepatocellular carcinoma. Onco Targets Ther. 9:1221–1230. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Steffens S, Janssen M, Roos FC, Becker F,
Schumacher S, Seidel C, Wegener G, Thüroff JW, Hofmann R, Stöckle
M, et al: Incidence and long-term prognosis of papillary compared
to clear cell renal cell carcinoma-a multicentre study. Eur J
Cancer. 48:2347–2352. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lan H, Zeng J, Chen G and Huang H:
Survival prediction of kidney renal papillary cell carcinoma by
comprehensive LncRNA characterization. Oncotarget. 8:110811–110829.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sourbier C, Liao PJ, Ricketts CJ, Wei D,
Yang Y, Baranes SM, Gibbs BK, Ohanjanian L, Spencer Krane L,
Scroggins BT, et al: Targeting loss of the Hippo signaling pathway
in NF2-deficient papillary kidney cancers. Oncotarget.
9:10723–10733. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chakraborty S, Tarantolo SR, Batra SK and
Hauke RJ: Incidence and prognostic significance of second primary
cancers in renal cell carcinoma. Am J Clin Oncol. 36:132–142. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
De P, Otterstatter MC, Semenciw R, Ellison
LF, Marrett LD and Dryer D: Trends in incidence, mortality, and
survival for kidney cancer in Canada, 1986–2007. Cancer Causes
Control. 25:1271–1281. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ke X, Zeng X, Wei X, Shen Y, Gan J, Tang H
and Hu Z: MiR-514a-3p inhibits cell proliferation and
epithelial-mesenchymal transition by targeting EGFR in clear cell
renal cell carcinoma. Am J Transl Res. 9:5332–5346. 2017.PubMed/NCBI
|
|
11
|
Akhtar M, Al-Bozom IA and Al Hussain T:
Papillary renal cell carcinoma (PRCC): An update. Adv Anat Pathol.
Nov 30–2018.(Epub ahead of print). doi:
10.1097/PAP.0000000000000220. PubMed/NCBI
|
|
12
|
Courthod G, Tucci M, Di Maio M and
Scagliotti GV: Papillary renal cell carcinoma: A review of the
current therapeutic landscape. Crit Rev Oncol Hematol. 96:100–112.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sleire L, Førde HE, Netland IA, Leiss L,
Skeie BS and Enger PØ: Drug repurposing in cancer. Pharmacol Res.
124:74–91. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45:W98–W102.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang Y, Huang JC, Cai KT, Yu XB, Chen YR,
Pan WY, He ZL, Lv J, Feng ZB and Chen G: Long noncoding RNA HOTTIP
promotes hepatocellular carcinoma tumorigenesis and development: A
comprehensive investigation based on bioinformatics, qRTPCR and
metaanalysis of 393 cases. Int J Oncol. 51:1705–1721. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
He Z, Tang F, Lu Z, Huang Y, Lei H, Li Z
and Zeng G: Analysis of differentially expressed genes, clinical
value and biological pathways in prostate cancer. Am J Transl Res.
10:1444–1456. 2018.PubMed/NCBI
|
|
17
|
Yang X, Zhu S, Li L, Zhang L, Xian S, Wang
Y and Cheng Y: Identification of differentially expressed genes and
signaling pathways in ovarian cancer by integrated bioinformatics
analysis. Onco Targets Ther. 11:1457–1474. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tripathi S, Pohl MO, Zhou Y,
Rodriguez-Frandsen A, Wang G, Stein DA, Moulton HM, DeJesus P, Che
J, Mulder LC, et al: Meta- and orthogonal integration of influenza
‘OMICs’ data defines a role for UBR4 in virus budding. Cell Host
Microbe. 18:723–735. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang J, Vasaikar S, Shi Z, Greer M and
Zhang B: WebGestalt 2017: A more comprehensive, powerful, flexible
and interactive gene set enrichment analysis toolkit. Nucleic Acids
Res. 45:W130–W137. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lamb J, Crawford ED, Peck D, Modell JW,
Blat IC, Wrobel MJ, Lerner J, Brunet JP, Subramanian A, Ross KN, et
al: The Connectivity Map: Using gene-expression signatures to
connect small molecules, genes, and disease. Science.
313:1929–1935. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Qu XA and Rajpal DK: Applications of
Connectivity Map in drug discovery and development. Drug Discov
Today. 17:1289–1298. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhong Y, Chen EY, Liu R, Chuang PY,
Mallipattu SK, Tan CM, Clark NR, Deng Y, Klotman PE, Ma'ayan A, et
al: Renoprotective effect of combined inhibition of
angiotensin-converting enzyme and histone deacetylase. J Am Soc
Nephrol. 24:801–811. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li C, Li X, Miao Y, Wang Q, Jiang W, Xu C,
Li J, Han J, Zhang F, Gong B, et al: SubpathwayMiner: A software
package for flexible identification of pathways. Nucleic Acids Res.
37:e1312009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Szklarczyk D, Santos A, von Mering C,
Jensen LJ, Bork P and Kuhn M: STITCH 5: Augmenting protein-chemical
interaction networks with tissue and affinity data. Nucleic Acids
Res. 44:D380–D384. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Szklarczyk D, Morris JH, Cook H, Kuhn M,
Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, et al:
The STRING database in 2017: Quality-controlled protein-protein
association networks, made broadly accessible. Nucleic Acids Res.
45:D362–D368. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Scardoni G, Tosadori G, Faizan M, Spoto F,
Fabbri F and Laudanna C: Biological network analysis with
CentiScaPe: Centralities and experimental dataset integration.
F1000Res. 3:1392014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Interactive human protein atlas launches.
Cancer Discov. 5:3392015. View Article : Google Scholar
|
|
28
|
Hsin KY, Matsuoka Y, Asai Y, Kamiyoshi K,
Watanabe T, Kawaoka Y and Kitano H: systemsDock: A web server for
network pharmacology-based prediction and analysis. Nucleic Acids
Res. 44:W507–W513. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Han L, Wang T, Wu J, Yin X, Fang H and
Zhang N: A facile route to form self-carried redox-responsive
vorinostat nanodrug for effective solid tumor therapy. Int J
Nanomedicine. 11:6003–6022. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Fernández-Rodríguez C, Salar A, Navarro A,
Gimeno E, Pairet S, Camacho L, Ferraro M, Serrano S, Besses C,
Bellosillo B, et al: Anti-tumor activity of the combination of
bendamustine with vorinostat in diffuse large B-cell lymphoma
cells. Leuk Lymphoma. 57:692–699. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Duvic M and Dimopoulos M: The safety
profile of vorinostat (suberoylanilide hydroxamic acid) in
hematologic malignancies: A review of clinical studies. Cancer
Treat Rev. 43:58–66. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zinzani PL, Bonthapally V, Huebner D,
Lutes R, Chi A and Pileri S: Panoptic clinical review of the
current and future treatment of relapsed/refractory T-cell
lymphomas: Cutaneous T-cell lymphomas. Crit Rev Oncol Hematol.
99:228–240. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Djamgoz MB, Coombes RC and Schwab A: Ion
transport and cancer: From initiation to metastasis. Philos Trans R
Soc Lond B Biol Sci. 369:201300922014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Guglietta S and Rescigno M:
Hypercoagulation and complement: Connected players in tumor
development and metastases. Semin Immunol. 28:578–586. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Samatov TR, Wicklein D and Tonevitsky AG:
L1CAM: Cell adhesion and more. Prog Histochem Cytochem. 51:25–32.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kourtidis A and Anastasiadis PZ: Bringing
together cell-to-cell adhesion and miRNA biology in cancer
research. Future Oncol. 12:1211–1214. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Brower V: Adhesion molecules, stem cells,
and the microenvironment in acute myeloid leukemia. J Natl Cancer
Inst. 108:djw1132016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Turaga SM and Lathia JD: Adhering towards
tumorigenicity: Altered adhesion mechanisms in glioblastoma cancer
stem cells. CNS Oncol. 5:251–259. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Hamidi H and Ivaska J: Every step of the
way: Integrins in cancer progression and metastasis. Nat Rev
Cancer. 18:533–548. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Tang Q, Zhang H, Kong M, Mao X and Cao X:
Hub genes and key pathways of non-small lung cancer identified
using bioinformatics. Oncol Lett. 16:2344–2354. 2018.PubMed/NCBI
|
|
41
|
Zhang Y, Zhang J, Shen Q, Yin W, Huang H,
Liu Y and Ni Q: High expression of Nectin-4 is associated with
unfavorable prognosis in gastric cancer. Oncol Lett. 15:8789–8795.
2018.PubMed/NCBI
|
|
42
|
Chang HY, Chang HM, Wu TJ, Chaing CY, Tzai
TS, Cheng HL, Raghavaraju G, Chow NH and Liu HS: The role of
Lutheran/basal cell adhesion molecule in human bladder
carcinogenesis. J Biomed Sci. 24:612017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zimpfer A, Maruschke M, Rehn S, Kundt G,
Litzenberger A, Dammert F, Zettl H, Stephan C, Hakenberg OW and
Erbersdobler A: Prognostic and diagnostic implications of
epithelial cell adhesion/activating molecule (EpCAM) expression in
renal tumours: A retrospective clinicopathological study of 948
cases using tissue microarrays. BJU Int. 114:296–302. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Alimbretov D, Askarova S, Umbayev B, Davis
T and Kipling D: Pharmacological targeting of cell cycle, apoptotic
and cell adhesion signaling pathways implicated in chemoresistance
of cancer cells. Int J Mol Sci. 19:E16902018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Shechter D, Dormann HL, Allis CD and Hake
SB: Extraction, purification and analysis of histones. Nat Protoc.
2:1445–1457. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Valenzuela-Fernández A, Cabrero JR,
Serrador JM and Sánchez-Madrid F: HDAC6: A key regulator of
cytoskeleton, cell migration and cell-cell interactions. Trends
Cell Biol. 18:291–297. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Kiweler N, Brill B, Wirth M, Breuksch I,
Laguna T, Dietrich C, Strand S, Schneider G, Groner B, Butter F, et
al: The histone deacetylases HDAC1 and HDAC2 are required for the
growth and survival of renal carcinoma cells. Arch Toxicol.
92:2227–2243. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Aggarwal R, Thomas S, Pawlowska N,
Bartelink I, Grabowsky J, Jahan T, Cripps A, Harb A, Leng J,
Reinert A, et al: Inhibiting histone deacetylase as a means to
reverse resistance to angiogenesis inhibitors: Phase I study of
abexinostat plus pazopanib in advanced solid tumor malignancies. J
Clin Oncol. 35:1231–1239. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Chun P: Therapeutic effects of histone
deacetylase inhibitors on kidney disease. Arch Pharm Res.
41:162–183. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Dokmanovic M, Clarke C and Marks PA:
Histone deacetylase inhibitors: Overview and perspectives. Mol
Cancer Res. 5:981–989. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Schmitt T, Mayer-Steinacker R, Mayer F,
Grünwald V, Schütte J, Hartmann JT, Kasper B, Hüsing J, Hajda J,
Ottawa G, et al: Vorinostat in refractory soft tissue
sarcomas-Results of a multi-centre phase II trial of the German
Soft Tissue Sarcoma and Bone Tumour Working Group (AIO). Eur J
Cancer. 64:74–82. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Pan CH, Chang YF, Lee MS, Wen BC, Ko JC,
Liang SK and Liang MC: Vorinostat enhances the cisplatin-mediated
anticancer effects in small cell lung cancer cells. BMC Cancer.
16:8572016. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Yoo C, Ryu MH, Na YS, Ryoo BY, Lee CW and
Kang YK: Vorinostat in combination with capecitabine plus cisplatin
as a first-line chemotherapy for patients with metastatic or
unresectable gastric cancer: Phase II study and biomarker analysis.
Br J Cancer. 114:1185–1190. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Li H, Wang X, Zhang C, Cheng Y, Yu M, Zhao
K, Ge W, Cai A, Zhang Y, Han F, et al: HDAC1-induced epigenetic
silencing of ASPP2 promotes cell motility, tumour growth and drug
resistance in renal cell carcinoma. Cancer Lett. 432:121–131. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Archin NM, Liberty AL, Kashuba AD,
Choudhary SK, Kuruc JD, Crooks AM, Parker DC, Anderson EM, Kearney
MF, Strain MC, et al: Administration of vorinostat disrupts HIV-1
latency in patients on antiretroviral therapy. Nature. 487:482–485.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Sung JA, Sholtis K, Kirchherr J, Kuruc JD,
Gay CL, Nordstrom JL, Bollard CM, Archin NM and Margolis DM:
Vorinostat renders the replication-competent latent reservoir of
human immunodeficiency virus (HIV) vulnerable to clearance by CD8 T
cells. EBioMedicine 23: 52–58, 2017. J Clin Invest. 127:3126–3135.
2017.PubMed/NCBI
|
|
57
|
Mota TM, Rasmussen TA, Rhodes A, Tennakoon
S, Dantanarayana A, Wightman F, Hagenauer M, Roney J, Spelman T,
Purcell DFJ, et al: No adverse safety or virological changes 2
years following vorinostat in HIV-infected individuals on
antiretroviral therapy. AIDS. 31:1137–1141. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Pili R, Liu G, Chintala S, Verheul H,
Rehman S, Attwood K, Lodge MA, Wahl R, Martin JI, Miles KM, et al:
Combination of the histone deacetylase inhibitor vorinostat with
bevacizumab in patients with clear-cell renal cell carcinoma: A
multicentre, single-arm phase I/II clinical trial. Br J Cancer.
116:874–883. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Schmeel LC, Schmeel FC, Blaum-Feder S and
Schmidt-Wolf IG: In vitro efficacy of naftifine against lymphoma
and multiple myeloma. Anticancer Res. 35:5921–5926. 2015.PubMed/NCBI
|
|
60
|
Bognar Z, Fekete K, Antus C, Hocsak E,
Bognar R, Tapodi A, Boronkai A, Farkas N, Gallyas F Jr, Sumegi B,
et al: Desethylamiodarone-A metabolite of amiodarone-Induces
apoptosis on T24 human bladder cancer cells via multiple pathways.
PLoS One. 12:e01894702017. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Chang YL, Liu ST, Wang YW, Lin WS and
Huang SM: Amiodarone promotes cancer cell death through elevated
truncated SRSF3 and downregulation of miR-224. Oncotarget.
9:13390–13406. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Felix F and Fontenele J: Valproic acid may
be tested in patients with H3F3A-mutated high-grade gliomas.
J Clin Oncol. 34:3104–3105. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Cha HY, Lee BS, Chang JW, Park JK, Han JH,
Kim YS, Shin YS, Byeon HK and Kim CH: Downregulation of Nrf2 by the
combination of TRAIL and Valproic acid induces apoptotic cell death
of TRAIL-resistant papillary thyroid cancer cells via suppression
of Bcl-xL. Cancer Lett. 372:65–74. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Xi W, Chen X, Sun J, Wang W, Huo Y, Zheng
G, Wu J, Li Y, Yang A and Wang T: Combined treatment with valproic
acid and 5-Aza-2′-deoxycytidine synergistically inhibits human
clear cell renal cell carcinoma growth and migration. Med Sci
Monit. 24:1034–1043. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Wei M, Mao S, Lu G, Li L, Lan X, Huang Z,
Chen Y, Zhao M, Zhao Y and Xia Q: Valproic acid sensitizes
metformin-resistant human renal cell carcinoma cells by
upregulating H3 acetylation and EMT reversal. BMC Cancer.
18:4342018. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Mao S, Lu G, Lan X, Yuan C, Jiang W, Chen
Y, Jin X and Xia Q: Valproic acid inhibits epithelialmesenchymal
transition in renal cell carcinoma by decreasing SMAD4 expression.
Mol Med Rep. 16:6190–6199. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Tajima S, Waki M, Doi W, Hayashi K,
Takenaka S, Fukaya Y and Kimura R: Acquired cystic
disease-associated renal cell carcinoma with a focal sarcomatoid
component: Report of a case showing more pronounced polysomy of
chromosomes 3 and 16 in the sarcomatoid component. Pathol Int.
65:89–94. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Hayes SA, Pandiri AR, Ton TV, Hong HH,
Clayton NP, Shockley KR, Peddada SD, Gerrish K, Wyde M, Sills RC,
et al: Renal cell carcinomas in vinylidene chloride-exposed male
B6C3F1 mice are characterized by oxidative stress and TP53 pathway
dysregulation. Toxicol Pathol. 44:71–87. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Malouf GG, Ali SM, Wang K, Balasubramanian
S, Ross JS, Miller VA, Stephens PJ, Khayat D, Pal SK, Su X, et al:
Genomic characterization of renal cell carcinoma with sarcomatoid
dedifferentiation pinpoints recurrent genomic alterations. Eur
Urol. 70:348–357. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Casuscelli J, Weinhold N, Gundem G, Wang
L, Zabor EC, Drill E, Wang PI, Nanjangud GJ, Redzematovic A,
Nargund AM, et al: Genomic landscape and evolution of metastatic
chromophobe renal cell carcinoma. JCI insight. 2:926882017.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Gao Y, Li J, Qiao N, Meng Q, Zhang M, Wang
X, Jia J, Yang S, Qu C, Li W, et al: Adrenomedullin blockade
suppresses sunitinib-resistant renal cell carcinoma growth by
targeting the ERK/MAPK pathway. Oncotarget. 7:63374–63387.
2016.PubMed/NCBI
|
|
72
|
Miyazaki A, Miyake H and Fujisawa M:
Molecular mechanism mediating cytotoxic activity of axitinib in
sunitinib-resistant human renal cell carcinoma cells. Clin Transl
Oncol. 18:893–900. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Ye H, Wang WG, Cao J and Hu XC: SPARCL1
suppresses cell migration and invasion in renal cell carcinoma. Mol
Med Rep. 16:7784–7790. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Ohnami S, Ohshima K, Nagashima T, Urakami
K, Shimoda Y, Saito J, Naruoka A, Hatakeyama K, Mochizuki T,
Serizawa M, et al: Comprehensive characterization of genes
associated with the TP53 signal transduction pathway in various
tumors. Mol Cell Biochem. 431:75–85. 2017. View Article : Google Scholar : PubMed/NCBI
|