Introduction
Glioma, the most common malignant primary brain
tumor in adults, is highly invasive and has a poor prognosis
(1). Of the 4 classes of glioma
classified by the World Health Organization (WHO), the highest
grade (grade IV) is glioblastoma multiforme (GBM) (2). Despite recent advances in our
understanding of the biology of GBM and extensive efforts to
develop new therapeutic options (3), the prognosis is dire, and the median
survival time of 15 months has not changed over the last 20 years
(4). Therefore, there is an urgent
need to identify novel diagnostic biomarkers and potential
therapeutic targets for this devastating disease.
MicroRNAs (miRNAs) are a class of small (~18–25
nucleotides) non-coding single-stranded RNAs (5), and are increasingly recognized to play
key regulatory roles in numerous physiological processes in plants
and animals (6). miRNAs are crucial
regulators of gene expression, and function by binding to partially
complementary sequences in the 3′ untranslated region (3′UTR) of
mRNAs, thereby preventing their translation and/or degradation
(7–10). Substantial evidence implicates
dysregulated expression and/or function of miRNAs in promoting or
inhibiting the development and progression of cancer, suggesting
they can act as tumor suppressors or oncogenes (11–13).
Among the cancer-related processes influenced by aberrant miRNA
expression are cell apoptosis, proliferation, invasion, and
resistance to chemotherapeutic agents (14–17).
One of the first miRNAs to be described was miR-21 originally
discovered in Caenorhabditis elegans, which is aberrantly
expressed in many human cancers and contributes to the malignant
phenotype by targeting critical tumor suppressor genes (18). Although there is increasing
recognition of the potential diagnostic and therapeutic utility of
miRNAs in cancer (19,20), relatively little is known about the
role of miRNAs in glioma development and progression.
Suppressor of zeste-12 (SUZ12) is a core protein of
the Polycomb repressive complex 2 (PRC2), which also includes EED,
EZH2, and RBBP4 and RBBP7 proteins. PRC2 catalyzes the
trimethylation of histone H3 lysine 27 (H3K27me3), which is
associated with repression of gene transcription (21). In human embryonic fibroblasts,
>1,000 genes are silenced by PRC2 activity (22). Moreover, aberrant expression of PRC2
has been revealed to contribute to various human diseases and
disorders, particularly cancer. In fact, many recent studies have
demonstrated that PRC2 is overexpressed in a variety of cancers,
where it plays a key role in preventing the expression of tumor
suppressor genes during cell transformation (23–25).
SUZ12 has been revealed to be aberrantly expressed in many types of
cancer, including bladder cancer (26), gastric carcinoma (27) and mantle cell lymphoma (28). However, the precise expression
pattern and function of SUZ12 in glioma remains unclear.
In the present study, it was revealed that the
expression of miR-767-5p was significantly downregulated in GBM
tissue samples and cell lines compared with their normal
counterparts. It was determined that ectopic expression of
miR-767-5p inhibited many cancer-related phenotypes of GBM cells.
In addition, SUZ12 was identified as a direct target of miR-767-5p,
and SUZ12 expression in GBM tissue was revealed to be negatively
correlated with miR-767-5p levels. Finally, it was determined that
miR-767-5p overexpression significantly suppressed tumor growth in
a mouse xenograft model of GBM. Collectively, our findings indicate
a crucial potential role for miR-767-5p in GBM.
Materials and methods
Human tissue samples
Human GBM samples (n=18) and normal brain tissues
(NBTs; n=8) were collected from patients at the Department of
Neurosurgery at the First Affiliated Hospital of Nanjing Medical
University, China. Fresh specimens were immediately frozen in
liquid nitrogen and stored at −80°C until analyzed.
Histopathological grading was based on the WHO criteria. None of
the GBM patients had undergone chemotherapy or radiotherapy prior
to surgery. The present study was approved by the Research Ethics
Committee of Nanjing Medical University, and written informed
consent was obtained from all patients.
Cell culture
The human GBM cell lines T98, A172, U87
(glioblastoma of unknown origin; STR profiling was performed),
LN229, U251 and U118 (derived from the U138MG astrocytoma cell
line; STR profiling was performed) were obtained from the Chinese
Academy of Sciences Cell Bank (Shanghai, China). All cells were
cultured in Dulbecco's modified Eagle's medium (DMEM) with high
glucose and sodium pyruvate, supplemented with 10% fetal bovine
serum (FBS) and antibiotics (100 U/ml penicillin and 100 ng/ml
streptomycin). Cells were maintained in a 5% CO2
atmosphere at 37°C. Normal human astrocytes (NHAs) were purchased
from Lonza (Walkersville, MD, USA) and cultured according to the
supplier's instructions. Human 293T cell lines were purchased from
the American Type Culture Collection (ATCC; Rockville, MD,
USA).
Lentiviral packaging and generation of
stable cell lines
A lentiviral packaging kit was purchased from
Shanghai GeneChem Co., Ltd. (Shanghai, China). The hsa-miR-767-5p
mimic and hsa-miR-negative control (miR-NC) sequences were
chemically synthesized by Guangzhou RiboBio Co., Ltd., Guangzhou,
China). Next, 293T cells were infected with the vectors, and
lentiviruses were collected from the supernatant according to the
manufacturer's instructions. U87 and U251 cells were infected with
lentiviruses for 24 h and selected with puromycin (1 µg/ml) to
establish stable cell lines. For SUZ12 overexpression, human SUZ12
cDNA was cloned into pcDNA3.1 to generate pCDNA3.1-SUZ12. Cells
were transfected with miR-767-5p, miR-NC, empty pCDNA3.1, or
pCNDA-SUZ12 using Lipofectamine 2000 (Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) according to the manufacturer's
protocol. Cells were used for experiments at 48 h after
transfection as indicated.
RNA extraction and quantitative
reverse-transcription PCR (RT-qPCR)
Total RNA was extracted from cells or tissues using
TRIzol (Invitrogen; Thermo Fisher Scientific, Inc.), following the
manufacturer's protocol. miR-767-5p levels were assessed by RT-qPCR
was performed using an ABI StepOne Plus System (Applied Biosystems;
Thermo Fisher Scientific, Inc.) with a Bulge-Loop™ miRNA RT-qPCR
Primer Kit (Guangzhou RiboBio Co., Ltd.) and primers purchased from
Guangzhou RiboBio Co., Ltd. miRNA levels were normalized to U6 mRNA
and quantified using the 2ΔΔCq method (29).
Protein extraction and
immunoblotting
Cells were harvested from 6-well plates and lysed in
RIPA lysis buffer (Shanghai Beyotime Biotechnology China, Shanghai,
China). Protein concentrations were determined using the
bicinchoninic acid assay (KenGen, Jiangsu, China). Western blotting
was performed as previously described (30). SUZ12 (cat. no. ab126577) were
purchased from obtained from Abcam (Cambridge, UK). p-ERK1/2 (cat.
no. 4370), total-ERK1/2 (cat. no. 4695), p-AKT (cat. no. 4060),
total-AKT (cat. no. 4691), were purchased from Cell Signaling
Technology (Beverly, MA, USA). β-actin (AA128) were obtained from
Beyotime Institute of Biotechnology. Secondary antibody mouse (cat.
no. A1902) and rabbit (cat. no. A0208) came from Beyotime Institute
of Biotechnology. In brief, proteins were separated by 10% SDS-PAGE
and transferred to nitrocellulose membranes (Thermo Fisher
Scientific, Inc.). After blocking in 5% non-fat milk for 2 h, the
membranes were incubated with primary antibodies against SUZ12
(dilution 1:100), p-ERK1/2 (dilution 1:2,000), total-ERK1/2
(dilution 1:1,000), p-AKT (dilution 1:2,000), total-AKT (dilution
1:1,000), and β-actin (1:1,000) overnight under 4°C. They were then
incubated with secondary antibodies for 2 h at room temperature.
Electrochemiluminescence detection system (Thermo Fisher
Scientific) was used for signal detection. Signals were examined by
densitometric scans using ImageJ software (version 1.51; available
at http://rsb.info.nih.gov/ij/) for
Pearson's correlation analysis.
miRNA target prediction
Potential miR-767-5p targets were identified using
the online predictive program miRBase Targets (http://www.diana.pcbi.upenn.edu/cgi-bin/miRGen/v3/Targets.cgi
#Results), TargetScan Release 7.0 (http://www.targetscan.org/).
Dual-luciferase reporter assay
The 3′UTR of SUZ12 mRNA containing the wild-type
sequence (WT) or containing mutations in the putative miR-767-5p
binding site (Mut) were amplified and cloned into the pmiRNA-Report
luciferase expression reporter vector (Shanghai GeneChem Co.,
Ltd.). Luciferase reporter assays were performed as previously
described (31). In brief, U87 and
U251 cells were seeded into 24-well plates for 24 h and then
co-transfected with SUZ12-WT or SUZ12-Mut luciferase vectors along
with miR-767-5p or miR-NC using Lipofectamine 2000 (Invitrogen;
Thermo Fisher Scientific, Inc.). Luciferase activities were
quantified at 24 h after transfection using the Promega
Dual-Luciferase Reporter Assay System (Promega Corp., Madison, WI,
USA).
Wound healing assay
Cell migration was examined using wound healing
assays conducted as previously described (32). Briefly, U87 and U251 glioma cells
were co-transfected with a SUZ12-pCDNA3.1 overexpression vector and
either miR-767-5p or miR-NC. After plating, the cell layer was
scratched and the cells were incubated for a further 24 h. Cells
were examined under a Leica light microscope (Leica Microsystems
GmbH, Wetzlar, Germany) and images of the wound area were captured
at 0 and 24 h after injury.
Invasion assay
Cell invasion was assessed using 24-well BD Matrigel
invasion chambers (BD Biosciences, Franklin Lakes, NJ, USA)
according to the manufacturer's instructions. Briefly,
3×104 cells in serum-free DMEM were added to the upper
chamber, and DMEM supplemented with 15% FBS was added to the lower
chamber as a chemoattractant. After 24 h, non-invading cells were
removed from the top well by scraping, and invaded cells in the
lower chamber were fixed with 4% paraformaldehyde for 15 min and
then stained with 0.1% crystal violet for 2 h. The wells were
photographed and the number of cells in 3 independent ×10
magnification fields was counted.
Cell proliferation and colony
formation assays
Cells in logarithmic growth phase were seeded at
5×103 cells/well in 96-well plates and cultured for the
indicated time-points. Cell proliferation was quantified using the
Cell Counting Kit-8 (CCK-8; Dojindo Laboratories, Kumamoto, Japan)
according to the manufacturer's instructions. For colony-forming
assays, cells were seeded at 2×102 cells/well in 6-well
plates and cultured for ~10 days. Cells were then fixed with 100%
methanol and stained with 0.1% crystal violet for 20 min. The
number of colonies with a diameter >0.5 mm was counted.
Cell cycle analysis and apoptosis
assay
For cell cycle analysis, cells were harvested,
centrifuged at 360 × g for 5 min, washed with phosphate-buffered
saline (PBS), and fixed in 75% ethanol at −20°C overnight. Fixed
cells were washed twice with PBS, stained as described for the Cell
Cycle Staining Kit (Hangzhou MultiSciences Biotech, Co., Ltd.,
(Hangzhou, China), incubated for 25 min in the dark, and analyzed
on a flow cytometer (Beckman Coulter, Inc., Brea, CA, USA).
Apoptosis was measured using an Apoptosis Detection Kit (BD
Biosciences) according to the manufacturer's protocol. Cells were
analyzed on a Gallios flow cytometer (Beckman Coulter, Inc.).
Xenograft experiments in vivo
Male BALB/c nude mice (n=12) at 4 weeks of age
(weighing ~20 g) were obtained from the Shanghai Laboratory Animal
Center (Shanghai, China). All mice were housed and maintained under
specific pathogen-free conditions in laminar flow cabinets. U87
cells transfected with lentiviruses encoding miR-767-5p or miR-NC
(5×105) were injected subcutaneously into the posterior
flanks of the mice (n=6/group) and tumor growth was monitored for
30 days. Tumor sizes were measured every 3 days using calipers, and
the volume (mm3) was calculated as
(lengthxwidth2)/2. The mice were sacrificed 30 days
after injection, and the tumors were excised, weighed and
photographed. Tumor samples were fixed with 4% paraformaldehyde,
embedded in paraffin, and processed for immunohistochemical (IHC)
staining as described below. All animal experiments were approved
by the Animal Management Rule of the Chinese Ministry of Health
(Document 55, 2001) and were conducted in accordance with the
approved guidelines and experimental protocols of Nanjing Medical
University.
Immunohistochemistry
IHC analysis of fresh brain tissue or excised tumor
xenografts was performed as previously described (33) using anti-SUZ12 (dilution 1:500) and
anti-Ki-67 (dilution 1:150; cat. no. ab156956) purchased form Abcam
(Cambridge, MA, USA).
Fluorescence in situ hybridization
(FISH)
miR-767-5p expression in GBM and NBT samples was
detected by FISH. A 5′-FAM-labeled miR-767-5p sequence
(5′-CATGCTCAGACAACCATGGTGCA-3′) was synthesized by GoodBio
Technology Co., Ltd. (Wuhan, China). FISH was performed according
to a protocol provided by BioSense (Guangzhou, China). In brief,
frozen tissues were fixed in 4% formaldehyde for 20 min, washed
twice for 3 min each with PBS, digested with proteinase K for 5
min, and washed twice again with PBS. After eliminating
auto-fluorescence and blocking endogenous biotin, the sections were
hybridized with the probes overnight at 42°C in a humid chamber.
The sections were then washed with 2X saline-sodium citrate (SSC)
for 10 min, 1X SSC for 10 min, and 0.5X SSC for 10 min at room
temperature. Finally, the sections were stained with
4′,6-diamidino-2-phenylindole (DAPI) for 15 min and examined with a
ZEISS LSM 700 Meta confocal microscope (Zeiss AG, Oberkochen,
Germany).
Statistical analysis
Data are presented as the mean ± standard deviation
(SD). Associations between miR-767-5p and SUZ12 levels in glioma
tissues were analyzed using Pearson's correlation analysis. For the
remaining experiments, and all data were analyzed by the Student's
t-test for pairwise comparison or one-way analysis of variance
(ANOVA) followed by Bonferroni test for multivariate analysis.
Differences were considered statistically significant at
P<0.05.
Results
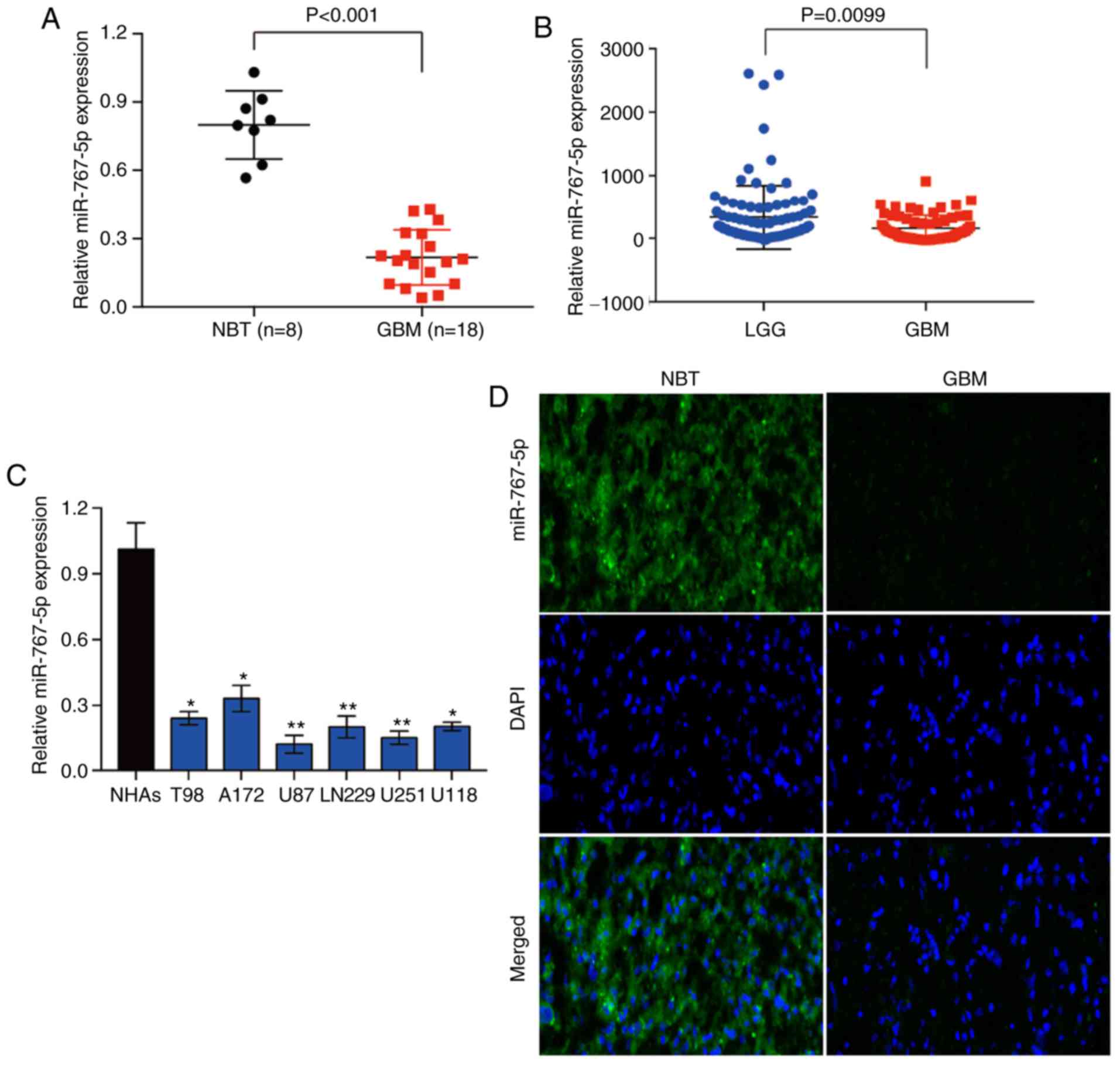
miR-767-5p expression is decreased in
human glioma specimens and cell lines
To determine whether miR-767-5p may play a role in
the development and/or progression of glioma, we first analyzed its
expression in 8 NBT samples and 18 histologically confirmed GBM
samples by RT-qPCR. The analysis revealed that miR-767-5p levels
were significantly lower in GBM tissues compared with NBTs
(Fig. 1A). To validate this
finding, miR-767-5p expression was evaluated in 158 glioma tissues
of different grades, based on data obtained from the Chinese Glioma
Genome Atlas (CGGA) database. Similarly, it was revealed that
miR-767-5p was significantly lower in GBM tissue compared with
low-grade glioma (grade II and III) (Fig. 1B). Next, the expression of
miR-767-5p was assessed in a panel of 6 glioma cell lines (T98,
A172, U87, LN229, U251 and U118) and normal human astrocytes
(NHAs). Consistent with the results of the tissue analyses,
miR-767-5p levels were significantly lower in all 6 glioma cell
lines compared with NHAs (Fig. 1C).
Finally, FISH was performed on sections of GBM and NBTs, which
confirmed the marked downregulation of miR-767-5p in GBM compared
with normal brain samples (Fig.
1D). Collectively, these data raise the possibility that the
loss of miR-767-5p expression in GBM may be involved in the
development and/or progression of glioma.
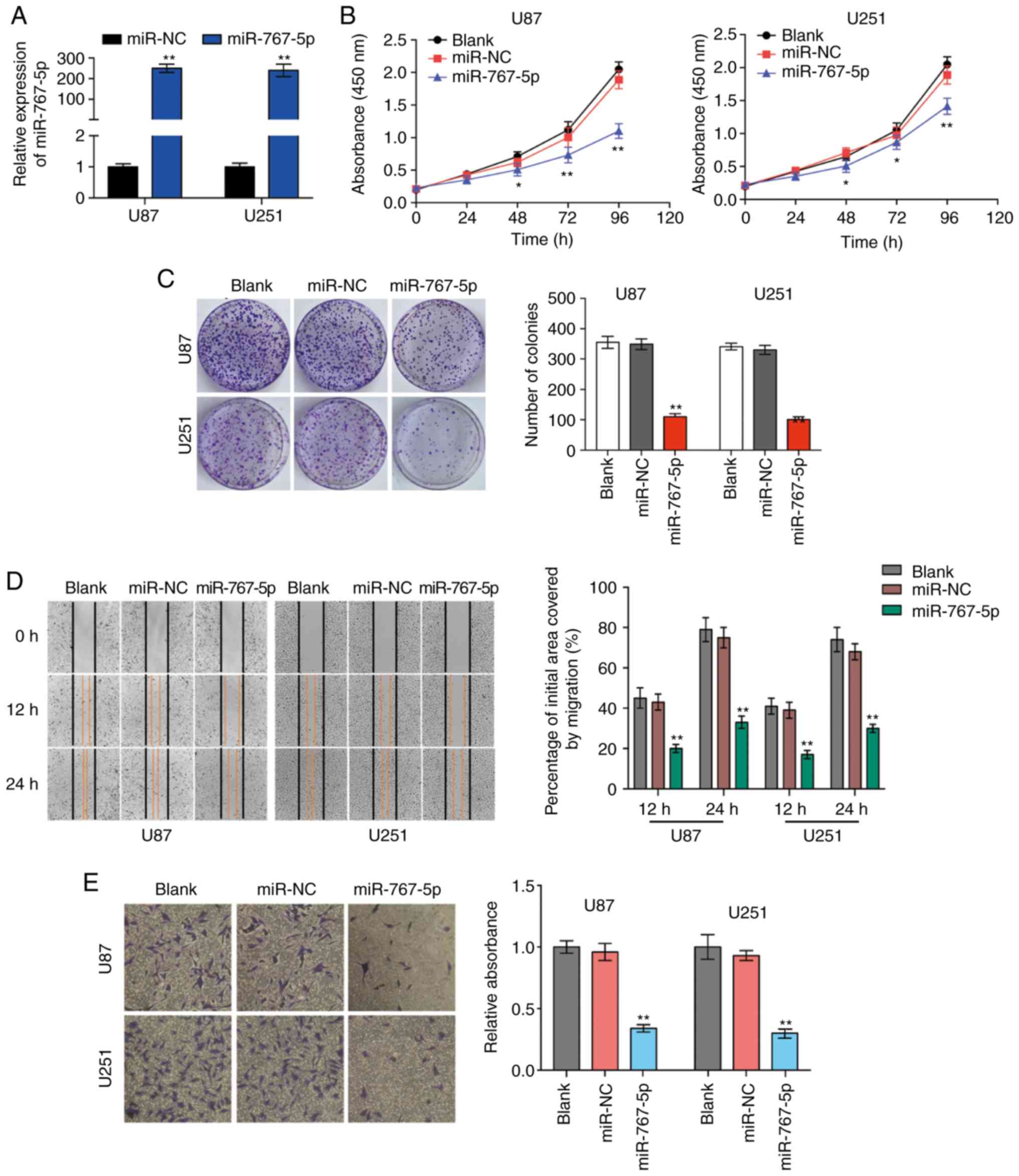
Overexpression of miR-767-5p
suppresses glioma cell proliferation and invasiveness and induces
apoptosis
To investigate the potential biological functions of
miR-767-5p in vitro, U87 and U251 cells were transfected
with lentiviral vectors encoding miR-767-5p or a control miRNA
sequence (miR-NC), and then the effects on various cancer-related
behaviors were analyzed. A specially constructed lentivirus for
miR-767-5p was transfected into U87 and U251 cells to alter the
expression level of miR-767-5p. RT-qPCR revealed that miR-767-5p
was significantly increased compared to negative control groups
(Fig. 2A). As revealed in Fig. 2B and C, the expression of the
miR-767-5p mimic significantly reduced a wide range of functions
compared with untransfected or miR-NC-transfected cells, including
proliferation (Fig. 2B) and colony
formation (Fig. 2C). In addition,
wound healing and Transwell invasion assays were performed to
evaluate GBM cell migration and invasion, respectively. Cells
overexpressing miR-767-5p revealed significantly reduced migratory
and invasive behaviors compared with un-transfected or
miR-NC-transfected cells (Fig. 2D and
E). Finally, the effects of miR-767-5p overexpression on GBM
cell cycle progression and apoptosis were evaluated. The proportion
of cells in G1 phase and G2/S phases was increased and decreased,
respectively, by expression of miR-767-5p compared with miR-NC
(Fig. 2F). Consistent with this
result, miR-767-5p overexpression resulted in an increase in the
number of apoptotic U87 and U251 cells compared with the control
cells (Fig. 2G). Furthermore, it
was revealed that miR-767-5p mainly affected early apoptosis of
cells, thus we primarily studied early apoptosis of cells in this
experiment. Collectively, these findings indicated that miR-767-5p
played a crucial role in regulating cell proliferation, colony
formation, migration, and survival of glioma cell lines.
SUZ12 is a specific target gene of
miR-767-5p and its expression is negatively correlated with
miR-767-5p levels in GBM tissue
To elucidate the mechanisms by which miR-767-5p may
inhibit GBM behavior, the bioinformatics analytical tool TargetScan
was used to identify mRNAs containing 3′UTR sequences complementary
to miR-767-5p. Of the potential targets identified, SUZ12 were
selected for further analysis. SUZ12 is a component of the PRC2
complex, which controls gene expression through its histone
methyltransferase activity and plays crucial roles in promoting
cancer cell growth and invasion. To determine whether miR-767-5p
does indeed interact with SUZ12 mRNA, we cloned the 3′UTR of SUZ12,
either WT or mutated in the putative miR-767-5p binding site (Mut)
into a luciferase reporter vector (Fig.
3A), which was transfected into U87 and U251 cells together
with miR-767-5p or miR-NC. The results revealed that
co-transfection with miR-767-5p significantly inhibited luciferase
activity driven by SUZ12-WT, but not by SUZ12-Mut (Fig. 3B), indicating that miR-767-5p may
regulate SUZ12 expression in GBM cells. Consistent with this, IHC
staining revealed strong expression of SUZ12 and the proliferation
marker Ki-67 in GBM tissues compared with NBTs (Fig. 3C). Furthermore, western blot
analysis revealed that SUZ12 expression in the GBM cell lines was
downregulated by overexpression of miR-767-5p (Fig. 3D) Notably, SUZ12 mRNA levels were
much higher in GBM tissues than NBTs (Fig. 3E), which was in contrast with the
pattern of miR-767-5p expression. In fact, Pearson's analysis
revealed a significant negative correlation between miR-767-5p and
SUZ12 mRNA levels in GBM tissues (Fig.
3F; r=−0.5847, P=0.0108). Collectively, these data indicated
that SUZ12 mRNA is a direct target of miR-767-5p in GBM.
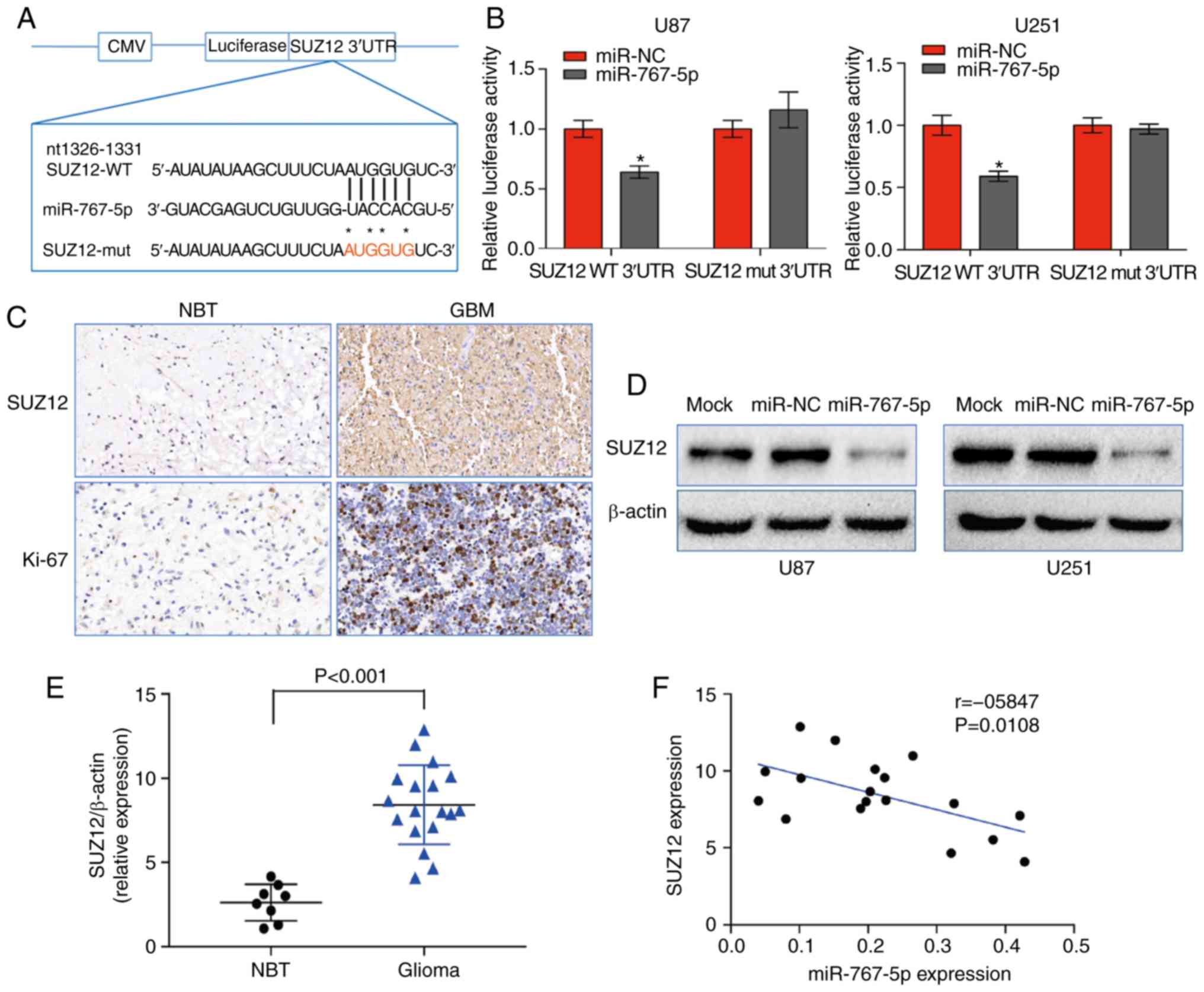 | Figure 3.miR-767-5p regulates the expression
of SUZ12 in GBM. (A) Predicted miR-767-5p binding site in the 3′UTR
of SUZ12 mRNA indicating the residues changed in the mutant
construct. (B) Relative luciferase activity driven by the WT or Mut
SUZ12 3′UTR in U87 and U251 cells co-transfected with miR-767-5p
mimic or a control sequence (miR-NC). Firefly luciferase activity
was normalized to Renilla luciferase. (C) Representative IHC
staining of SUZ12 and Ki-67 in GBM and NBT. (D) Western blot
analysis of SUZ12 protein levels in U87 and U251 cells expressing
miR-767-5p or miR-NC. β-actin was probed as a loading control. (E)
Relative expression of SUZ12 determined by western blot analysis of
18 GBM and 8 NBT specimens. SUZ12 levels were normalized to
β-actin. (F) Pearson's correlation analysis of SUZ12 and miR-767-5p
expression in 18 GBM tissues (Spearman correlation analysis,
r=−0.5847, P=0.0108). Data are presented as the mean ± SD of three
independent experiments. *P<0.05. SUZ12, suppressor of zeste-12;
GBM, glioblastoma multiforme; 3′UTR, 3′ untranslated region; WT,
wild-type; Mut, mutated; IHC, immunohistochemistry; NBT, normal
brain tissue; SD, standard deviation. |
Exogenous expression of SUZ12
overcomes the inhibitory effects of miR-767-5p on GBM cells
Previous research has revealed that SUZ12 plays a
pivotal role in the proliferation and migration of many types of
human cancers. To determine whether SUZ12 may mediate the effects
of miR-767-5p on GBM cells, we assessed whether ectopic expression
of SUZ12 could reverse the effects of miR-767-5p overexpression in
U87 and U251 cells. RT-qPCR demonstrated that SUZ12 was increased
compared to negative control groups (Fig. 4A). As revealed in Fig. 4, overexpression of SUZ12 partially
or completely reversed the miR-767-5p-induced effects on cell
proliferation (Fig. 4B), colony
formation (Fig. 4C), migration
(Fig. 4D), invasion (Fig. 4E), cell cycle arrest (Fig. 4F), and apoptosis (Fig. 4G). Immunofluorescence staining of
U87 and U251 cells confirmed that SUZ12 protein levels were
markedly downregulated by transfection with miR-767-5p compared
with miR-NC (Fig. 4H). Finally,
whether miR-767-5p and SUZ12 modulation affected two major
signaling pathways in GBM cells was examined. In fact, ERK1/2 and
AKT pathway activation was suppressed by the miR-767-5p mimic, as
demonstrated by a specific reduction of the phosphorylated
(activated) forms of ERK1/2 and AKT (Fig. 4I). Notably, these miR-767-5p effects
could be rescued by co-expression of SUZ12 (Fig. 4I). Thus, our data revealed that
SUZ12 mediated the effects of miR-767-5p, at least in part, on
glioma cell cancer-related behaviors.
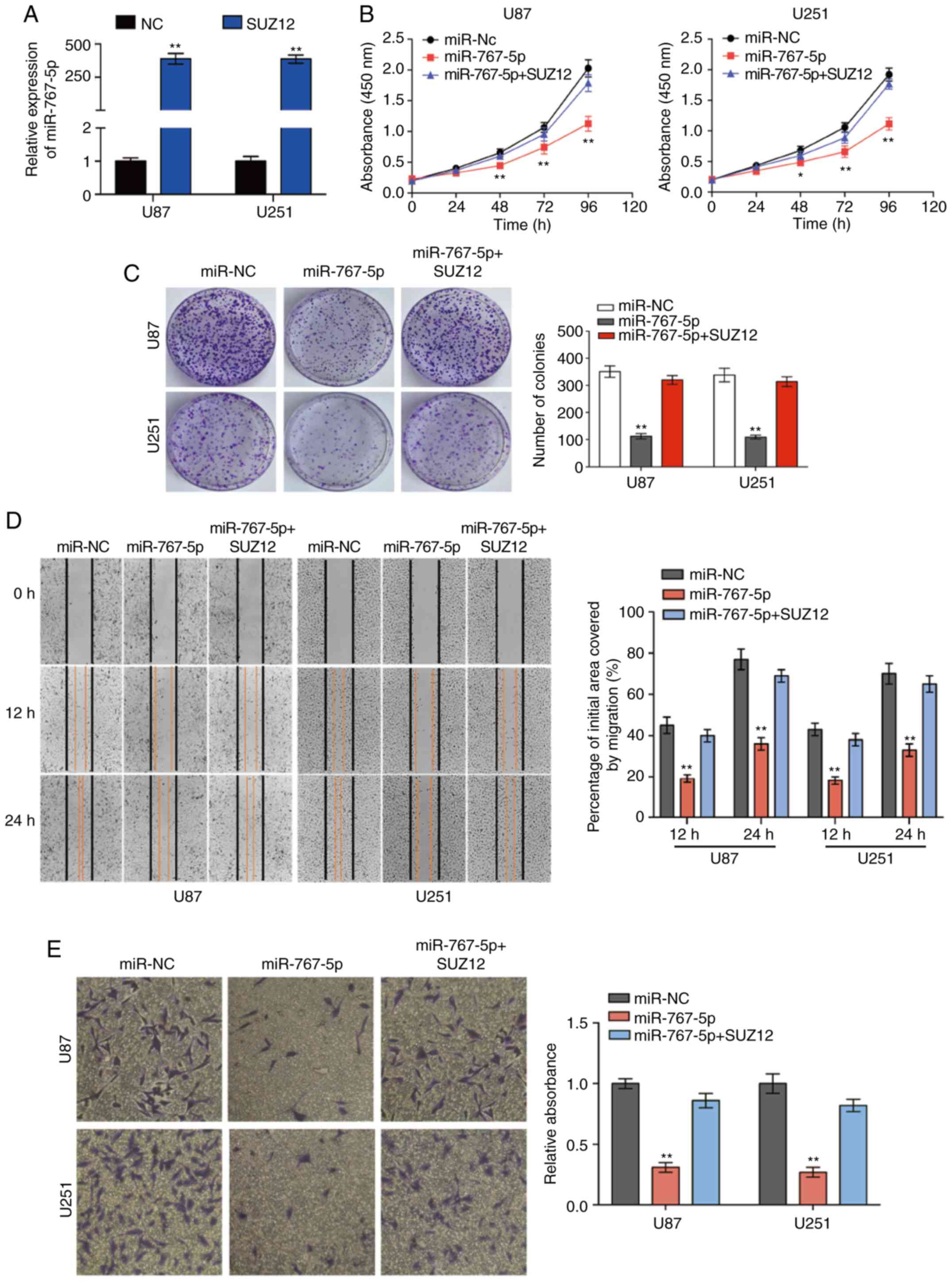 | Figure 4.SUZ12 reverses the inhibitory effects
of miR-767-5p on GBM cell lines. (A) The expression of SUZ12 in U87
and U251 cells was calculated by RT-qPCR after transfection. (B-I)
For all assays, U87 and U251 cells were transfected with miR-767-5p
mimic, negative control sequence (miR-NC), or miR-767-5p and a
SUZ12 overexpression plasmid. (B) CCK-8 cell proliferation assay.
(C and D) Representative images (left) and quantification (right)
of (C) colony-forming assays, (D) wound healing assays, and (E)
Transwell invasion assays. (F) Flow cytometric analysis of the cell
cycle analyzed 48 h after transfection representative histograms
(left) and quantification (right) of the percentage of cells in G1,
G2, and S phases of the cell cycle. (G) Representative dot plots
(left) and quantification (right) of an Annexin V-FITC/PI apoptosis
assay. (H) Representative images of immunofluorescence staining of
SUZ12 in U87 and U251 cells. Nuclei were stained with DAPI. (H)
Representative images of immunofluorescence staining of SUZ12 in
U87 and U251 cells. Nuclei were stained with DAPI. (I) Western blot
analysis of SUZ12, phosphorylated (p)-ERK1/2, total ERK1/2, p-AKT,
and total AKT levels (left) and quantification of relative p-AKT
and p-ERK levels (right). β-actin was probed as an internal
control. Data are presented as the mean ± SD of n=3. *P<0.05,
**P<0.01, miR-767-5p vs. miR-767-5p+SUZ12. SUZ12, suppressor of
zeste-12; GBM, glioblastoma multiforme; CCK-8, Cell Counting
Kit-8. |
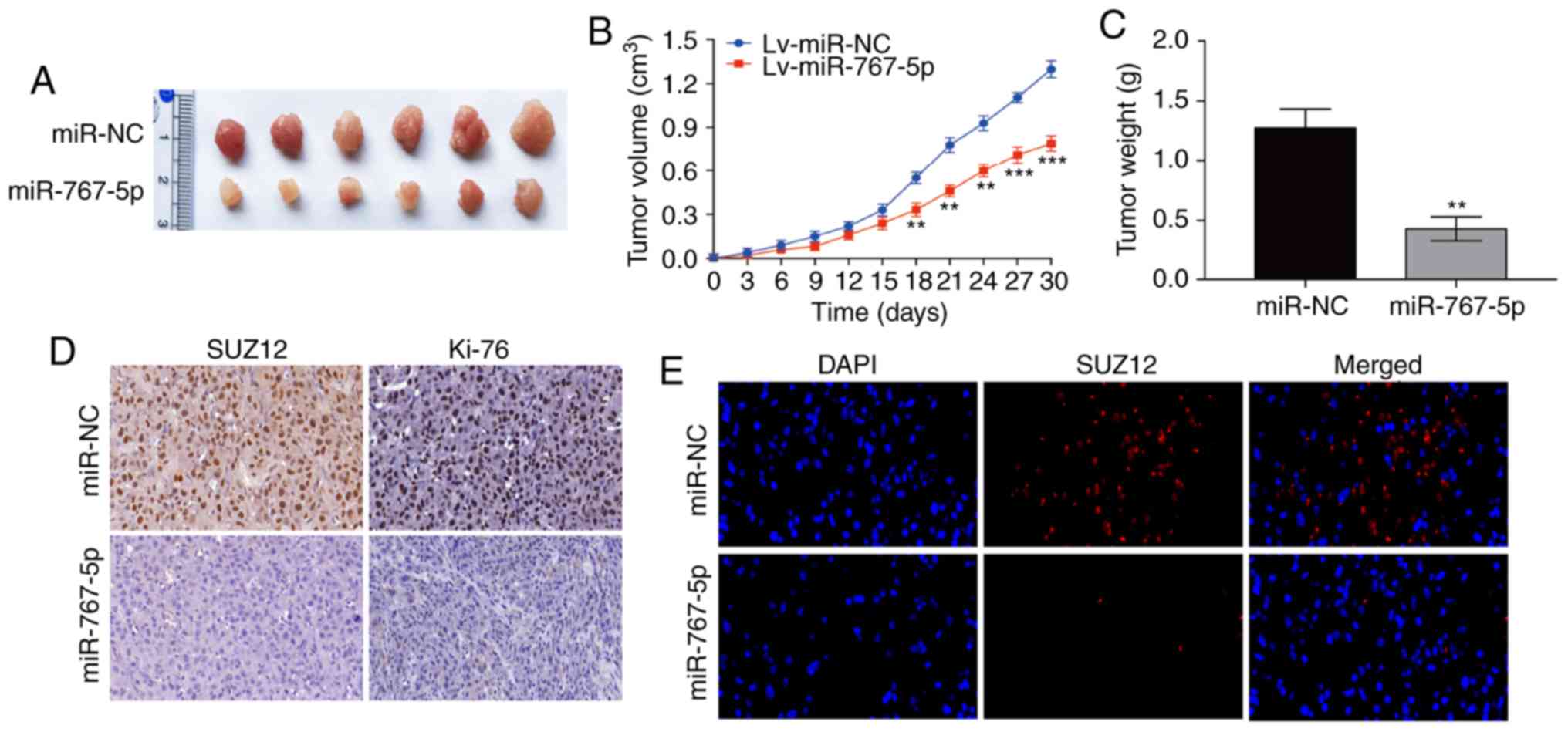
Overexpression of miR-767-5p
suppresses tumor growth in vivo
Having established the pivotal role played by
miR-767-5p in regulating GBM functions in vitro, its effects
on tumor growth in vivo were next examined, using a mouse
xenograft model. For this, U87 cells stably expressing miR-767-5p
or miR-NC were injected subcutaneously into nude mice, and tumor
growth was monitored over the next 30 days. As revealed in Fig. 5A-C, tumors derived from
miR-767-5p-expressing U87 cells grew significantly slower and
formed significantly smaller tumors than the miR-NC-expressing
cells. miR-767-5p-overexpressing tumors excised after 30 days
contained markedly reduced levels of SUZ12 and Ki-67 proteins
compared with the miR-NC-expressing tumors, as determined by both
IHC (Fig. 5D) and
immunofluorescence (Fig. 5E)
staining. These results indicated that overexpression of miR-767-5p
suppressed GBM growth in vivo, potentially via the effects
on SUZ12.
Discussion
The miRNA class of small non-coding RNAs is
increasingly recognized to play important roles in tumor activation
or suppression via regulation of many genes involved in cancer
progression and metastasis (34,35).
Deregulated miRNA expression has been revealed in many tumors,
including the most common and malignant glioblastoma subtypes
(36). Notably, the aberrant
expression of a single miRNA can have profound implications for the
expression of various transcripts that play an important role in
human disease. Multiple miRNAs have been implicated in several
crucial processes in glioma, such as cell proliferation, apoptosis,
and invasion (37–39). In the present study, the biological
role of miR-767-5p and its target gene SUZ12 were explored in
glioma biology.
The present study revealed that miR-767-5p levels
were significantly reduced in glioma compared with NBT based on
data from the CGGA database and direct analysis of tissue samples.
It was also revealed that miR-767-5p expression was significantly
lower in glioma cell lines than in NHAs, and forced expression of
miR-767-5p significantly inhibited cell growth, migration, and
invasion, and induced cell cycle arrest and apoptosis. Notably, the
inhibitory effect of miR-767-5p was partially or completely
abolished by ectopic expression of its target gene SUZ12. Western
blot analysis and luciferase reporter assays confirmed that
miR-767-5p negatively regulates the expression of SUZ12 by binding
to the SUZ12 3′UTR in glioma cells, leading to downregulation of
SUZ12 protein. Our study thus describes the first experimental
validation of a miR-767-5p target gene. The clinical relevance of
our findings was also confirmed by demonstrating the ability of
miR-767-5p overexpression to suppress the growth of human GBM
tumors in nude mice. To the best of our knowledge, this is the
first evidence that miR-767-5p is a tumor suppressive miRNA.
SUZ12 is a component of PRC2, a crucial regulator of
multiple cellular functions. A recent study revealed that SUZ12
protein is overexpressed in a variety of cancers, including ovarian
(40) and colorectal cancer
(41), and mantle cell lymphoma
(28). PRC2 is known to regulate
the expression of various oncogenes. While the molecular mechanism
of SUZ12 upregulation observed in glioma was previously unclear,
there is growing evidence that miRNAs may be involved, consistent
with our findings in the present study. For example, SUZ12
expression was regulated by miR-200b/c, and overexpression of this
miRNA inhibited cholangiocarcinoma tumorigenesis and metastasis
(42). Our data in the present
study are the first to reveal that miR-767-5p specifically targets
SUZ12 in glioma. SUZ12 levels were correlated inversely with
miR-767-5p in glioma tumor tissues, and forced SUZ12 overexpression
overcame the inhibitory effects of miR-767-5p in GBM cells.
Finally, downregulation of SUZ12 by miR-767-5p inhibited the
phosphorylation of ERK1/2 and AKT, suggesting a potential molecular
mechanism for its activity. Collectively, our data provide the
first evidence that miR-767-5p suppresses glioma cell growth and
metastasis by inhibiting SUZ12 translation.
We propose that miR-767-5p and SUZ12 could be useful
novel prognostic markers as well as potential therapeutic targets
for GBM. Collectively, these findings provide important new
insights into the molecular mechanisms underlying glioma
development and reveal potential new approaches to its
treatment.
Acknowledgements
Not applicable.
Funding
This present study was supported by the Jiangsu
Provincial Department of Health General Program [H201506 (EA15)],
and the Program for Advanced Talents within Six Industries of
Jiangsu Province [2015-WSN-023 (IB15)].
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
XL and JialZ proposed and designed this research.
JialZ, SX and JX conducted the experiments. YL was responsible for
the collection and analysis of the patients' data. JianZ and JieZ
participated in the design of the study and assisted in drafting
the manuscript. XL and JialZ wrote this manuscript. XL, JialZ, SX,
JX, YL, JieZ and JianZ reviewed and edited the manuscript. All
authors have read and approved the manuscript and agreed to be
responsible for all aspects of the study to ensure proper
investigation and resolution of the accuracy or integrity of any
part of the work.
Ethics approval and consent to
participate
The present study was approved by the Research
Ethics Committee of Nanjing Medical University, and written
informed consent was obtained from all patients. All animal
experiments were approved by the Animal Management Rule of the
Chinese Ministry of Health (Document 55, 2001) and were conducted
in accordance with the approved guidelines and experimental
protocols of Nanjing Medical University.
Patient consent for publication
Informed consent was obtained from all individual
participants included in the study.
Competing interests
The authors declare that they have no conflict of
interest.
References
|
1
|
Ohgaki H and Kleihues P: Genetic
alterations and signaling pathways in the evolution of gliomas.
Cancer Sci. 100:2235–2241. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Louis DN, Ohgaki H, Wiestler OD, Cavenee
WK, Burger PC, Jouvet A, Scheithauer BW and Kleihues P: The 2007
WHO classification of tumours of the central nervous system. Acta
Neuropathol. 114:97–109. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ng SS, Cheung YT, An XM, Chen YC, Li M, Li
GH, Cheung W, Sze J, Lai L, Peng Y, et al: Cell cycle-related
kinase: A novel candidate oncogene in human glioblastoma. J Natl
Cancer Inst. 99:936–948. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Stupp R, Mason WP, van den Bent MJ, Weller
M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn
U, et al: Radiotherapy plus concomitant and adjuvant temozolomide
for glioblastoma. N Engl J Med. 352:987–996. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lai EC: Micro RNAs are complementary to
3′UTR sequence motifs that mediate negative post-transcriptional
regulation. Nat Genet. 30:363–364. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Huntzinger E and Izaurralde E: Gene
silencing by microRNAs: Contributions of translational repression
and mRNA decay. Nat Rev Genet. 12:99–110. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Fu LL, Wen X, Bao JK and Liu B:
MicroRNA-modulated autophagic signaling networks in cancer. Int J
Biochem Cell Biol. 44:733–736. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
van Jaarsveld MT, Helleman J, Berns EM and
Wiemer EA: MicroRNAs in ovarian cancer biology and therapy
resistance. Int J Biochem Cell Biol. 42:1282–1290. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Janga SC and Vallabhaneni S: MicroRNAs as
post-transcriptional machines and their interplay with cellular
networks. Adv Exp Med Biol. 722:59–74. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Paulin R, Courboulin A, Barrier M and
Bonnet S: From oncoproteins/tumor suppressors to microRNAs, the
newest therapeutic targets for pulmonary arterial hypertension. J
Mol Med (Berl). 89:1089–1101. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen CZ: MicroRNAs as oncogenes and tumor
suppressors. N Engl J Med. 353:1768–1771. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gabriely G, Yi M, Narayan RS, Niers JM,
Wurdinger T, Imitola J, Ligon KL, Kesari S, Esau C, Stephens RM, et
al: Human glioma growth is controlled by microRNA-10b. Cancer Res.
71:3563–3572. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang CZ, Zhang JX, Zhang AL, Shi ZD, Han
L, Jia ZF, Yang WD, Wang GX, Jiang T, You YP, et al: miR-221 and
miR-222 target PUMA to induce cell survival in glioblastoma. Mol
Cancer. 9:2292010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Di Leva G, Garofalo M and Croce CM:
MicroRNAs in cancer. Annu Rev Pathol. 9:287–314. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cho WC: OncomiRs: The discovery and
progress of microRNAs in cancers. Mol Cancer. 6:602007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Esquela-Kerscher A and Slack FJ:
Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer.
6:259–269. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Volinia S, Calin GA, Liu CG, Ambs S,
Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et
al: A microRNA expression signature of human solid tumors defines
cancer gene targets. Proc Natl Acad Sci USA. 103:2257–2261. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chan JA, Krichevsky AM and Kosik KS:
MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells.
Cancer Res. 65:6029–6033. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang V and Wu W: MicroRNA-based
therapeutics for cancer. BioDrugs. 23:15–23. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chase A and Cross NC: Aberrations of EZH2
in cancer. Clin Cancer Res. 17:2613–2618. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bracken AP, Dietrich N, Pasini D, Hansen
KH and Helin K: Genome-wide mapping of polycomb target genes
unravels their roles in cell fate transitions. Genes Dev.
20:1123–1136. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Iliopoulos D, Lindahl-Allen M, Polytarchou
C, Hirsch HA, Tsichlis PN and Struhl K: Loss of miR-200 inhibition
of Suz12 leads to polycomb-mediated repression required for the
formation and maintenance of cancer stem cells. Mol Cell.
39:761–772. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Villa R, Pasini D, Gutierrez A, Morey L,
Occhionorelli M, Viré E, Nomdedeu JF, Jenuwein T, Pelicci PG,
Minucci S, et al: Role of the polycomb repressive complex 2 in
acute promyelocytic leukemia. Cancer Cell. 11:513–525. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Herranz N, Pasini D, Díaz VM, Francí C,
Gutierrez A, Dave N, Escrivà M, Hernandez-Muñoz I, Di Croce L,
Helin K, et al: Polycomb complex 2 is required for E-cadherin
repression by the Snail1 transcription factor. Mol Cell Biol.
28:4772–4781. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Fan Y, Shen B, Tan M, Mu X, Qin Y, Zhang F
and Liu Y: TGF-β-induced upregulation of malat1 promotes bladder
cancer metastasis by associating with suz12. Clin Cancer Res.
20:1531–1541. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Cui Y, Chen J, He Z and Xiao Y: SUZ12
depletion suppresses the proliferation of gastric cancer cells.
Cell Physiol Biochem. 31:778–784. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Martin-Perez D, Sanchez E, Maestre L,
Suela J, Vargiu P, Di Lisio L, Martínez N, Alves J, Piris MA and
Sánchez-Beato M: Deregulated expression of the polycomb-group
protein SUZ12 target genes characterizes mantle cell lymphoma. Am J
Pathol. 177:930–942. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang J, Zhang J, Zhang J, Qiu W, Xu S, Yu
Q, Liu C, Wang Y, Lu A, Zhang J and Lu X: MicroRNA-625 inhibits the
proliferation and increases the chemosensitivity of glioma by
directly targeting AKT2. Am J Cancer Res. 7:1835–1849.
2017.PubMed/NCBI
|
|
31
|
Henriksen JR, Haug BH, Buechner J, Tømte
E, Løkke C, Flaegstad T and Einvik C: Conditional expression of
retrovirally delivered anti-MYCN shRNA as an in vitro model system
to study neuronal differentiation in MYCN-amplified neuroblastoma.
BMC Dev Biol. 11:12011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang G, Mao W and Zheng S: MicroRNA-183
regulates Ezrin expression in lung cancer cells. FEBS Lett.
582:3663–3668. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wang YY, Sun G, Luo H, Wang XF, Lan FM,
Yue X, Fu LS, Pu PY, Kang CS, Liu N and You YP: miR-21 modulates
hTERT through a STAT3-dependent manner on glioblastoma cell growth.
CNS Neurosci Ther. 18:722–728. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Møller HG, Rasmussen AP, Andersen HH,
Johnsen KB, Henriksen M and Duroux M: A systematic review of
microRNA in glioblastoma multiforme: Micro-modulators in the
mesenchymal mode of migration and invasion. Mol Neurobiol.
47:131–144. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Huang Q, Gumireddy K, Schrier M, le Sage
C, Nagel R, Nair S, Egan DA, Li A, Huang G, Klein-Szanto AJ, et al:
The microRNAs miR-373 and miR-520c promote tumour invasion and
metastasis. Nat Cell Biol. 10:202–210. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Bou Kheir T, Futoma-Kazmierczak E,
Jacobsen A, Krogh A, Bardram L, Hother C, Grønbæk K, Federspiel B,
Lund AH and Friis-Hansen L: miR-449 inhibits cell proliferation and
is down-regulated in gastric cancer. Mol Cancer. 10:292011.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Mott JL, Kobayashi S, Bronk SF and Gores
GJ: mir-29 regulates Mcl-1 protein expression and apoptosis.
Oncogene. 26:6133–6140. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Li H, Cai Q, Wu H, Vathipadiekal V, Dobbin
ZC, Li T, Hua X, Landen CN, Birrer MJ, Sánchez-Beato M and Zhang R:
SUZ12 promotes human epithelial ovarian cancer by suppressing
apoptosis via silencing HRK. Mol Cancer Res. 10:1462–1472. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Liu YL, Gao X, Jiang Y, Zhang G, Sun ZC,
Cui BB and Yang YM: Expression and clinicopathological significance
of EED, SUZ12 and EZH2 mRNA in colorectal cancer. J Cancer Res Clin
Oncol. 141:661–669. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Peng F, Jiang J, Yu Y, Tian R, Guo X, Li
X, Shen M, Xu M, Zhu F, Shi C, et al: Direct targeting of
SUZ12/ROCK2 by miR-200b/c inhibits cholangiocarcinoma
tumourigenesis and metastasis. Br J Cancer. 109:3092–3104. 2013.
View Article : Google Scholar : PubMed/NCBI
|