Introduction
Colorectal cancer (CRC) is one of the most commonly
diagnosed types of malignancy, with ~134,490 new cases diagnosed
worldwide (1,2). In 2016, ~49,190 CRC-associated
mortalities were reported worldwide (2). Distant metastasis of cancer is a
predominant cause of CRC-associated mortality (3). The 5-year survival rate of metastatic
CRC is as low as ~10%. In the past few decades, several regulators
of CRC metastasis have been identified, including HNRNPLL and PGE2
(4,5). HNRNPLL has been revealed to modulate
alternative splicing of CD44 during the epithelial-mesenchymal
transition (EMT), which leads to suppression of CRC metastasis
(4). PGE2 induced an expansion of
CRC stem cells to promote liver metastases in mice by activating
NF-κB (5). However, to the best of
our knowledge, the mechanisms underlying CRC metastasis remain
unclear.
MEIS proteins, including MEIS1, MEIS2 and MEIS3,
serve crucial roles in regulating the neural crest and limb
development (6,7). MEIS proteins interacts with HOX or PBX
proteins to form a homeoprotein-DNA complex. MEIS2, a member of the
MEIS protein family, has been implicated in the pathogenesis of
human cancer (8,9). MEIS2 has been revealed to be
overexpressed in neuroblastoma cells and to promote neuroblastoma
cell proliferation and tumorigenicity (10). In addition, MEIS2 was upregulated
and required for AML1-ETO-positive AML growth (11). Conversely, a high expression of
MEIS2 has been associated with an improved prognosis for patients
with ovarian cancer (12). A recent
study demonstrated that the protein expression level of MEIS2 was
associated with a lack of biochemical recurrence and progression to
clinically metastatic disease in prostate cancer (9). However, the functional roles of MEIS2
in CRC, particularly in CRC metastasis, remain unclear.
The present study aimed to investigate the role of
MEIS2 in the regulation of CRC metastasis using in vivo and
in vitro experiments. Furthermore, public datasets were
analyzed to evaluate the prognostic value of MEIS2 in CRC. The
present study provided novel information that supports the
potential clinical use of MEIS2 as a prognostic marker for CRC.
Materials and methods
Cell culture
HCT116 cells were obtained from The Cell Bank of
Type Culture Collection of the Chinese Academy of Sciences
(Shanghai, China) and cultured in RPMI-1640 medium (HyClone; GE
Healthcare, Chicago, IL, USA) containing 10% fetal bovine serum
(FBS; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA). The
HCT116 cells were cultured at 37°C in a humidified incubator with
5% CO2.
Lentiviral constructs and
transfections
The MEIS2 shRNA sequences were obtained from
Shanghai GeneChem Co., Ltd. (Shanghai, China). Recombinant
lentiviral vectors carrying MEIS2 shRNA were constructed according
to the manufacturer's protocol. The MEIS2 shRNA sequences were
designed using an online system (http://rnaidesigner.thermofisher.com/rnaiexpress/) and
purchased from Shanghai GeneChem Co., Ltd. The sequence of MEIS2
shRNA-1 was
5′-CCGGCCCATGATTGACCAGTCAAATTTCAAGAGAATTTGACTGGTCAATCATGGGTTTTTG-3′
and the sequence of MEIS2 shRNA-2 was
CCGGCCCATGATTGACCAGTCAAATTTCAAGAGAATTTGACTGGTCAATCATGGGTTTTTG.
Recombinantlentiviral vectors carrying MEIS2 shRNAs were
constructed with standard molecular techniques. 293T cells were
transfected with the recombinant vectors to generate lentiviruses.
Concentrated lentiviruses were transfected in HCT116 cells at a
multiplicity of infection (MOI) of 40 in RPMI-1640 medium (HyClone;
GE Healthcare) without FBS. The expression of MEIS2 in HCT116
knockdown cells was validated by quantitative real-time polymerase
chain reaction (qRT-PCR).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from cells using TRIzol
reagent (Invitrogen; Thermo Fisher Scientific, Inc.). Subsequently,
cDNA was synthesized using the RevertAid First Strand cDNA
Synthesis kit (Promega Corp., Madison, WI, USA), according to the
manufacturer's protocol. qPCR was performed using the iQ™
SYBR-Green SuperMix (Bio-Rad Laboratories, Inc., Hercules, CA,
USA). The following primers were used for qPCR: CEBPA forward,
5′-CCAGAAAGCTAGGTCGTGGGT-3′ and reverse,
5′-TGGACTGATCGTGCTTCGTGT-3′; JUN forward,
5′-ATGGTCAGGTTATACTCCTCCTC-3′ and reverse,
5′-CACATGCCACTTGATACAATCC-3′; TGFBR2 forward,
5′-TGGCTGTATGGAGAAAGA-3′ and reverse, 5′-GTCAGGATTGCTGGTGTT-3′;
GAPDH forward, 5′-TGACTTCAACAGCGACACCCA-3′ and reverse,
5′-CACCCTGTTGCTGTAGCCAAA-3′; MDM2 forward,
5′-GAATCATCGGACTCAGGTACATC-3′ and reverse,
5′-TCTGTCTCACTAATTGCTCTCCT-3′; CDKN1A forward,
5′-CTGTCTTGTACCCTTGTGCCT-3′ and reverse,
5′-GGTAGAAATCTGTCATGCTGGT-3′; and TGFBR2 forward,
5′-CGATAACTTCTGCCACCGAT-3′ and reverse, 5′-AGGGTTATGGTCAGCGAGAT-3′.
The 2−ΔΔCq method was used to calculate the relative
expression levels of the target genes (13).
Wound healing assay
HCT116 cells were seeded into a 6-well plate. Once
the confluency of the cells reached ~80%, a scratch was created in
a monolayer of the cells using a sterile micropipette tip. The
detached cells were then washed with phosphate-buffered saline
(PBS). The extent of wound healing was observed at 0, 24 and 48 h.
Images were captured from 5 random fields using an inverted
microscope. Triplicate wells for each condition were examined.
Transwell assay
Transwell assays were performed using 8-µm pore size
Transwell® plates (Corning Inc., Corning, NY, USA). The
invasion assay was performed using Matrigel® (BD
Biosciences, San, Jose, CA, USA), which was used to pre-coat
Transwell® plates. A total of 50,000 HCT116 cells,
cultured in RPMI-1640 with 2% FBS, were seeded into the upper well.
In addition, RPMI-1640 with 10% FBS was added to the bottom well.
Following incubation for 72 h the number of invading cells was
counted.
In vivo tumor metastasis assays
A total of 4×106 MEIS2-knockdown or
negative control HCT116 cells were transplanted into ten 5-week-old
female nude mice (from the Shanghai Slake Laboratory Animal Co.,
Ltd., Shanghai, China) weighing 15–20 g through the lateral tail
vein. The mice were housed at a temperature of 20–26 °C, a relative
humidity of 40–70 % and light/dark cycle of 12/12 h.
Luciferase-expressing HCT116 cells were constructed.
MEIS2-knockdown and control luciferase-expressing HCT116 cells were
then injected into the mice and monitored using an IVIS system
(IVIS; PerkinElmer, Inc., Waltham, MA, USA). The animal was
sacrificed when the tumor reached 1 cm in diameter. The mice were
sacrificed 7 weeks after injection with CO2 (with the
flow rate of CO2 euthanasia displacing ≤30% of the
chamber volume/min). To detect the effect of MEIS2 on tumor
metastasis, the lungs were collected and hematoxylin and eosin
(H&E) staining was performed. All in vivo study
protocols were approved by the Shanghai Medical Experimental Animal
Care Commission (Approval ID: ShCI-14-008).
Western blot analysis
The CRC cells were rinsed with PBS, and lysates were
prepared using RIPA buffer (Sigma-Aldrich; Merck KGaA, Darmstadt,
Germany). Total protein concentrations were determined using a BCA
protein concentration assay kit (Beyotime Institute of
Biotechnology, Haimen, China). A quantity of 50 µg protein sample
was loaded to each lane before electrophoresis began. Proteins were
separated by 10% sodium dodecyl sulfate-polyacrylamide gel
electrophoresis (SDS-PAGE) and transferred to polyvinylidene
difluoride (PVDF) membranes using a Bio-Rad System (Bio-Rad
Laboratories, Inc.). Blotted membranes were firstly moved to
blocking buffer containing 5% non-fat dry milk (diluted in TBST) at
room temperature for 1 h. Western blot analysis was performed with
E-cadherin (diluted 1:1,000; cat. no. 14472; Cell Signaling
Technologie), Snail (diluted 1:1,000; cat. no. 3895; Cell Signaling
Technologies), MEIS2 (diluted 1:1,000; cat. no. ab73164; Abcam,
Cambridge, MA, USA), Twist (diluted 1:1,000; cat. no. ab50581;
Abcam), JUN (diluted 1:1,000; cat. no. ab32137; Abcam), CEBPA
(diluted 1:1,000; cat. no. 8178; Cell Signaling Technologies), MDM2
(diluted 1:1,000; cat. no. ab38618; Abcam), TGFBR2 (diluted
1:1,000; cat. no. ab61213; Abcam) and CDKN1A (diluted 1:1,000; cat.
no. 2947; Cell Signaling Technologies), and mouse anti-GAPDH
(diluted 1:1,000; cat. no. c-25778; Santa Cruz Biotechnology, Inc.,
Dallas, TX, USA) at 4°C overnight and then incubated with 2 mg/ml
HRP-conjugated anti-rabbit IgG (diluted 1:5,000; cat. no. A9169;
Sigma-Aldrich; Merck KGaA) for 1 h at room temperature. After
washing, ECL Western Blotting reagent (Millipore; Merck KGaA) was
applied for the detection. The Quantity One software package
(Bio-Rad Laboratories, Inc.) was used for quantitation of the
signal intensities.
Microarray and expression
datasets
Total RNA was extracted using TRIzol reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) and was quantified by
the NanoDrop ND-2000 (Thermo Fisher Scientific, Inc.). Global
expression of mRNAs in 3 MCM10 shCtrl samples and 3 shMCM10 were
examined using the GeneChip PrimeView Human Gene Expression Array
(Thermo Fisher Scientific, Inc.). The raw data of the mRNA
expression profiles were downloaded and analyzed by R language
software. Background correction, quartile data normalization, and
probe summarization were applied for the original data. The limma
method in Bioconductor (http://www.bioconductor.org/) was used to identify
genes which were differentially expressed between two groups; the
significance of DEGs was calculated by t-test and was represented
by a P-value. The threshold set for upregulated and downregulated
genes was a fold-change ≥1.5.
Microarray data
The gene expression profiles of MEIS2 in CRC by
analyzing a series of public datasets, including The Cancer Genome
Atlas (TCGA), GSE17536 (14),
GSE17537 (14) and GSE41258
(15) datasets. TCGA dataset
included 10 normal colon samples and 367 CRC samples. GSE17536
dataset included 177 patients with CRC from the Moffitt Cancer
Center (Tampa, FL, USA) were used as the independent dataset.
GSE17537 dataset included 55 CRC patients from Vanderbilt Medical
Center (Nashville, TN, USA). GSE41258 dataset included 54 normal
colon samples, 13 normal liver samples, 7 normal lung samples and
186 primary CRC samples. Moreover, we analyzed the protein levels
of MEIS2 in CRC by analyzing Human Protein Atlas database
(https://www.proteinatlas.org/).
Functional enrichment analysis
The Database for Annotation, Visualization, and
Integrated Discovery (DAVID) online tool (version 6.8; david.ncifcrf.gov) was applied to perform Gene
Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG)
analysis. Adjusted P<0.05 was considered to indicate statistical
significance.
Statistical analysis
SPSS 13.0 software (SPSS, Inc., Chicago, IL, USA)
was used to perform statistical analysis. Each experiment was
performed 3 times. Student's t-test was used to calculate the
statistical significance between 2 groups. For >2 groups,
one-way analysis of variance followed by Newman-Keuls post hoc test
was used. P<0.05 was considered to indicate a statistically
significant difference.
Results
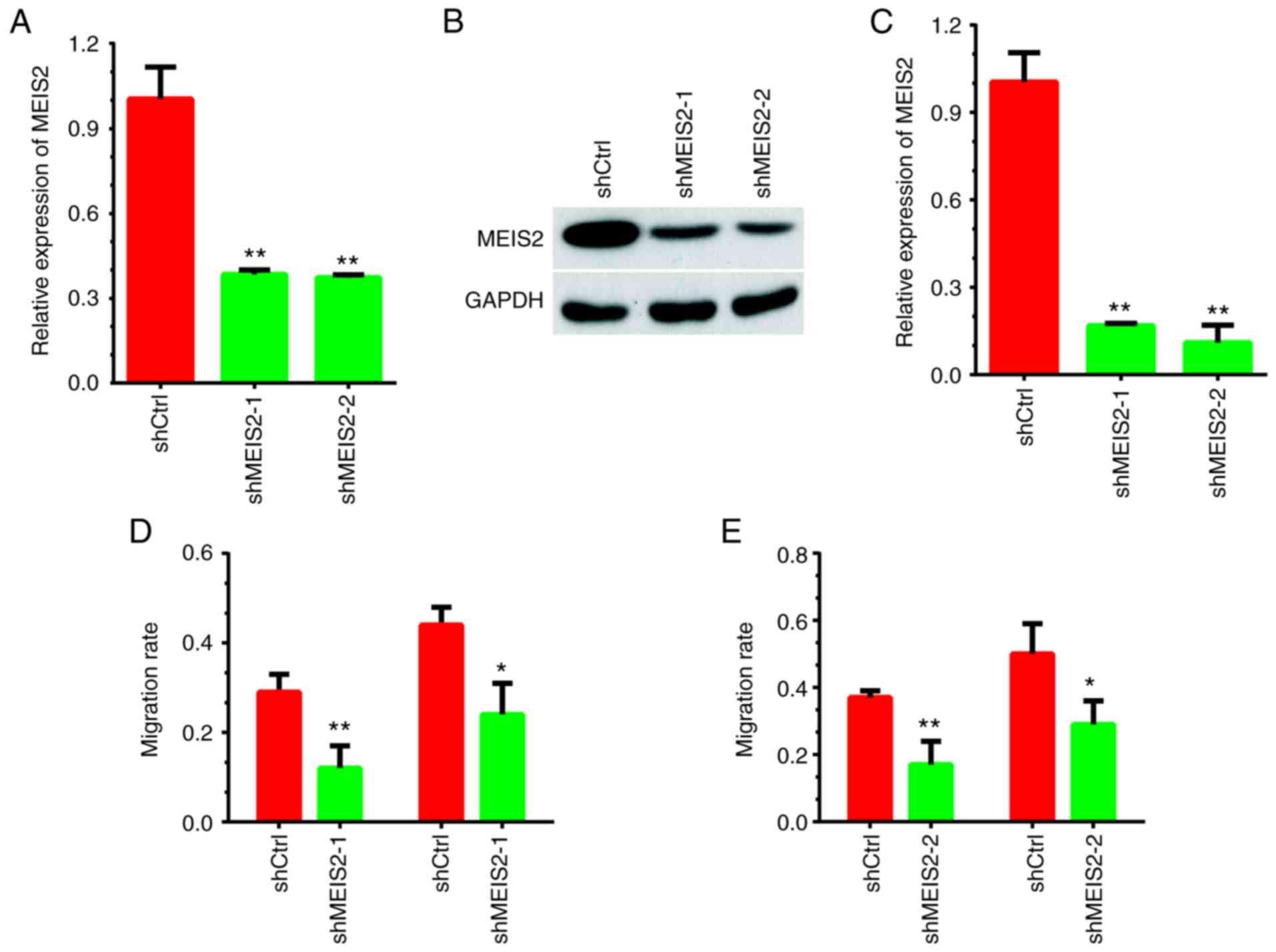
Knockdown of MEIS2 suppresses cell
migration in CRC
The present study performed loss-of-function assays
by silencing MEIS2 expression to investigate its role in CRC. As
presented in Fig. 1A-C,
shRNA-mediated silencing of MEIS2 led to a significant decrease in
both mRNA and protein expression levels of MEIS2 in HCT116 cells.
Subsequently, the effects of MEIS2 on cell migration were examined
by wound healing assay. Compared with the negative control group,
MEIS2-knockdown significantly suppressed the number of HCT116 cells
migrating toward the wound area (Fig.
1D-E and S1).
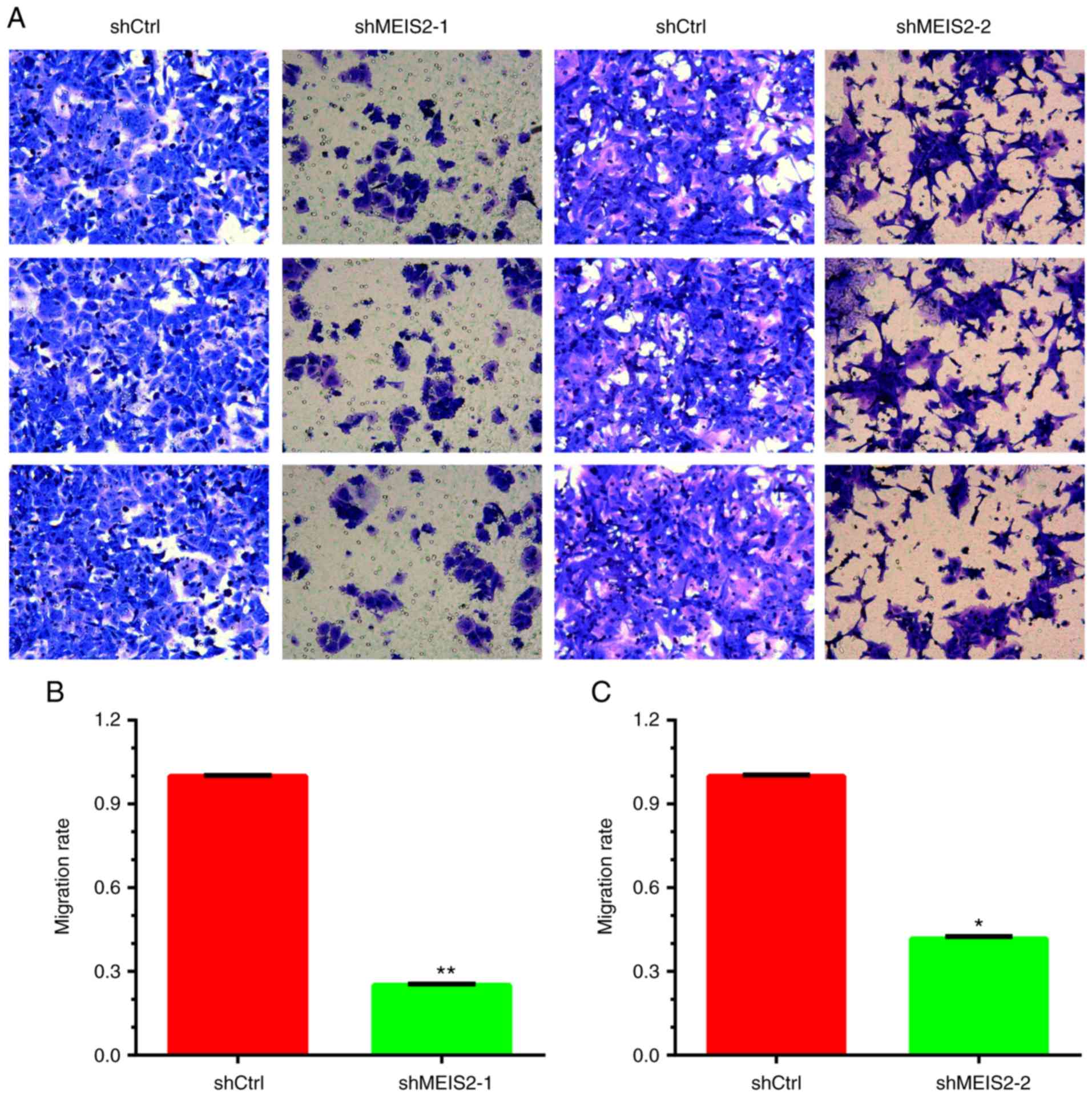
Furthermore, a Transwell assay was performed to
investigate the role of MEIS2 in the regulation of migration. It
was revealed that the number of migrating HCT116 cells decreased by
~70 and 60% in the shRNA-1 (Fig. 2A and
B) and shRNA-2 (Fig. 2A and C)
knockdown groups, respectively, compared with the negative control
group.
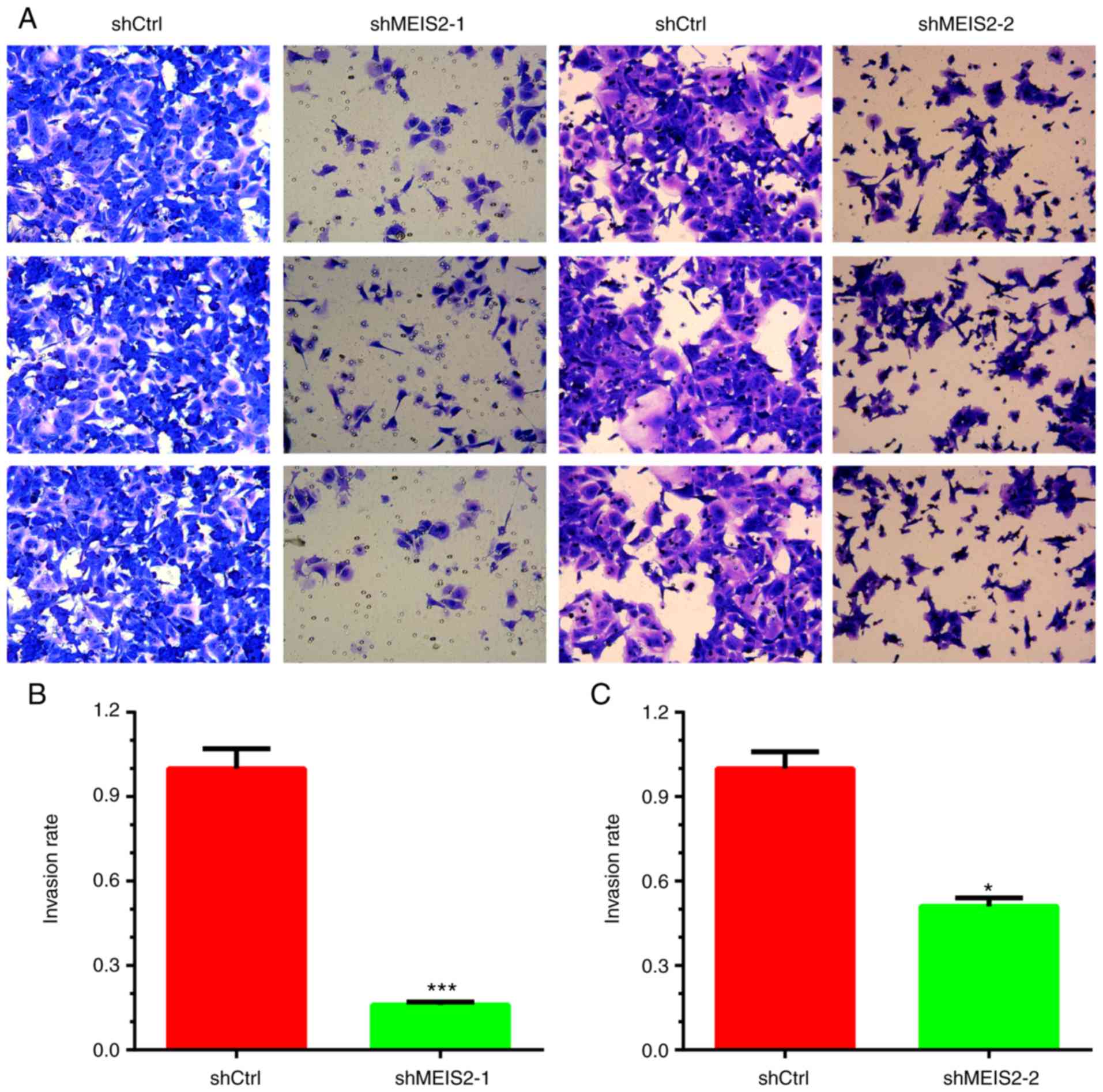
Knockdown of MEIS2 inhibits cell
invasion in CRC
The invasive ability of cells transfected with MEIS2
shRNAs was assessed by Matrigel cell invasion assay. The results
indicated that knockdown of MEIS2 significantly suppressed the
invasion of CRC cells (P<0.05). The number of invading HCT116
cells decreased by ~80 and 45% in the shRNA-1 (Fig. 3A and B) and shRNA-2 (Fig. 3A and C) knockdown groups,
respectively, compared with the negative control group.
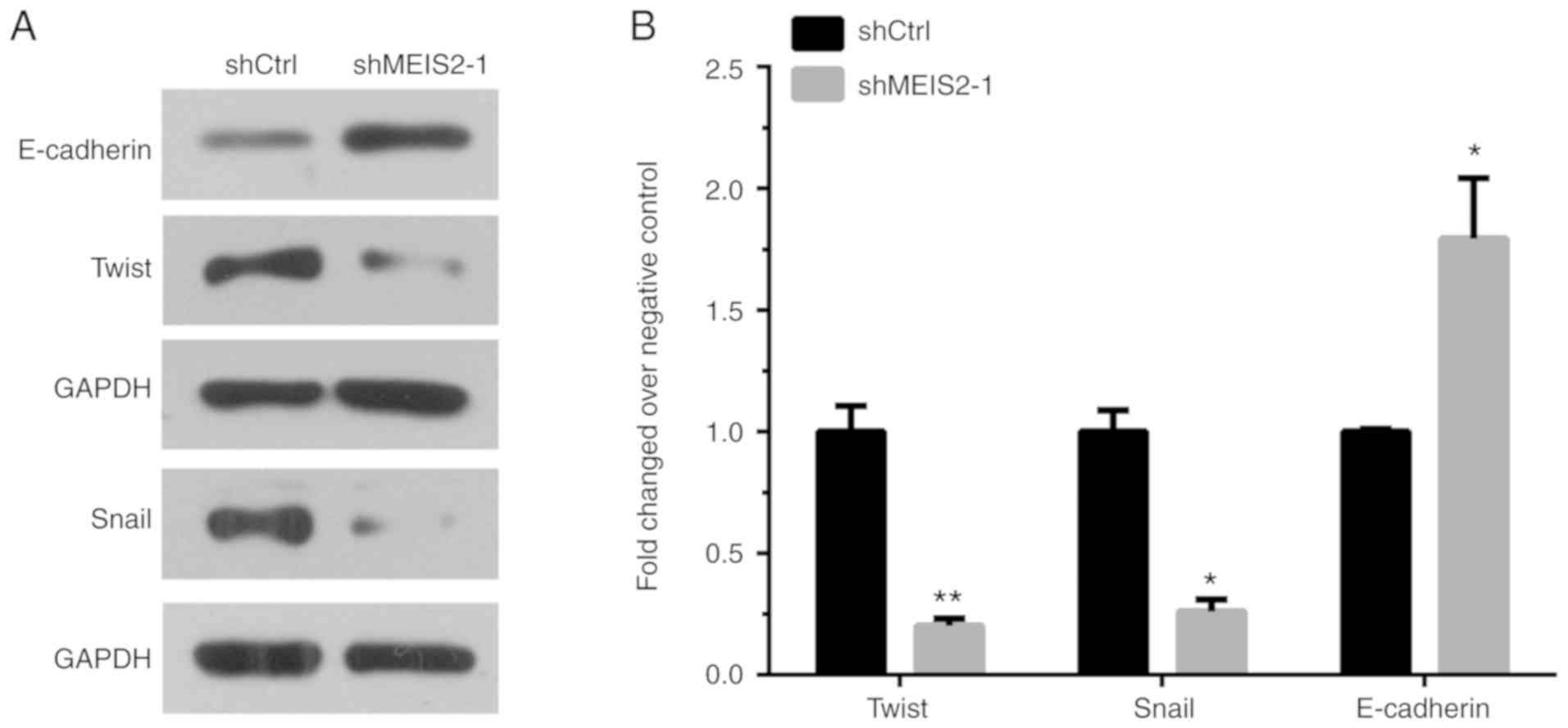
Knockdown of MEIS2 inhibits EMT in
CRC
To investigate whether MEIS2 promotes metastasis by
regulating EMT, the protein levels of E-cadherin, Twist and Snail
were detected following MEIS2 knockdown in CRC cells. Western blot
analysis revealed that the expression level of E-cadherin
increased, while Twist and Snail levels decreased in
MEIS2-knockdown HCT116 cells compared with the control groups
(Fig. 4A and B).
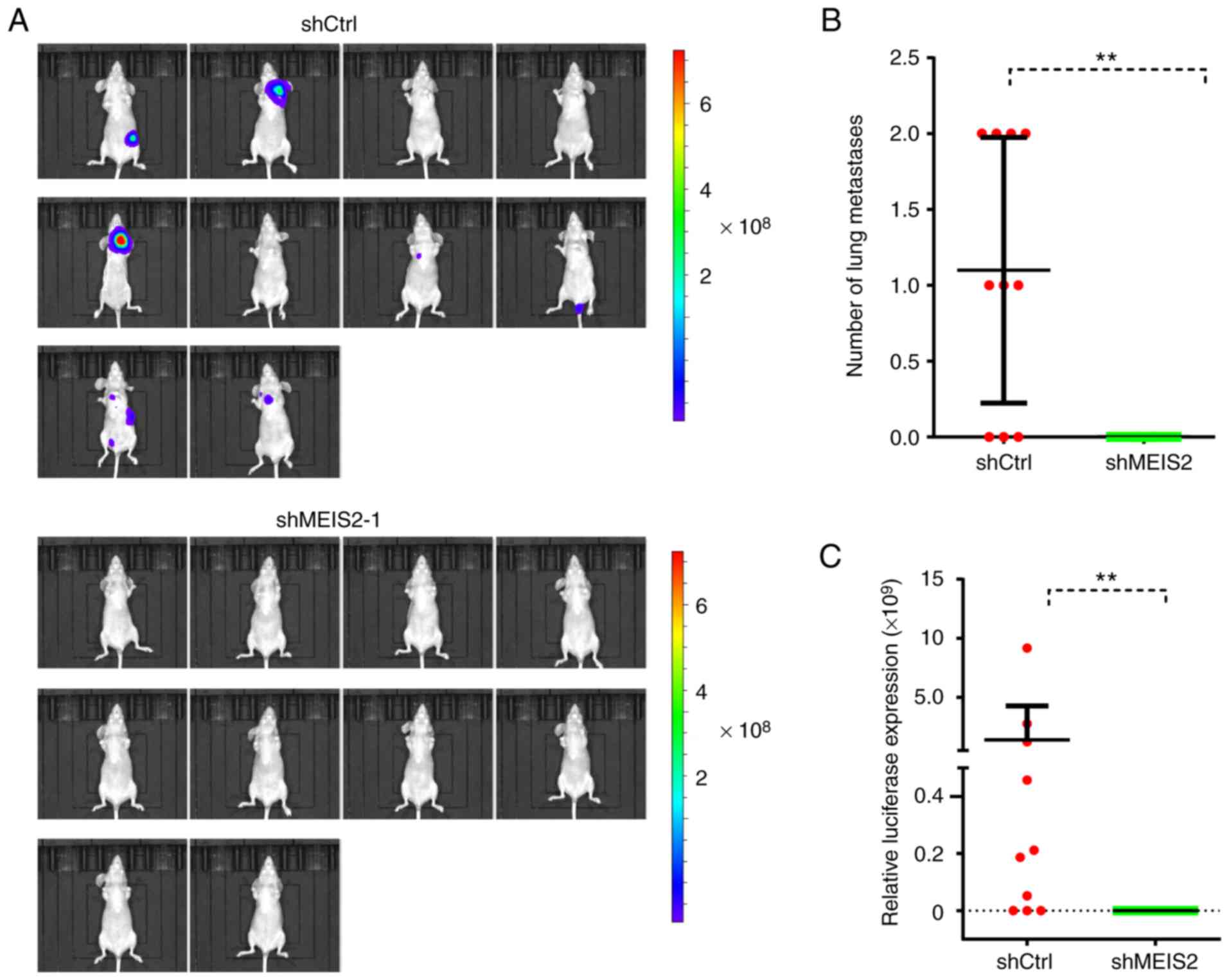
Knockdown of MEIS2 inhibits CRC
metastasis in vivo
Since it was demonstrated that MEIS2 knockdown
inhibited cell motility in vitro, the effect of MEIS2 on CRC
metastasis was further validated in vivo. The results
demonstrated that MEIS2-knockdown significantly inhibited CRC
metastasis in vivo. The luciferase signaling in the
MEIS2-knockdown group was significantly decreased compared with the
control groups (Fig. 5A and C).
Furthermore, histological analysis was performed to
confirm that MEIS2-knockdown inhibits the formation of lung
metastasis. It was revealed that the number of lung metastasis
nodules was markedly lower in the MEIS2-knockdown group compared
with the control group (Fig.
5B).
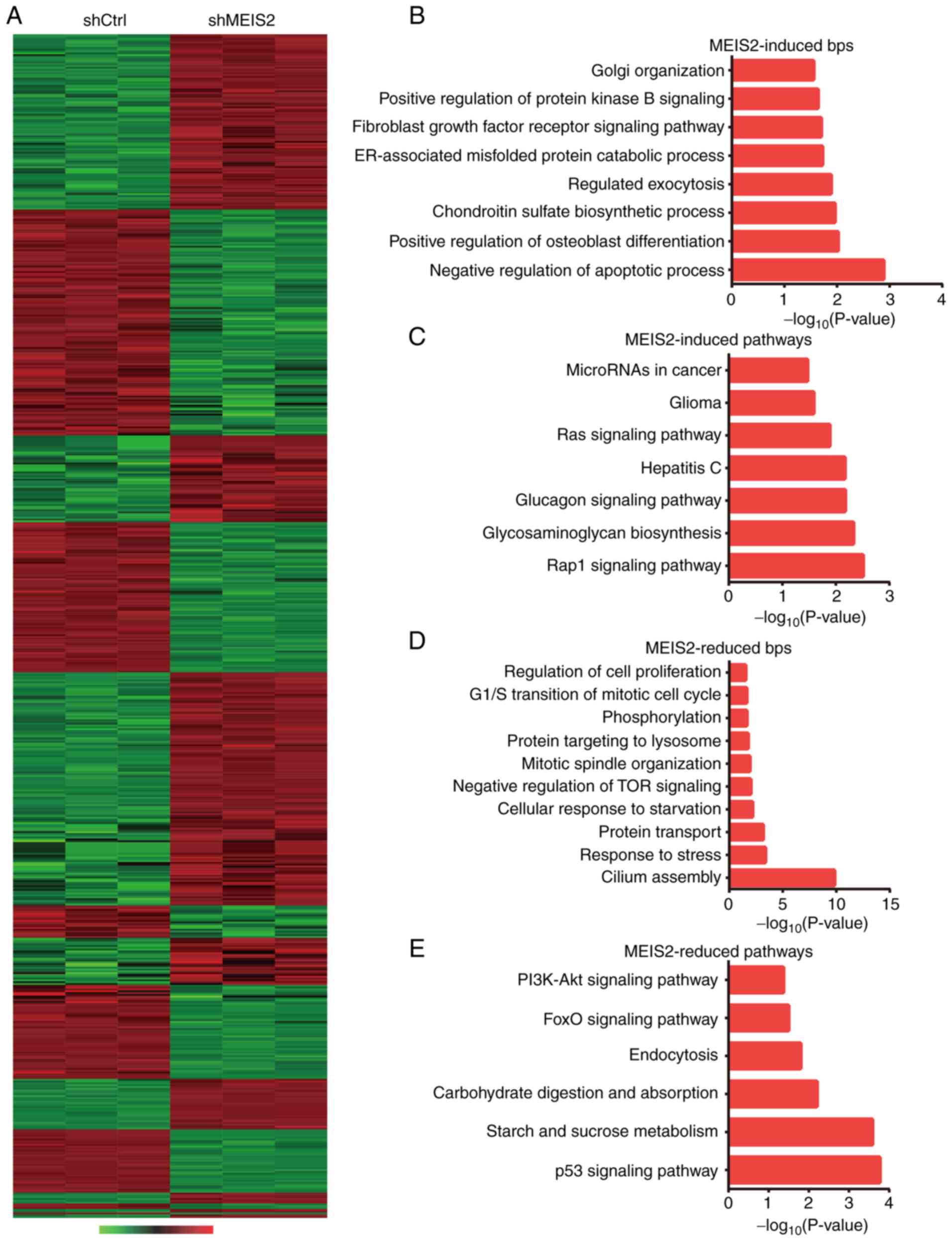
Microarray analysis reveals targets of
MEIS2 in CRC
Previous studies have indicated that MEIS2 is
involved in the regulation of gene transcription by interacting
with HOX and PBX proteins to form protein-DNA complex. However, the
targets of MEIS2 in certain human cancer types, including CRC,
remain unknown. The present study performed microarray analysis,
which identified 338 upregulated and 317 downregulated genes
following MEIS2-knockdown (Fig.
6A). The top 10 upregulated genes are presented in Table I and the top 10 downregulated genes
are revealed in Table II.
Bioinformatics analyses, including Gene Ontology (GO) and Kyoto
Encyclopedia of Genes and Genomes pathway analysis, were
subsequently performed.
 | Table I.The top 10 upregulated genes after
MEIS2 knockdown in CRC cell line. |
Table I.
The top 10 upregulated genes after
MEIS2 knockdown in CRC cell line.
| Entrez | Gene symbol | Fold-change | logFC | Regulation | P-value | FDR |
|---|
| 5899 | RALB | 2.42 | 1.27 | Upregulation | 4.87E-14 | 3.46E-11 |
| 10769 | PLK2 | 2.46 | 1.3 | Upregulation | 2.03E-11 | 2.97E-09 |
| 23639 | LRRC6 | 2.52 | 1.33 | Upregulation | 1.69E-11 | 2.63E-09 |
| 9270 | ITGB1BP1 | 2.57 | 1.36 | Upregulation | 5.17E-10 | 3.39E-08 |
| 23568 | ARL2BP | 2.71 | 1.44 | Upregulation | 5.11E-17 | 7.51E-13 |
| 5824 | PEX19 | 2.75 | 1.46 | Upregulation | 1.08E-13 | 6.16E-11 |
| 4193 | MDM2 | 2.85 | 1.51 | Upregulation | 8.80E-09 | 3.14E-07 |
| 5887 | RAD23B | 2.93 | 1.55 | Upregulation | 4.03E-16 | 1.57E-12 |
| 51200 | CPA4 | 3.51 | 1.81 | Upregulation | 5.73E-17 | 7.51E-13 |
| 5906 | RAP1A | 22.75 | 4.50 | Upregulation | 6.09E-27 | 2.39E-22 |
 | Table II.The top 10 downregulated genes after
MEIS2 knockdown in CRC cell line. |
Table II.
The top 10 downregulated genes after
MEIS2 knockdown in CRC cell line.
| Entrez | Gene symbol | Fold-change | logFC | Regulation | P-value | FDR |
|---|
| 9802 | DAZAP2 | −2.83 | −1.50 | Downregulation | 1.29E-16 | 1.01E-12 |
| 81839 | VANGL1 | −2.81 | −1.49 | Downregulation | 5.55E-14 | 3.63E-11 |
| 7504 | XK | −2.77 | −1.47 | Downregulation | 9.71E-13 | 2.75E-10 |
| 8519 | IFITM1 | −2.74 | −1.45 | Downregulation | 2.53E-16 | 1.34E-12 |
| 154807 | VKORC1L1 | −2.66 | −1.41 | Downregulation | 2.01E-12 | 4.95E-10 |
| 5880 | RAC2 | −2.53 | −1.34 | Downregulation | 5.26E-16 | 1.72E-12 |
| 9474 | ATG5 | −2.53 | −1.34 | Downregulation | 1.45E-12 | 3.80E-10 |
| 79801 | SHCBP1 | −2.50 | −1.32 | Downregulation | 1.03E-13 | 5.95E-11 |
| 5160 | PDHA1 | −2.42 | −1.27 | Downregulation | 2.55E-12 | 5.77E-10 |
| 55591 | VEZT | −2.39 | −1.25 | Downregulation | 1.43E-09 | 7.27E-08 |
As anticipated, it was revealed that the
downregulated genes following MEIS2-knockdown were associated with
negative regulation of apoptotic process, chondroitin sulfate
biosynthetic process, regulated exocytosis, ER-associated misfolded
protein catabolic process, protein kinase B signaling, Golgi
organization, Rap1 signaling pathway and glycosaminoglycan
biosynthesis. Furthermore, upregulated genes following
MEIS2-knockdown were identified to be involved in regulating cilium
assembly, response to stress, protein transport, cellular response
to starvation, negative regulation of TOR signaling, mitotic
spindle organization, p53 signaling pathway, FoxO signaling
pathway, carbohydrate digestion and absorption, endocytosis, and
PI3K-Akt signaling pathway (Fig.
6B-E).
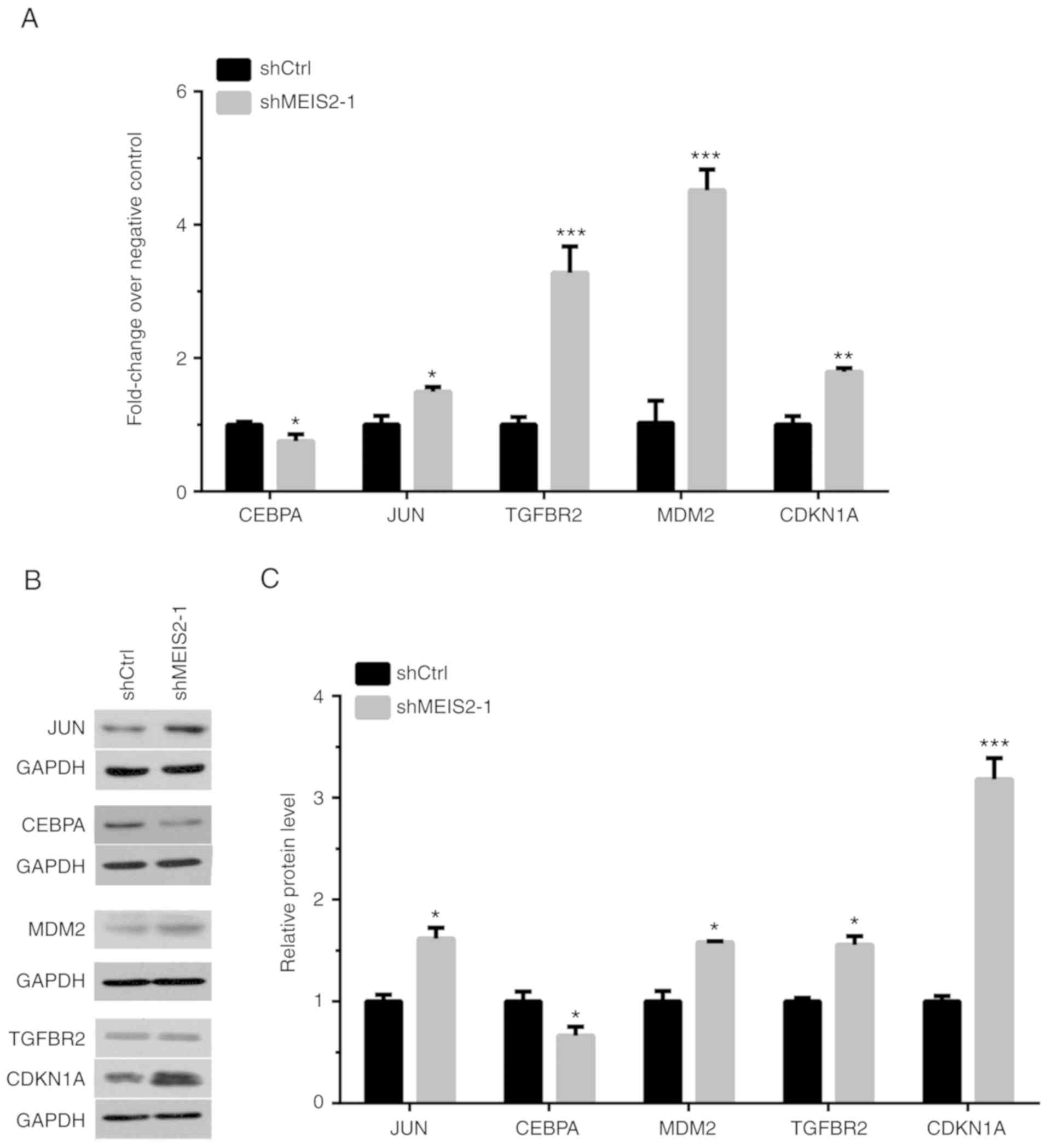
To further validate the microarray analysis results,
the expression levels of several key pathway regulators were
detected using RT-qPCR following MEIS2-knockdown in CRC cells. As
presented in Fig. 7A, it was
revealed that CEBPA was downregulated, and JUN, TGFBR2, MDM2 and
CDKN1A were upregulated following MEIS2-knockdown in HCT116 cells.
Western blot analysis also revealed similar results (Fig. 7B and C).
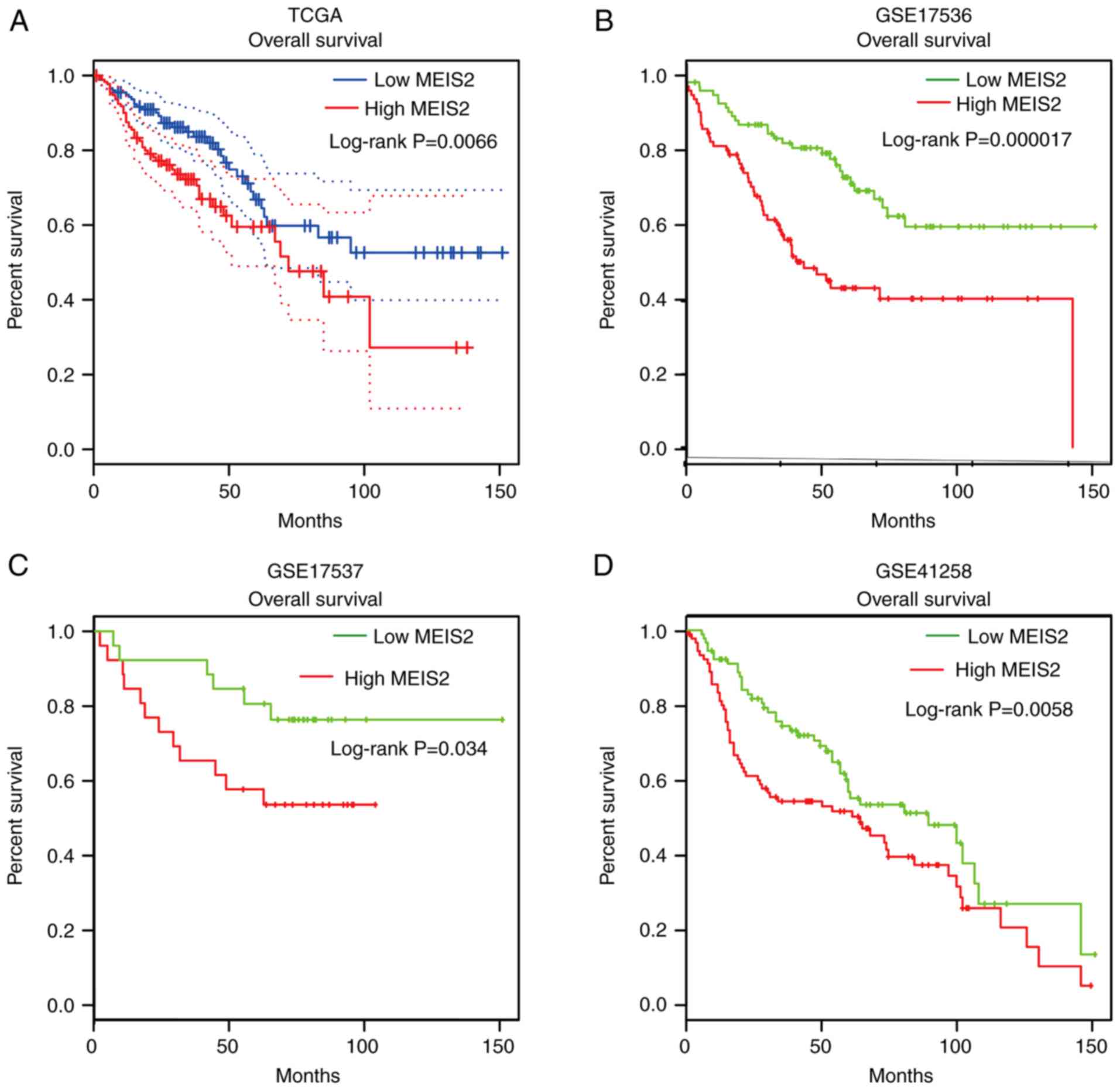
Higher MEIS2 expression is associated
with shorter overall survival time in CRC
Next, the protein levels of MEIS2 in CRC tissue and
normal colorectal tissue were analyzed using the Human Protein
Atlas database. As revealed in Fig.
S2, it was demonstrated that the MEIS2 protein levels in CRC
samples were high. In addition, the MEIS2 protein levels in normal
colon and rectum samples were also high. Furthermore, the present
study analyzed whether the dysregulation of MEIS2 was correlated
with overall survival time in CRC by analyzing a series of public
datasets, including The Cancer Genome Atlas (TCGA), GSE17536
(14), GSE17537 (14) and GSE41258 (15) datasets. Analysis of TCGA data
revealed that a high expression level of MEIS2 was correlated with
a shorter overall survival time for patients with CRC (Fig. 8A). Subsequently, the Gene Expression
Omnibus datasets, including GSE17536, GSE17537 and GSE41258, were
further analyzed to validate the aforementioned analysis. A similar
result was observed, where the overall survival time was shorter in
the MEIS2-high group compared with the MEIS2-low group (Fig. 8B-D). These results indicated that
the dysregulation of MEIS2 could serve as a novel biomarker for
CRC.
Discussion
MEIS2 has been identified to be involved in the
tumorigenesis of human cancer. Previous studies have demonstrated
that MEIS2 served crucial roles in cancer proliferation, and was
dysregulated in neuroblastoma (10), AML1-ETO-positive AML (11), and ovarian cancer (12). A recent study regarding prostate
cancer revealed that MEIS2 may be associated with the progression
of metastasis, since tumor expression of MEIS2 was correlated with
clinically metastatic disease (8,9).
However, to the best of our knowledge, the roles of MEIS2 in the
regulation of CRC metastasis progression remain unknown. For the
first time, the present study determined the effect of MEIS2 on CRC
metastasis using in vivo and in vitro assays.
Distant metastasis of cancer is a key cause of
CRC-associated mortality. In the past few decades, several genes
have been reported to be regulators of CRC metastasis. For example,
Qi et al (16) revealed that
BOP1, CKS2 and NFIL3 served as new targets of the Wnt pathway and
influenced CRC metastasis in mice. miR-224 has also been revealed
to act as a promoter of metastasis by suppressing SMAD4 in CRC
(17). However, to the best of our
knowledge, the detailed mechanisms underlying CRC metastasis remain
to be further investigated. The present study demonstrated that
silencing of MEIS2 significantly suppressed cell metastasis.
Furthermore, it was determined that MEIS2-knockdown suppressed EMT
progression by inducing E-cadherin expression and reducing Twist
and Snail expression. In addition, this result was validated using
an in vivo model by transplanting HCT116 cells into nude
mice through the tail vein. In summary, these analyses indicated
that MEIS2 acted as a promoter of metastasis in CRC.
To investigate the detailed mechanisms of MEIS2 in
CRC progression, the present study performed microarray analysis to
identify MEIS2 targets. GO analysis revealed that MEIS2 was
significantly associated with regulating the apoptotic process,
protein kinase B signaling, the Rap1 pathway, TOR signaling, the
FoxO pathway, the PI3K/Akt pathway, mitotic spindle organization
and the p53 pathway. A number of studies have indicated that these
pathways serve crucial roles in CRC progression. For example,
PI3K-Akt signaling has been identified to be involved in regulating
cell growth, cell apoptosis and cell metastasis in CRC (18–21).
Furthermore, in the present study, RT-qPCR and western blot
analysis demonstrated that the expression levels of TGFBR2, CDKN1A,
JUN and MDM2 increased, and the level of CEBPA decreased following
knockdown of MEIS2 in HCT116 cells. Numerous studies have reported
that these genes serve key roles in a number of human cancer types,
including CRC. For example, TGFBR2, a key member of the TGF-β
signaling pathway, has been revealed to act as a suppressor of
metastasis, since downregulation of TGFBR2 promoted migration and
invasion in CRC (22,23). CDKN1A, a widely studied cell cycle
regulator, was involved in regulating CRC proliferation (24,25).
JUN, a core member of the AP-1 complex, regulated CRC progression
via transcriptional regulation of various targets, including miR-22
(26–28).
Notably, the 5-year survival rate of metastatic CRC
is as low as ~10%. In the past few decades, numerous studies have
aimed to identify biomarkers for CRC. Several genes have been
identified to be dysregulated and associated with tumor progression
in CRC. For example, serum CNPY2 isoform 2 was revealed to be
upregulated in tumor samples and served as a novel biomarker for
early detection of CRC (29).
miR-6852 was downregulated and correlated with an improved
prognosis for patients with CRC (30). However, there remains an urgent
requirement to identify new biomarkers for CRC. By analyzing Human
Protein Atlas database, it was revealed that the MEIS2 protein
levels in CRC samples were high. In addition, MEIS2 protein levels
in normal colon and rectum samples were also high. By analyzing a
series of public datasets, including GSE17536, GSE17537, GSE41258
and TCGA datasets, it was revealed that a high MEIS2 expression
level was associated with a poor prognosis for patients with CRC.
These results revealed that MEIS2 may not regulate CRC
tumorigenisis but participated in regulation CRC progression.
In summary, to the best of our knowledge, the
present study demonstrated for that first time that MEIS2 acted as
a promoter of metastasis in CRC. Using in vivo and in
vitro experiments it was revealed that knockdown of MEIS2
significantly suppressed CRC migration, invasion and EMT.
Furthermore, microarray and bioinformatics analyses were performed
to investigate the underlying mechanisms of MEIS2 in the regulation
of CRC metastasis. In addition, it was identified that MEIS2 was
associated with a shorter overall survival time for patients with
CRC. In conclusion, the present study demonstrated that MEIS2 may
serve as a novel biomarker for CRC.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was financially supported by the
Natural Science Foundation of Zhejiang Province (LY18H160041,
LY15H160050, LY15H030014 and LY17H160064), the Funding Project of
Health and Family Planning Commission of Zhejiang Province
(2018KY217 and 2016KYA020), the Funding Project Administration of
Traditional Chinese Medicine of Zhejiang Province (2018ZA009), and
the Funding Project of CSCO, Chinese Society of Clinical Oncology
(Y-MX2016-047).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
RC, ST and XH conceived the experiments and drafted
the manuscript. ZW, BC and HY conducted the experiments. QD, BZ,
XM, WP, YT and QY analyzed and interpreted the data. All authors
read and approved the manuscript and agree to be accountable for
all aspects of the research in ensuring that the accuracy or
integrity of any part of the work are appropriately investigated
and resolved.
Ethics approval and consent to
participate
All in vivo study protocols were approved by
the Shanghai Medical Experimental Animal Care Commission (Approval
ID: ShCI-14-008).
Patient consent for publication
Not applicable.
Competing interests
The authors state that they have no competing
interests.
References
|
1
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Roshan MH, Tambo A and Pace NP: The role
of testosterone in colorectal carcinoma: Pathomechanisms and open
questions. EPMA J. 7:222016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Calon A, Espinet E, Palomo-Ponce S,
Tauriello DV, Iglesias M, Céspedes MV, Sevillano M, Nadal C, Jung
P, Zhang XH, et al: Dependency of colorectal cancer on a
TGF-beta-driven program in stromal cells for metastasis initiation.
Cancer Cell. 22:571–584. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sakuma K, Sasaki E, Kimura K, Komori K,
Shimizu Y, Yatabe Y and Aoki M: HNRNPLL, a newly identified
colorectal cancer metastasis suppressor, modulates alternative
splicing of CD44 during epithelial-mesenchymal transition. Gut.
67:1103–1111. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yan G, Zhao H, Zhang Q, Zhou Y, Wu L, Lei
J, Wang X, Zhang J, Zhang X, Zheng L, et al: A
RIPK3-PGE2 circuit mediates myeloid-derived suppressor
cell-potentiated colorectal carcinogenesis. Cancer Res.
78:5586–5599. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Geerts D, Schilderink N, Jorritsma G and
Versteeg R: The role of the MEIS homeobox genes in neuroblastoma.
Cancer Lett. 197:87–92. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Salzberg A, Elias S, Nachaliel N, Bonstein
L, Henig C and Frank D: A Meis family protein caudalizes neural
cell fates in Xenopus. Mech Dev. 80:3–13. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bhanvadia RR, VanOpstall C, Brechka H,
Barashi NS, Gillard M, McAuley EM, Vasquez JM, Paner G, Chan WC,
Andrade J, et al: MEIS1 and MEIS2 expression and prostate cancer
progression: A role for HOXB13 binding partners in metastatic
disease. Clin Cancer Res. 24:3668–3680. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jeong JH, Park SJ, Dickinson SI and Luo
JL: A constitutive intrinsic inflammatory signaling circuit
composed of miR-196b, Meis2, PPP3CC, and p65 drives prostate cancer
castration resistance. Mol Cell. 65:154–167. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zha Y, Xia Y, Ding J, Choi JH, Yang L,
Dong Z, Yan C, Huang S and Ding HF: MEIS2 is essential for
neuroblastoma cell survival and proliferation by transcriptional
control of M-phase progression. Cell Death Dis. 5:e14172014.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Vegi NM, Klappacher J, Oswald F, Mulaw MA,
Mandoli A, Thiel VN, Bamezai S, Feder K, Martens JHA, Rawat VPS, et
al: MEIS2 is an oncogenic partner in AML1-ETO-positive AML. Cell
Rep. 16:498–507. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Crijns AP, de Graeff P, Geerts D, Ten Hoor
KA, Hollema H, van der Sluis T, Hofstra RM, de Bock GH, de Jong S,
van der Zee AG, et al: MEIS and PBX homeobox proteins in ovarian
cancer. Eur J Cancer. 43:2495–2505. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2−ΔΔCT method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Smith JJ, Deane NG, Wu F, Merchant NB,
Zhang B, Jiang A, Lu P, Johnson JC, Schmidt C, Bailey CE, et al:
Experimentally derived metastasis gene expression profile predicts
recurrence and death in patients with colon cancer.
Gastroenterology. 138:958–968. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sheffer M, Bacolod MD, Zuk O, Giardina SF,
Pincas H, Barany F, Paty PB, Gerald WL, Notterman DA and Domany E:
Association of survival and disease progression with chromosomal
instability: A genomic exploration of colorectal cancer. Proc Natl
Acad Sci USA. 106:7131–7136. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Qi J, Yu Y, Akilli Ozturk O, Holland JD,
Besser D, Fritzmann J, Wulf-Goldenberg A, Eckert K, Fichtner I and
Birchmeier W: New wnt/beta-catenin target genes promote
experimental metastasis and migration of colorectal cancer cells
through different signals. Gut. 65:1690–1701. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang Z, Yang J, Di J, Cui M, Xing J, Wu F,
Wu W, Yang H, Zhang C, Yao Z, et al: Downregulated USP3 mRNA
functions as a competitive endogenous RNA of SMAD4 by sponging
miR-224 and promotes metastasis in colorectal cancer. Sci Rep.
7:42812017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yang L, Liu Y, Wang M, Qian Y, Dai X, Zhu
Y, Chen J, Guo S and Hisamitsu T: Celastrus orbiculatus extract
triggers apoptosis and autophagy via PI3K/Akt/mTOR inhibition in
human colorectal cancer cells. Oncol Lett. 12:3771–3778. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Qian DC, Xiao X, Byun J, Suriawinata, Her
SC, Amos CI and Barth RJ Jr: PI3K/Akt/mTOR signaling and plasma
membrane proteins are implicated in responsiveness to adjuvant
dendritic cell vaccination for metastatic colorectal cancer. Clin
Cancer Res. 23:399–406. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xu L, Zhang Y, Wang H, Zhang G, Ding Y and
Zhao L: Tumor suppressor miR-1 restrains epithelial-mesenchymal
transition and metastasis of colorectal carcinoma via the MAPK and
PI3K/AKT pathway. J Transl Med. 12:2442014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Pandurangan AK: Potential targets for
prevention of colorectal cancer: A focus on PI3K/Akt/mTOR and Wnt
pathways. Asian Pac J Cancer Prev. 14:2201–2205. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Fricke F, Lee J, Michalak M, Warnken U,
Hausser I, Suarez- Carmona M, Halama N, Schnölzer M, Kopitz J and
Gebert J: TGFBR2-dependent alterations of exosomal cargo and
functions in DNA mismatch repair-deficient HCT116 colorectal cancer
cells. Cell Commun Signal. 15:142017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lee J, Katzenmaier EM, Kopitz J and Gebert
J: Reconstitution of TGFBR2 in HCT116 colorectal cancer cells
causes increased LFNG expression and enhanced
N-acetyl-D-glucosamine incorporation into Notch1. Cell Signal.
28:1105–1113. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ding J, Li J, Wang H, Tian Y, Xie M, He X,
Ji H, Ma Z, Hui B, Wang K and Ji G: Long noncoding RNA CRNDE
promotes colorectal cancer cell proliferation via epigenetically
silencing DUSP5/CDKN1A expression. Cell Death Dis. 8:e29972017.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Dunlop MG, Dobbins SE, Farrington SM,
Jones AM, Palles C, Whiffin N, Tenesa A, Spain S, Broderick P, Ooi
LY, et al: Common variation near CDKN1A, POLD3 and SHROOM2
influences colorectal cancer risk. Nat Genet. 44:770–776. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liu Y, Chen X, Cheng R, Yang F, Yu M, Wang
C, Cui S, Hong Y, Liang H, Liu M, et al: The Jun/miR-22/HuR
regulatory axis contributes to tumourigenesis in colorectal cancer.
Mol Cancer. 17:112018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bae JA, Yoon S, Park SY, Lee JH, Hwang JE,
Kim H, Seo YW, Cha YJ, Hong SP, Kim H, et al: An unconventional
KITENIN/ErbB4-mediated downstream signal of EGF upregulates c-jun
and the invasiveness of colorectal cancer cells. Clin Cancer Res.
20:4115–4128. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Takeda K, Kinoshita I, Shimizu Y, Ohba Y,
Itoh T, Matsuno Y, Shichinohe T and Dosaka-Akita H:
Clinicopathological significance of expression of p-c-Jun, TCF4 and
beta-Catenin in colorectal tumors. BMC Cancer. 8:3282008.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Peng J, Ou Q, Pan Z, Zhang R, Zhao Y, Deng
Y, Lu Z, Zhang L, Li C, Zhou Y, et al: Serum CNPY2 isoform 2
represents a novel biomarker for early detection of colorectal
cancer. Aging. 10:1921–1931. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Cui BH and Hong X: miR-6852 serves as a
prognostic biomarker in colorectal cancer and inhibits tumor growth
and metastasis by targeting TCF7. Exp Ther Med. 16:879–885.
2018.PubMed/NCBI
|