Introduction
Papillary renal cell carcinoma (PRCC) is the second
most prevalent subtype of renal cell carcinoma (RCC) and it
represents 15–20% of all kidney neoplasms (1–4).
Compared with patients with clear RCC (CCRCC), those with PRCC
generally have a more favorable outcome following surgical
treatment and these patients are less likely to exhibit distant
metastasis and recurrence (5–7).
However, as incidents of RCC continue to increase, representing
2–3% of all adult malignancies, PRCC still receives attention as
the second most common subtype of RCC (8–10).
Although many genes have been demonstrated to be involved in the
development of PRCC in recent years (11), the underlying molecular mechanism of
PRCC still remains uncertain. Regarding the PRCC treatment, several
agents, such as mechanistic target of rapamycin suppressor and
anti-vascular endothelial growth factor agents, are suitable
options for patients with progressive and metastatic PRCC,
following the support of clinical trials consisting of all RCC
subtypes (12). However, specific
evidence from patients affected by PRCC is insufficient and
controversial, and results from the limited amount of small samples
(12). Therefore, it is necessary
to investigate the underlying mechanisms of PRCC and search for
additional ignored drugs for its treatment.
Although it is difficult to support expenditure in
researching novel oncologic therapeutic drugs, the clinical
application of a newly-discovered drug typically requires long-term
trials to ensure its safety and tolerance in the human body
(13). Therefore, the repurposing
of known drugs is a feasible drug development strategy, which
provides substantial advantages in meeting the high demands of
better therapeutic agents in anti-PRCC treatment by searching for
more suppressors with known safety but ignored oncologic
chemotherapy.
In the present study, the genes that are expressed
differently in cases of patients with PRCC and non-PRCC controls
were screened using The Cancer Genome Atlas (TCGA) data. A
bioinformatics analysis, including Gene Ontology (GO) analysis and
Kyoto Encyclopedia of Genes (KEGG) enrichment pathway, was
performed to explore the underlying mechanism in PRCC. Next, the
Connectivity map (Cmap), which is a database that contains
>6,900 expression profiles and 1,309 compounds, was used to
search for potential drugs for PRCC treatment based on differently
expressed genes (DEGs). Then, the hub genes of DEGs in PRCC were
selected following the construction of protein-protein interaction
(PPI) network analysis, and molecular docking tests between query
drugs and hub genes were further performed to validate the
prospective application in PRCC treatment.
Materials and methods
Identification of differently
expressed genes based on TCGA data
The TCGA database, which comprises 33 cancer types
and >10,000 samples, has been widely used to investigate the
underlying mechanism in human cancers. GEPIA (14), which is an online tool based on the
TCGA data, can be used for DEGs, correlation, survival, and
co-expressed genes analyses in various types of cancer. In the
present study, GEPIA was used to analyze the DEGs between PRCC
tissues and non-PRCC adjacent tissues with one-way analysis of
variance (ANOVA) method and Tukey's test, and genes with a q-value
<0.05 and ǀlog2fold-change (FC) ǀ>2 were secerned and
considered as the significant DEGs.
Functional annotation and KEGG pathway
enrichment analysis
For the investigation of the underlying mechanism of
PRCC, gene functional annotation and KEGG pathways analyses were
performed to explore how aforementioned DEGs function in the onset
and development of PRCC (15–17).
MetaScape (18) (http://metascape.org), which was updated in 2018, is a
web-based tool that provides gene functional annotation and
enrichment analysis. In the present study, GO analysis was
performed for the gene function annotation by MetaScape, and
another tool, Webgestalt (ORA method) (19) (http://www.webgestalt.org), was applied for KEGG
pathway enrichment analysis to illustrate which pathways may
contribute to the occurrence of PRCC.
Prediction of potential drugs for PRCC
treatment based on DEGs by Cmap and Drug Pair Seeker
The Cmap database (20,21)
uses gene-expression signatures to predict small molecular
compounds for a specific disease. In the present study, the DEGs of
PRCC were divided into 2 groups: Upregulated and downregulated
genes. Upregulated and downregulated genes were subsequently
uploaded to the Cmap in the ‘query’ page, and searches for small
molecule drugs that may treat PRCC were performed. Scores ranging
from −1-1 represented the correlation between the drug and DEGs.
The more negatively correlated drugs indicate greater correlation
with the uploaded DEGs and are more likely to be used for PRCC
treatment. In the present study, drugs with a score of ≤0.75 were
considered as candidate drugs for PRCC treatment. Additionally,
Drug Pair Seeker (DPS, version 1.4.0, http://www.maayanlab.net/DPS/) was also utilized to
predict which drug from Old AFFY Cmap data could be correlated with
the query drug together to reverse the direction of gene expression
(22).
Construction of the drug-pathway
network
For the exploration of the associations between
candidate agents and pathways, the expression profiles for each of
the candidate agents were downloaded and the genes affected by
candidate drugs were obtained from the Cmap database. The Cmap
incorporates 6,100 instances for 1,309 small molecular agents, and
each instance includes gene expression profiles of control and
corresponding treatment for a certain agent. For each instance,
expression profiles of treatment and control were matched for the
previously listed candidate drugs according to descriptions of the
annotation file. Then, DEGs were identified between control and
treatment with log2FC >1 or ≤1 (ORA method) for each candidate
drug, and these DEGs were considered as genes that were affected by
certain drugs. Finally, these affected genes for each above
candidate molecules were entered into SubpathwayMiner (3,23) (an
R package for identifying subpathways depend on the KEGG database)
to identify significantly enriched subpathways, and a subpathway
with false discovery rate (FDR) <0.1 was considered as
statistically significant.
Construction of drug-target
networks
To further explore the potential mechanism of the
top 10 prospective drugs, the SMILE structure of these drugs was
obtained from the DrugBank database (https://www.drugbank.ca/). Next, the STITCH database
(24) was applied to identify
targets of these drugs, and then drug-target networks were
construed to show the interactions between the top 10 drugs and
their corresponding targets.
Further exploration of query drugs for
PRCC treatment
Many types of dysregulated genes are involved in
tumorigenesis. To elucidate which genes may serve as key roles in
such a complex connection network, a PPI network was constructed
for the DEGs of PRCC using the STRING database (25), followed by the identification of the
hub genes by CentiScape (26), a
plugin of Cytoscape, which may have a key role in the gene
regulation network. Next, the expression levels of hub genes were
determined by TCGA data in GEPIA. Finally, immunohistochemical
(IHC) results of complement C3 (C3) and annexin 1 (ANXA1) in
various types of cancers were acquired from the Human Protein Atlas
(27) (version 18, https://www.proteinatlas.org/; ANXA1, https://www.proteinatlas.org/ENSG00000135046-ANXA1/pathology;
C3, http://wwwhttps://www.proteinatlas.org/ENSG00000125730-C3/pathology),
from which the prognostic value of C3 and ANXA1 based on TCGA data
(ANXA1, https://www.proteinatlas.org/ENSG00000135046-ANXA1/pathology/tissue/renal+cancer/KIRP;
C3, http://www.proteinatlas.org/ENSG00000125730-C3/pathology/tissue/renal+cancer/KIRP),
as well as IHC images in renal cancer and normal kidney were also
obtained (C3 protein in normal kidney, https://www.proteinatlas.org/ENSG00000125730-C3/tissue/kidney#;
C3 protein in renal cancer, https://www.proteinatlas.org/ENSG00000125730-C3/pathology/tissue/renal+cancer#;
ANXA1 protein in normal kidney, https://www.proteinatlas.org/ENSG00000135046-ANXA1/tissue/kidney#;
ANXA1 protein in renal cancer, http://www.proteinatlas.org/ENSG00000135046-ANXA1/pathology/tissue/renal+cancer#).
To further investigate the potential application of
query drugs in PRCC treatment, molecular docking study, as
determined by systemsDock (28), a
web-online tool for network pharmacology-based prediction and
analyses, was performed to simulate the drug-protein interactions
between the query drugs and hub genes. This tool provides a
high-precision docking simulation and docking pattern map to
systematically illustrate the ligand selectivity and the
interaction ability between a ligand and proteins, as well as to
elucidate how a specific ligand acts on a complex protein. The
interaction ability between the query drug and proteins are
assessed by docking scores. A drug that interacts well with ANXA1
(PDB code: 1HM6) and C3 (PDB code: 1GHQ; docking score >4) may
have a better anticancer application prospect in PRCC as it
suppresses the gene regulation network by inhibiting hub genes. A
flow chart detailing the experimental design of the present study
is presented in Fig. S1.
Statistical analysis
To analyze the genes that are differently expressed
in PRCC and non-PRCC adjacent tissues, the one-way ANOVA method and
Tukey's test was applied and genes with a q-value <0.05 and
ǀlog2FCǀ >2 were selected as the significant DEGs. For GO and
KEGG pathway analysis, the overrepresentation enrichment analysis
method was used, in which a GO term or pathway with P<0.05 was
significant. Following the determination of hub genes among the
above DEGs, Kaplan-Meier survival curves were performed to explore
their prognostic value in patients with PRCC. Furthermore, the
one-way ANOVA method and Tukey's test was also utilized to identify
the genes affected by each candidate drug (log2FC >1 or ≤1).
Thereafter, these affected genes were used to explore significantly
enriched subpathways (FDR<0.1) affected by candidate agents with
SubpathwayMiner tool.
Results
GO analysis and KEGG pathway
enrichment analysis using DEGs of PRCC
Altogether, 627 DEGs were identified from TCGA data,
including 161 upregulated and 466 downregulated genes (Fig. S2). The GO analysis includes three
categories (biological process, molecular function, and cellular
component), and the 10 significant enrichment terms were displayed
for each category (Table I). From
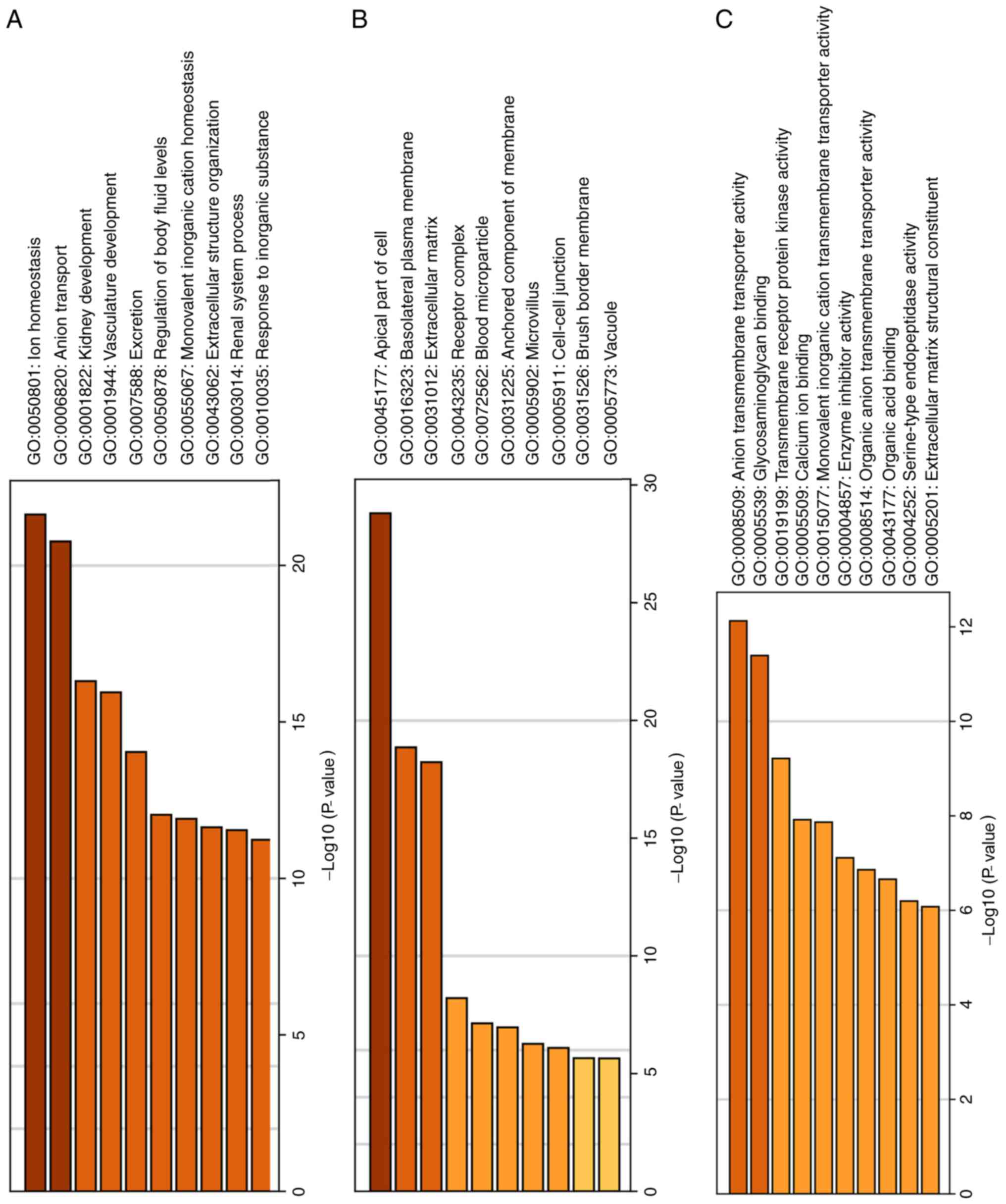
the biological process (Fig. 1A),
it was observed that DEGs were predominantly associated with anion
transport, ion hemostasis and kidney development. For the cellular
component (Fig. 1B), these DEGs
were enriched in the apical part of the cell, extracellular matrix
and basolateral plasma membrane. In molecular function (Fig. 1C), these DEGs were associated with
glycosaminoglycan binding, anion transmembrane transporter
activity, and calcium ion binding. Regarding the KEGG pathway
(Fig. 2; Table II), the results demonstrated that
DEGs are significantly associated with complement and coagulation
cascades, cell adhesion molecules and mineral absorption.
 | Table I.Significant GO terms for each GO
category enriched by MetaScape. |
Table I.
Significant GO terms for each GO
category enriched by MetaScape.
| Categories | GO ID | GO terms | Gene numbers | Log10
(P-value) |
|---|
| Biological
processes | GO:0006820 | Anion
transport | 58 | −17.90430 |
|
| GO:0050801 | Ion
homeostasis | 65 | −17.56780 |
|
| GO:0001822 | Kidney
development | 35 | −15.00420 |
|
| GO:0072358 | Cardiovascular
system development | 59 | −14.05610 |
|
| GO:0007588 | Excretion | 18 | −13.69620 |
|
| GO:0055067 | Monovalent
inorganic cation homeostasis | 23 | −11.67040 |
|
| GO:0050878 | Regulation of body
fluid levels | 43 | −11.62370 |
|
| GO:0007169 | Transmembrane
receptor protein tyrosine kinase signaling pathway | 50 | −10.33970 |
|
| GO:0048871 | Multicellular
organismal homeostasis | 33 | −10.24460 |
|
| GO:0043062 | Extracellular
structure organization | 35 | −9.96938 |
| Cellular
components | GO:0045177 | Apical part of
cell | 55 | −26.49700 |
|
| GO:0016323 | Basolateral plasma
membrane | 36 | −19.19540 |
|
| GO:0031012 | Extracellular
matrix | 46 | −11.64710 |
|
| GO:0009986 | Cell surface | 50 | −9.54270 |
|
| GO:0031225 | Anchored component
of membrane | 17 | −6.36750 |
|
| GO:0005911 | Cell-cell
junction | 28 | −5.48420 |
|
| GO:0072562 | Blood
microparticle | 17 | −5.43264 |
|
| GO:0031526 | Brush border
membrane | 9 | −5.35366 |
|
| GO:0005902 | Microvillus | 11 | −5.14557 |
|
| GO:0000323 | Lytic vacuole | 37 | −2.91416 |
| Molecular
functions | GO:0005539 | Glycosaminoglycan
binding | 27 | −11.27660 |
|
| GO:0008509 | Anion transmembrane
transporter activity | 34 | −10.93000 |
|
| GO:0019199 | Transmembrane
receptor protein kinase activity | 15 | −8.91288 |
|
| GO:0015081 | Sodium ion
transmembrane transporter activity | 20 | −8.45459 |
|
| GO:0004857 | Enzyme inhibitor
activity | 30 | −7.03172 |
|
| GO:0005509 | Calcium ion
binding | 40 | −5.9180 |
|
| GO:0004252 | Serine-type
endopeptidase activity | 21 | −5.86497 |
|
| GO:0019825 | Oxygen binding | 9 | −5.83118 |
|
| GO:0033293 | Monocarboxylic acid
binding | 10 | −5.32175 |
|
| GO:0019838 | Growth factor
binding | 14 | −5.29116 |
 | Table II.Significant KEGG pathways enriched by
Webgestalt. |
Table II.
Significant KEGG pathways enriched by
Webgestalt.
| Pathway ID | KEGG pathway | Counts | P-value |
|---|
| hsa04610 | Complement and
coagulation cascades | 15 | <0.00001 |
| hsa04514 | Cell adhesion
molecules | 18 | <0.00010 |
| hsa04978 | Mineral
absorption | 10 | <0.00001 |
| hsa04966 | Collecting duct
acid secretion | 7 | <0.00001 |
| hsa04960 |
Aldosterone-regulated sodium
reabsorption | 8 | 0.00010 |
| hsa04670 | Leukocyte
transendothelial migration | 14 | 0.00018 |
| hsa05323 | Rheumatoid
arthritis | 10 | 0.00250 |
| hsa04270 | Vascular smooth
muscle contraction | 12 | 0.00255 |
| hsa00590 | Arachidonic acid
metabolism | 8 | 0.00261 |
| hsa05110 | Vibrio cholerae
infection | 7 | 0.00336 |
| hsa00260 | Glycine, serine and
threonine metabolism | 6 | 0.00417 |
| hsa00010 |
Glycolysis/Gluconeogenesis | 8 | 0.00426 |
| hsa04020 | Calcium signaling
pathway | 15 | 0.00480 |
| hsa04976 | Bile secretion | 8 | 0.00608 |
| hsa00980 | Metabolism of
xenobiotics by cytochrome P450 | 8 | 0.00779 |
| hsa04015 | Rap1 signaling
pathway | 16 | 0.00842 |
| hsa04971 | Gastric acid
secretion | 8 | 0.00843 |
| hsa04961 | Endocrine and other
factor-regulated calcium reabsorption | 6 | 0.00929 |
| hsa00350 | Tyrosine
metabolism | 5 | 0.01079 |
| hsa04614 | Renin-angiotensin
system | 4 | 0.01113 |
The drugs of repurposing for PRCC
treatment
From the prediction of the Cmap dataset, 60
candidate drugs (after removing duplicates) that scored ≤-0.75 were
considered as potential drugs for PRCC treatment (Table SI). The information of the 10 drugs
with significant scores is listed in Table III and their 2D molecular
structures, as provided by Drugbank (https://www.drugbank.ca/), are displayed in Fig. S3.
 | Table III.Information of the 10 prospective
drugs with significant scores for papillary renal cell carcinoma
treatment. |
Table III.
Information of the 10 prospective
drugs with significant scores for papillary renal cell carcinoma
treatment.
| Drug name | Molecular
formula | Classification | Clinical
application |
|---|
| Pinacidil |
C13H19N5 | Membrane transport
modulators |
Antihypertension |
| Ciclosporin |
C62H111N11O12 |
Immunosuppressant | Transplant
rejection, rheumatoid arthritis, and severe psoriasis |
| Naftifine |
C21H21N | Antifungal
agent | Tinea pedis, tinea
cruris, and tinea corporis |
| Vorinostat |
C14H20N2O3 | Histone deacetylase
suppressor | Cutaneous T-cell
lymphoma |
| Metacycline |
C22H22N2O8 | Tetracycline
antibiotic agent | Acute bacterial
exacerbations of chronic bronchitis |
| Sulfacetamide |
C8H10N2O3S | Sulfonamide
antibiotic agent | Bacterial
vaginitis, keratitis, acute conjunctivitis, and blepharitis |
|
Chlorprothixene |
C18H18ClNS | Antipsychotic
agent | Psychotic
disorders, and acute mania |
| Amiodarone |
C25H29I2NO3 | Anti-Arrhythmia
agent | Frequently
recurring ventricular fibrillation and tachycardia |
| Noretynodrel |
C20H26O2 | Hormonal agent | Gynecological
disorders, and contraceptives |
| Valproic acid |
C8H16O2 | Anti-epileptic
Agent | Epilepsy, mania,
and migraine headache |
Construction of drug-pathway and
drug-target network for candidate drugs
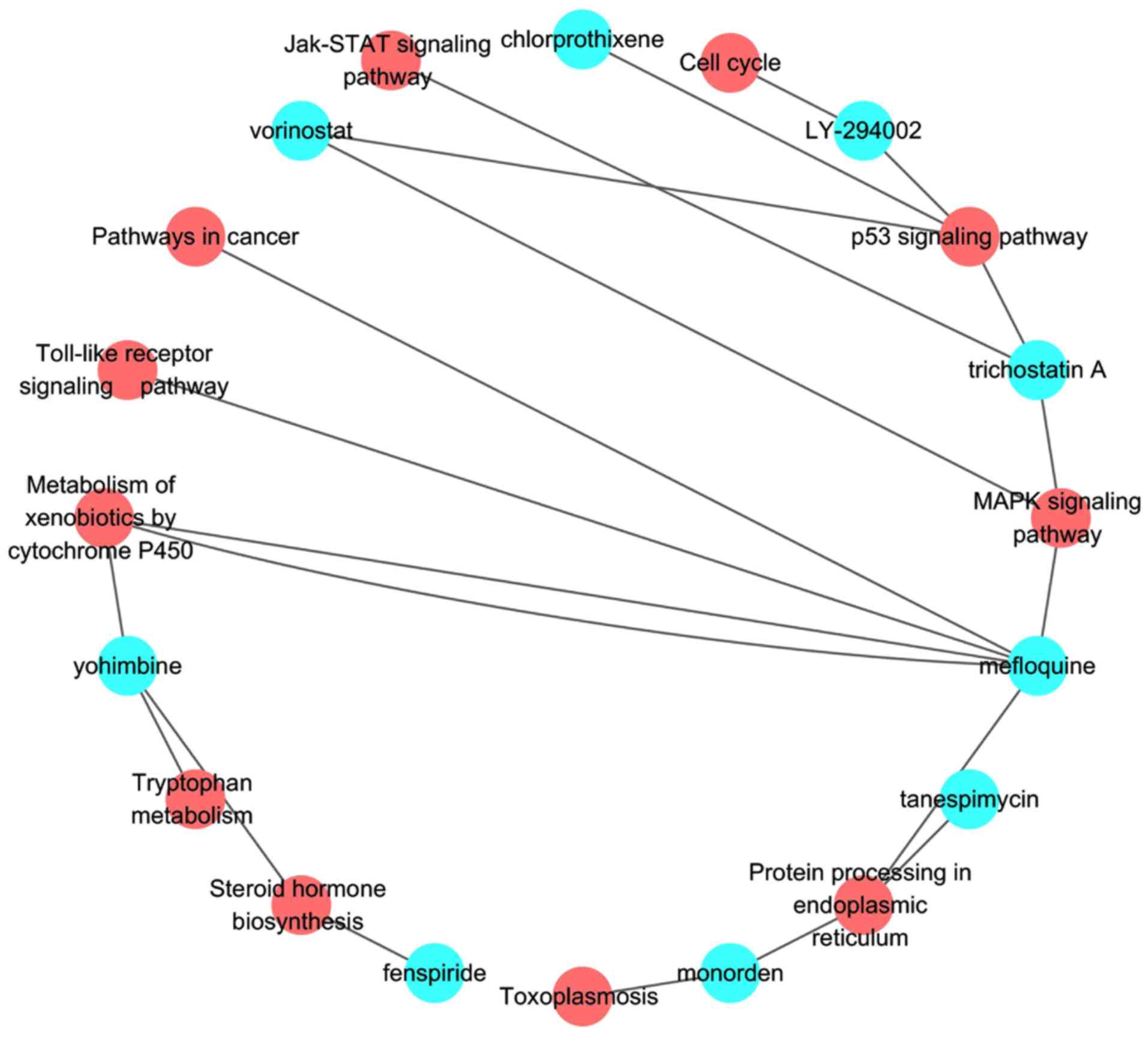
In total, 8 small molecular components among 60
candidate drugs are significantly associated with 9 metabolic
pathways (Table SII and Fig. 3). For the 10 significant drugs, the
corresponding pathways of vorinostat are p53 and MAPK signing
pathway, and the p53 signaling pathway is affected by
chlorprothixene. Then, the targets were predicted for the top 10
drugs using the STITCH database; however the targets of pinacidil,
ciclosporin, and metacycline were not available. Therefore, only 7
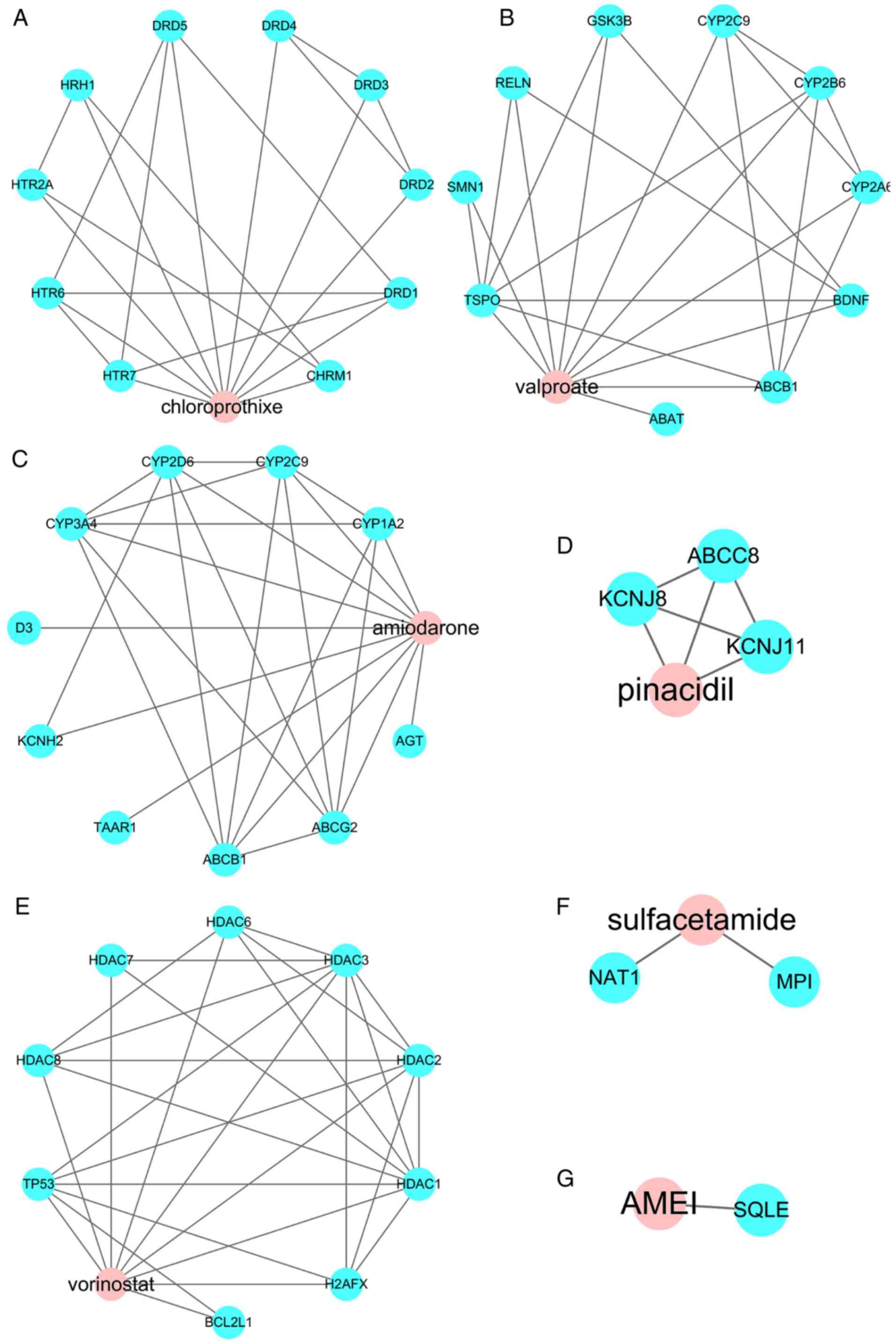
drug-target networks are presented (Fig. 4).
Molecular docking study and drug
pairing prediction for vorinostat
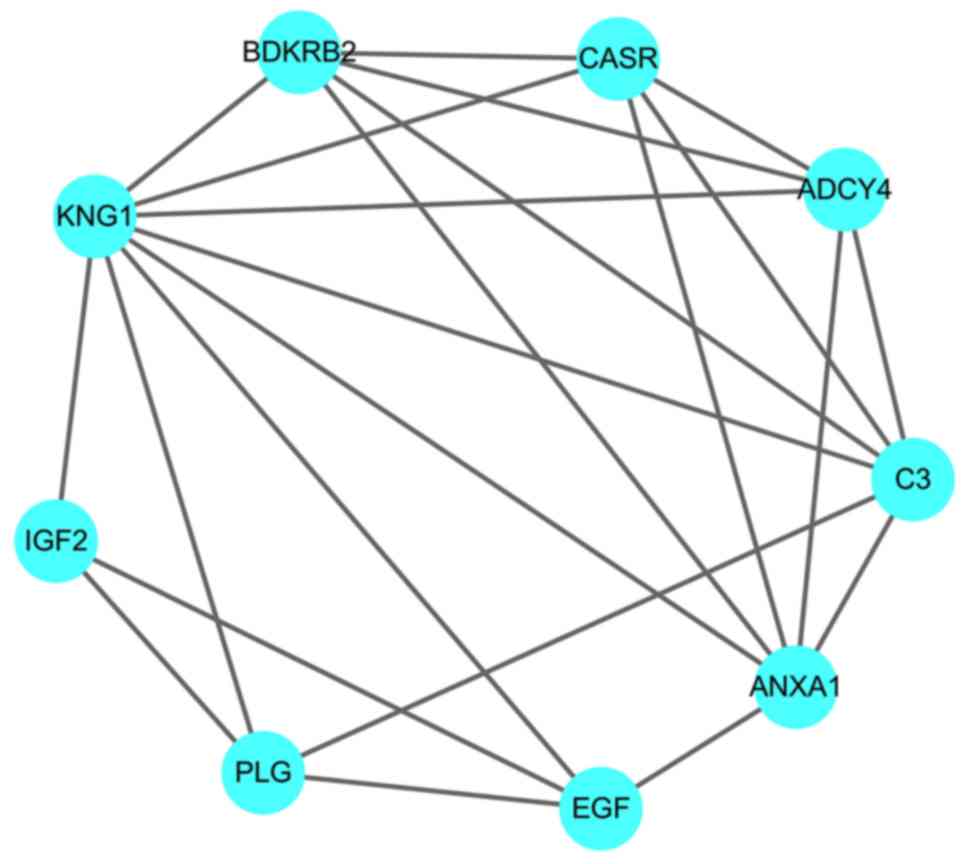
Regarding hub genes in the regulation network of
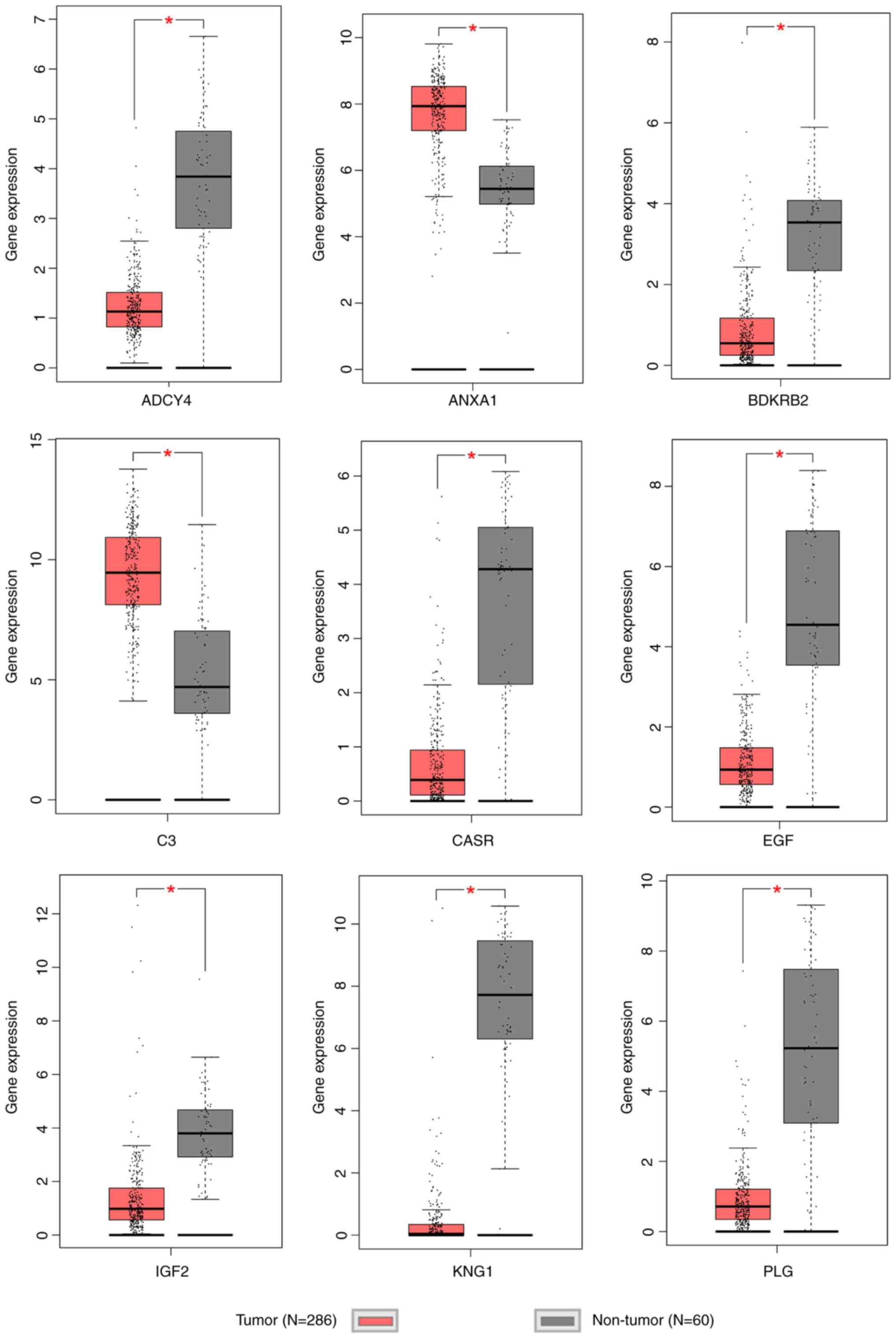
DEGs, 9 genes (BDKRB2, C3, PLG, EGF, IGF2, KNG1, CASR, ANXA1 and
ADCY4) were selected as hub genes due to their centrality degree
≥15 (Fig. 5). Among these 9 hub
genes, 7 genes are significantly downregulated and 2 genes (C3 and
ANXA1) are significantly upregulated (Fig. 6), which suggests that C3 and ANXA1
may serve as potential therapeutic targets in the chemotherapy of
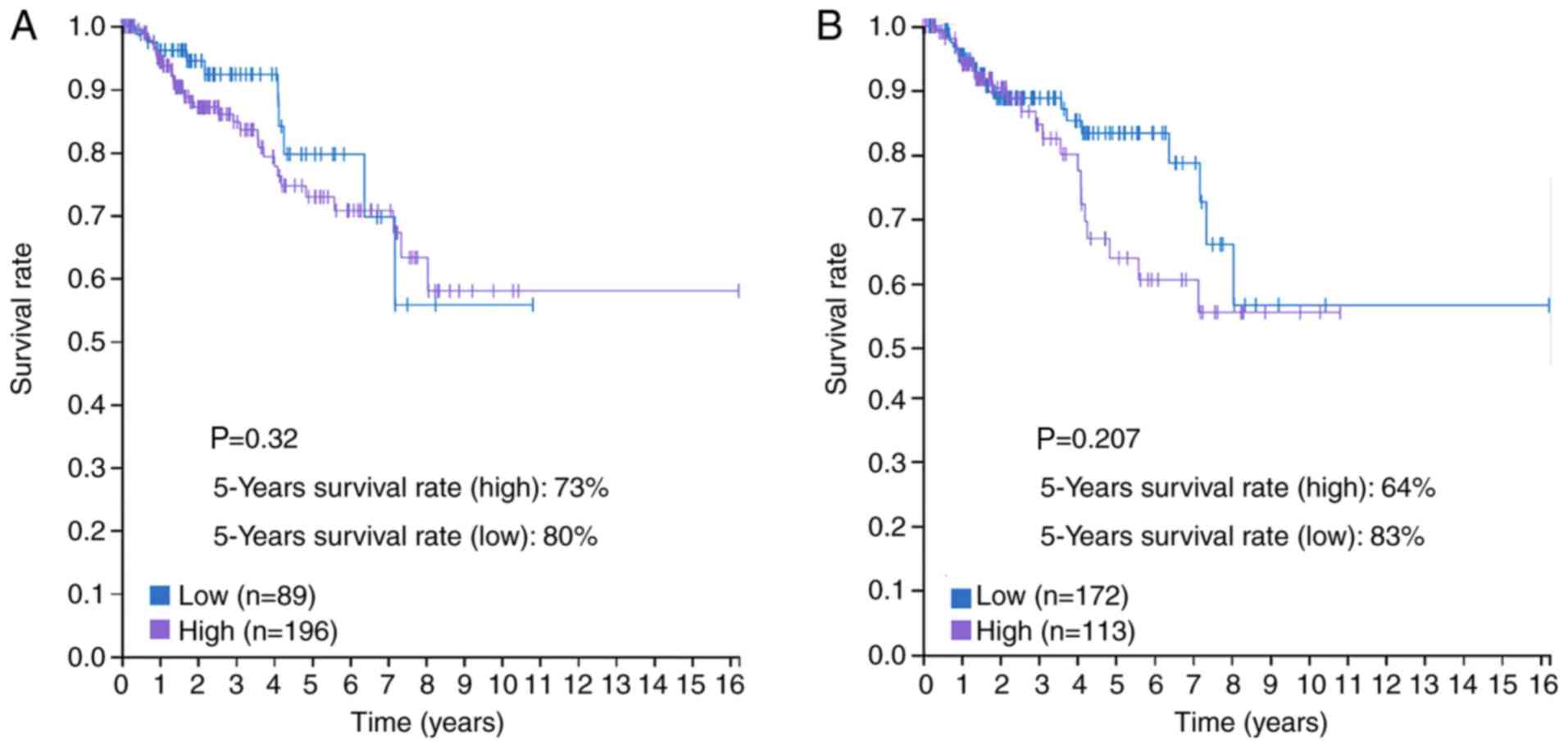
PRCC. The prognostic value of C3 and ANXA1, as determined by TCGA
data, is presented in Fig. 7.
Furthermore, the validation of protein levels for C3 and ANXA1 in
various types of tumors is presented in Figs. 8 and 9. Distinctly positive C3 protein was
observed in tumors' stromal and the majority of malignant cells
displayed weak-to-moderate cytoplasmic immunoreactivity. Similarly,
most malignant cells displayed moderate-to-strong cytoplasmic and
nuclear positivity of ANXA1 protein except breast cancers and
malignant lymphomas. Regarding renal cancer tissues, 7 of 13
(53.8%) renal cancer tissues exhibited high/medium C3 protein
expression and 9 of 12 (75%) renal cancer tissues exhibited
high/medium ANXA1 protein expression. Nevertheless, the expression
difference of C3 and ANXN1 protein in renal cancer and non-cancer
kidney is not well demonstrated due to the limited controls. Among
the top 10 agents, vorinostat was reported to be closely correlated
with cell cycle and had been repurposing for the patients with
progressive cutaneous T-cell lymphoma (29–32).
Consequently, the potential chemotherapy effect of vorinostat in
PRCC patients seems to be a feasible investigation. Notably, the
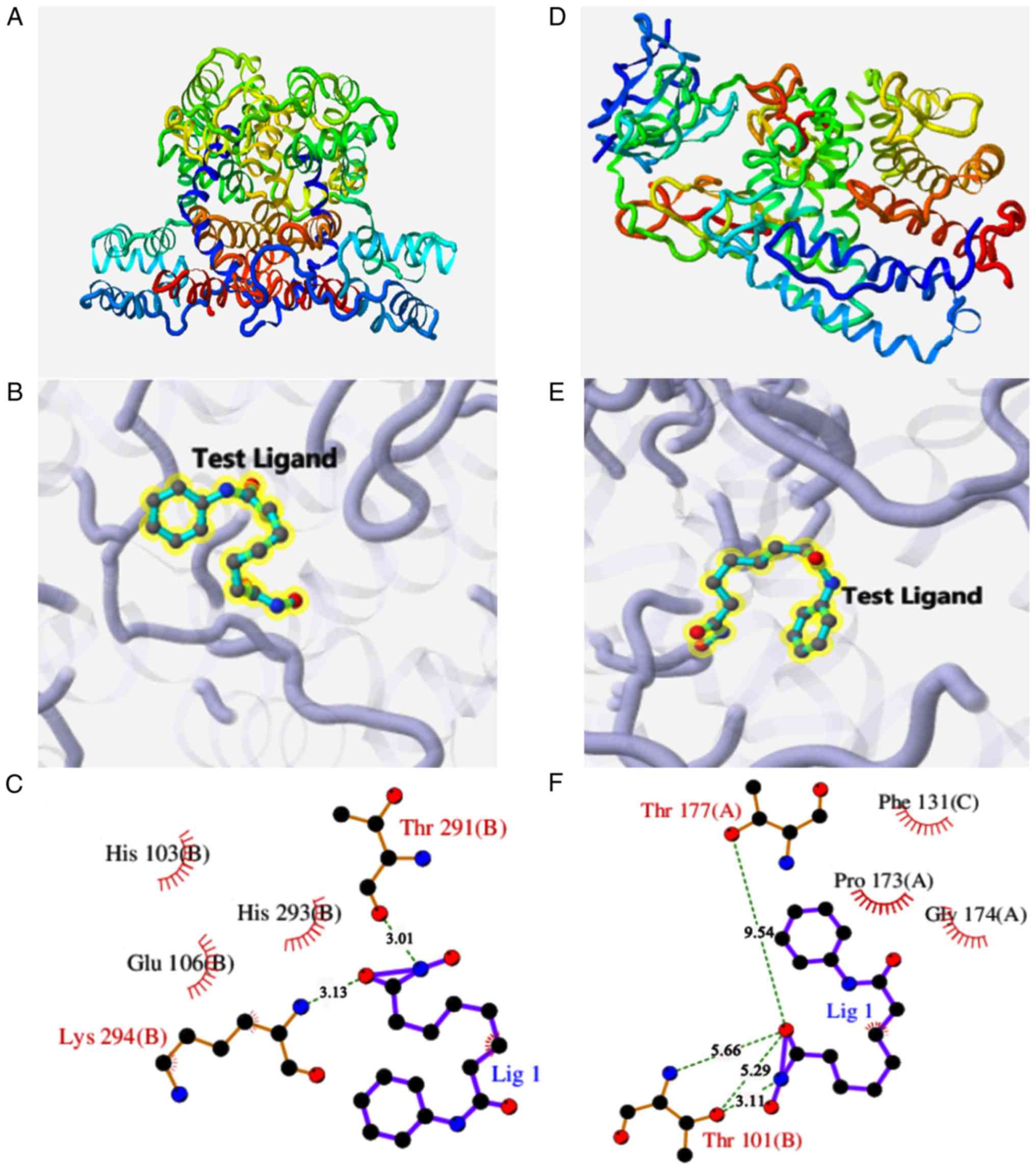
molecular docking tests indicate that vorinostat can interact well
with ANXA1 and C3 proteins, and that the docking scores for ANXA1
(PDB code: 1HM6; Fig. 10A-C) and
C3 (PDB code: 1GHQ; Fig. 10D-F)
are 4.866 and 4.634, respectively. Furthermore, the docking results
suggest that vorinostat is a potentially prospective agent to treat
and reverse PRCC by interfering with gene regulation network
through targeting C3 and ANXN1. In addition, the DPS program
computationally-predicted which drugs would improve the reversal
effects of gene expression changes when combined with vorinostat in
PRCC treatment. Via this method, a total of 10 drugs (including
propofol and sulfamonomethoxine) were revealed to have latent
synergistic effects when combined with vorinostat (Table IV).
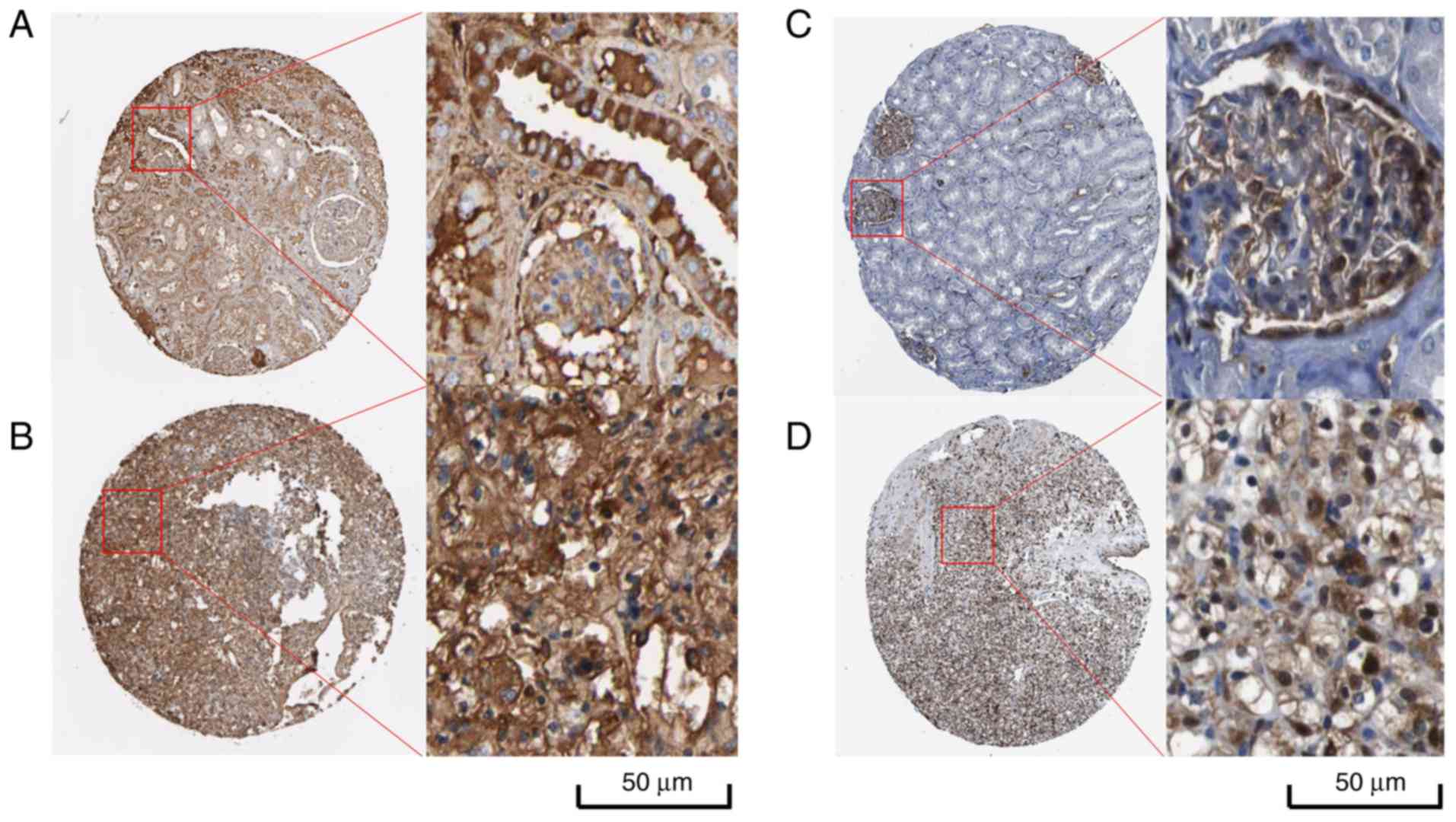 | Figure 9.IHC results of C3 and ANXN1 protein
levels in normal kidney and renal cancer from Protein Atlas
(26). (A) IHC result of C3 protein
in normal kidney. Staining. medium; intensity, moderate; quantity,
75–25%; location, cytoplasmic/membranous. (B) IHC result of C3
protein in renal cancer. Staining, high; intensity, strong;
quantity, 75–25%; location, cytoplasmic/membranous. (C) IHC result
of ANXA1 protein in normal kidney tissue. Staining, high;
intensity, strong; quantity, 75–25%; location,
cytoplasmic/membranous/nuclear. (D) IHC result of ANXA1 protein in
renal cancer. Staining, high; intensity, strong; quantity, 75–25%;
location, cytoplasmic/membranous/nuclear. IHC, immunohistochemical;
C3, complement C3; ANXA1, annexin 1. |
 | Table IV.Computationally-predicted drugs that
may improve the reversal effects of gene expression changes when
combined with vorinostat in Drug Pair Seeker. |
Table IV.
Computationally-predicted drugs that
may improve the reversal effects of gene expression changes when
combined with vorinostat in Drug Pair Seeker.
| Drug 1 | Drug 2 | Total coverage | Total
conflicts | Drug 1
coverage | Drug 1
conflicts | Drug 2
coverage | Drug 2
conflicts |
|---|
|
Vorinostat-4444 | Propofol-3048 | 61 | 20 | 21 | 8 | 42 | 12 |
|
Vorinostat-4444 |
Sulfamonomethoxine-2742 | 59 | 18 | 21 | 8 | 38 | 10 |
|
Vorinostat-4444 |
Methazolamide-2733 | 58 | 18 | 21 | 8 | 38 | 10 |
|
Vorinostat-4444 |
Phthalylsulfathiazole-5249 | 56 | 16 | 21 | 8 | 35 | 8 |
|
Vorinostat-4444 | Lobeline-1770 | 49 | 11 | 21 | 8 | 35 | 8 |
|
Vorinostat-4444 |
Parbendazole-3881 | 59 | 20 | 21 | 8 | 41 | 12 |
|
Vorinostat-4444 | Glipizide-6645 | 56 | 17 | 21 | 8 | 36 | 9 |
|
Vorinostat-1220 | Propofol-3048 | 57 | 19 | 16 | 7 | 42 | 17 |
|
Vorinostat-4444 |
Azacitidine-3348 | 57 | 19 | 21 | 8 | 38 | 13 |
|
Vorinostat-4444 |
Rimexolone-5092 | 59 | 21 | 21 | 8 | 40 | 14 |
Discussion
In the present study, the DEGs of PRCC were
identified using TCGA data and a bioinformatics analysis including
GO analysis and KEGG pathway was performed to investigate the
underlying mechanisms of PRCC. The identified DEGs were also used
to search for potential drugs using the Cmap dataset for the
treatment of PRCC. Subsequently, the potential application of query
drugs in PRCC was further explored with the drug pathway network,
drug-target network and a molecular docking test.
According to the GEPIA tool, 627 genes in total were
considered as DEGs in PRCC, among which 161 were upregulated genes
and 466 were downregulated genes. The GO functional annotation was
performed based on these 627 DEGs by MetaScape and it demonstrated
that these DEGs were mainly associated with anion transport, ion
homeostasis, kidney development, and anion transmembrane
transporter activity, which is consistent with other findings that
suggest that ion transport has an essential role in tumor
development and metastasis by altering substantially normal
biological processes (33). The
results from the KEGG enrichment pathway, as determined by
WebGestalt, also reveal how these DEGS function in PRCC. Among the
top 20 metabolic pathways, several significant pathways, such as
complement and coagulation cascades and cell adhesion molecules
(CAMs), are associated with human tumors (34,35).
Cancer migration originates from the disruption of cell adhesion
interaction between cancer and normal cells/matrix, followed by an
increased cell adhesion activity that interacts with other tissue.
Therefore, CAMs is deemed to be a crucial pathway in the
development and metastasis of human cancers (36–39).
As previously reported, CAMs greatly contributes to migration and
invasion in lung cancer, gastric cancer, and bladder cancer
(40–42). In a previous study by Zimpfer et
al (43), the overexpression of
CAMs was located in 126 of 155 patients with PRCC and is clearly
associated with higher grade and worse prognosis in PRCC patients.
However, the majority of previous studies focused on the
investigation of CCRCC, and to date there have been no published
studies that investigate how CAMs pathway functions in PRCC based
on the molecular mechanism. Therefore, more experiments are
required to determine the importance of CAMs in PRCC, which may
serve as an ignored therapeutic target in PRCC chemotherapy
(44).
To identify more potential drugs for PRCC treatment,
60 candidate drugs were obtained from the prediction of the Cmap
dataset depending on DEGs of PRCC. Among the top 10 drugs,
vorinostat was particularly interesting and it is considered to be
the most promising drug in PRCC treatment for detailed
discussion.
Vorinostat, a histone deacetylase (HDAC) suppressor,
has been widely applied for therapy in progressive cutaneous T-cell
lymphoma via blocking cell cycle and/or inducing cell apoptosis
that results from the accumulation of acetylated histone (29–32).
In biology, DNA is wrapped around histones and its expression
relies on the regulation of acetyltransferases and deacetylases
(45). HDACs are a group of enzymes
in eukaryotic nuclei that help histone deacetylation, and
accordingly allow histones to assemble and transform DNA into
bioactive units (46). It was
reported that HDACs (HDAC1 and HDAC2) are required for cell growth
and survival in RCC tumors (47).
The inhibition of HDACs may reverse resistance to angiogenesis
inhibitors and enhance oncologic chemotherapy responses in advanced
RCC (48). A growing volume of
evidence has suggested the incorporation of HDACs in the
development of renal tumors, illustrating its decrease or
suppression as a prospective therapeutic method to restrain renal
tumors (49,50). Recent studies suggest that
vorinostat possesses antitumor activity against soft tissue
sarcomas, gastric and lung cancer, and even RCC (51–54).
In addition, the anti-virus effect of vorinostat in patients with
HIV is also reported (55–57). For the safety of vorinostat in
clinical application, a clinical trial published in 2017 suggested
that the combination of bevacizumab and vorinostat is relatively
safe and tolerated in patients with CCRCC (58). Chemotherapy effects of vorinostat
for patients with PRCC, however, are still not confirmed by
clinical trials. Regarding other drugs, such as naftifine,
amiodarone and valproic acid, the antitumor effect of these drugs
in human cancers has also been reported in recent years (59–66).
In the present study, the results of drug prediction
in Cmap suggest that vorinostat had a relatively low connectivity
score, which indicates a high inverse correlation between
vorinostat and DEGs of PRCC. From the prediction of drug targets,
it was observed that vorinostat is directly targeted to TP53, and
there have been a number of published studies, that argue that the
mutation of TP53 greatly contributes to the tumorigenesis and
development of RCC (67–69). The current study also observed that
vorinostat exerts a significant influence in regulating the p53 and
MAPK signaling pathway. Previous studies have indicated that both
p53 and MAPK signaling pathway are clearly associated with various
cellular functions, including apoptosis, cell growth, migration and
induction of aging, and serve as key pathways for tumorigenesis and
progression in kidney cancers (68,70–74).
Therefore, vorinostat may possess an antitumor activity by
inhibiting p53 and MAPK signaling pathway.
For a better investigation of the specific molecular
mechanism and potential application of vorinostat in anti-PRCC
activity, a PPI network was constructed to search for hub genes in
the gene regulation network of PRCC. From the PPI network, 9 genes
are considered to be hub genes in the DEGs of PRCC, in which 2
genes, C3 and ANXN1, are significantly upregulated. Notably, there
was no statistical difference in survival curves for C3 and ANXN1
to support their role in the prognosis of PRCC patients. However,
these 2 genes are considered as oncologic therapeutic targets in
the PRCC treatment as their key roles of hub genes in the
regulation network of DEGs, therefore, the suppression of these 2
genes may interfere with a series of interactions between the DEGs,
thereby inhibiting the development and progression of PRCC and then
help to treat patients with PRCC. A target drug performs its
effects on cancer cells via interaction with the specific proteins
encoded by oncogenic or key genes, therefore, molecular docking
tests were performed to investigate the drug-protein interactions
between vorinostat and these two proteins (C3 and ANXN1). The
results provided by systemDock precisely simulate their interaction
patterns and illustrate how vorinostat acts on C3 and ANXN1
proteins in the human body. Usually, the binding ability of small
molecules and proteins are evaluated by docking scores.
Surprisingly, the results as observed from docking tests
demonstrate that vorinostat can recognize and interact with both C3
and ANXN1 proteins (docking tests >4), which suggests that
vorinostat has a considerably prospective performance in PRCC
treatment by suppressing the regulation network of DEGs through
inhibiting C3 and ANXN1 proteins. In the future, more experimental
evidence and long-term clinical trials are required to validate the
effects of vorinostat in PRCC treatment.
Some limitations in the present study remain to be
answered. First, the hub genes verified from the PPI network should
be further validated in vitro to observe their specific role
in PRCC. Second, the effect of potential drugs from Cmap prediction
for PRCC treatment should also be further investigated using
experimental evidence.
In summary, disregarding the above limitations, 627
DEGs have been identified in PRCC based on TCGA data in the current
study, and the underlying mechanism of PRCC has been further
investigated by GO and KEGG pathway analysis. Furthermore, to the
best of our knowledge, the present study was the first to predict
60 candidate drugs for PRCC treatment based on DEGs by integrating
Cmap dataset, in which vorinostat was considered to be the most
prospective drug and exhibited significant anti-PRCC activity by
inhibiting the regulation network of DEGs by targeting C3 and
ANXA1.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the Fund of
Promoting Project of Basic Capacity for Young and Middle-aged
University Teachers in Guangxi (grant no. KY2016LX034) and the
Medical Excellence Award Funded by the Creative Research
Development Grant from the First Affiliated Hospital of Guangxi
Medical University.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
GC, HBY, and SHL involved in the conception and
design of the study, as well as designed the figures and tables.
JSP, PL, XDW, and ZKL contributed to the statistical analysis, as
well as wrote and corrected the manuscript. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
PRCC
|
papillary renal cell carcinoma
|
|
TCGA
|
The Cancer Genome Atlas
|
|
DEGs
|
differently expressed genes
|
|
GO
|
Gene Ontology
|
|
KEGG
|
Kyoto Encyclopedia of Genes and
Genomes
|
|
Cmap
|
Connectivity Map
|
|
CCRCC
|
clear renal cell carcinoma
|
|
CAMs
|
cell adhesion molecules
|
References
|
1
|
Wu X, Liu D, Gao X, Xie F, Tao D, Xiao X,
Wang L, Jiang G and Zeng F: Inhibition of BRD4 suppresses cell
proliferation and induces apoptosis in renal cell carcinoma. Cell
Physiol Biochem. 41:1947–1956. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wang C, Cai L, Liu J, Wang G, Li H, Wang
X, Xu W, Ren M, Feng L, Liu P, et al: MicroRNA-30a-5p inhibits the
growth of renal cell carcinoma by modulating GRP78 expression. Cell
Physiol Biochem. 43:2405–2419. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wang J, Li M, Wang Y and Liu X:
Integrating subpathway analysis to identify candidate agents for
hepatocellular carcinoma. Onco Targets Ther. 9:1221–1230. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Steffens S, Janssen M, Roos FC, Becker F,
Schumacher S, Seidel C, Wegener G, Thüroff JW, Hofmann R, Stöckle
M, et al: Incidence and long-term prognosis of papillary compared
to clear cell renal cell carcinoma-a multicentre study. Eur J
Cancer. 48:2347–2352. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lan H, Zeng J, Chen G and Huang H:
Survival prediction of kidney renal papillary cell carcinoma by
comprehensive LncRNA characterization. Oncotarget. 8:110811–110829.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sourbier C, Liao PJ, Ricketts CJ, Wei D,
Yang Y, Baranes SM, Gibbs BK, Ohanjanian L, Spencer Krane L,
Scroggins BT, et al: Targeting loss of the Hippo signaling pathway
in NF2-deficient papillary kidney cancers. Oncotarget.
9:10723–10733. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chakraborty S, Tarantolo SR, Batra SK and
Hauke RJ: Incidence and prognostic significance of second primary
cancers in renal cell carcinoma. Am J Clin Oncol. 36:132–142. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
De P, Otterstatter MC, Semenciw R, Ellison
LF, Marrett LD and Dryer D: Trends in incidence, mortality, and
survival for kidney cancer in Canada, 1986–2007. Cancer Causes
Control. 25:1271–1281. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ke X, Zeng X, Wei X, Shen Y, Gan J, Tang H
and Hu Z: MiR-514a-3p inhibits cell proliferation and
epithelial-mesenchymal transition by targeting EGFR in clear cell
renal cell carcinoma. Am J Transl Res. 9:5332–5346. 2017.PubMed/NCBI
|
|
11
|
Akhtar M, Al-Bozom IA and Al Hussain T:
Papillary renal cell carcinoma (PRCC): An update. Adv Anat Pathol.
Nov 30–2018.(Epub ahead of print). doi:
10.1097/PAP.0000000000000220. PubMed/NCBI
|
|
12
|
Courthod G, Tucci M, Di Maio M and
Scagliotti GV: Papillary renal cell carcinoma: A review of the
current therapeutic landscape. Crit Rev Oncol Hematol. 96:100–112.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sleire L, Førde HE, Netland IA, Leiss L,
Skeie BS and Enger PØ: Drug repurposing in cancer. Pharmacol Res.
124:74–91. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45:W98–W102.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang Y, Huang JC, Cai KT, Yu XB, Chen YR,
Pan WY, He ZL, Lv J, Feng ZB and Chen G: Long noncoding RNA HOTTIP
promotes hepatocellular carcinoma tumorigenesis and development: A
comprehensive investigation based on bioinformatics, qRTPCR and
metaanalysis of 393 cases. Int J Oncol. 51:1705–1721. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
He Z, Tang F, Lu Z, Huang Y, Lei H, Li Z
and Zeng G: Analysis of differentially expressed genes, clinical
value and biological pathways in prostate cancer. Am J Transl Res.
10:1444–1456. 2018.PubMed/NCBI
|
|
17
|
Yang X, Zhu S, Li L, Zhang L, Xian S, Wang
Y and Cheng Y: Identification of differentially expressed genes and
signaling pathways in ovarian cancer by integrated bioinformatics
analysis. Onco Targets Ther. 11:1457–1474. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tripathi S, Pohl MO, Zhou Y,
Rodriguez-Frandsen A, Wang G, Stein DA, Moulton HM, DeJesus P, Che
J, Mulder LC, et al: Meta- and orthogonal integration of influenza
‘OMICs’ data defines a role for UBR4 in virus budding. Cell Host
Microbe. 18:723–735. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang J, Vasaikar S, Shi Z, Greer M and
Zhang B: WebGestalt 2017: A more comprehensive, powerful, flexible
and interactive gene set enrichment analysis toolkit. Nucleic Acids
Res. 45:W130–W137. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lamb J, Crawford ED, Peck D, Modell JW,
Blat IC, Wrobel MJ, Lerner J, Brunet JP, Subramanian A, Ross KN, et
al: The Connectivity Map: Using gene-expression signatures to
connect small molecules, genes, and disease. Science.
313:1929–1935. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Qu XA and Rajpal DK: Applications of
Connectivity Map in drug discovery and development. Drug Discov
Today. 17:1289–1298. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhong Y, Chen EY, Liu R, Chuang PY,
Mallipattu SK, Tan CM, Clark NR, Deng Y, Klotman PE, Ma'ayan A, et
al: Renoprotective effect of combined inhibition of
angiotensin-converting enzyme and histone deacetylase. J Am Soc
Nephrol. 24:801–811. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li C, Li X, Miao Y, Wang Q, Jiang W, Xu C,
Li J, Han J, Zhang F, Gong B, et al: SubpathwayMiner: A software
package for flexible identification of pathways. Nucleic Acids Res.
37:e1312009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Szklarczyk D, Santos A, von Mering C,
Jensen LJ, Bork P and Kuhn M: STITCH 5: Augmenting protein-chemical
interaction networks with tissue and affinity data. Nucleic Acids
Res. 44:D380–D384. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Szklarczyk D, Morris JH, Cook H, Kuhn M,
Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, et al:
The STRING database in 2017: Quality-controlled protein-protein
association networks, made broadly accessible. Nucleic Acids Res.
45:D362–D368. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Scardoni G, Tosadori G, Faizan M, Spoto F,
Fabbri F and Laudanna C: Biological network analysis with
CentiScaPe: Centralities and experimental dataset integration.
F1000Res. 3:1392014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Interactive human protein atlas launches.
Cancer Discov. 5:3392015. View Article : Google Scholar
|
|
28
|
Hsin KY, Matsuoka Y, Asai Y, Kamiyoshi K,
Watanabe T, Kawaoka Y and Kitano H: systemsDock: A web server for
network pharmacology-based prediction and analysis. Nucleic Acids
Res. 44:W507–W513. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Han L, Wang T, Wu J, Yin X, Fang H and
Zhang N: A facile route to form self-carried redox-responsive
vorinostat nanodrug for effective solid tumor therapy. Int J
Nanomedicine. 11:6003–6022. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Fernández-Rodríguez C, Salar A, Navarro A,
Gimeno E, Pairet S, Camacho L, Ferraro M, Serrano S, Besses C,
Bellosillo B, et al: Anti-tumor activity of the combination of
bendamustine with vorinostat in diffuse large B-cell lymphoma
cells. Leuk Lymphoma. 57:692–699. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Duvic M and Dimopoulos M: The safety
profile of vorinostat (suberoylanilide hydroxamic acid) in
hematologic malignancies: A review of clinical studies. Cancer
Treat Rev. 43:58–66. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zinzani PL, Bonthapally V, Huebner D,
Lutes R, Chi A and Pileri S: Panoptic clinical review of the
current and future treatment of relapsed/refractory T-cell
lymphomas: Cutaneous T-cell lymphomas. Crit Rev Oncol Hematol.
99:228–240. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Djamgoz MB, Coombes RC and Schwab A: Ion
transport and cancer: From initiation to metastasis. Philos Trans R
Soc Lond B Biol Sci. 369:201300922014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Guglietta S and Rescigno M:
Hypercoagulation and complement: Connected players in tumor
development and metastases. Semin Immunol. 28:578–586. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Samatov TR, Wicklein D and Tonevitsky AG:
L1CAM: Cell adhesion and more. Prog Histochem Cytochem. 51:25–32.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kourtidis A and Anastasiadis PZ: Bringing
together cell-to-cell adhesion and miRNA biology in cancer
research. Future Oncol. 12:1211–1214. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Brower V: Adhesion molecules, stem cells,
and the microenvironment in acute myeloid leukemia. J Natl Cancer
Inst. 108:djw1132016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Turaga SM and Lathia JD: Adhering towards
tumorigenicity: Altered adhesion mechanisms in glioblastoma cancer
stem cells. CNS Oncol. 5:251–259. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Hamidi H and Ivaska J: Every step of the
way: Integrins in cancer progression and metastasis. Nat Rev
Cancer. 18:533–548. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Tang Q, Zhang H, Kong M, Mao X and Cao X:
Hub genes and key pathways of non-small lung cancer identified
using bioinformatics. Oncol Lett. 16:2344–2354. 2018.PubMed/NCBI
|
|
41
|
Zhang Y, Zhang J, Shen Q, Yin W, Huang H,
Liu Y and Ni Q: High expression of Nectin-4 is associated with
unfavorable prognosis in gastric cancer. Oncol Lett. 15:8789–8795.
2018.PubMed/NCBI
|
|
42
|
Chang HY, Chang HM, Wu TJ, Chaing CY, Tzai
TS, Cheng HL, Raghavaraju G, Chow NH and Liu HS: The role of
Lutheran/basal cell adhesion molecule in human bladder
carcinogenesis. J Biomed Sci. 24:612017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zimpfer A, Maruschke M, Rehn S, Kundt G,
Litzenberger A, Dammert F, Zettl H, Stephan C, Hakenberg OW and
Erbersdobler A: Prognostic and diagnostic implications of
epithelial cell adhesion/activating molecule (EpCAM) expression in
renal tumours: A retrospective clinicopathological study of 948
cases using tissue microarrays. BJU Int. 114:296–302. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Alimbretov D, Askarova S, Umbayev B, Davis
T and Kipling D: Pharmacological targeting of cell cycle, apoptotic
and cell adhesion signaling pathways implicated in chemoresistance
of cancer cells. Int J Mol Sci. 19:E16902018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Shechter D, Dormann HL, Allis CD and Hake
SB: Extraction, purification and analysis of histones. Nat Protoc.
2:1445–1457. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Valenzuela-Fernández A, Cabrero JR,
Serrador JM and Sánchez-Madrid F: HDAC6: A key regulator of
cytoskeleton, cell migration and cell-cell interactions. Trends
Cell Biol. 18:291–297. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Kiweler N, Brill B, Wirth M, Breuksch I,
Laguna T, Dietrich C, Strand S, Schneider G, Groner B, Butter F, et
al: The histone deacetylases HDAC1 and HDAC2 are required for the
growth and survival of renal carcinoma cells. Arch Toxicol.
92:2227–2243. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Aggarwal R, Thomas S, Pawlowska N,
Bartelink I, Grabowsky J, Jahan T, Cripps A, Harb A, Leng J,
Reinert A, et al: Inhibiting histone deacetylase as a means to
reverse resistance to angiogenesis inhibitors: Phase I study of
abexinostat plus pazopanib in advanced solid tumor malignancies. J
Clin Oncol. 35:1231–1239. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Chun P: Therapeutic effects of histone
deacetylase inhibitors on kidney disease. Arch Pharm Res.
41:162–183. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Dokmanovic M, Clarke C and Marks PA:
Histone deacetylase inhibitors: Overview and perspectives. Mol
Cancer Res. 5:981–989. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Schmitt T, Mayer-Steinacker R, Mayer F,
Grünwald V, Schütte J, Hartmann JT, Kasper B, Hüsing J, Hajda J,
Ottawa G, et al: Vorinostat in refractory soft tissue
sarcomas-Results of a multi-centre phase II trial of the German
Soft Tissue Sarcoma and Bone Tumour Working Group (AIO). Eur J
Cancer. 64:74–82. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Pan CH, Chang YF, Lee MS, Wen BC, Ko JC,
Liang SK and Liang MC: Vorinostat enhances the cisplatin-mediated
anticancer effects in small cell lung cancer cells. BMC Cancer.
16:8572016. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Yoo C, Ryu MH, Na YS, Ryoo BY, Lee CW and
Kang YK: Vorinostat in combination with capecitabine plus cisplatin
as a first-line chemotherapy for patients with metastatic or
unresectable gastric cancer: Phase II study and biomarker analysis.
Br J Cancer. 114:1185–1190. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Li H, Wang X, Zhang C, Cheng Y, Yu M, Zhao
K, Ge W, Cai A, Zhang Y, Han F, et al: HDAC1-induced epigenetic
silencing of ASPP2 promotes cell motility, tumour growth and drug
resistance in renal cell carcinoma. Cancer Lett. 432:121–131. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Archin NM, Liberty AL, Kashuba AD,
Choudhary SK, Kuruc JD, Crooks AM, Parker DC, Anderson EM, Kearney
MF, Strain MC, et al: Administration of vorinostat disrupts HIV-1
latency in patients on antiretroviral therapy. Nature. 487:482–485.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Sung JA, Sholtis K, Kirchherr J, Kuruc JD,
Gay CL, Nordstrom JL, Bollard CM, Archin NM and Margolis DM:
Vorinostat renders the replication-competent latent reservoir of
human immunodeficiency virus (HIV) vulnerable to clearance by CD8 T
cells. EBioMedicine 23: 52–58, 2017. J Clin Invest. 127:3126–3135.
2017.PubMed/NCBI
|
|
57
|
Mota TM, Rasmussen TA, Rhodes A, Tennakoon
S, Dantanarayana A, Wightman F, Hagenauer M, Roney J, Spelman T,
Purcell DFJ, et al: No adverse safety or virological changes 2
years following vorinostat in HIV-infected individuals on
antiretroviral therapy. AIDS. 31:1137–1141. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Pili R, Liu G, Chintala S, Verheul H,
Rehman S, Attwood K, Lodge MA, Wahl R, Martin JI, Miles KM, et al:
Combination of the histone deacetylase inhibitor vorinostat with
bevacizumab in patients with clear-cell renal cell carcinoma: A
multicentre, single-arm phase I/II clinical trial. Br J Cancer.
116:874–883. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Schmeel LC, Schmeel FC, Blaum-Feder S and
Schmidt-Wolf IG: In vitro efficacy of naftifine against lymphoma
and multiple myeloma. Anticancer Res. 35:5921–5926. 2015.PubMed/NCBI
|
|
60
|
Bognar Z, Fekete K, Antus C, Hocsak E,
Bognar R, Tapodi A, Boronkai A, Farkas N, Gallyas F Jr, Sumegi B,
et al: Desethylamiodarone-A metabolite of amiodarone-Induces
apoptosis on T24 human bladder cancer cells via multiple pathways.
PLoS One. 12:e01894702017. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Chang YL, Liu ST, Wang YW, Lin WS and
Huang SM: Amiodarone promotes cancer cell death through elevated
truncated SRSF3 and downregulation of miR-224. Oncotarget.
9:13390–13406. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Felix F and Fontenele J: Valproic acid may
be tested in patients with H3F3A-mutated high-grade gliomas.
J Clin Oncol. 34:3104–3105. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Cha HY, Lee BS, Chang JW, Park JK, Han JH,
Kim YS, Shin YS, Byeon HK and Kim CH: Downregulation of Nrf2 by the
combination of TRAIL and Valproic acid induces apoptotic cell death
of TRAIL-resistant papillary thyroid cancer cells via suppression
of Bcl-xL. Cancer Lett. 372:65–74. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Xi W, Chen X, Sun J, Wang W, Huo Y, Zheng
G, Wu J, Li Y, Yang A and Wang T: Combined treatment with valproic
acid and 5-Aza-2′-deoxycytidine synergistically inhibits human
clear cell renal cell carcinoma growth and migration. Med Sci
Monit. 24:1034–1043. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Wei M, Mao S, Lu G, Li L, Lan X, Huang Z,
Chen Y, Zhao M, Zhao Y and Xia Q: Valproic acid sensitizes
metformin-resistant human renal cell carcinoma cells by
upregulating H3 acetylation and EMT reversal. BMC Cancer.
18:4342018. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Mao S, Lu G, Lan X, Yuan C, Jiang W, Chen
Y, Jin X and Xia Q: Valproic acid inhibits epithelialmesenchymal
transition in renal cell carcinoma by decreasing SMAD4 expression.
Mol Med Rep. 16:6190–6199. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Tajima S, Waki M, Doi W, Hayashi K,
Takenaka S, Fukaya Y and Kimura R: Acquired cystic
disease-associated renal cell carcinoma with a focal sarcomatoid
component: Report of a case showing more pronounced polysomy of
chromosomes 3 and 16 in the sarcomatoid component. Pathol Int.
65:89–94. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Hayes SA, Pandiri AR, Ton TV, Hong HH,
Clayton NP, Shockley KR, Peddada SD, Gerrish K, Wyde M, Sills RC,
et al: Renal cell carcinomas in vinylidene chloride-exposed male
B6C3F1 mice are characterized by oxidative stress and TP53 pathway
dysregulation. Toxicol Pathol. 44:71–87. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Malouf GG, Ali SM, Wang K, Balasubramanian
S, Ross JS, Miller VA, Stephens PJ, Khayat D, Pal SK, Su X, et al:
Genomic characterization of renal cell carcinoma with sarcomatoid
dedifferentiation pinpoints recurrent genomic alterations. Eur
Urol. 70:348–357. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Casuscelli J, Weinhold N, Gundem G, Wang
L, Zabor EC, Drill E, Wang PI, Nanjangud GJ, Redzematovic A,
Nargund AM, et al: Genomic landscape and evolution of metastatic
chromophobe renal cell carcinoma. JCI insight. 2:926882017.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Gao Y, Li J, Qiao N, Meng Q, Zhang M, Wang
X, Jia J, Yang S, Qu C, Li W, et al: Adrenomedullin blockade
suppresses sunitinib-resistant renal cell carcinoma growth by
targeting the ERK/MAPK pathway. Oncotarget. 7:63374–63387.
2016.PubMed/NCBI
|
|
72
|
Miyazaki A, Miyake H and Fujisawa M:
Molecular mechanism mediating cytotoxic activity of axitinib in
sunitinib-resistant human renal cell carcinoma cells. Clin Transl
Oncol. 18:893–900. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Ye H, Wang WG, Cao J and Hu XC: SPARCL1
suppresses cell migration and invasion in renal cell carcinoma. Mol
Med Rep. 16:7784–7790. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Ohnami S, Ohshima K, Nagashima T, Urakami
K, Shimoda Y, Saito J, Naruoka A, Hatakeyama K, Mochizuki T,
Serizawa M, et al: Comprehensive characterization of genes
associated with the TP53 signal transduction pathway in various
tumors. Mol Cell Biochem. 431:75–85. 2017. View Article : Google Scholar : PubMed/NCBI
|