Introduction
Hepatocellular carcinoma (HCC) has one of the
highest rates of incidence and mortality of all types of cancer
worldwide, exhibiting as well as a low complete resection rate and
a high postoperative recurrence rate (1–3), which
are primarily driven by a high invasiveness and intrahepatic and/or
extrahepatic metastasis (4).
Fundamental research has demonstrated that epithelial-mesenchymal
transition (EMT) serves a crucial role in the metastasis of HCC
(5). Therefore, assessing the
mechanism of HCC cell EMT is important for improving the prognosis
of patients with HCC and for lengthening their survival.
As a zinc-finger protein, Snail1 binds to the E-box
of the E-cadherin promoter and reduces the transcription of
E-cadherin to promote EMT (6).
Snail1 also indirectly activates vimentin and α-SMA to facilitate
EMT (6). MicroRNAs (miRNAs) and
long non-coding RNAs (lncRNAs) serve important roles in the
regulation of cellular processes (7). It has been revealed that lncRNA
influences the occurrence and development of tumors by regulating
the expression of miRNAs, causing the dysfunction of protein
encoding genes if expressed abnormally (8). The expression profile of lncRNAs in
certain tumor cells differ from that in normal cells, which may
contribute to tumor development (9). Therefore, assessing the interaction
between miRNAs and lncRNAs may further elucidate cell structural
and regulatory networks, and provide scientific and clinical value.
AB209371 is the lncRNA with the most differential expression among
primary HCC and metastases that we found through screening and
sequencing of the transcriptome. Furthermore, the level of AB209371
was found to decrease from metastases to primary HCC and then to
adjacent normal tissues, suggesting that AB209371 may be involved
in HCC metastases. Our present study demonstrates for the first
time that AB209371 silencing suppresses EMT of HCC cells by
restoring the negative regulation of Snail1 by hsa-miR199a-5p,
providing a solid theoretic base for using lncRNA as target of
genetic therapy for metastasis and migration of HCC as well as
offering a reasonable explanation for the inactivation of miRNA in
different tumors or tumors at different stages.
Materials and methods
Cell culture
MHCC97-H (SCSP-528) and HCC-LM3 (HCCC-9810) HCC cell
lines were obtained from the Type Collection of the Chinese Academy
of Sciences (Shanghai, China) and maintained in RPMI-1640 medium
(cat. no. 11875093; Invitrogen; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS;
cat. no. 10100147 Invitrogen; Thermo Fisher Scientific, Inc.).
293TN cells (CRL-3216) were purchased from the American Type
Culture Collection (Manassas, VA, USA) and maintained in Dulbecco's
Minimum Essential medium (DMEM; cat. no. 10569044; Invitrogen;
Thermo Fisher Scientific, Inc.) supplemented with 10% FBS. All
adherent cells were passaged via 0.25% trypsin digestion and
incubated in an atmosphere of 5% CO2 at 37°C.
Assessment of AB209371,
hsa-miR199a-5p, Snail1 protein and EMT markers in primary and
metastatic HCC tissue, and adjacent normal tissue
Primary HCC tissue, metastasis tissue and adjacent
normal tissue (2 cm from the primary site) were obtained from 20
patients at the Department of General Surgery, Yancheng City No. 1
People's Hospital (Yancheng, China) from December 2016 to March
2017. Whether patients had received any preoperative routine
chemotherapy or not was not considered for enrollment. Detailed
patient information is presented in Table I. Informed consent was obtained from
all patients prior to enrollment and the ethical approval was
obtained from the Ethics Committee of Yancheng City No. 1 People's
Hospital. RNA was extracted from tissues using the Trizol reagent
(cat. no. 15596018; Invitrogen; Thermo Fisher Scientific, Inc.) and
reverse transcription-quantitative polymerase chain reaction
(RT-qPCR) was performed to assess AB209371, hsa-miR199a-5p and
Snail1 mRNA. Total protein was extracted from tissues using the
T-PER tissue protein extraction reagent (cat. no. 78510; Pierce;
Thermo Fisher Scientific, Inc.) and used for the detection of
Snail1 and EMT markers, including E-cadherin, vimentin, α-smooth
muscle actin (α-SMA) and matrix metallopeptidase-9 (MMP-9), via
western blotting.
 | Table I.Clinicopathological features of 20
patients with metastatic hepatocellular carcinoma. |
Table I.
Clinicopathological features of 20
patients with metastatic hepatocellular carcinoma.
| Number | Sex | Age | TNM stage | Metastasis
type |
|---|
| 1 | M | 39 | T2NXM1 | Bone |
| 2 | F | 72 | T3N0M1 | Lung |
| 3 | M | 59 | T1NXM1 | Bone |
| 4 | M | 49 | T3N0M1 | Head |
| 5 | M | 68 | T3NXM1 | Colon |
| 6 | F | 71 | T3N1M1 | Lung |
| 7 | M | 66 | T1N1M1 | Colon |
| 8 | M | 51 | T3NXM1 | Colon |
| 9 | M | 62 | T2N0M1 | Lung |
| 10 | M | 66 | T3NXM1 | Lung |
| 11 | F | 65 | T3N1M1 | Stomach |
| 12 | M | 47 | T4N0M1 | Bone |
| 13 | F | 59 | T3N1M1 | Lung |
| 14 | M | 45 | T2N1M1 | Stomach |
| 15 | F | 62 | T3N0M1 | Stomach |
| 16 | M | 55 | T2NXM1 | Bone |
| 17 | F | 60 | T3N0M1 | Head |
| 18 | M | 58 | T3NXM1 | Lung |
| 19 | M | 62 | T2N0M1 | Lung |
| 20 | F | 55 | T4N1M1 | Lung |
Construction of vectors
A small interfering RNA (siRNA) sequence
(5′-GGTAACAGATGGAAATCCG-3′) with complementary binding to AB209371
was selected (each; Sangon Biotech Co., Ltd., Shanghai, China). The
oligonucleotide templates of siRNA were chemically synthesized and
cloned into the linear pSIH1-cop green fluorescent protein (GFP)
siRNA Vector (cat. no. SI501A-1; System Biosciences LLC, Palo Alto,
CA, USA) which was obtained through digestion by BamHI and
EcoRI (Takara Biotechnology Co., Ltd., Dalian, China) and
purification by agarose gel electrophoresis. An invalid siRNA
sequence (5′-GAAGCCAGATCCAGCTTCC-3′) was used as a negative control
(NC). The recombinant vectors were named pSIH1-siRNA-AB209371 and
pSIH1-NC, respectively.
The coding sequence of human Snail1 (NM_005985.3)
was amplified using the primers
5′-GGAATTCGCCACCATGCCGCGCTCTTTCCTCG-3′ and
5′-CGGGATCCTCAGCGGGGACATCCTGAGCAGC-3′, which contain an
EcoRI cutting site and kozak sequence (5′-GCCACC-3′) and a
BamHI cutting site, respectively, with the cDNA prepared by
reverse transcription of RNA isolated from 293TN cells with a M-MLV
Reverse Transcriptase kit (cat. no. 2640; Takara Biotechnology Co.,
Ltd.) in accordance with the manufacturer's protocol. Takara
Ex Tap (cat. no. RR001Q; Takara Biotechnology Co., Ltd.) was
utilized for the PCR procedure with the following thermocycling
conditions: 35 cycles of denaturation at 94°C for 30 sec, annealing
at 55°C for 30 sec and elongation at 72°C for 45 sec. The PCR
product was digested and cloned into the pCDH-CMV-MCS-EF1α-copGFP
Cloning and Expression Lentivector (cat. no. CD511B-1; System
Biosciences LLC). The recombinant vector was named pcDH-Snail1.
Luciferase reporter assay
The present study utilized TargetScan 7.1
(http://www.targetscan.org/) to predict
whether a hsa-miR199a-5p binding site exists within the
3′-untranslated region (UTR) of human Snail1 mRNA. The same tool
was used to predict the binding sites of hsa-miR199a-5p on
AB209371. Primers that targeted the 3′-UTR of the Snail1 gene were
designed such that flanking XbaI restriction sites were
introduced into the 158 bp PCR product containing the
hsa-miR199a-5p target site (5′-ACACTGG-3′). The primer sequences
were 5′-GCTCTAGATCTGACCGATGTGTCTC-3′ (Forward) and
5′-GCTCTAGAGAATATCAAACTGTACAT-3′ (Reverse), respectively. The PCR
reaction was performed using Takara Ex Taq (cat. no. RR001A;
Takara Bio, Inc., Otsu, Japan) under the following conditions: 35
cycles of denaturation 94°C for 30 sec, annealing at 55°C for 30
sec and elongation at 72°C for 10 sec. The PCR product was digested
with XbaI (cat. no. 1093S; Takara Bio, Inc.) and cloned into
the pGL3-promoter luciferase reporter vector (cat. no. E1761;
Promega Corporation, Madison, WI, USA) to generate the vector
pGL3-wild-type (wt)-Snail1. The hsa-miR199a-5p target site in the
pGL3-wt-Snail1 vector was mutated from 5′-ACACTGG-3′ to
5′-ACATCGG-3′ to construct the mutated reporter vector,
pGL3-mt-Snail1 using a Site Directed Mutagenesis kit (cat. no.
630701; Takara Bio, Inc.). The products of the vectors were
confirmed by DNA sequencing on an ABI 3700 DNA sequencer (Applied
Biosystems; Thermo Fisher Scientific, Inc.). Endotoxin free DNA was
prepared in all cases. The hsa-miR199a-5p mimic
(5′-CCCAGUGUUCAGACUACCUGUUCtt-3′), the hsa-miR199a-5p inhibitor
(5′-GAACAGGUAGUCUGAACACUGGGtt-3′) and NC
(5′-CCCAGUGUUCAGACUACCUGUUCtt-3′) were all chemically synthesized
by Invitrogen (Thermo Fisher Scientific, Inc.).
The number of viable 293TN cells in logarithmic
phase growth were counted using a hemocytometer in conjunction with
trypan blue staining at room temperature for 5 min. Viable cells
were then seeded into 6-well plates (2×105 cells/well)
and maintained in DMEM supplemented with 10% FBS at 37°C for 24 h
in a 5% CO2 atmosphere. The transfection of plasmid DNA
and RNA was performed using Lipofectamine 2000 (cat. no. 11668027;
Invitrogen; Thermo Fisher Scientific, Inc.). Transfection of cells
with 100 ng pGL-TK (cat. no. E6921; Promega Corporation) served as
a reference for luciferase detection. Luciferase activity was
measured using the dual luciferase reporter assay kit (cat. no.
E1901; Promega Corporation) 48 h following transfection in
accordance with the manufacturer's protocol the results of which
were normalized to that of Renilla luciferase. The
luciferase method was also used for observing the inhibition of
hsa-miR489-3p function by AB209371 in MHCC97-H cells. The plasmid
transfection and luciferase activity assay were the same as that in
experiment of validation the target site.
Lentivirus packaging
One day prior to transfection, 293TN cells were
seeded into 10 cm dishes (cat. no. 430167; Corning, Inc., Corning,
NY, USA). pSIH1-siRNA-AB209371 (2 µg) and pPACK Packaging Plasmid
mix (10 µg; cat. no. LV500A-1; System Biosciences LLC) were
co-transfected using Lipofectamine 2000 in accordance with the
manufacturer's protocol. The medium (DMEM with 10% FBS) was
replaced with DMEM with 1% FBS. A total of 48 h later, the
supernatant was harvested, cleared via centrifugation at 5,000 × g
at 4°C for 5 min and passed through a 0.45 µm polyvinylidene
fluoride membrane (cat. no. IPVH00010; EMD Millipore, Billerica,
MA, USA). The titer of virus was determined via gradient dilution.
The packaged lentiviruses were named as Lv-siRNA-AB209371.
Recombinant lentivirus Lv-NC, Lv-Snail1 and Lv-miR199a-3p was
packaged under the same conditions.
Genetic intervention using a
lentiviral approach
The experiment included 5 groups: A control group
(without viral infection), an NC group (infected with Lv-NC), an
AB209371 silencing group (infected with Lv-siRNA-AB209371), an
Snail1 expression group (infected with Lv-Snail1) and an miR199a-5p
expression group (infected with Lv-miR199a-5p). MHCC97-H and
HCC-LM3 cells in logarithmic phase were seeded into 6-well plates
at a density of 5×105 cells/well. Following one day,
viral solution was added at a multiplicity of infection of 10.
Infection efficiency was assessed by observing and analyzing the
level of GFP fluorescence 72 h following infection using a
fluorescence inverted microscope (IX71; Olympus Corporation, Tokyo,
Japan). Infection rate was estimated by dividing the number of
cells expressing GFP by the total number of cells in each view. For
statistics, five views were randomly selected and the mean was
calculated. Total RNA and protein were isolated from the cells and
subjected to RT-qPCR for AB209371 and hsa-miR199a-5p determination,
and western blotting for Snail1 protein expression.
Effect of silencing AB209371 on
hsa-miR199a-5p, Snail1 protein and EMT
Cells were divided into 9 groups: A control group
(without virus infection or induction), a model group (induced by
TGF-β1), an NC combined model group (infected with Lv-NC and
induced by TGF-β1), an AB209371 silencing model group (infected
with Lv-siRNA-AB209371 and induced by TGF-β1), a Snail1 expression
model group (infected with Lv-Snail1 and induced by TGF-β1), an
AB209371 silencing and Snail1 expression model group (infected with
Lv-Snail1 and Lv-siRNA-AB209371, and induced by TGF-β1), a
hsa-miR199a-5p expression model group (infected with Lv-miR199a-5p
and induced by TGF-β1); a hsa-miR199a-5p expression AB209371
silencing model group (infected with Lv-miR199a-5p and
Lv-siRNA-AB209371 and induced by TGF-β1) and a hsa-miR199a-5p
expression AB209371 silencing Snail1 expression model group
(infected with Lv-miR199a-5p, Lv-siRNA-AB209371 and Lv-Snail1, and
induced by TGF-β1). MHCC97-H and HCC-LM3 cells in the logarithmic
phase were seeded into 6-well plates at a density of
5×105 cells/well and viral solution was added following
1 day of normal culture (37°C with 5% CO2) following. An
EMT model was established by adding 10 ng/ml of TGF-β1 protein
(cat. no. ab50036; Abcam, Cambridge, UK) and incubated under normal
conditions (37°C with 5% CO2) for 48 h. Total RNA was
isolated from the cells and subjected to RT-qPCR for AB209371 and
hsa-miR199a-5p determination, and protein was isolated and
subjected to western blotting for Snail1, E-cadherin, Vimentin and
α-SMA protein expression.
Cellular proliferation and migration
assay
MHCC97-H and HCC-LM3 cells were divided into four
groups: A control group (without virus infection or induction), a
model group (induced by TGF-β1), an NC combined model group
(infected with Lv-NC and induced by TGF-β1) and an AB209371
silencing combined model group (infected with Lv-siRNA-AB209371 and
induced by TGF-β1). A total of 48 h following infection, cells were
trypsinized and seeded into 96-well plates at a density of
1×104 cells/well. Cells were cultured under normal
conditions (37°C with 5% CO2) and cell proliferation was
determined using a Cell Counting Kit-8 (CCK-8) assay at 12, 24, 48
and 72 h time points. A total of 10 µl CCK-8 solution (cat. no.
CK-04; Dojindo Molecular Technologies, Inc., Kumamoto, Japan) was
added and cells were cultured under normal conditions (37°C with 5%
CO2) for an additional 4 h prior to the measurement of
absorbance at 450 nm.
Cell migration experiments were performed using the
QCMTM 24-well Fluorimetric Cell Migration Assay kit (cat. no.
ECM508; Chemicon International; Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol. The kit uses an insert
polycarbonate membrane with an 8-µm pore size. The insert was
coated with a thin layer of EC Matrix™ (part of the
migration assay kit) that occluded membrane pores and blocked the
migration of non-invasive cells. RPMI-1640 medium (500 µl)
supplemented with 10% FBS was used as chemoattractant. Cells that
migrated and invaded the underside of the membrane were fixed with
4% paraformaldehyde at room temperature for 24 h. Invading cells
were stained at room temperature for 10 min with 0.5% crystal
violet (dilution ratio, 1:10) and counted via fluorescence and
reported as relative fluorescence units (RFUs). The grouping and
cell treatments were the same as aforementioned in the cell
viability assay. A total of 72 h following lentiviral infection,
cells were trypsinized. Viable cells were counted via 0.4% trypan
blue (dilution ratio, 1:10) staining at room temperature for 5 min
using a fluorescence inverted microscope (IX73; Olympus
Corporation, Tokyo, Japan; Amplification factor, 1:160), seeded
into the upper chamber of the transwell equipment at a density of
5×105 cells/well, incubated under normal conditions
(37°C and 5% CO2) for 48 h, with RPMI-1640 medium
containing 20% FBS plated in the lower chamber. The experiment was
performed with MHCC97-H and HCC-LM3 cells at the same time.
RT-qPCR
Total RNA of tissues and HCC cell lines were
isolated using the Trizol Reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. Samples
were then reverse transcribed into cDNA using the M-MLV Reverse
Transcriptase kit (cat. no. 2640; Takara Biotechnology Co., Ltd.)
and oligo(dT)18 primer. The following specific primers were used:
AB209371 forward, 5′-TTCCAGTGACTCCACGTGC-3′ and reverse,
5′-AACTTTGGGCCTGTGCCGAAGGGT-3′; Snail1 forward,
5′-TTACCTTCCAGCAGCCCTACG-3′ and reverse,
5′-AGGTCAGCTCTGCCACCCTGG-3′; β-actin forward
5′-CCTGTACGCCAACACAGTGC-3′ and reverse, 5′-ATACTCCTGCTTGCTGATCC-3′.
The lengths of the amplified products were 219, 183 and 211 bp,
respectively. RT-qPCR was performed using a SYBR Premix Ex
Taq kit (cat. no. RR420A; Takara Biotechnology Co., Ltd.) and the
TP800 System (Takara Biotechnology Co., Ltd.). cDNA from 200 ng
total RNA was used as the template. The thermocycling conditions
for PCR were as follows: 40 cycles of denaturation at 95°C for 10
sec, annealing at 60°C for 20 sec and elongation at 72°C for 20
sec. The mRNA levels of Snail1 and AB209371 were normalized to the
expression of endogenous β-actin using the 2−∆∆Cq method
(10). For each sample, triplicate
determinations were performed and the mean values were utilized for
further analysis. To assess hsa-miR199a-5p, total RNA (2 µg) was
used for cDNA preparation using the M-MLV reverse transcription kit
(cat. no. 2640; Takara Biotechnology Co., Ltd.) in accordance with
the manufacturer's protocol. The specific primers used were as
follows: U6 snRNA: 5′-TACCTTGCGAAGTGCTTAAAC-3′ and hsa-miR199a-5p:
5′-GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACGCTGCC-3′. The
following PCR primers were utilized: U6 forward,
5′-GTGACATCACATATACGGCAGC-3′ and reverse,
5′-GTCGTATCCAGTGCGTGTCGTG-3′; hsa-miR199a-5p forward,
5′-GTGCTCGCTTCGGCAGCACAT-3′ and reverse,
5′-TACCTTGCGAAGTGCTTAAAC-3′. Takara SYBR Premix Ex Tap was utilized
for the PCR procedure with the following thermocycling conditions:
40 cycles of denaturation at 95°C for 10 sec, annealing at 60°C for
20 sec and extension at 72°C for 20 sec. The relative level of
hsa-miR199a-5p was analyzed using the 2−∆∆Cq method,
with all values being normalized to that of U6 (10).
Detection of protein contents in cells
and tissues
Total protein was extracted from HCC cells or
tissues using the M-PER mammalian protein extraction reagent (cat.
no. 78501; Pierce; Thermo Fisher Scientific, Inc.) and the T-PER
tissue protein extraction reagent (cat. no. 78510; Pierce; Thermo
Fisher Scientific, Inc.), respectively. Protein was determined
using the bicinchoninic acid protein assay kit (cat. no. A53227;
Pierce; Thermo Fisher Scientific, Inc.). Equal quantities of
protein (15 µg/lane) were then loaded onto 11% SDS-PAGE gels and
transferred onto nitrocellulose membranes. Membranes were
subsequently blocked with 5% bovine serum albumin (cat. no. 37520;
Thermo Fisher Scientific, Inc.) in tris-buffered saline at and room
temperature for 1 h and probed with monoclonal antibodies against
human Snail1 (cat. no. sc-271977; 1:400), Vimentin (cat. no.
sc-6260; 1:300), α-SMA (cat. no. sc-53015; 1:200), E-cadherin (cat.
no. sc-8426; 1:450), MMP-9 (cat. no. sc-13520; 1:400) and β-actin
(cat. no. sc-81178; 1:1,000; all, Santa Cruz Biotechnology, Inc.,
Dallas, TX, USA) at 4°C for 12 h. Samples were then treated with
secondary horseradish peroxidase-conjugated anti-mouse and rabbit
antibodies (cat. nos. sc-516102 and sc-2370; Santa Cruz
Biotechnology, Inc.; 1:3,000) at 4°C for 4 h. Following washing,
the bands were detected via chemiluminescence (Pierce; Thermo
Fisher Scientific, Inc.) and imaged with X-ray films (Kodak,
Rochester, NY, USA). β-actin was used as an endogenous reference
for normalization. Relative optical densities were analyzed using
image processing software Total Lab v1.10 (Total Laboratory
Services Ltd., Blandford Forum, UK).
Statistical analysis
Data are presented as the mean ± standard deviations
of three independent experiments. All statistical data were
analyzed using SPSS GradPack version 20.0 statistical software (IBM
Corp., Armonk, NY, USA) and GraphPad Prism 7.0 software (GraphPad
Software, Inc., La Jolla, CA, USA). Comparisons between groups were
made using the Student's t-test. P<0.05 was considered to
indicate a statistically significant difference.
Results
Assessment of AB209371,
hsa-miR199a-5p, Snail1 mRNA and protein levels in primary HCC and
metastatic HCC tissues
The relative levels of AB209371 and hsa-miR199a-p in
primary and metastatic HCC tissues were significantly increased
compared with adjacent normal tissue (P<0.01 and P<0.05).
Furthermore, compared with primary HCC tissues, these were
expressed at higher levels in HCC metastatic tissue (P<0.05;
Fig. 1A). Additionally, Snail1
protein was expressed at higher levels in primary and metastatic
HCC tissue compared with adjacent normal tissue (P<0.01), with
an increased expression in HCC metastatic tissue compared with that
of primary HCC tissue (P<0.05). The results of Snail1 mRNA
assessment demonstrated that there were no marked differences
between the four groups of tissue (Fig.
1B). The present study detected EMT-associated proteins in
primary and metastatic HCC tissue, and in adjacent tissue. The
results revealed an increase in vimentin, α-SMA and MMP-9 in
primary and metastatic HCC tissue compared with adjacent tissues
(P<0.01 in vimentin and α-SMA, P<0.05 in MMP-9), with the
increase in metastatic tissue being more significant than the
primary HCC tissue (P<0.01 in vimentin and α-SMA, P<0.05 in
MMP-9). The reverse trend was observed in E-cadherin in the three
tissues (Fig. 1C).
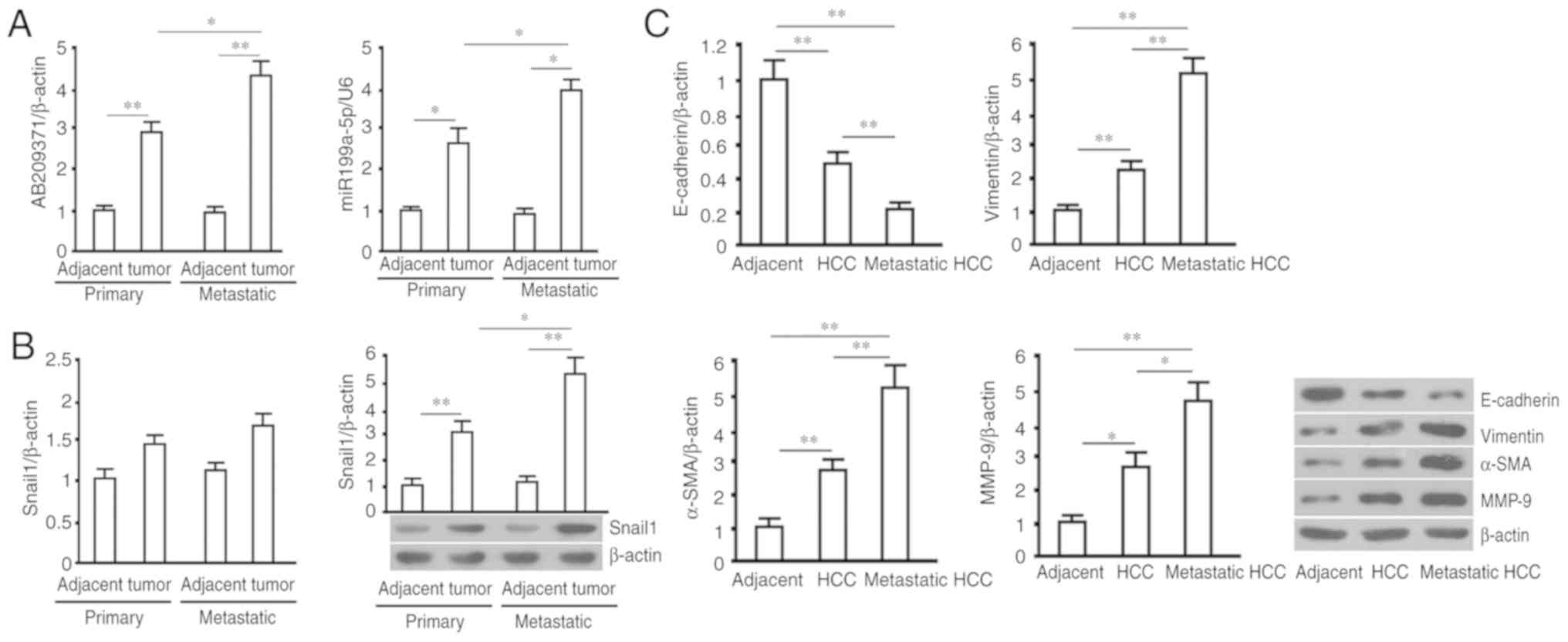 | Figure 1.RT-qPCR (AB209371, hsa-miR199a-5p and
Smail1 mRNA) and western blot analysis (Sanil1, E-cadherin,
Vimentin, α-SMA and MMP-9) was performed using primary and
metastatic HCC tumors, and adjacent normal tissue. (A) Relative
levels of AB209371 and hsa-miR199a-5p, with β-actin and U6 as
internal controls. (B) Snail1 mRNA and mRNA and protein levels
determined via RT-qPCR and western blotting, respectively. β-actin
was utilized as the internal control. (C) Protein levels of
E-cadherin (116 kDa), Vimentin (55 kDa), α-SMA (37 kDa) and MMP-9
(92 kDa) were determined via western blotting and subsequent
quantification with β-actin (43 kDa) as the internal control. All
data are expressed as the mean ± standard deviations (sample size,
n=20). *P<0.05 and **P<0.01. RT-qPCR, reverse
transcription-quantitative polymerase chain reaction; miR, miRNA;
α-SMA, α-smooth muscle actin; MMP-9, matrix metallopeptidase-9;
HCC, hepatocellular carcinoma. |
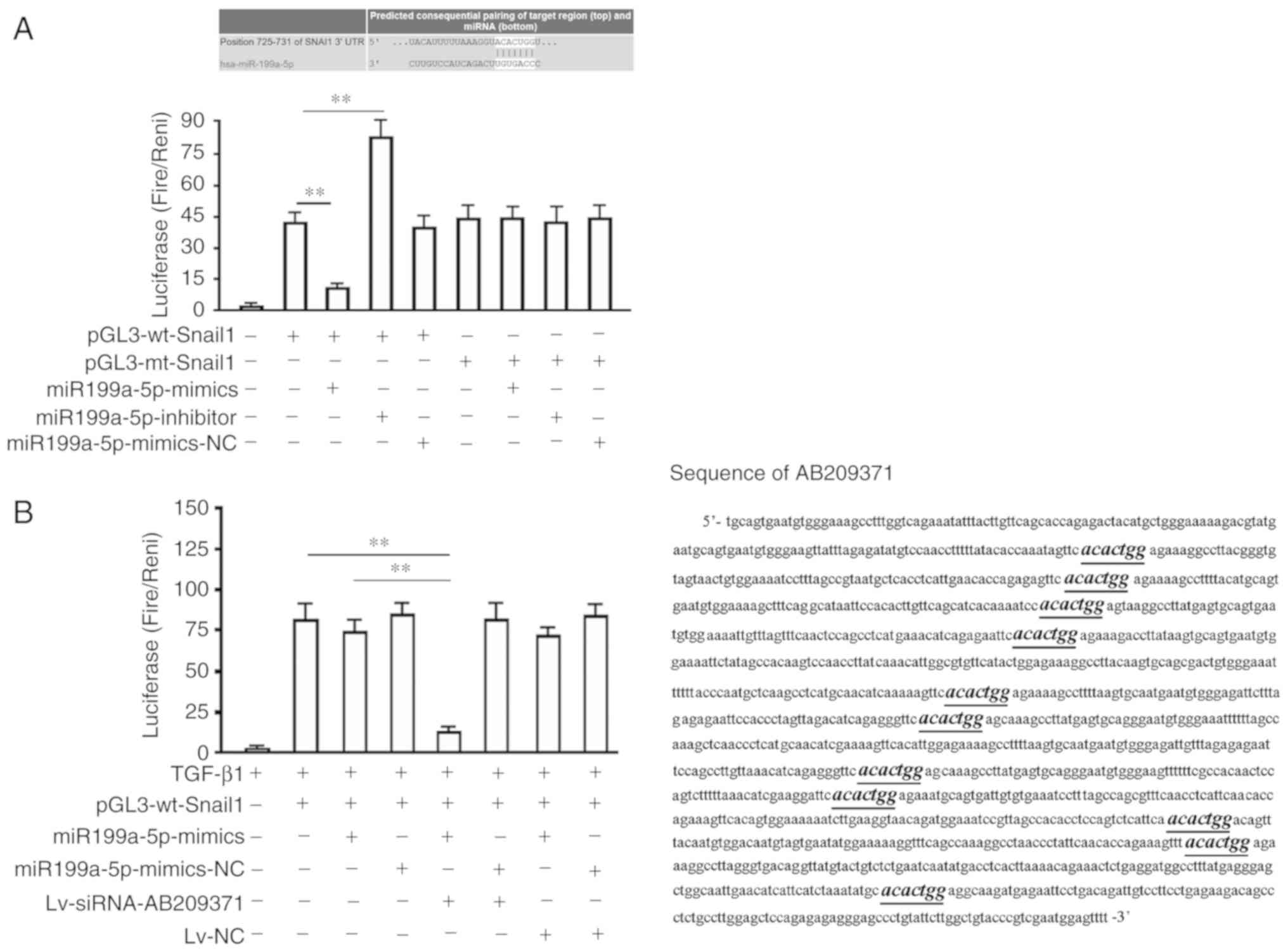
hsa-miR199a-5p binds to the Smail1
3′-UTR, which is influenced by AB209371
Bioinformatics analysis identified a seven-based
hsa-miR199a-5p seed sequence in the 3miR199a-5Snail1 mRNA. The
present study therefore constructed luciferase reporter vectors to
verify whether this site represents a valid hsa-miR199a-5p target.
The reporter vectors contained wild-type Snail1 three vectors in
which the hsa-miR199a-5p target site within the 3′-UTR had been
mutated. Each reporter construct expressed luciferase at a high
level when compared with the untransfected group. However,
miR199a-5p mimics significantly inhibited the luciferase activity
of cells transfected with the reporter vector encoding the
wild-type 3ithin the 3′-UTR had been mutated (P<0.01; Fig. 2A). Furthermore, the miR199a-5p
inhibitor significantly increased the luciferase activity of
pGL3-wt-Snail1 transfected cells (39.55±3.01 vs. 81.22±10.85;
P<0.01; Fig. 2A). Conversely, in
cells transfected with the reporter vector encoding the mutated
hsa-miR199a-5p target site (pGl3-mt-Snail1), neither the miR199a-5p
mimics nor the miR199a-5p inhibitor demonstrated any marked effect
on the luciferase activity. Additionally, the co-transfection of
miR199a-3p-NC (non-targeting control) had no effect on the
luciferase activity of either of vector (Fig. 2A). miR199a-5p mimics were also
revealed to lose their ability to inhibit luciferase expressed by
the wt luciferase reporter vector in MHCC97-H cells (Fig. 2B). However, the inhibitive ability
of mimics was regained following AB209371 silencing. In addition,
the theoretical binding sites of hsa-miR199a-5p and AB209371 were
predicted using the bioinformatics software ‘TargetScan’. The
results revealed that there were 11 theoretical binding sites of
hsa-miR199a-5p on AB209371, which were distributed along the length
of the 700 bp sequence (Fig
2B).
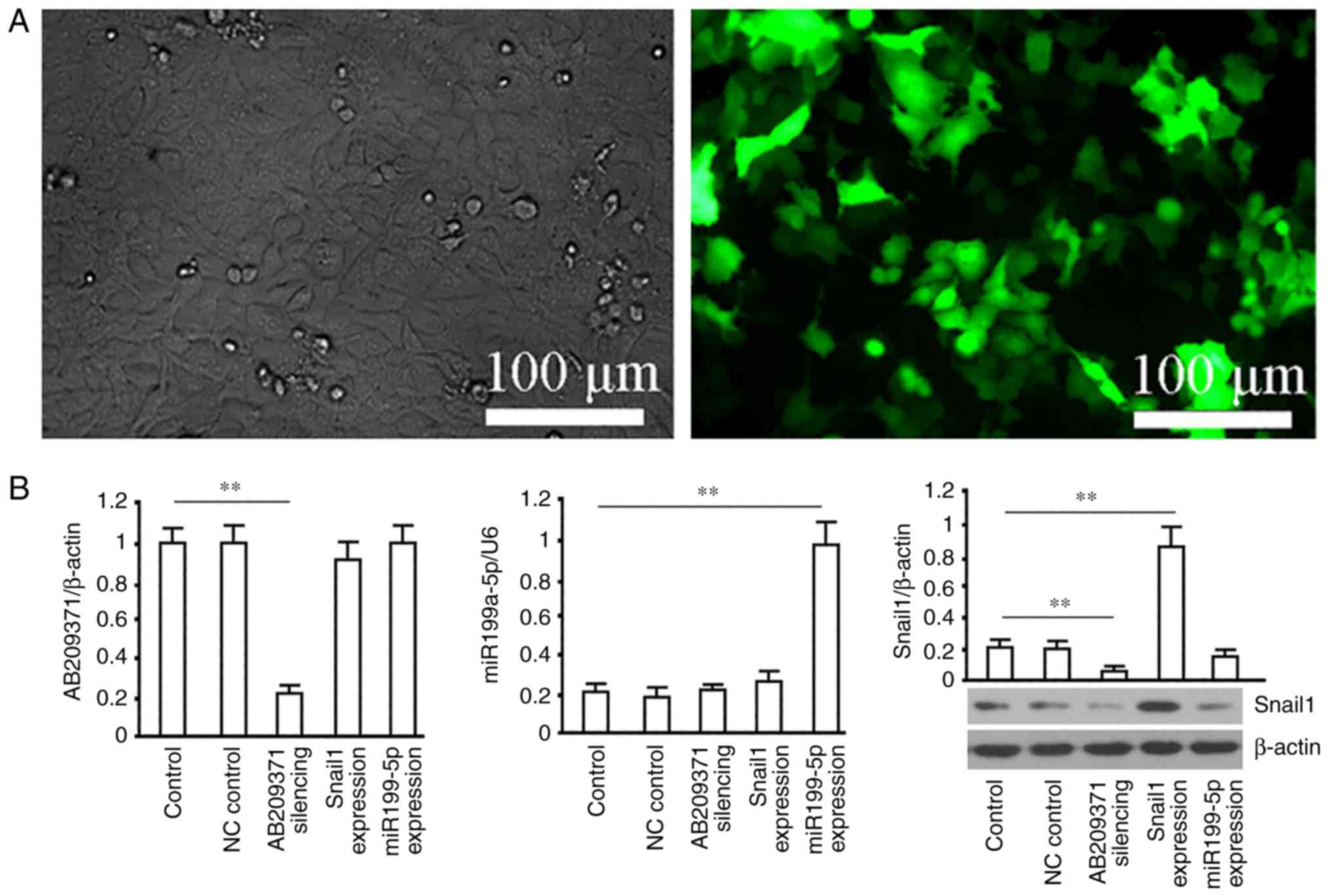
Effect of silencing AB209371 using the
lentiviral approach on hsa-miR199a-5p and Snail1 protein in HCC
cells
Recombinant lentiviruses (Lv-NC, Lv-siRNA-AB209371,
Lv-Snail1 and Lv-miR199a-5p) were used to infect MHCC97-H cells.
The results revealed that GFP was detected in the majority of cells
72 h following infection and the proportion of GFP-expressing cells
indicated that the efficiency of gene delivery was >95% in
MHCC97-H cells (Fig. 3A).
Furthermore, AB209371 was significantly decreased following
Lv-siRNA-AB209371 transfection (P<0.01 vs. the control group).
However, no change was observed in cells infected with Lv-Snail1 or
Lv-miR199a-5p. Additionally, miR199a-5p was significantly increased
by Lv-miR199a-5p (P<0.01 vs. the control group), but no change
was observed in cells transfected with Lv-Snail1 or Lv-AB209371.
Snail1 protein levels were also significantly increased following
Lv-Snail1 transfection and decreased following Lv-siRNA-AB209371
transfection (P<0.01 vs. the control group). No change was
detected in cells infected with Lv-miR199a-5p (Fig 3B). The results from another HCC cell
line, HCC-LM3, were the same as those for MHCC97-H (data not
shown).
AB209371 silencing inhibits EMT in
TGF-β1 induced HCC cells
To verify that AB209371 silencing inhibits EMT in
HCC cells by restoring the negative regulation of Snail1 by
hsa-miR199a-5p, an in vitro EMT model was constructed using
10 ng/ml TGF-β1 for induction. The results revealed that, following
exposure to 10 ng/ml TGF-β1 for 48 h, vimentin and α-SMA levels
were increased, and E-cadherin was decreased in MHCC97-H cells,
clearly indicating EMT (Fig. 4A).
An RT-qPCR assay and western blotting also revealed that AB209371
silencing reduced the TGF-β1-induced increase in AB209371 and
Snail1 protein (P<0.01), but had no effect on hsa-miR199a-5p.
Furthermore, the overexpression of hsa-miR199a-5p increased
hsa-miR199a-5p and Snail1 protein levels in TGF-β1-induced MHCC97-H
cells, but had no marked effect on AB209371. Additionally, the
overexpression of Snail1 increased the expression of TGF-β1 induced
Snail1 in MHCC97-H cells (P<0.05), but did not affect AB209371
and hsa-miR199a-5p (Fig. 4A and B).
Western blotting for EMT marker proteins revealed that the
overexpression of hsa-miR199a-5p had no significant effect on the
EMT of MHCC97-H cells induced by TGF-β1. However, the
overexpression of hsa-miR199a-5p combined with AB209371 silencing
significantly inhibited EMT. This affect was completely reversed by
overexpression of Snail1 (Fig.
4A).
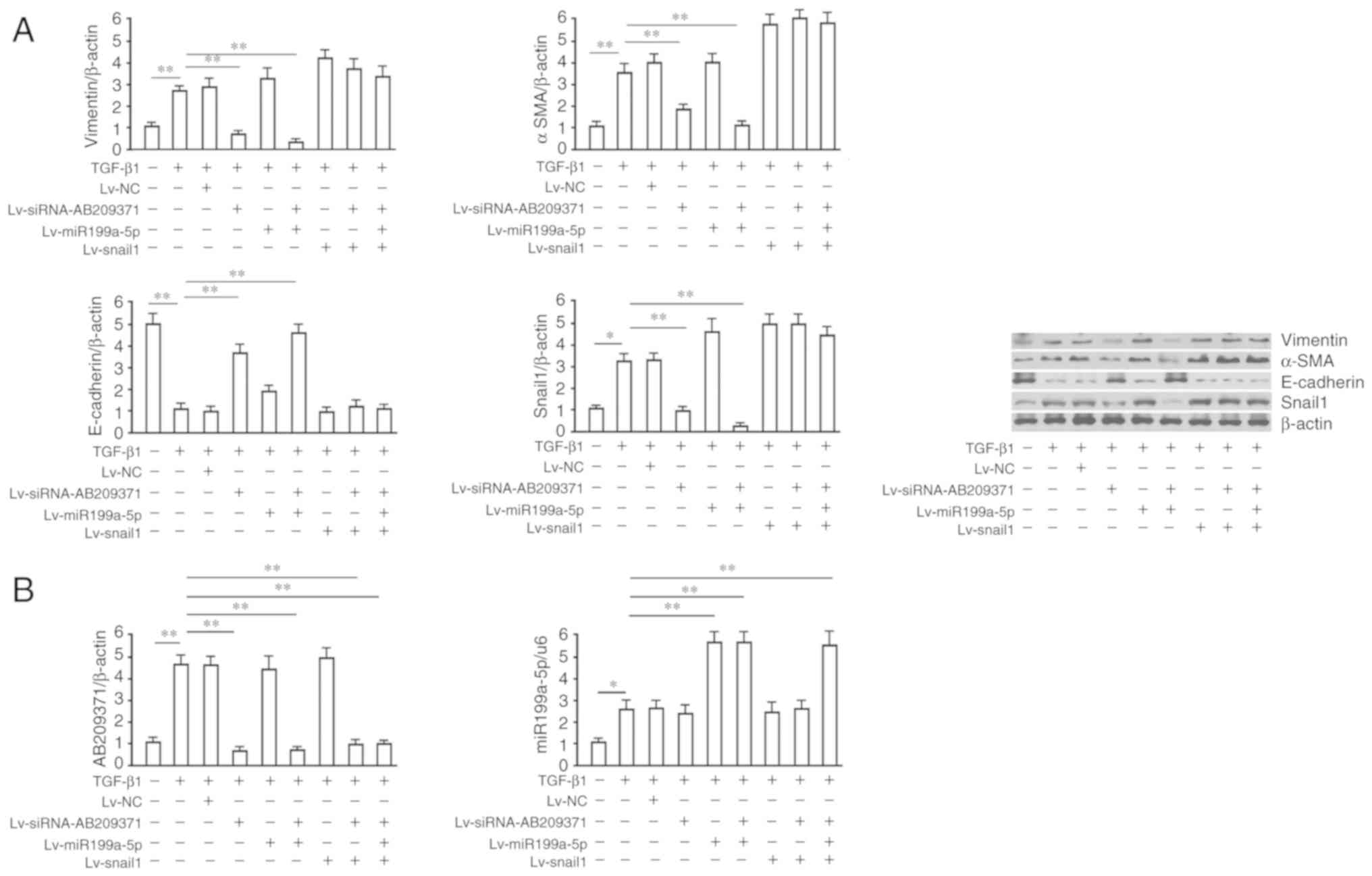 | Figure 4.AB209371 silencing inhibits
hepatocellular carcinoma cell EMT induced by TGF-β1 via the
restoration of Snail1 inhibition by hsa-miR199-5p. (A) Western
blotting of EMT markers, E-cadherin (116 kDa), Vimentin (55 kDa),
α-SMA (37 kDa) and Snail1 (34 kDa) protein. A total of 24 h
following MHCC97-H transfection with recombinant viruses, cells
were induced with TGF-β1 (10 ng/ml) for a further 48 h. β-actin was
utilized as a loading control. (B) Determination of AB209371 and
hsa-miR199a-5p levels via reverse transcription-quantitative
polymerase chain reaction. Data are representative of at least
three independent experiments and are presented as the mean ±
standard deviation. **P<0.01 and *P<0.05. EMT,
epithelial-mesenchymal transition; TGF-β1, transforming growth
factor-β1; miR, miRNA; α-SMA, α-smooth muscle actin; NC, negative
control; siRNA, small interfering RNA. |
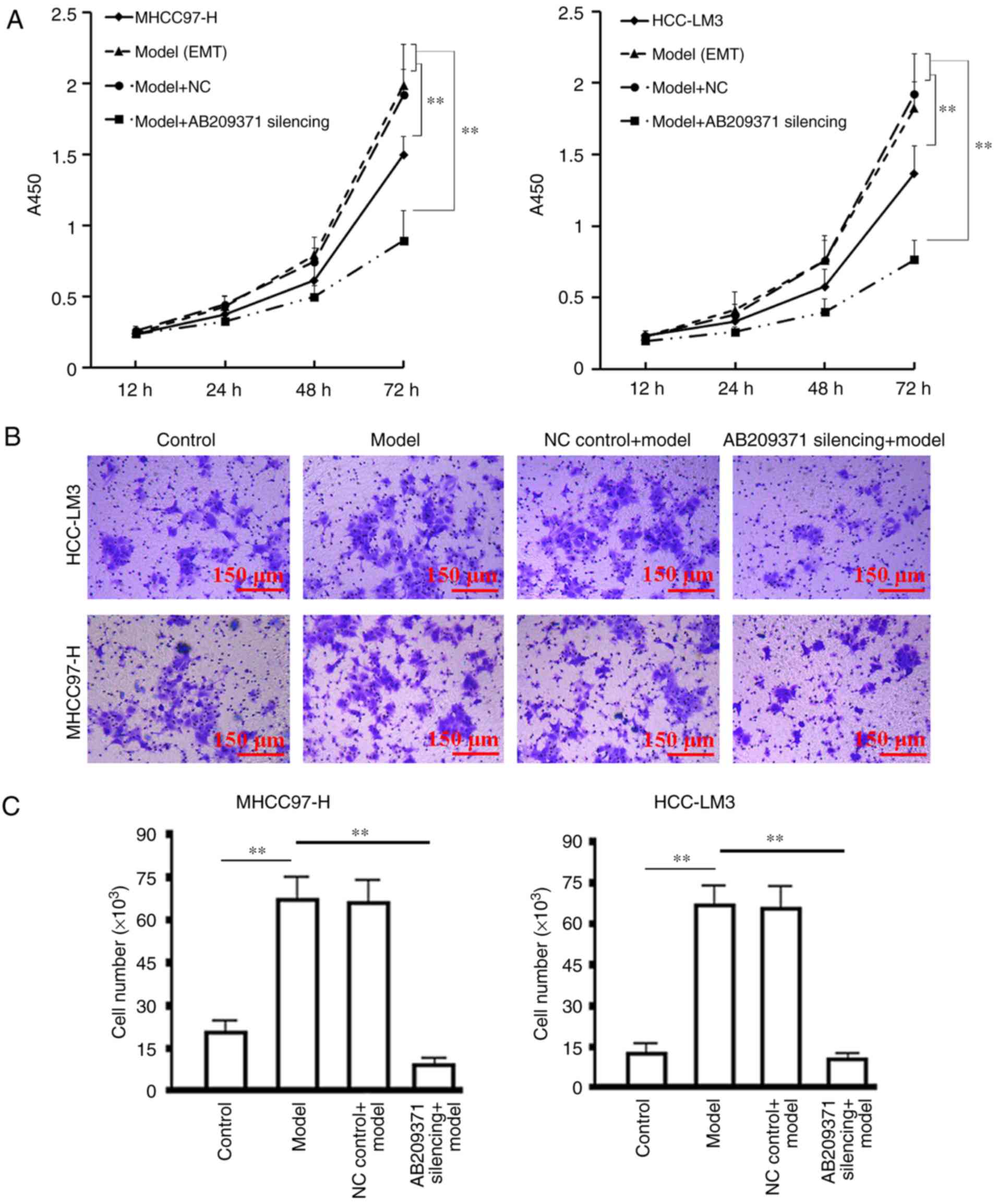
Effect of AB209371 silencing on the
proliferation and migration of HCC cells induced by TGF-β1
The results of the cell proliferation assay revealed
that TGF-β1 induction significantly enhanced cell proliferation
compared with the control group (P<0.01). Furthermore, AB209371
silencing effectively inhibited the proliferation of MHCC97-H and
HCC-LM3 cells induced by TGF-β1 in the logarithmic growth phase
(P<0.01 vs. the model or NC control model group; Fig. 5A). The migration assay demonstrated
that TGF-β1 induction significantly enhanced the migration of the
two cell lines compared with the control group (P<0.01).
Furthermore, AB209371 silencing effectively inhibited cell
migration (P<0.01 vs. all other groups induced by TGF-β1), which
was demonstrated by the decreased number of cells that passed
through the basement membrane (Fig.
5B).
Discussion
LncRNA is a class of natural DNA transcriptional
products comprising >200 bases (11). LncRNA has also been determined to
serve an important role in the proliferation and regulation of
certain types of tumor (12,13).
Studies have revealed that lncRNA and miRNA interact to regulate
tumor metastasis. LncRNA can also regulate miRNAs by serving as an
endogenous miRNA inhibitor that suppresses miRNA function and thus
affects the malignant biological behavior of cancer cells (14). LncRNAs participate in the promotion
of HCC initiation and metastasis progression, as revealed by
previous studies (15,16). The invasion and metastasis of cancer
involves cancer cells breaking away from the primary tumor site,
transferring to discrete tissues or remote organs and proliferating
into cancer cells of the same type. This process is dependent on
the interaction of cancer cells and the internal environment, which
may promote their survival, growth angiogenesis, invasion and
metastasis (17,18). Therefore, EMT is important for the
invasion and metastasis of cancer cells (19–21).
The EMT of HCC cells is closely associated with HCC metastasis and
post-operative metastasis (22,23).
However, the lncRNAs that are involved in the EMT of HCC cells are
yet to be fully determined. Furthermore, to the best of our
knowledge, the biological role and underlying mechanism of AB209371
in the EMT of HCC has not yet been reported.
As a transcription factor, Snail1 functions by
binding to the E-box element of the 5′-CANNTG-3′ core base sequence
(N=A, T, C or G) in the promoter region of many tumor-associated
genes (24,25). Snail1 can regulate the EMT of
various tumors by promoting or inhibiting the transcription of
E-cadherin, interleukin-8 and cluster of differentiation 147
(26,27). To verify whether Snail1 contributes
to HCC metastasis, the present study tested primary and metastatic
HCC samples, and adjacent normal tissues. The results revealed that
the expression of Snail1 protein was significantly increased in
primary or metastatic HCC tissues compared with adjacent tissues.
The results also indicated that the underlying cause of this
increased expression was the decrease in hsa-miR199a-5p content.
The present study therefore hypothesized that a further decrease in
hsa-miR199a-5p may further increase Snail1 protein expression,
resulting in EMT. The present study hypothesizes that there is a
mechanism which may explain these results and the results which
demonstrated the weakened negative regulation of hsa-miR199a-5p on
the Snail1 protein in the process of metastasis of HCC. The results
of the present study indicate that the inactivation process of
hsa-miR199a-5p starts from the initial stage of EMT or HCC
metastasis. Based on data from previous studies, the present study
hypothesizes that lncRNA may be the key factor (28), that is, a group of miRNAs are
elevated and the binding of has-miR199a-5p to the 3′-UTR region of
the Snail1 gene is inhibited by competitive binding. Screening was
carried out based on two aspects. The present study hypothesized
that the expression of hsa-miR199a-5p and Snail1 protein in HCC and
its metastatic tissues revealed the following: i) hsa-miR199a-5p
was elevated in metastatic tissues of HCC and may not effectively
inhibit Snail1 protein expression; ii) the increased expression of
Snail1 protein in HCC metastasis may be due to post-transcriptional
regulation inactivation (with no change in mRNA). Based on these
two facts combined with existing theories that lncRNA regulates
miRNA to serve as endogenous miRNA inhibitor to suppress miRNA
function (8), the present study
hypothesizes that the inactivation of hsa-miR199a-5p may be caused
by an increase in the content of a lncRNA and that this lncRNA must
have two sets of characteristics: i) Its content in the metastatic
tissues of HCC is higher than that in primary HCC; ii) it
effectively inhibits the binding of hsa-miR199a-5p to the 3′-UTR of
the Snail1 gene.
The experiments of the present study revealed that
AB209371 was increased in primary and metastatic HCC compared with
adjacent normal tissues, and was also increased to a greater degree
in metastatic HCC tissues compared with primary HCC tissues,
indicating that there may be an association among AB209371, EMT and
HCC metastasis. The results of bioinformatics indicated that there
was a 7-base seed region of hsa-miR199a-5p on the 3′-UTR of Snail1
and that there were 11 seeding regions on the 800-base AB209371. As
a result, the present study hypothesized that elevated AB209371
bound to hsa-miR199a-5p as a miRNA inhibitor, disabling the
negative regulation of Snail1 by hsa-miR199a-5p, meaning that
Snail1 expression was increased, resulting in in HCC EMT. LncRNAs,
as a competing endogenous RNAs, interact with miRNAs to regulate
target genes and as such serve important roles in the EMT of HCC
cells and the metastasis of HCC (29,30).
The present study concluded that AB209371 silencing restores the
negative regulation of Snail1 by hsa-miR199a-5p to inhibit cancer.
This conclusion was based on the following results: i)
hsa-miR199a-5p negatively regulated Snail1 by binding to its
3′-UTR; ii) AB209371 silencing inhibited EMT in HCC, which was
reversed by the overexpression of Snail1; iii) the overexpression
of hsa-miR199a-5p exhibited no marked effect on HCC EMT, but did
inhibit EMT when AB209371 was silenced. Considering that Snail1
overexpressed by the lentiviral system did not contain the
wild-type 3′-UTR, it would not be affected by miRNA. Therefore, the
present study hypothesizes that a hsa-miR199a-5p/Snail1 pathway
regulates HCC EMT. The upregulation of hsa-miR199a-5p is considered
to be an intrinsic antagonistic response to HCC metastasis and
elevated AB209371 may be the reason why the inhibition of HCC
metastasis is unable to occur. The present study established an EMT
model of HCC cell lines via induction with TGF-β1 to clarify the
effect of AB209371 on the EMT of HCC cells. The present study
confirmed that AB209371 silencing effectively restored
hsa-miR199a-5p inhibition of hepatoma cell EMT and HCC metastasis.
Furthermore, AB209371 silencing in combination with hsa-miR199a-5p
overexpression was an effective method for preventing HCC
metastasis by inhibiting the EMT of HCC cells. These results may
aid the development of clinical personalized treatments for
patients with HCC in the future. In addition, hsa-miR199a-5p was
determined to negatively regulate Snail1 protein expression in
several groups of stem cells and was inactivated following
TGF-β1-induction. This outcome further confirms that TGF-β1
induction leads to an increase in the AB209371 content of HCC
cells, which may be the key to hsa-miR199a-5p inactivation.
Although the present study systematically assessed
the associations of AB209371, hsa-miR199a-5p and Snail1, further
study is required to determine how the elevation of AB209371 in HCC
cells occurs. However, The present study demonstrated that lncRNA
may serve as a link between the EMT of HCC cells and the metastasis
of HCC. The present study demonstrated for the first time that
AB209371 silencing suppresses the EMT of HCC cells by restoring the
negative regulation of Snail1 by hsa-miR199a-5p. These results may
lay a solid theoretic base for the use of lncRNA as a target of
genetic therapy for HCC metastasis and invasion.
Acknowledgements
The authors would like to thank Professor HaiXin
Qian for his guidance in the design and implementation of the
present study.
Funding
The present study was supported by the Jiangsu
Provincial Scientific Research of 333 Project (grant no.
BRA2018251) and The Yancheng Medical Science Development Foundation
(grant no. YK2017007).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
RF, CX, XW and WC supervised the present study. CX,
XW, HY, XC, XS and YM performed the experiments. RF, CX, XW and WC
collected the data, performed data analysis and wrote the
manuscript. All authors read and approved the manuscript and agree
to be accountable for all aspects of the research in ensuring that
the accuracy or integrity of any part of the work are appropriately
investigated and resolved.
Ethics approval and consent to
participate
The study was reviewed and approved by the Yancheng
City First People's Hospital Institutional Review Board (Yancheng,
China).
Patient consent for publication
Written informed consent was obtained from all
patients for the publication of this article and accompanying
datasets.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Mulcahy MF: Management of hepatocellular
cancer. Curr Treat Options Oncol. 6:423–435. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bosch FX, Ribes J, Díaz M and Cléries R:
Primary HCC: Worldwide incidence and trends. Gastroenterology. 127
(Suppl 1):S5–S16. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
El-Serag HB: Hepatocellular carcinoma. N
Engl J Med. 365:1118–1127. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yang H, Lin M, Xiong F, Yang Y, Nie X,
McNutt MA and Zhou R: Combined lysosomal protein transmembrane 4
beta-35 and argininosuccinate synthetase expression predicts
clinical outcome in hepatocellular carcinoma patients. Surg Today.
41:810–817. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chen JS, Li HS, Huang JQ, Zhang LJ, Chen
XL, Wang Q, Lei J, Feng JT, Liu Q and Huang XH: Down-regulation of
Gli-1 inhibits hepatocellular carcinoma cell migration and
invasion. Mol Cell Biochem. 393:283–291. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kaufhold S and Bonavida B: Central role of
Snail1 in the regulation of EMT and resistance in cancer: A target
for therapeutic intervention. J Exp Clin Cancer Res. 33:622014.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Abdelmohsen K and Gorospe M: Noncoding RNA
control of cellular enescence. Wiley Interdiscip Rev RNA.
6:615–629. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Alaei-Mahabadi B and Larsson E: Limited
evidence for evolutionarily conserved targeting of long non-coding
RNAs by microRNAs. Silence. 4:42013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Schmitt AM and Chang HY: Long Noncoding
RNAs in Cancer Pathways. Cancer Cell. 29:452–463. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2−ΔΔCT method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu H, Luo J, Luan S, He C and Li Z: Long
non-coding RNAs involved in cancer metabolic reprogramming. Cell
Mol Life Sci. 76:495–504. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bergmann JH and Spector DL: Long
non-coding RNAs: Modulators of nuclear structure and function. Curr
Opin Cell Biol. 26:10–18. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Huang JF, Guo YJ, Zhao CX, Yuan SX, Wang
Y, Tang GN, Zhou WP and Sun SH: Hepatitis B virus X protein
(HBx)-related long noncoding RNA (lncRNA) down-regulated expression
by HBx (Dreh) inhibits hepatocellular carcinoma metastasis by
targeting the intermediate filament protein vimentin. Hepatology.
57:1882–1892. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Noh JH and Gorospe M: AKTions by
cytoplasmic lncRNA CASC9 promote hepatocellular carcinoma
survival. Hepatology. 68:1675–1677. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wu LL, Cai WP, Lei X, Shi KQ, Lin XY and
Shi L: NRAL mediates cisplatin resistance in hepatocellular
carcinoma via miR-340-5p/Nrf2 axis. J Cell Commun Signal.
13:99–112. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tsang WP, Wong TW, Cheung AH, Co CN and
Kwok TT: Induction of drug resistance and transformation in human
cancer cells by the noncoding RNA CUDR. RNA. 13:890–898. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kulesa PM and McLennan R: Neural crest
migration: Trailblazing ahead. F1000Prime Rep. 7(02)2015.PubMed/NCBI
|
|
19
|
Maritzen T, Schachtner H and Legler DF: On
the move: Endocytic trafficking in cell migration. Cell Mol Life
Sci. 72:2119–2134. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Deng B, Zhang S, Miao Y, Zhang Y, Wen F
and Guo K: Down-regulation of Frizzled-7 expression inhibits
migration, invasion, and epithelial-mesenchymal transition of
cervical cancer cell lines. Med Oncol. 32:1022015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yuan W, Yuan Y, Zhang T and Wu S: Role of
Bmi-1 in regulation of ionizing irradiation-induced
epithelial-mesenchymal transition and migration of breast cancer
cells. PLoS One. 10:e01187992015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bao YX, Cao Q, Yang Y, Mao R, Xiao L,
Zhang H, Zhao HR and Wen H: Expression and prognostic significance
of golgiglycoprotein73 (GP73) with epithelial-mesenchymal
transition (EMT) related molecules in hepatocellular carcinoma
(HCC). Diagn Pathol. 8:1972013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang H, Song Y, Zhou C, Bai Y, Yuan D,
Pan Y and Shao C: Blocking endogenous H2S signaling
attenuated radiation-induced long-term metastasis of residual HepG2
cells through inhibition of EMT. Radiat Res. 190:374–384. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Nieto MA: The snail super family of
zinc-finger transcription factors. Nat Rev Mol Cell Biol.
3:155–166. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
25
|
Cheng CW, Wu PE, Yu JC, Huang CS, Yue CT,
Wu CW and Shen CY: Mechanisms of inactivation of E-cadherin in
breast carcinoma: Modification of the two-hit hypothesis of turn
out suppressor gene. Oncngene. 20:3814–3823. 2001. View Article : Google Scholar
|
|
26
|
Argast GM, Krueger JS, Thomson S,
Sujka-Kwok I, Carey K, Silva S, O'Connor M, Mercado P, Mulford IJ,
Young GD, et al: Inducible expression of TGFβ, Snail and Zeb1
recapitulates EMT in vitro and in vivo in a NSCLC model. Clin Exp
Metastasis. 28:593–614. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Barnett P, Arnold RS, Mezencev R, Chung
LW, Zayzafoon M and Odero-Marah V: Snail-mediated regulation of
reactive oxygen species in ARCa P human prostate cancer cells.
Biochem Biophys Res Commun. 404:34–39. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lanzafame M, Bianco G, Terracciano LM, Ng
CKY and Piscuoglio S: The role of long non-coding RNAs in
hepatocarcinogenesis. Int J Mol Sci. 19:E6822018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chen L, Wang W, Cao L, Li Z and Wang X:
Long non-coding RNA CCAT1 acts as a competing endogenous RNA to
regulate cell growth and differentiation in acute myeloid leukemia.
Mol Cells. 39:330–336. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Karreth FA and Pandolfi PP: ceRNA
cross-talk in cancer: When ce-bling rivalries go awry. Cancer
Discov. 3:1113–1121. 2013. View Article : Google Scholar : PubMed/NCBI
|