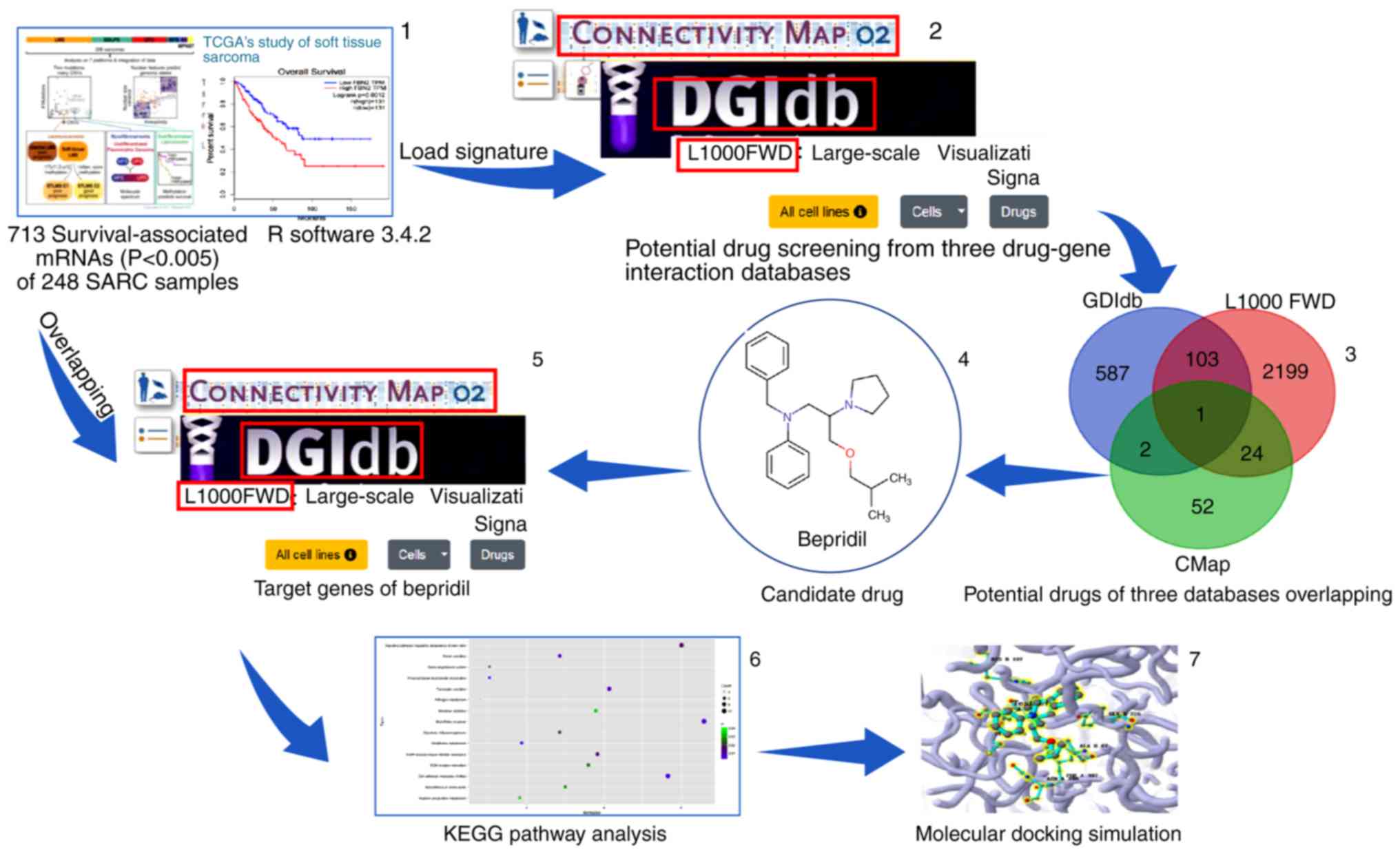
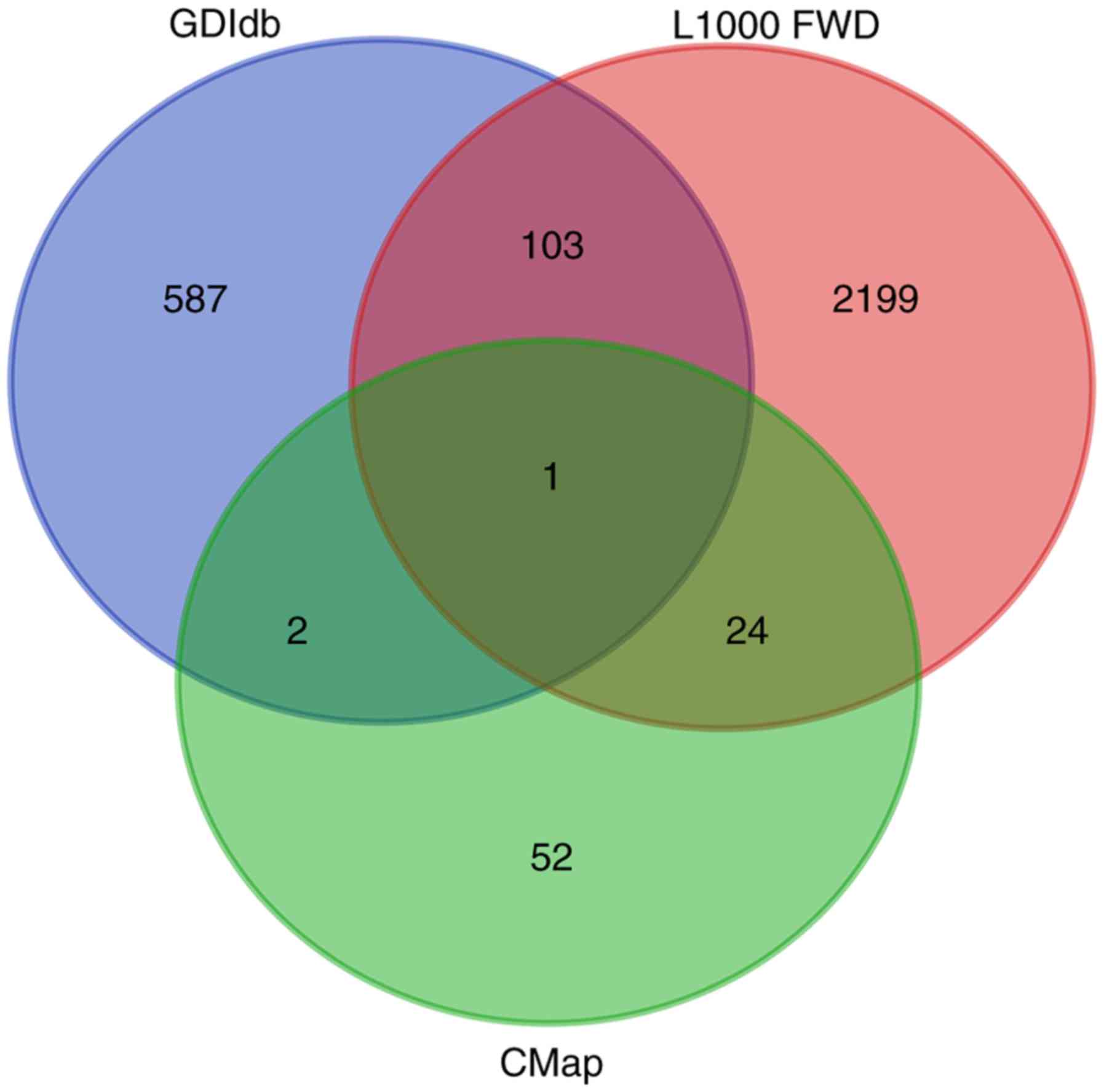
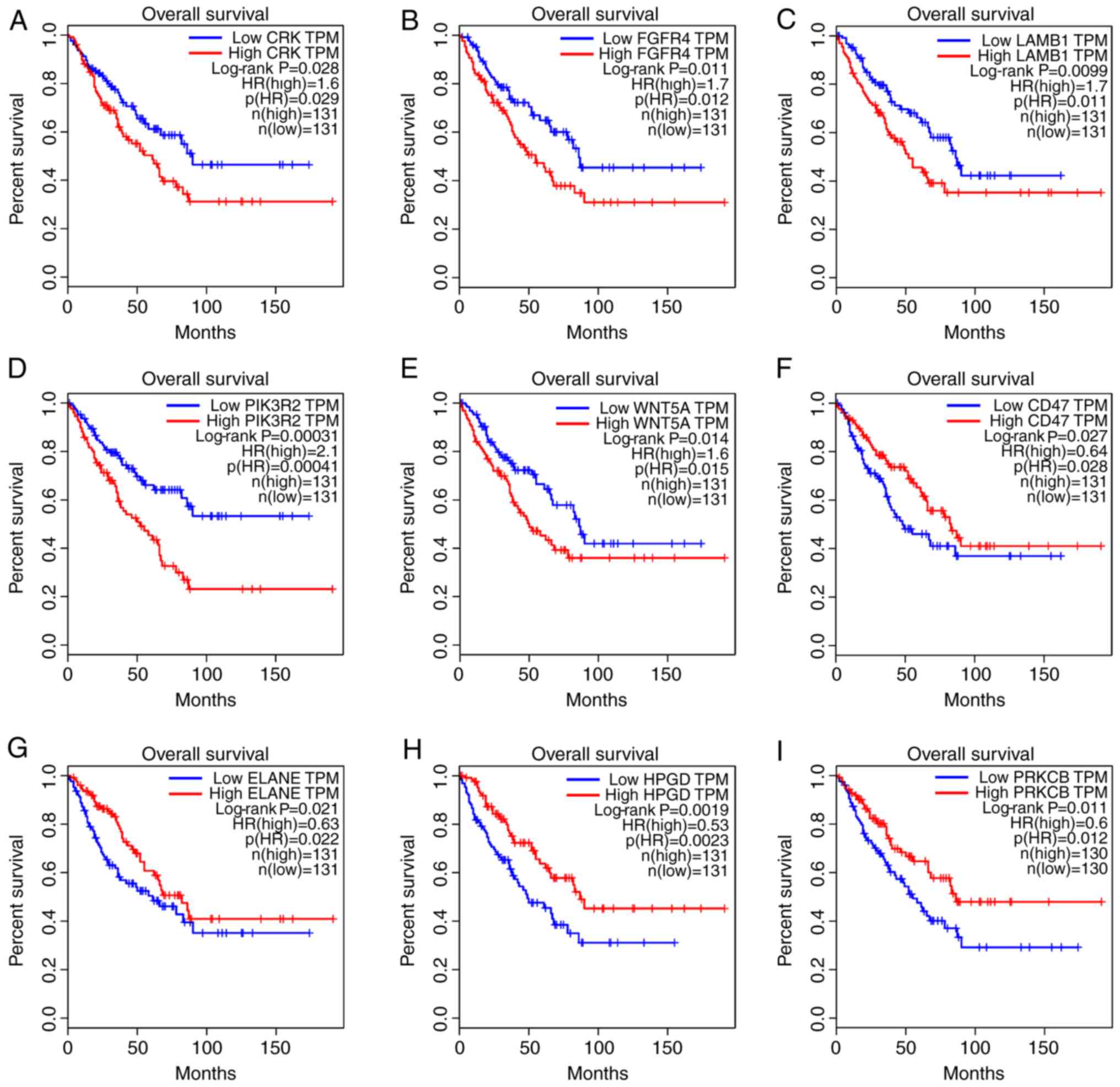
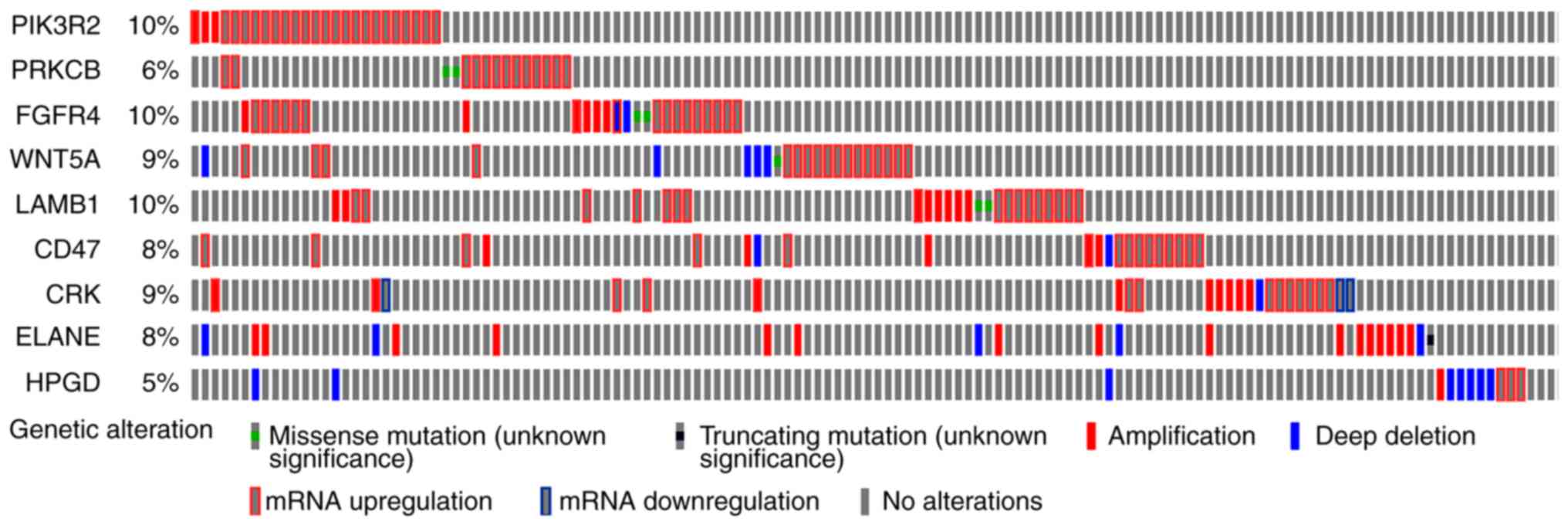
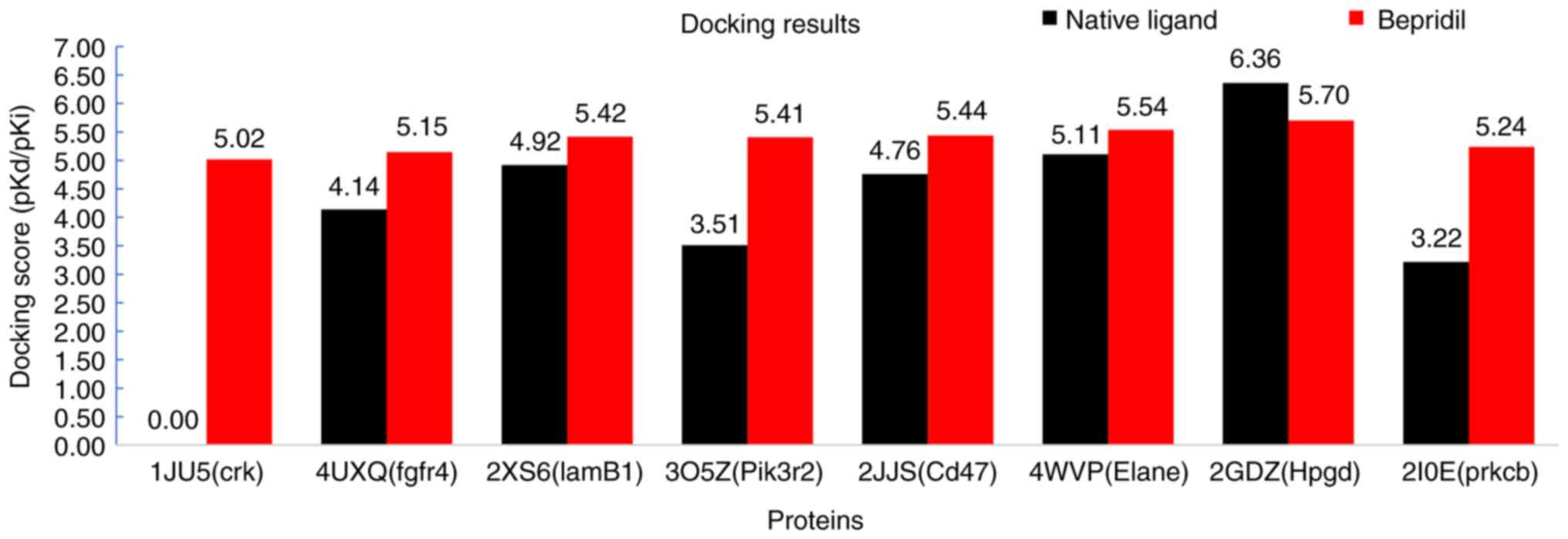
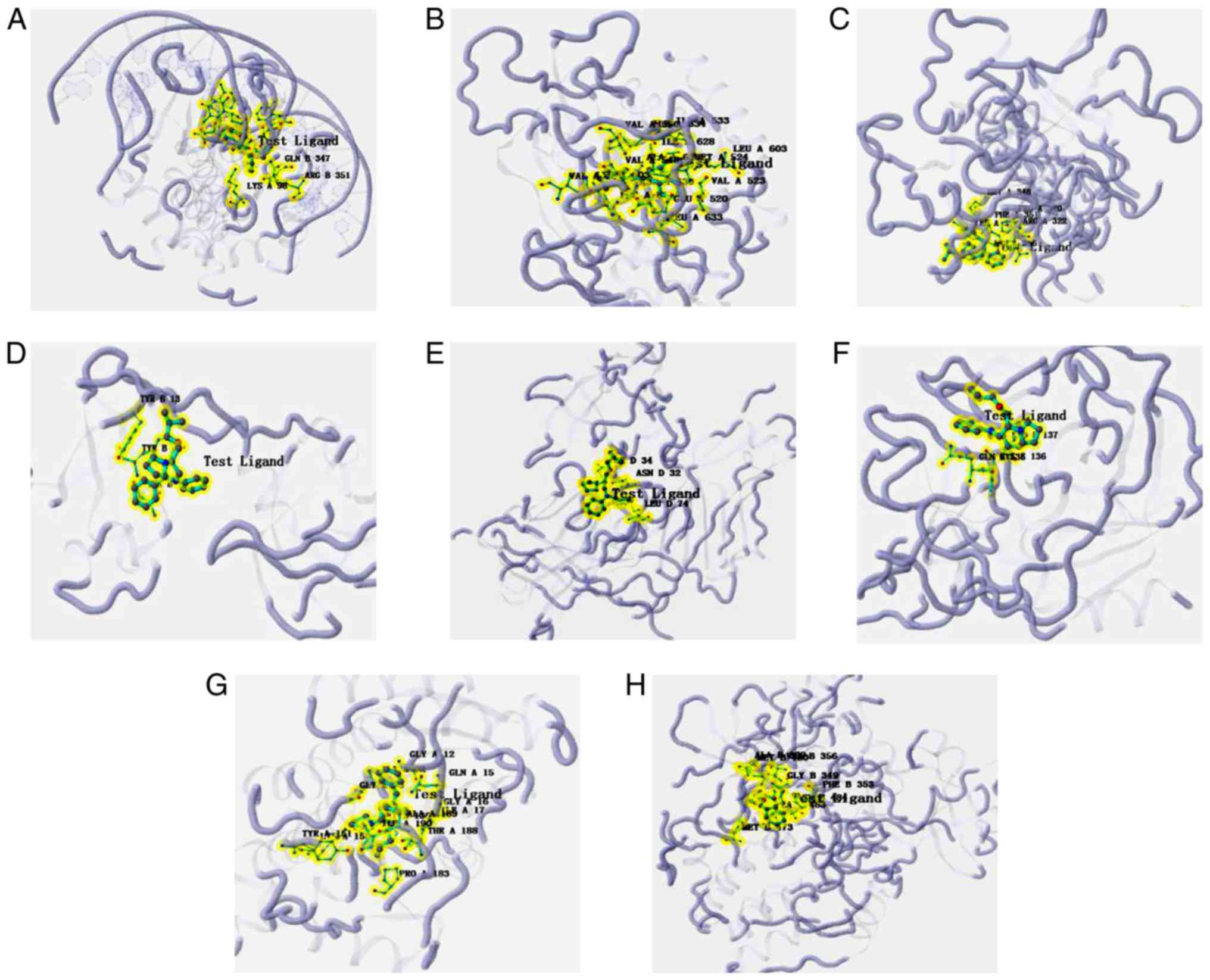
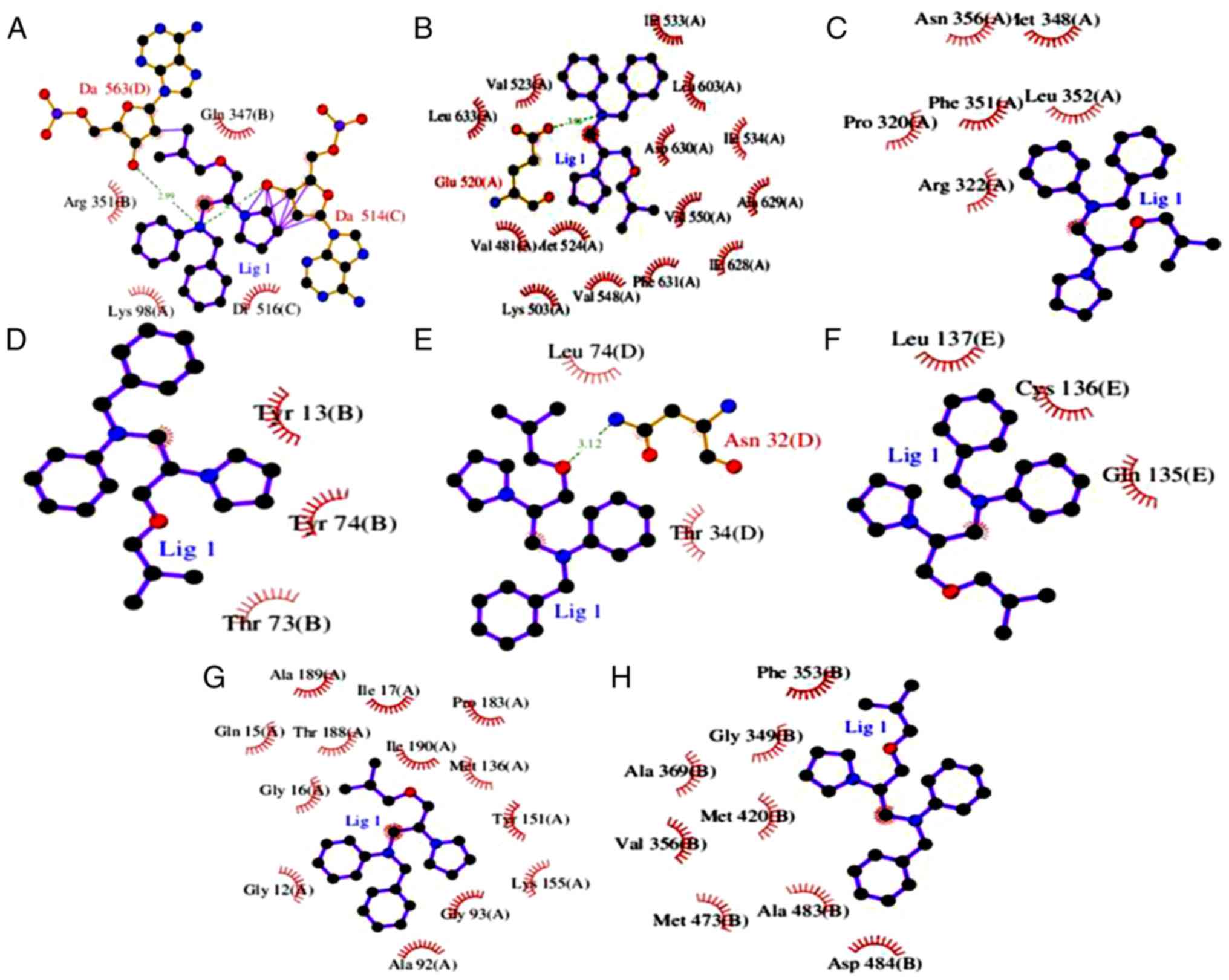
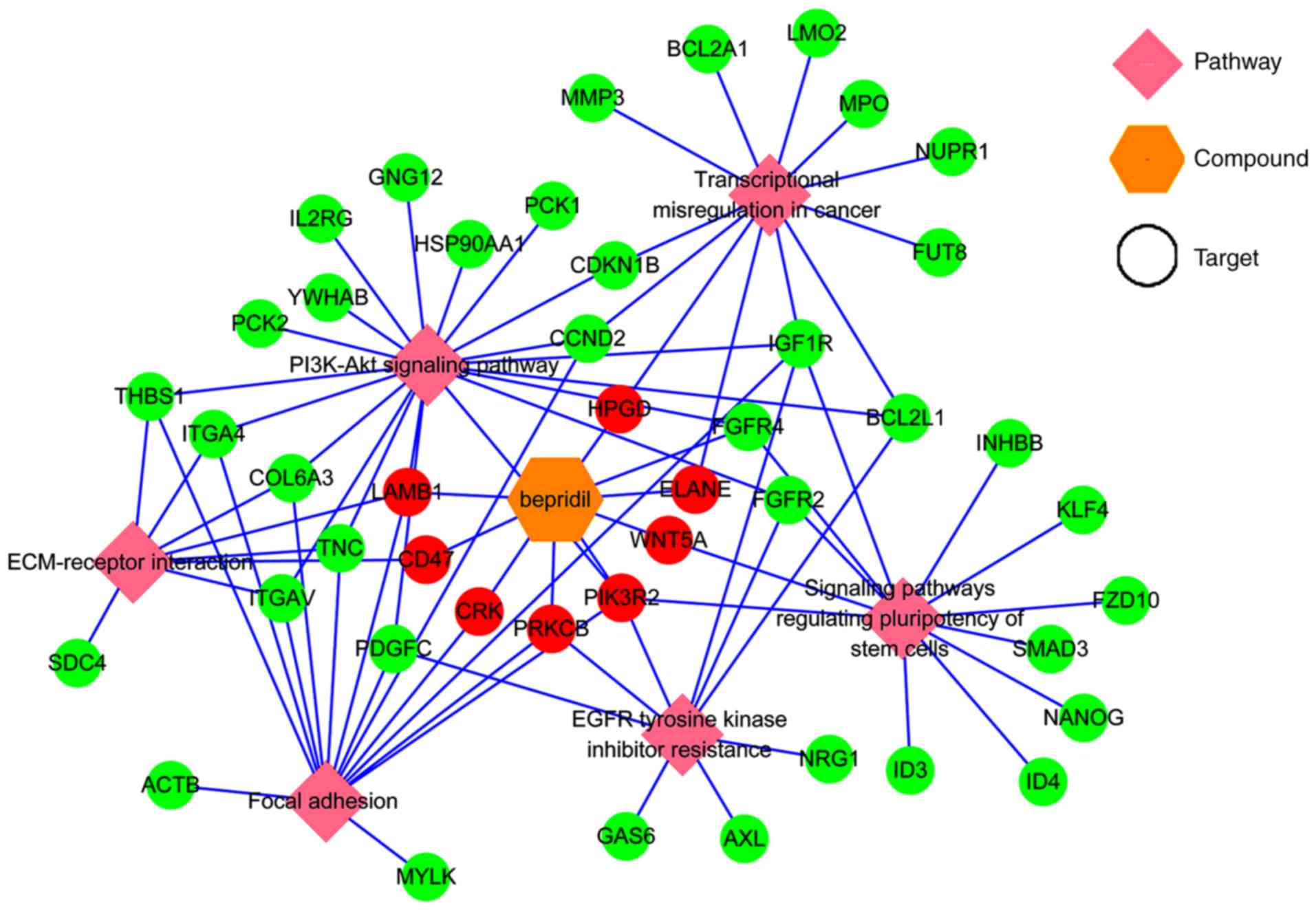
|
1
|
Hoang NT, Acevedo LA, Mann MJ and Tolani
B: A review of soft-tissue sarcomas: Translation of biological
advances into treatment measures. Cancer Manag Res. 10:1089–1114.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Burningham Z, Hashibe M, Spector L and
Schiffman JD: The epidemiology of sarcoma. Clin Sarcoma Res.
2:142012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Koliou P, Karavasilis V, Theochari M,
Pollack SM, Jones RL and Thway K: Advances in the treatment of soft
tissue sarcoma: Focus on eribulin. Cancer Manag Res. 10:207–216.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bourcier K and Italiano A: Newer
therapeutic strategies for soft-tissue sarcomas. Pharmacol Ther.
188:118–123. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Recine F, Bongiovanni A, Riva N, Fausti V,
De Vita A, Mercatali L, Liverani C, Miserocchi G, Amadori D and
Ibrahim T: Update on the role of trabectedin in the treatment of
intractable soft tissue sarcomas. Onco Targets Ther. 10:1155–1164.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Oliveira IM, Borges A, Borges F and Simoes
M: Repurposing ibuprofen to control Staphylococcus aureus
biofilms. Eur J Med Chem. 166:197–205. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Qu XA and Rajpal DK: Applications of
connectivity map in drug discovery and development. Drug Discov
Today. 17:1289–1298. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Cotto KC, Wagner AH, Feng YY, Kiwala S,
Coffman AC, Spies G, Wollam A, Spies NC, Griffith OL and Griffith
M: DGIdb 3.0: A redesign, and expansion of the drug-gene
interaction database. Nucleic Acids Res. 46:D1068–D1073. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang Z, Lachmann A, Keenan AB and Ma'ayan
A: L1000FWD: Fireworks visualization of drug-induced transcriptomic
signatures. Bioinformatics. 34:2150–2152. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lamb J, Crawford ED, Peck D, Modell JW,
Blat IC, Wrobel MJ, Lerner J, Brunet JP, Subramanian A, Ross KN, et
al: The connectivity map: Using gene-expression signatures to
connect small molecules, genes, and disease. Science.
313:1929–1935. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hurle MR, Yang L, Xie Q, Rajpal DK,
Sanseau P and Agarwal P: Computational drug repositioning: From
data to therapeutics. Clin Pharmacol Ther. 93:335–341. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhong Y, Chen EY, Liu R, Chuang PY,
Mallipattu SK, Tan CM, Clark NR, Deng Y, Klotman PE, Ma'ayan A, et
al: Renoprotective effect of combined inhibition of
angiotensin-converting enzyme, and histone deacetylase. J Am Soc
Nephrol. 24:801–811. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Karube K, Tsuzuki S, Yoshida N, Arita K,
Kato H, Katayama M, Ko YH, Ohshima K, Nakamura S, Kinoshita T, et
al: Comprehensive gene expression profiles of NK cell neoplasms
identify vorinostat as an effective drug candidate. Cancer Lett.
333:47–55. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Dyle MC, Ebert SM, Cook DP, Kunkel SD, Fox
DK, Bongers KS, Bullard SA, Dierdorff JM and Adams CM:
Systems-based discovery of tomatidine as a natural small molecule
inhibitor of skeletal muscle atrophy. J Biol Chem. 289:14913–14924.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gao L, Zhao G, Fang JS, Yuan TY, Liu AL
and Du GH: Discovery of the neuroprotective effects of alvespimycin
by computational prioritization of potential anti-Parkinson agents.
FEBS J. 281:1110–1122. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chen YT, Xie JY, Sun Q and Mo WJ: Novel
drug candidates for treating esophageal carcinoma: A study on
differentially expressed genes, using connectivity mapping, and
molecular docking. Int J Oncol. 54:152–166. 2019.PubMed/NCBI
|
|
18
|
Drullion C, Marot G, Martin N, Desle J,
Saas L, Salazar-Cardozo C, Bouali F, Pourtier A, Abbadie C and
Pluquet O: Pre-malignant transformation by senescence evasion is
prevented by the PERK and ATF6alpha branches of the Unfolded
protein response. Cancer Lett. 438:187–196. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tang X, Xu Y, Lu L, Jiao Y, Liu J, Wang L
and Zhao H: Identification of key candidate genes, and small
molecule drugs in cervical cancer by bioinformatics strategy.
Cancer Manag Res. 10:3533–3549. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Christinat A and Leyvraz S: Role of
trabectedin in the treatment of soft tissue sarcoma. Onco Targets
Ther. 2:105–113. 2009.PubMed/NCBI
|
|
21
|
Demetri GD, von Mehren M, Jones RL,
Hensley ML, Schuetze SM, Staddon A, Milhem M, Elias A, Ganjoo K,
Tawbi H, et al: Efficacy and safety of trabectedin or dacarbazine
for metastatic liposarcoma or leiomyosarcoma after failure of
conventional chemotherapy: Results of a Phase III randomized
multicenter clinical trial. J Clin Oncol. 34:786–793. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Xie L, Guo W, Wang Y, Yan T, Ji T and Xu
J: Apatinib for advanced sarcoma: Results from multiple
institutions' off-label use in China. BMC Cancer. 18:3962018.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhu B, Li J, Xie Q, Diao L, Gai L and Yang
W: Efficacy and safety of apatinib monotherapy in advanced bone and
soft tissue sarcoma: An observational study. Cancer Biol Ther.
19:198–204. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li S, Chen X, Liu X, Yu Y, Pan H, Haak R,
Schmidt J, Ziebolz D and Schmalz G: Complex integrated analysis of
lncRNAs-miRNAs-mRNAs in oral squamous cell carcinoma. Oral Oncol.
73:1–9. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Brum AM, van de Peppel J, Nguyen L, Aliev
A, Schreuders- Koedam M, Gajadien T, van der Leije CS, van Kerkwijk
A, Eijken M, van Leeuwen JPTM, et al: Using the connectivity map to
discover compounds influencing human osteoblast differentiation. J
Cell Physiol. 233:4895–4906. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang J, Vasaikar S, Shi Z, Greer M and
Zhang B: WebGestalt 2017: A more comprehensive, powerful, flexible,
and interactive gene set enrichment analysis toolkit. Nucleic Acids
Res. 45:W130–W137. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yoav Benjamini and Yosef Hochberg:
Controlling the false discovery rate: A practical and powerful
approach to multiple testing. J R Stat Soc Series B. 57:289–300.
1995.
|
|
28
|
Vilar S, Sobarzo-Sanchez E, Santana L and
Uriarte E: Molecular docking and drug discovery in beta-Adrenergic
receptors. Curr Med Chem. 24:4340–4359. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Eswari JS, Dhagat S, Kaser S and Tiwari A:
Homology modeling and molecular docking studies of bacillomycin and
iturin synthetases with novel ligands for the production of
therapeutic lipopeptides. Curr Drug Discov Technol. 15:132–141.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hsin KY, Matsuoka Y, Asai Y, Kamiyoshi K,
Watanabe T, Kawaoka Y and Kitano H: systemsDock: A web server for
network pharmacology-based prediction and analysis. Nucleic Acids
Res. 44:W507–W513. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Jain AN: Surflex: Fully automatic flexible
molecular docking using a molecular similarity-based search engine.
J Med Chem. 46:499–511. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zsoldos Z, Reid D, Simon A, Sadjad BS and
Johnson AP: eHiTS: An innovative approach to the docking and
scoring function problems. Curr Protein Pept Sci. 7:421–435. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ma F, Takanari H, Masuda K, Morishima M
and Ono K: Short- and long-term inhibition of cardiac
inward-rectifier potassium channel current by an antiarrhythmic
drug bepridil. Heart Vessels. 31:1176–1184. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Vitiello PP, Cardone C, Martini G,
Ciardiello D, Belli V, Matrone N, Barra G, Napolitano S, Della
Corte C, Turano M, et al: Receptor tyrosine kinase-dependent PI3K
activation is an escape mechanism to vertical suppression of the
EGFR/RAS/MAPK pathway in KRAS-mutated human colorectal cancer cell
lines. J Exp Clin Cancer Res. 38:412019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ge W, Wang SH, Sun B, Zhang YL, Shen W,
Khatib H and Wang X: Melatonin promotes Cashmere goat (Capra
hircus) secondary hair follicle growth: A view from integrated
analysis of long non-coding and coding RNAs. Cell Cycle.
17:1255–1267. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Doi T, Yang JC, Shitara K, Naito Y, Cheng
AL, Sarashina A, Pronk LC, Takeuchi Y and Lin CC: Phase I study of
the focal adhesion kinase inhibitor BI 853520 in Japanese and
Taiwanese patients with advanced or metastatic solid tumors. Target
Oncol. Feb 6–2019.(Epub ahead of print). doi:
10.1007/s11523-019-00620-0. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Paik H, Chung AY, Park HC, Park RW, Suk K,
Kim J, Kim H, Lee K and Butte AJ: Repurpose terbutaline sulfate for
amyotrophic lateral sclerosis using electronic medical records. Sci
Rep. 5:85802015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Dudley JT, Sirota M, Shenoy M, Pai RK,
Roedder S, Chiang AP, Morgan AA, Sarwal MM, Pasricha PJ and Butte
AJ: Computational repositioning of the anticonvulsant topiramate
for inflammatory bowel disease. Sci Transl Med. 3:96ra762011.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Musa A, Ghoraie LS, Zhang SD, Glazko G,
Yli-Harja O, Dehmer M, Haibe-Kains B and Emmert-Streib F: A review
of connectivity map, and computational approaches in
pharmacogenomics. Brief Bioinform. 19:506–523. 2018.PubMed/NCBI
|
|
40
|
Gaspar T, Kis B, Snipes JA, Lenzsér G,
Mayanagi K, Bari F and Busija DW: Neuronal preconditioning with the
antianginal drug, bepridil. J Neurochem. 102:595–608. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
van Kalken CK, van der Hoeven JJ, de Jong
J, Giaccone G, Schuurhuis GJ, Maessen PA, Blokhuis WM, van der
Vijgh WJ and Pinedo HM: Bepridil in combination with anthracyclines
to reverse anthracycline resistance in cancer patients. Eur J
Cancer. 27:739–744. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Lee YS, Sayeed MM and Wurster RD:
Intracellular Ca2+ mediates the cytotoxicity induced by
Bepridil, and benzamil in human brain tumor cells. Cancer Lett.
88:87–91. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Baldoni S, Del Papa B, Dorillo E, Aureli
P, De Falco F, Rompietti C, Sorcini D, Varasano E, Cecchini D, Zei
T, et al: Bepridil exhibits anti-leukemic activity associated with
NOTCH1 pathway inhibition in chronic lymphocytic leukemia. Int J
Cancer. 143:958–970. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Li C, Zeng X, Liu Z, Li F, Wang K and Wu
B: BDNF VAL66MET polymorphism elevates the risk of bladder cancer
via MiRNA-146b in Micro-Vehicles. Cell Physiol Biochem. 45:366–377.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Gong XH, Chen C, Hou P, Zhu SC, Wu CQ,
Song CL, Ni W, Hu JF, Yao DK, Kang JH, et al: Overexpression of
miR-126 inhibits the activation, and migration of HSCs through
targeting CRK. Cell Physiol Biochem. 33:97–106. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Watanabe T, Tsuda M, Tanaka S, Ohba Y,
Kawaguchi H, Majima T, Sawa H and Minami A: Adaptor protein Crk
induces Src-dependent activation of p38 MAPK in regulation of
synovial sarcoma cell proliferation. Mol Cancer Res. 7:1582–1592.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Matsumoto R, Tsuda M, Wang L, Maishi N,
Abe T, Kimura T, Tanino M, Nishihara H, Hida K, Ohba Y, et al:
Adaptor protein CRK induces epithelial-mesenchymal transition, and
metastasis of bladder cancer cells through HGF/c-Met feedback loop.
Cancer Sci. 106:709–717. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Kumar S, Lu B, Davra V, Hornbeck P,
Machida K and Birge RB: Crk tyrosine phosphorylation regulates
PDGF-BB-inducible Src activation, and breast tumorigenicity, and
metastasis. Mol Cancer Res. 16:173–183. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Xu M, Chen S, Yang W, Cheng X, Ye Y, Mao
J, Wu X, Huang L and Ji J: FGFR4 links glucose metabolism, and
chemotherapy resistance in breast cancer. Cell Physiol Biochem.
47:151–160. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Katoh M and Nakagama H: FGF receptors:
Cancer biology, and therapeutics. Med Res Rev. 34:280–300. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Schelch K, Kirschner MB, Williams M, Cheng
YY, van Zandwijk N, Grusch M and Reid G: A link between the
fibroblast growth factor axis, and the miR-16 family reveals
potential new treatment combinations in mesothelioma. Mol Oncol.
12:58–73. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Lin Q, Lim HS, Lin HL, Tan HT, Lim TK,
Cheong WK, Cheah PY, Tang CL, Chow PK and Chung MC: Analysis of
colorectal cancer glyco-secretome identifies laminin β-1 (LAMB1) as
a potential serological biomarker for colorectal cancer.
Proteomics. 15:3905–3920. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Qi L, Sun K, Zhuang Y, Yang J and Chen J:
Study on the association between PI3K/AKT/mTOR signaling pathway
gene polymorphism, and susceptibility to gastric cancer. J BUON.
22:1488–1493. 2017.PubMed/NCBI
|
|
54
|
Wu XJ, Zhao ZF, Kang XJ, Wang HJ, Zhao J
and Pu XM: MicroRNA-126-3p suppresses cell proliferation by
targeting PIK3R2 in Kaposi's sarcoma cells. Oncotarget.
7:36614–36621. 2016.PubMed/NCBI
|
|
55
|
Kobayashi Y, Kadoya T, Amioka A, Hanaki H,
Sasada S, Masumoto N, Yamamoto H, Arihiro K, Kikuchi A and Okada M:
Wnt5a-induced cell migration is associated with the aggressiveness
of estrogen receptor-positive breast cancer. Oncotarget.
9:20979–20992. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Wang L, Yao M, Fang M, Zheng WJ, Dong ZZ,
Pan LH, Zhang HJ and Yao DF: Expression of hepatic Wnt5a, and its
clinicopathological features in patients with hepatocellular
carcinoma. Hepatobiliary Pancreat Dis Int. 17:227–232. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Ye X, Wang X, Lu R, Zhang J, Chen X and
Zhou G: CD47 as a potential prognostic marker for oral leukoplakia,
and oral squamous cell carcinoma. Oncol Lett. 15:9075–9080.
2018.PubMed/NCBI
|
|
58
|
Makaryan V, Zeidler C, Bolyard AA, Skokowa
J, Rodger E, Kelley ML, Boxer LA, Bonilla MA, Newburger PE,
Shimamura A, et al: The diversity of mutations, and clinical
outcomes for ELANE-associated neutropenia. Curr Opin Hematol.
22:3–11. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Vainio P, Gupta S, Ketola K, Mirtti T,
Mpindi JP, Kohonen P, Fey V, Perälä M, Smit F, Verhaegh G, et al:
Arachidonic acid pathway members PLA2G7, HPGD, EPHX2, and
CYP4F8 identified as putative novel therapeutic targets in
prostate cancer. Am J Pathol. 178:525–536. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Qi X, Wang Y, Hou J and Huang Y: A Single
nucleotide polymorphism in HPGD gene is associated with
prostate cancer risk. J Cancer. 8:4083–4086. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Pereira C, Queiros S, Galaghar A, Sousa H,
Pimentel-Nunes P, Brandão C, Moreira-Dias L, Medeiros R and
Dinis-Ribeiro M: Genetic variability in key genes in prostaglandin
E2 pathway (COX-2, HPGD, ABCC4, and
SLCO2A1), and their involvement in colorectal cancer
development. PLoS One. 9:e920002014. View Article : Google Scholar : PubMed/NCBI
|