Cervical cancer, one of the most common malignant
tumors affecting the female population, brings a serious problem to
affect public women's health (1).
The morbidity and mortality associated with cervical cancer has
significantly decreased over the past three decades. However,
cervical cancer remains the third most commonly diagnosed type of
cancer among women worldwide and some patients with this disease
have a poor prognosis (2–4). Therefore, novel and effective
therapeutic strategies for cervical cancer are urgently required.
Accumulating evidence has indicated that the development of
cervical cancer is a multi-step process, including the abnormal
expression of oncogenes and tumor suppressor genes (5–7).
Despite advances in cervical cancer, the precise molecular
mechanisms of carcinogenesis and progression underlying cervical
cancer are only partly understood (5). Therefore, the identification of the
specific molecular biomarkers and detailed underlying mechanisms,
which may contribute to the development of novel diagnostic and
treatment strategies for patients with cervical cancer is
critical.
Long non-coding RNAs (lncRNAs) are a distinct
subclass of RNA transcripts that more than 200 nucleotides in
length (8). An increasing number of
individual lncRNAs have been determined to exert a multitude of
effects within cells, such as modulating gene expression at the
epigenetic, transcriptional, post-transcriptional and translational
levels under both physiological and pathological conditions
(9–13). The aberrant expression of lncRNAs
has been linked to tumor initiation and progression (14–18).
Cancer RNA-Seq Nexus (CRN) is a database of
phenotype-specific transcriptome profiling in cancer cells. Using
this database, we systematically obtained RNA-seq datasets
concerning cervical cancer tissues from The Cancer Genome Atlas
(TCGA), Sequence Read Archive (SRA) and NCBI Gene Expression
Omnibus (GEO) (28). The expression
data of AFAP1-AS1 in cervical cancer was downloaded from this
database. The DNA methylation data of cervical cancer was
downloaded from the MethHC database (http://methhc.mbc.nctu.edu.tw/php/index.php), data
which came from TCGA (29). The
data used to draw the survival curve were all from the UCSC XENA
database (http://xena.ucsc.edu/).
The HeLa cell line (ATCC no. CCL-2) is the most
widely usedcervical cancer cell line. Due to its strong ability to
proliferate, invade and migrate, we selected it for further
research. The HeLa cells were purchased from the American Type
Culture Collection (ATCC; Manassas, VA, USA) and cultured in
RPMI-1640 medium supplemented with 10% fetal bovine serum (FBS;
Thermo Fisher Scientific, Inc., Waltham, MA, USA), penicillin and
streptomycin (both from Sigma-Aldrich; Merck KGaA, Darmstadt,
Germany) in a humidified incubator with 5% CO2 at 37°C.
For gene knockdown, the cells were seeded and cultured overnight.
Subsequently, 20 nM AFAP1-AS1 siRNA or scramble control (NC) siRNA
(Guangzhou RiboBio Co., Ltd., Guangzhou, China) were transfected
into the cervical cancer cells using Lipofectamine 3000 (Thermo
Fisher Scientific, Inc.) according to the manufacturer's
instructions. The two siRNAs used in this manuscript had no
off-target effect and exhibited the optimal knockdown efficiency in
previous studies by us, as well as other groups (19,30).
The cycling conditions of PCR were as follows: First, 10 min at
95°C. Denaturation at 95°C for 15 sec, annealing at 60°C for 30
sec, and elongation at 70°C for 10 sec. After 40 cycles, the PCR
tubes were incubated 5 min at 70°C. The sequences were as follows:
AFAP1-AS1 siRNA1, 5′-GGGCTTCAATTTACAAGCATT-3′ and AFAP1-AS1 siRNA2,
5′-CCTATCTGGTCAACACGTATT-3′. The abovementioned nucleotide
sequences were synthesized by Guangzhou RiboBio Co., Ltd.
The cervical cancer cells were seeded and cultured
in 6-well culture plates following transfection for 24 h. When the
cells grew to 90% confluence, a 10 µl tip was used to create a
scratch. Images were captured (magnification, ×20) at different
time points (0, 24 and 48 h) using a microscope (The Cell Culture
Laboratory Solution CKX53; Olympus, Tokyo, Japan). The ocular ruler
was performed to measure the gap width in each group at the
identified time point. All the wounds in the experimental group had
the same width at the 0 h (31–33).
The HeLa cells were harvested after being
transfected with the siRNAs for 36 h and total RNA was extracted
using TRIzol reagent (Invitrogen/Thermo Fisher Scientific, Inc.)
according to the manufacturer's instructions. The RNA was reversed
into cDNA using the 5X All-In-One RT Master Mix Reagent kit (Abm
Canada Inc., Milton, ON, Canada) according to the kit protocol.
cDNA was subjected to RT-qPCR using the SYBR Premix Ex Taq II kit
(Takara Bio, Inc., Shiga, Japan). Reactions were carried out on the
CFX96 Real-Time PCR Detection System (185–5196; Bio-Rad
Laboratories, Inc., Hercules, CA, USA). Fold changes of lncRNA were
calculated using the 2−ΔΔCq method, with β-actin as an
internal control (41). The
sequences of the primers were as follows: AFAP1-AS1 forward,
5′-AATGGTGGTAGGAGGGAGGA-3′ and reverse, 5′-CACACAGGGGAATGAAGAGG-3′;
β-actin forward, 5′-TCACCAACTGGGACGACATG-3′ and reverse,
5′-GTCACCGGAGTCCATCACGAT-3′.
Cell cycle distribution was detected by flow
cytometry. When the cells were transfected with the siRNAs for 48
h, the cells were collected and fixed in 75% ethanol at −20°C
overnight. The fixed cells were then washed thrice with
phosphate-buffered saline (PBS) before being incubated at room
temperature with RNase A for 20 min. These cells were stained using
a propidium iodide (PI) staining kit (BD Biosciences) and incubated
in the dark for 30 min at 4°C. A Beckman flow cytometer (Beckman
Coulter, Inc.) was used for cell cycle analysis. Three independent
experiments were conducted.
Total protein in the HeLa cells was extracted using
RIPA extraction reagent (Beyotime Institute of Biotechnology,
Shanghai, China) supplemented with a protease inhibitor cocktail
(Roche Diagnostics, Basel, Switzerland). Protein from the HeLa
cells was quantified by the BCA method. To separate the protein,
30–50 µg protein per lane was loaded and electrophoresed in 10%
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
(SDS-PAGE). Following electrophoresis, the protein bands were
transferred onto PVDF membranes. The membranes were blocked with 5%
skim milk for 1 h at room temperature and then incubated with
primary anti-Rho GDP-dissociation inhibitor 1 (RHOGDI, 1:1,000;
cat. no. 14282-1-AP), anti-Rac Family Small GTPase 2 (RAC2,
1:1,000; cat. no. 10735-1-AP), anti-RAB1B (1:800; cat. no.
17824-1-AP), anti-RAB11A (1:800; cat. no. 20229-1-AP),
anti-profilin 1 (PFN1, 1:1,000; cat. no. 11680-1-AP) and anti-LIM
and SH3 protein 1 (LASP1, 1:1,000; cat. no. 10515-1-AP) antibodies
(ProteinTech, Inc., Rosemont, IL, USA) or E-cadherin (1:1,000; cat.
no. 3195), Zonula occludens-1 (ZO-1, 1:1,000; cat. no. 8193),
Vimentin (1:1,000; cat. no. 5741), β-catenin antibodies (1:1,000;
cat. no. 8480) (Cell Signaling Technology, Inc., Danvers, MA, USA)
overnight at 4°C. The following day, the membranes were incubated
with a horseradish peroxidase-conjugated secondary antibody (goat
anti-mouse IgG-HRP; cat. no. SC-2005; and goat anti-rabbit IgG-HRP,
cat. no. SC-2005; 1:2,000; Santa Cruz Biotechnology, Inc., Santa
Cruz, CA, USA) for 30 min at 37°C. Immunoreactive bands were
visualized using an ECL detection reagent (Amersham; GE Healthcare
Life Sciences, Chalfont, UK). β-actin (1:1,000; cat. no. 4970; Cell
Signaling Technology, Inc.) expression was used as a housekeeping
gene control.
All the experimental data are represented as the
means ± standard error (SE) and processed using SPSS statistical
software, version 19.0 (SPSS, Inc., Chicago, IL, USA). Overall
survival (OS) was analyzed using the Kaplan-Meier method and the
results of the analysis were considered significant if log-rank
test yielded a value of P<0.05. Significance between 2 groups
was evaluated by a Student's t-test. Pearson's correlation analysis
was used to assess the correlation between AFAP1-AS1 expression and
its methylation. For the comparison of multiple groups, analysis of
variance (ANOVA) with Dunnett's post hoc t-test was performed. A
P-value <0.05 was considered to indicate a statistically
significant difference.
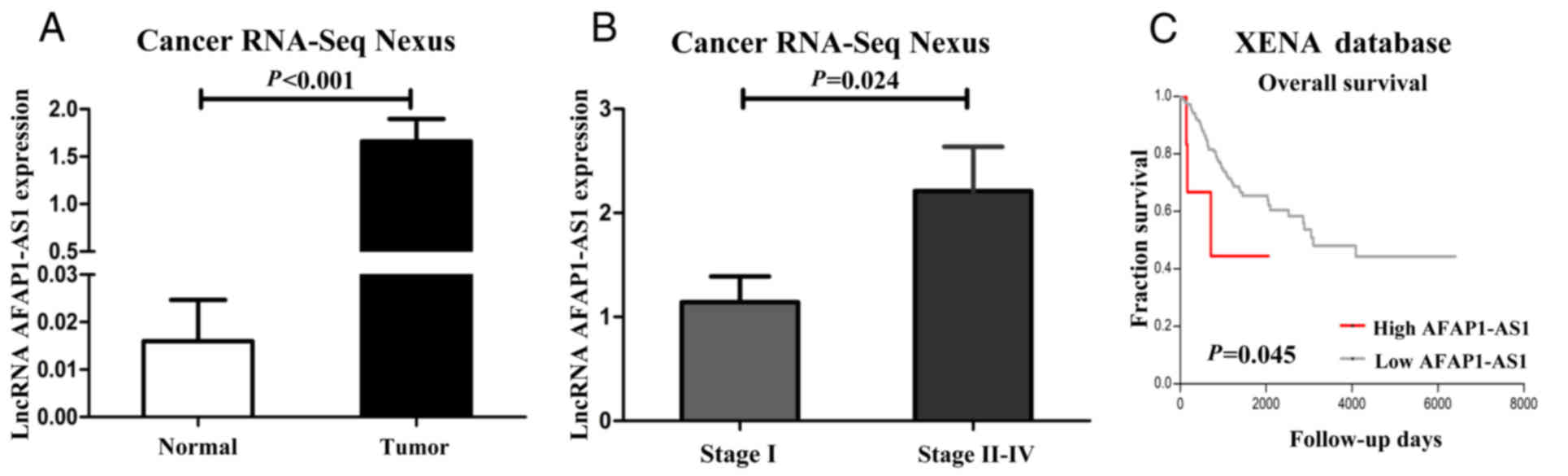
Firstly, the expression level of AFAP1-AS1 in
cervical cancer samples was analyzed using TCGA data from the CRN
database. The data suggested that AFAP1-AS1 was more abundant in
the cervical cancer tissues compared to the normal tissues
(P<0.001; Fig. 1A). Moreover, a
high AFAP1-AS1 expression was positively associated with the TNM
stage (P<0.05; Fig. 1B). To
validate whether the expression levels of AFAP1-AS1 were associated
with the OS of patients with cervical cancer, we analyzed the
patient outcome data from the UCSC XENA database. This analysis
revealed that compared to patients with a low expression of
AFAP1-AS1, patients with a high AFAP1-AS1 expression had a shorter
survival time (P<0.05; Fig.
1C).
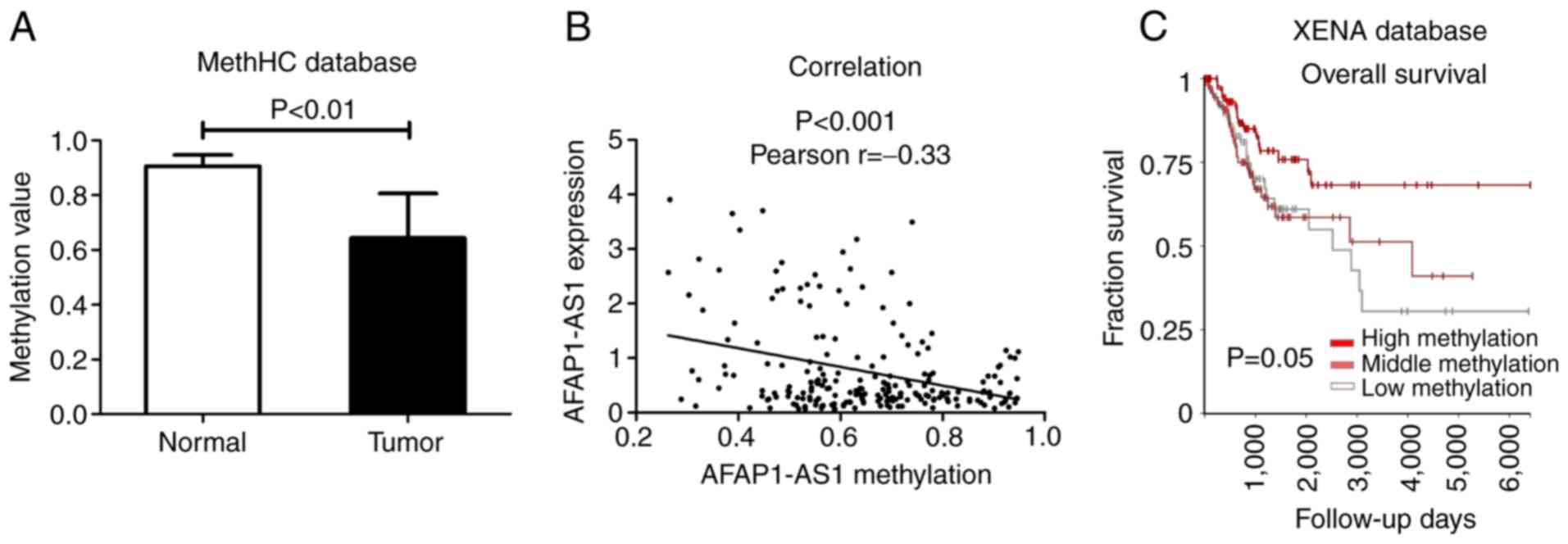
DNA methylation is an important part of epigenetic
inheritance and is involved in the transcriptional expression of
lncRNAs (42,43). In the present study, to investigate
whether the expression of AFAP1-AS1 is influenced by DNA
methylation, we downloaded the methylation data of AFAP1-AS1 in
cervical cancer from the human pan-cancer method database, MethCH.
We found that the average methylation level of the AFAP1-AS1
promoter region was lower in the cervical cancer samples when
compared with the non-tumor cervical cancer samples (P<0.01;
Fig. 2A). In addition, the results
of Pearson's correlation analysis revealed that the expression
level of AFAP1-AS1 negatively correlated with its methylation level
(r=−0.33, P<0.001; Fig. 2B). The
survival analysis was constructed using the UCSC XENA database, and
we identified that the hypomethylation of AFAP1-AS1 was associated
with a poor overall survival of patients with cervical cancer
(P=0.05; Fig. 2C). These data
suggested that the high expression of AFAP1-AS1 may be related to
the hypomethylation of its promoter region. In addition, the
hypomethylation of the AFAP1-AS1 promoter region and the high
expression levels of AFAP1-AS1 may act as independent prognostic
indicators of patients with cervical cancer.
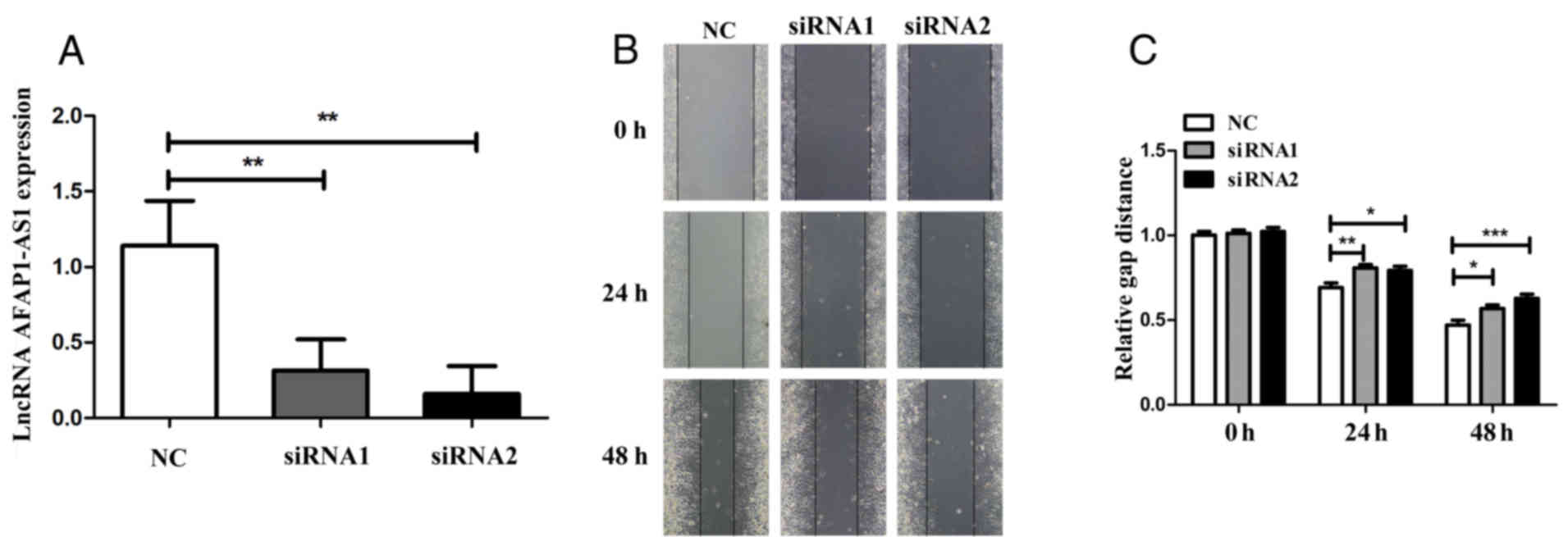
To elucidate the role of AFAP1-AS1 in cervical
cancer, the AFAP1-AS1 expression was downregulated by transfection
of the cervical cancer cell line, HeLa, with two validated siRNAs
targeting AFAP1-AS1 (siRNA1 and siRNA2). We examined the knockdown
efficiency of these two siRNAs by RT-qPCR which yielded
satisfactory results. AFAP1-AS1 expression was knocked down by at
least 65% in the HeLa cells (both siRNAs, P<0.01; Fig. 3A). The functional experiments were
performed to examine the phenotypic alterations induced by the
silencing of AFAP1-AS1 in the cervical cancer cells. The scratch
test results revealed that the migratory capacity of the
AFAP1-AS1-silenced cells was suppressed at 24 and 48 h following
transfection with siRNA compared to the si-control group (24 h,
siRNA1 vs. control, P<0.01; siRNA2 vs. control, P<0.05; 48 h,
siRNA1 vs. control, P<0.05; siRNA2 vs. control, P<0.001;
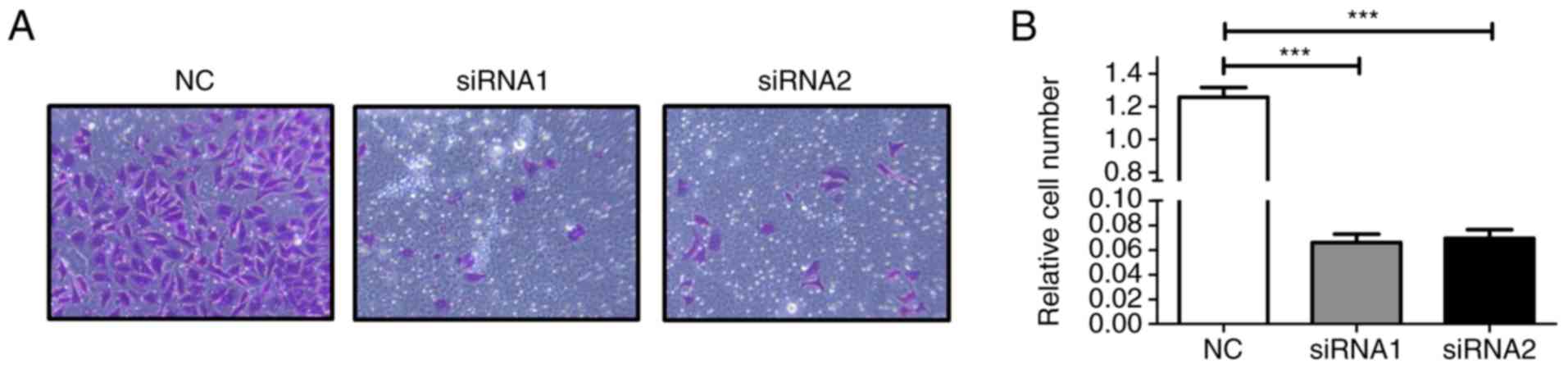
Fig. 3B and C). Moreover, as shown
in Fig. 4, the number of invaded
cells was also decreased in the AFAP1-AS1 siRNA-transfected cancer
cells compared with the si-control group (both siRNAs, P<0.001;
Fig. 4). However, we noted that
there was no significant association between the expression of
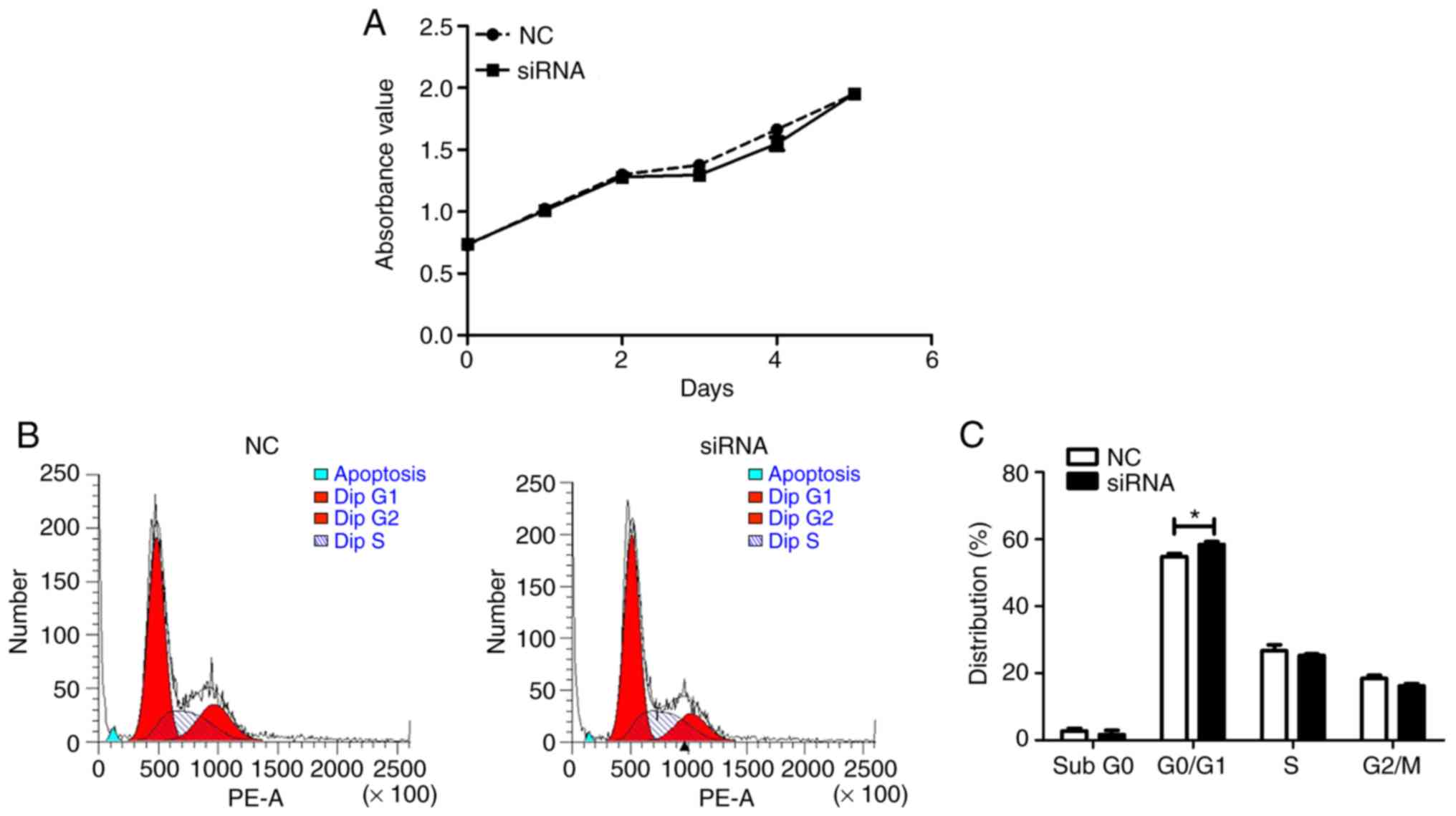
AFAP1-AS1 and cell viability and cell apoptosis. By contrast,
AFAP1-AS1 knockdown slightly, yet significantly increased the
number of cells in the G0/G1 phase in the present study (P<0.05;
Fig. 5).
To investigate the potential mechanisms of AFAP1-AS1
regarding its promoting effect on the migration and invasion of
cervical cancer cells, we examined the expression levels of several
key molecules of the Rho/Rac signaling pathways by western blot
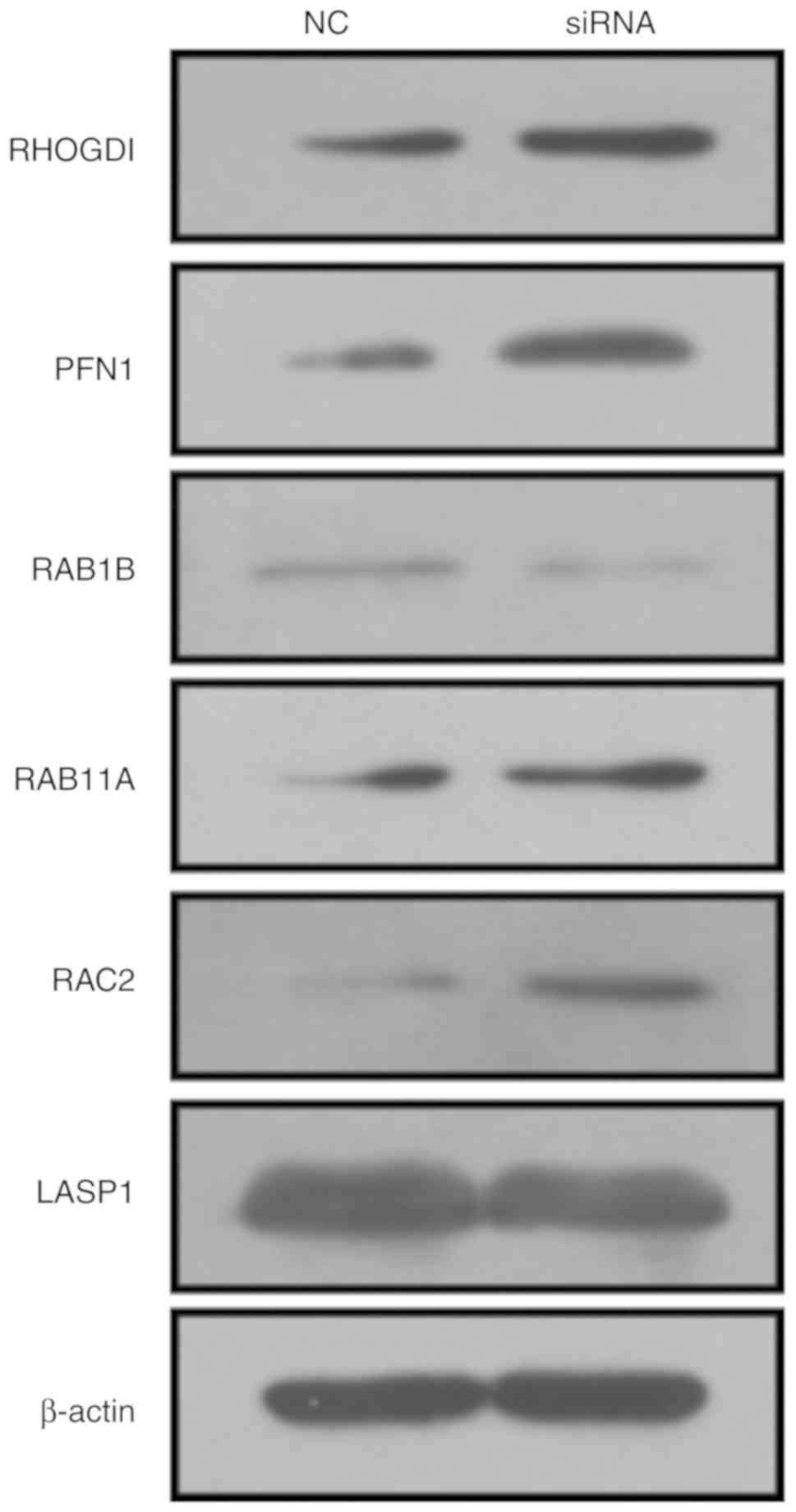
analysis. The results suggested that the knockdown of AFAP1-AS1
increased the expression of RHOGDI, PFN1, RAB11A and RAC2, while it
decreased the expression of RAB1B and LASP1 (Fig. 6).
In order to further investigate the mechanisms
underlying the role of AFAP1-AS1 in cervical cancer development, we
also detected changes in the protein levels of some EMT-related
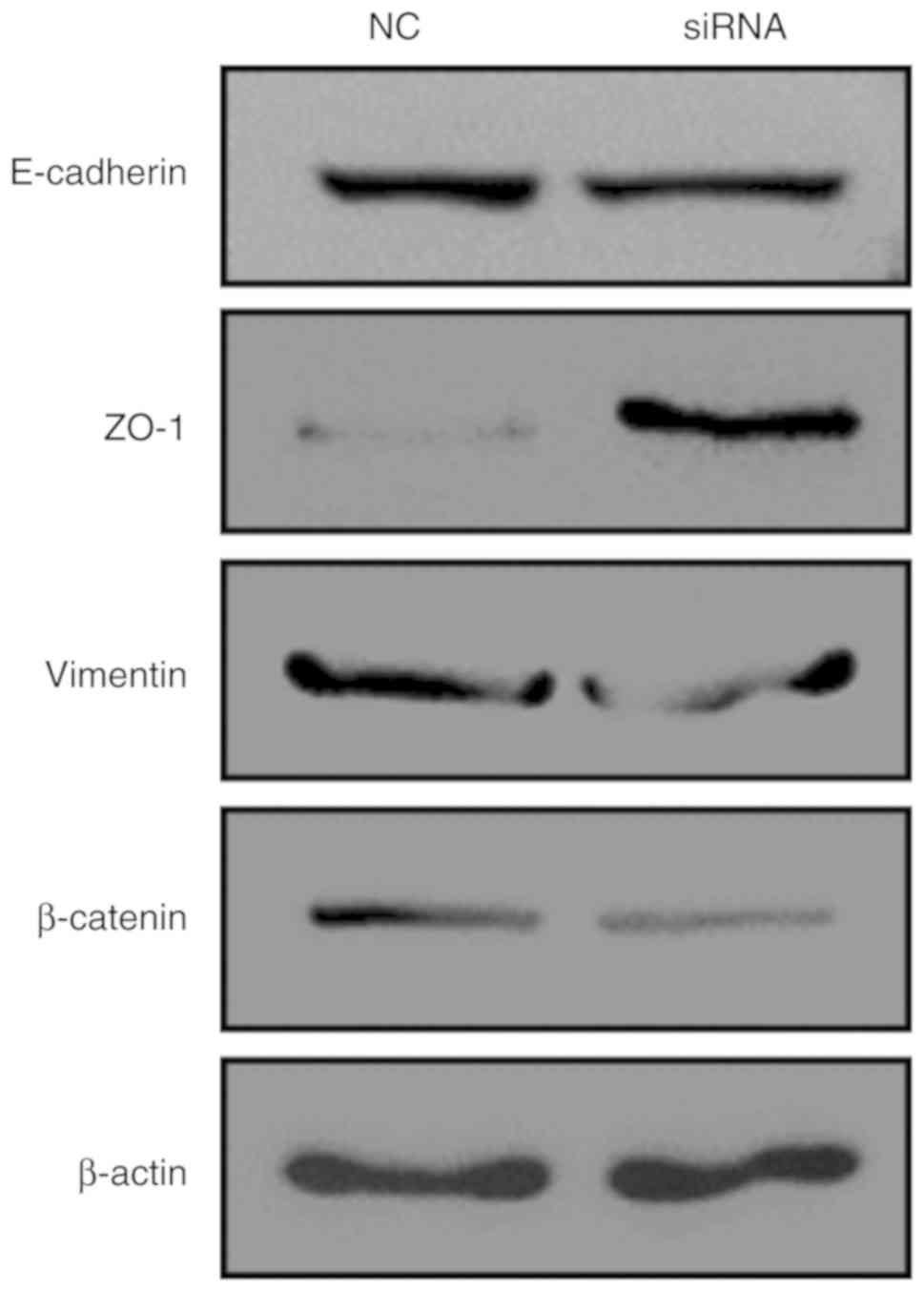
genes. The results revealed that following the knockdown of
AFAP1-AS1, the expression of ZO-1 increased, while the expression
of Vimentin and β-catenin decreased. However, E-cadherin expression
was not markedly altered (Fig.
7).
In recent years, an increasing number of studies
have reported that non-coding RNAs, including lncRNAs, miRNAs and
circular RNAs (circRNAs) act as tumor suppressors or promoters in
tumorigenesis and pathogenesis (44–46).
Accumulating evidence indicates that the abnormal expression of
lncRNAs is associated with tumorigenesis and progression (43,47–52).
The TCGA database includes genome, transcriptome, epigenetic,
proteomic and clinical phenotypic data of 34 tumors, which is one
of the most authoritative cancer databases worldwide (53–55).
Based on the TCGA database, researchers developed the MethCH, XENA
and CRN databases. These provide numerous transcriptome, epigenetic
and clinical phenotypic data for data mining about human malignant
tumors. The mining of these data is a frequently used and
convenient method for the exploration of gene or lncRNA expression
and function (43).
In the present study, we combined the MethCH, XENA
and CRN databases to investigate whether the transcription level
and methylation level of lncRNA AFAP1-AS1 differ in cervical
cancer. We found that AFAP1-AS1 was upregulated and hypomethylated
in cervical cancer tissues compared to normal tissues, which is
consistent with the findings of a previous study showing that the
increased expression of AFAP1-AS1 was due to its promoter
demethylation in esophageal carcinoma (30). Moreover, AFAP1-AS1 upregulation and
hypomethylation were associated with a shorter survival time of
patients with cervical cancer. In addition, some studies have
revealed that lncRNA AFAP1-AS1 is enriched in several tumors and
functions as an oncogene (19,20,30,56,57).
Thus, we hypothesized that AFAP1-AS1 may also function as an
oncogene in cervical cancer. Subsequent functional experiments
confirmed that cell migration and invasion were significantly
suppressed following the knockdown of AFAP1-AS1 in cervical cancer
cells, while no effect on cell proliferation or cell apoptosis
in vitro were observed. However, AFAP1-AS1 knockdown
slightly affected cell cycle distribution.
The Rho/Rac GTPase family is an important member of
the Ras superfamily. It regulates cytoskeletal remodeling by
affecting the polymerization and stability of microtubules and
microfilaments. The Rho/Rac GTPase family has been reported to
regulate cell adhesion and motility (58–60).
Several studies have found that Rho/Rac GTPase family members are
involved in the migration and invasion of various tumors, including
colorectal cancer and ovarian cancer (61,62).
Moreover, two recent studies indicated that lncRNA AFAP1-AS1 was
involved in the regulation of Rho/Rac GTPase family members
(19,56). In the present study, we determined
that the RHOGDI, PFN1, RAB11A and RAC2 protein levels were
elevated, while the RAB1B and LASP1 protein levels were suppressed
after the silencing of AFAP1-AS1 in HeLa cells. These results were
consistent with those of previous studies on lung cancer and
nasopharyngeal carcinoma (19,20).
However, the change in RAC2 protein levels was contrary to the
findings of a previous study on liver cancer (56). This suggests that AFAP1-AS1 may
modulate the Rho/Rac pathway through different mechanisms in
different types of tumors.
EMT is a vital malignant phenotype in tumor cell
migration and invasion which promotes tumor development (48,63). A
number of studies have indicated that lncRNAs play important roles
in the EMT process in tumors (64–69).
In the present study, to identify whether AFAP1-AS1 regulates the
EMT process in cervical cancer as well, the protein levels of some
EMT-related genes were measured by western blot analysis. The data
suggested that AFAP1-AS1 knockdown downregulated the EMT-related
genes vimentin and β-catenin, while it upregulated ZO-1 in cervical
cancer cells. Our data indicated that AFAP1-AS1 may function as a
tumor promoter by affecting the EMT process in HeLa cells.
β-catenin is an important member of the Wnt/β-catenin signaling
pathway. We demonstrated that AFAP1-AS1 knockdown decreased the
protein levels of β-catenin. Nevertheless, it is not clear whether
the Wnt/β-catenin signaling pathway suggests a downstream of
AFAP1-AS1 in cervical cancer. This question warrants further
investigation. In addition, only one cell line was used in the
present study to explore the function and mechanism of AFAP1-AS1 in
cervical cancer. The lack of multiple cell lines in the present
study was a limitation, and that the results should be confirmed in
further experimental models.
Taken together, the findings of the present study
revealed that AFAP1-AS1 was significantly hypomethylated and
upregulated in cervical cancer. And hypomethylation and
upregulation of AFAP1-AS1 are both associated with a poor outcome
of patients with cervical cancer. We further confirmed that
AFAP1-AS1 knockdown suppressed the migration and invasion of HeLa
cells. The inhibition of AFAP1-AS1 negatively regulated the Rho/Rac
pathway and EMT-related gene protein products. The identified
lncRNA AFAP1-AS1 may be a novel target for cervical cancer
therapy.
The authors would like to thank Dr Wei Wang
(Department of Pathology, Affiliated Hospital of Jining Medical
University, Jining, Shandong, China) for providing excellent
technical assistance.
The present study was supported by grants from The
National Natural Science Foundation of China (nos. 81572787,
81672683, 81672993, 81672688, 81702907, 81772901, 81772928,
81803025 and 81872278), the Overseas Expertise Introduction Project
for Discipline Innovation (111 Project, no. 111-2-12), the Natural
Science Foundation of Hunan Province (nos. 2016JC2035, 2017SK2105,
2018JJ3704, 2018JJ3815, 2018SK21210 and 2018SK21211), the
Foundation from the Changsha Science and Technology Board
(kq1706045) and Chinese Anti-Cancer Association, the Fundamental
Research Funds for the Central Universities of Central South
University (no. 1053320171023) and the Special Fund of Clinical
Medicine of Chinese Medical Association (no. 17020280697).
All data generated or analyzed during the present
study are included in this published article or are available from
the corresponding author on reasonable request.
HB and ZG mainly performed the experiments, analyzed
the data and wrote the paper. ZL, LS, CG, XL, QL, WZ, KC and MZ
helped with the experiments and analyzed the data. BX, XL, QL and
LF helped with the data acquisition. BX, XL, WX, LF, ZZ, SZ and FX
helped with the paper writing. GL, WX, ZZ, SZ and FX carried out
the experiment design and manuscript drafting. All authors read and
approved the manuscript and agree to be accountable for all aspects
of the research in ensuring that the accuracy or integrity of any
part of the work are appropriately investigated and resolved.
All patient data analyzed in the present study were
downloaded from online databases. Thus, no ethics approval was
required.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Torre LA, Islami F, Siegel RL, Ward EM and
Jemal A: Global Cancer in Women: Burden and Trends. Cancer
Epidemiol Biomarkers Prev. 26:444–457. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wei F, Wu Y, Tang L, Xiong F, Guo C, Li X,
Zhou M, Xiang B, Li X, Li G, et al: Trend analysis of cancer
incidence and mortality in China. Sci China Life Sci. 60:1271–1275.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Deng X, Xiong F, Li X, Xiang B, Li Z, Wu
X, Guo C, Li X, Li Y, Li G, et al: Application of atomic force
microscopy in cancer research. J Nanobiotechnology. 16:1022018.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Cancer Genome Atlas Research Network and
Albert Einstein College of Medicine; Analytical Biological
Services; Barretos Cancer Hospital; Baylor College of Medicine;
Beckman Research Institute of City of Hope; Buck Institute for
Research on Aging; Canada's Michael Smith Genome Sciences Centre;
Harvard Medical School; Helen F. Graham Cancer Center &
Research Institute at Christiana Care Health Services, ; et al
Integrated genomic and molecular characterization of cervical
cancer. Nature. 543:378–384. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Xie G, Wang Z, Chen Y, Zhang S, Feng L,
Meng F and Yu Z: Dual blocking of PI3K and mTOR signaling by
NVP-BEZ235 inhibits proliferation in cervical carcinoma cells and
enhances therapeutic response. Cancer Lett. 388:12–20. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Liao S, Xiao S, Chen H, Zhang M, Chen Z,
Long Y, Gao L, Zhu G, He J, Peng S, et al: CD38 enhances the
proliferation and inhibits the apoptosis of cervical cancer cells
by affecting the mitochondria functions. Mol Carcinog.
56:2245–2257. 2017. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gong Z, Zhang S, Zhang W, Huang H, Li Q,
Deng H, Ma J, Zhou M, Xiang J, Wu M, et al: Long non-coding RNAs in
cancer. Sci China Life Sci. 55:1120–1124. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tang Y, Wang J, Lian Y, Fan C, Zhang P, Wu
Y, Li X, Xiong F, Li X, Li G, et al: Linking long non-coding RNAs
and SWI/SNF complexes to chromatin remodeling in cancer. Mol
Cancer. 16:422017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yang L, Tang Y, Xiong F, He Y, Wei F,
Zhang S, Guo C, Xiang B, Zhou M, Xie N, et al: LncRNAs regulate
cancer metastasis via binding to functional proteins. Oncotarget.
9:1426–1443. 2018.PubMed/NCBI
|
|
11
|
Wang JP, Tang YY, Fan CM, He Y, Wei F,
Zhang S, Guo C, Xiang B, Zhou M, Xie N, et al: The role of exosomal
non-coding RNAs in cancer metastasis. Oncotarget. 9:12487–12502.
2018.PubMed/NCBI
|
|
12
|
Fan C, Tang Y, Wang J, Xiong F, Guo C,
Wang Y, Zhang S, Gong Z, Wei F, Yang L, et al: Role of long
non-coding RNAs in glucose metabolism in cancer. Mol Cancer.
16:1302017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gong Z, Yang Q, Zeng Z, Zhang W, Li X, Zu
X, Deng H, Chen P, Liao Q, Xiang B, et al: An integrative
transcriptomic analysis reveals p53 regulated miRNA, mRNA, and
lncRNA networks in nasopharyngeal carcinoma. Tumour Biol.
37:3683–3695. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Fan C, Wang J, Tang Y, Wang Y, Xiong F,
Zhang S, Li X, Xiang B, Wu X, Guo C, et al: Long non-coding RNA
LOC284454 promotes migration and invasion of nasopharyngeal
carcinoma via modulating the Rho/Rac signaling pathway.
Carcinogenesis. Oct 30–2018.(Epub ahead of print). doi:
10.1093/carcin/bgy143. View Article : Google Scholar
|
|
15
|
He Y, Jing Y, Wei F, Tang Y, Yang L, Luo
J, Yang P, Ni Q, Pang J, Liao Q, et al: Long non-coding RNA PVT1
predicts poor prognosis and induces radioresistance by regulating
DNA repair and cell apoptosis in nasopharyngeal carcinoma. Cell
Death Dis. 9:2352018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yu J, Liu Y, Guo C, Zhang S, Gong Z, Tang
Y, Yang L, He Y, Lian Y, Li X, et al: Upregulated long non-coding
RNA LINC00152 expression is associated with progression and poor
prognosis of tongue squamous cell carcinoma. J Cancer. 8:523–530.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yu J, Liu Y, Gong Z, Zhang S, Guo C, Li X,
Tang Y, Yang L, He Y, Wei F, et al: Overexpression long non-coding
RNA LINC00673 is associated with poor prognosis and promotes
invasion and metastasis in tongue squamous cell carcinoma.
Oncotarget. 8:16621–16632. 2017.PubMed/NCBI
|
|
18
|
Wang Y, Xue D, Li Y, Pan X, Zhang X, Kuang
B, Zhou M, Li X, Xiong W, Li G, et al: The long noncoding RNA
MALAT-1 is a novel biomarker in various cancers: A meta-analysis
based on the GEO database and literature. J Cancer. 7:991–1001.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bo H, Gong Z, Zhang W, Li X, Zeng Y, Liao
Q, Chen P, Shi L, Lian Y, Jing Y, et al: Upregulated long
non-coding RNA AFAP1-AS1 expression is associated with progression
and poor prognosis of nasopharyngeal carcinoma. Oncotarget.
6:20404–20418. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zeng Z, Bo H, Gong Z, Lian Y, Li X, Li X,
Zhang W, Deng H, Zhou M, Peng S, et al: AFAP1-AS1, a long noncoding
RNA upregulated in lung cancer and promotes invasion and
metastasis. Tumour Biol. 37:729–737. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang Y, Mo Y, Yang X, Zhou R, Wu Z, He Y,
Yang X, Zhong Y, Du Y, Zhou H, et al: Long non-coding RNA AFAP1-AS1
is a novel biomarker in various cancers: A systematic review and
meta-analysis based on the literature and GEO datasets. Oncotarget.
8:102346–102360. 2017.PubMed/NCBI
|
|
22
|
Tang Y, He Y, Shi L, Yang L, Wang J, Lian
Y, Fan C, Zhang P, Guo C, Zhang S, et al: Co-expression of
AFAP1-AS1 and PD-1 predicts poor prognosis in nasopharyngeal
carcinoma. Oncotarget. 8:39001–39011. 2017.PubMed/NCBI
|
|
23
|
Lian Y, Xiong F, Yang L, Bo H, Gong Z,
Wang Y, Wei F, Tang Y, Li X, Liao Q, et al: Long noncoding RNA
AFAP1-AS1 acts as a competing endogenous RNA of miR-423-5p
to facilitate nasopharyngeal carcinoma metastasis through
regulating the Rho/Rac pathway. J Exp Clin Cancer Res. 37:2532018.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bo H, Fan L, Li J, Liu Z, Zhang S, Shi L,
Guo C, Li X, Liao Q, Zhang W, et al: High expression of lncRNA
AFAP1-AS1 promotes the progression of colon cancer and predicts
poor prognosis. J Cancer. 9:4677–4683. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Guo JQ, Li SJ and Guo GX: Long noncoding
RNA AFAP1-AS1 promotes cell proliferation and apoptosis of gastric
cancer cells via PTEN/p-AKT pathway. Dig Dis Sci. 62:2004–2010.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Han X, Wang L, Ning Y, Li S and Wang Z:
Long non-coding RNA AFAP1-AS1 facilitates tumor growth and promotes
metastasis in colorectal cancer. Biol Res. 49:362016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Deng J, Liang Y, Liu C, He S and Wang S:
The up-regulation of long non-coding RNA AFAP1-AS1 is associated
with the poor prognosis of NSCLC patients. Biomed Pharmacother.
75:8–11. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li JR, Sun CH, Li W, Chao RF, Huang CC,
Zhou XJ and Liu CC: Cancer RNA-Seq Nexus: A database of
phenotype-specific transcriptome profiling in cancer cells. Nucleic
Acids Res. 44:D944–951. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Huang WY, Hsu SD, Huang HY, Sun YM, Chou
CH, Weng SL and Huang HD: MethHC: A database of DNA methylation and
gene expression in human cancer. Nucleic Acids Res. 43:D856–D861.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wu W, Bhagat TD, Yang X, Song JH, Cheng Y,
Agarwal R, Abraham JM, Ibrahim S, Bartenstein M, Hussain Z, et al:
Hypomethylation of noncoding DNA regions and overexpression of the
long noncoding RNA, AFAP1-AS1, in Barrett's esophagus and
esophageal adenocarcinoma. Gastroenterology. 144:956–966 e954.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wei F, Tang L, He Y, Wu Y, Shi L, Xiong F,
Gong Z, Guo C, Li X, Liao Q, et al: BPIFB1 (LPLUNC1) inhibits
radioresistance in nasopharyngeal carcinoma by inhibiting VTN
expression. Cell Death Dis. 9:4322018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Song Y, Li X, Zeng Z, Li Q, Gong Z, Liao
Q, Li X, Chen P, Xiang B, Zhang W, et al: Epstein-Barr virus
encoded miR-BART11 promotes inflammation-induced carcinogenesis by
targeting FOXP1. Oncotarget. 7:36783–36799. 2016.PubMed/NCBI
|
|
33
|
Zeng Z, Huang H, Huang L, Sun M, Yan Q,
Song Y, Wei F, Bo H, Gong Z, Zeng Y, et al: Regulation network and
expression profiles of Epstein-Barr virus-encoded microRNAs and
their potential target host genes in nasopharyngeal carcinomas. Sci
China Life Sci. 57:315–326. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yi M, Wang W, Chen S, Peng Y, Li J, Cai J,
Zhou Y, Peng Q, Ban Y, Zeng Z, et al: Dual-functionality of RASSF1A
overexpression in A375 cells is mediated by activation of
IL-6/STAT3 regulatory loop. Mol Biol Rep. 45:1277–1287. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zou G, Ren B, Liu Y, Fu Y, Chen P, Li X,
Luo S, He J, Gao G, Zeng Z, et al: Inhibin B suppresses anoikis
resistance and migration through the transforming growth factor-β
signaling pathway in nasopharyngeal carcinoma. Cancer Sci.
109:3416–3427. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
He R, Liu P, Xie X, Zhou Y, Liao Q, Xiong
W, Li X, Li G, Zeng Z and Tang H: circGFRA1 and GFRA1 act as ceRNAs
in triple negative breast cancer by regulating miR-34a. J Exp Clin
Cancer Res. 36:1452017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Liao Q, Zeng Z, Guo X, Li X, Wei F, Zhang
W, Li X, Chen P, Liang F, Xiang B, et al: LPLUNC1 suppresses
IL-6-induced nasopharyngeal carcinoma cell proliferation via
inhibiting the Stat3 activation. Oncogene. 33:2098–2109. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Yang Y, Liao Q, Wei F, Li X, Zhang W, Fan
S, Shi L, Li X, Gong Z, Ma J, et al: LPLUNC1 inhibits
nasopharyngeal carcinoma cell growth via down-regulation of the MAP
kinase and cyclin D1/E2F pathways. PLoS One. 8:e628692013.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Liang F, Li Q, Li X, Li Z, Gong Z, Deng H,
Xiang B, Zhou M, Li X, Li G, et al: TSC22D2 interacts with PKM2 and
inhibits cell growth in colorectal cancer. Int J Oncol.
49:1046–1056. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Li Q, Chen P, Zeng Z, Liang F, Song Y,
Xiong F, Li X, Gong Z, Zhou M, Xiang B, et al: Yeast two-hybrid
screening identified WDR77 as a novel interacting partner of
TSC22D2. Tumour Biol. 37:12503–12512. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2−ΔΔCT method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zhou Y, Zhang X and Klibanski A: MEG3
noncoding RNA: A tumor suppressor. J Mol Endocrinol. 48:R45–R53.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wang Z, Yang B, Zhang M, Guo W, Wu Z, Wang
Y, Jia L, Li S; Cancer Genome Atlas Research Network, ; Xie W and
Yang D: lncRNA epigenetic landscape analysis identifies
EPIC1 as an oncogenic lncRNA that interacts with MYC and
promotes cell-cycle progression in cancer. Cancer Cell.
33:706–720.e9. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Wang Y, Mo Y, Gong Z, Yang X, Yang M,
Zhang S, Xiong F, Xiang B, Zhou M, Liao Q, et al: Circular RNAs in
human cancer. Mol Cancer. 16:252017. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zhong Y, Du Y, Yang X, Mo Y, Fan C, Xiong
F, Ren D, Ye X, Li C, Wang Y, et al: Circular RNAs function as
ceRNAs to regulate and control human cancer progression. Mol
Cancer. 17:792018. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zhou R, Wu Y, Wang W, Su W, Liu Y, Wang Y,
Fan C, Li X, Li G, Li Y, et al: Circular RNAs (circRNAs) in cancer.
Cancer Lett. 425:134–142. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ,
Tao QF, Liu F, Pan W, Wang TT, Zhou CC, et al: A long noncoding RNA
activated by TGF-β promotes the invasion-metastasis cascade in
hepatocellular carcinoma. Cancer Cell. 25:666–681. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
He B, Li W, Wu Y, Wei F, Gong Z, Bo H,
Wang Y, Li X, Xiang B, Guo C, et al: Epstein-Barr virus-encoded
miR-BART6-3p inhibits cancer cell metastasis and invasion by
targeting long non-coding RNA LOC553103. Cell Death Dis.
7:e23532016. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Gong Z, Zhang S, Zeng Z, Wu H, Yang Q,
Xiong F, Shi L, Yang J, Zhang W, Zhou Y, et al: LOC401317, a
p53-regulated long non-coding RNA, inhibits cell proliferation and
induces apoptosis in the nasopharyngeal carcinoma cell line HNE2.
PLoS One. 9:e1106742014. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Zeng Z, Fan S, Zhang X, Li S, Zhou M,
Xiong W, Tan M, Zhang W and Li G: Epstein-Barr virus-encoded small
RNA 1 (EBER-1) could predict good prognosis in nasopharyngeal
carcinoma. Clin Transl Oncol. 18:206–211. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Xu K, Xiong W, Zhou M, Wang H, Yang J, Li
X, Chen P, Liao Q, Deng H, Li X, et al: Integrating ChIP-sequencing
and digital gene expression profiling to identify BRD7 downstream
genes and construct their regulating network. Mol Cell Biochem.
411:57–71. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Zhang W, Huang C, Gong Z, Zhao Y, Tang K,
Li X, Fan S, Shi L, Li X, Zhang P, et al: Expression of LINC00312,
a long intergenic non-coding RNA, is negatively correlated with
tumor size but positively correlated with lymph node metastasis in
nasopharyngeal carcinoma. J Mol Histol. 44:545–554. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Tu C, Zeng Z, Qi P, Li X, Guo C, Xiong F,
Xiang B, Zhou M, Liao Q, Yu J, et al: Identification of genomic
alterations in nasopharyngeal carcinoma and nasopharyngeal
carcinoma-derived Epstein-Barr virus by whole genome sequencing.
Carcinogenesis. 39:1517–1528. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Tu C, Zeng Z, Qi P, Li X, Yu Z, Guo C,
Xiong F, Xiang B, Zhou M, Gong Z, et al: Genome-wide analysis of 18
Epstein-Barr viruses isolated from primary nasopharyngeal carcinoma
biopsy specimens. J Virol. 91(pii): e00301–17. 2017.PubMed/NCBI
|
|
55
|
Xiao K, Yu Z, Li X, Li X, Tang K, Tu C, Qi
P, Liao Q, Chen P, Zeng Z, et al: Genome-wide analysis of
Epstein-Barr virus (EBV) integration and strain in C666-1 and Raji
cells. J Cancer. 7:214–224. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Zhang JY, Weng MZ, Song FB, Xu YG, Liu Q,
Wu JY, Qin J, Jin T and Xu JM: Long noncoding RNA AFAP1-AS1
indicates a poor prognosis of hepatocellular carcinoma and promotes
cell proliferation and invasion via upregulation of the RhoA/Rac2
signaling. Int J Oncol. 48:1590–1598. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Lu X, Zhou C, Li R, Deng Y, Zhao L and
Zhai W: Long noncoding RNA AFAP1-AS1 promoted tumor growth and
invasion in cholangiocarcinoma. Cell Physiol Biochem. 42:222–230.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Tang Y, He Y, Zhang P, Wang J, Fan C, Yang
L, Xiong F, Zhang S, Gong Z, Nie S, et al: LncRNAs regulate the
cytoskeleton and related Rho/ROCK signaling in cancer metastasis.
Mol Cancer. 17:772018. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Wang M, Zhao J, Zhang L, Wei F, Lian Y, Wu
Y, Gong Z, Zhang S, Zhou J, Cao K, et al: Role of tumor
microenvironment in tumorigenesis. J Cancer. 8:761–773. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Yi M, Cai J, Li J, Chen S, Zeng Z, Peng Q,
Ban Y, Zhou Y, Li X, Xiong W, et al: Rediscovery of NF-κB signaling
in nasopharyngeal carcinoma: How genetic defects of NF-κB pathway
interplay with EBV in driving oncogenesis? J Cell Physiol.
233:5537–5549. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Gou WF, Zhao Y, Lu H, Yang XF, Xiu YL,
Zhao S, Liu JM, Zhu ZT, Sun HZ, Liu YP, et al: The role of RhoC in
epithelial-to-mesenchymal transition of ovarian carcinoma cells.
BMC Cancer. 14:4772014. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Makrodouli E, Oikonomou E, Koc M, Andera
L, Sasazuki T, Shirasawa S and Pintzas A: BRAF and
RAS oncogenes regulate Rho GTPase pathways to mediate
migration and invasion properties in human colon cancer cells: A
comparative study. Mol Cancer. 10:1182011. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Zhou Y, Liao Q, Li X, Wang H, Wei F, Chen
J, Yang J, Zeng Z, Guo X, Chen P, et al: HYOU1, regulated by
LPLUNC1, is up-regulated in nasopharyngeal carcinoma and associated
with poor prognosis. J Cancer. 7:367–376. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Wei F, Wu Y, Tang L, He Y, Shi L, Xiong F,
Gong Z, Guo C, Li X, Liao Q, et al: BPIFB1 (LPLUNC1) inhibits
migration and invasion of nasopharyngeal carcinoma by interacting
with VTN and VIM. Br J Cancer. 118:233–247. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Yang L, Tang Y, He Y, Wang Y, Lian Y,
Xiong F, Shi L, Zhang S, Gong Z, Zhou Y, et al: High expression of
LINC01420 indicates an unfavorable prognosis and modulates cell
migration and invasion in nasopharyngeal carcinoma. J Cancer.
8:97–103. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Fan C, Tang Y, Wang J, Xiong F, Guo C,
Wang Y, Xiang B, Zhou M, Li X, Wu X, et al: The emerging role of
Epstein-Barr virus encoded microRNAs in nasopharyngeal carcinoma. J
Cancer. 9:2852–2864. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Zhang Y, Xia M, Jin K, Wang S, Wei H, Fan
C, Wu Y, Li X, Li X, Li G, et al: Function of the c-Met receptor
tyrosine kinase in carcinogenesis and associated therapeutic
opportunities. Mol Cancer. 17:452018. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Tang L, Wei F, Wu Y, He Y, Shi L, Xiong F,
Gong Z, Guo C, Li X, Deng H, et al: Role of metabolism in cancer
cell radioresistance and radiosensitization methods. J Exp Clin
Cancer Res. 37:872018. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Wang YA, Li XL, Mo YZ, Fan CM, Tang L,
Xiong F, Guo C, Xiang B, Zhou M, Ma J, et al: Effects of tumor
metabolic microenvironment on regulatory T cells. Mol Cancer.
17:1682018. View Article : Google Scholar : PubMed/NCBI
|