Introduction
Breast cancer is the most frequently diagnosed
cancer and accounts for 30% of all new malignancies diagnosed in
females (1). Although advances in
screening and treatment have improved survival rates of patients
with breast cancer, currently, breast cancer remains the leading
cause of cancer-related death among females worldwide (2). Approximately 90% of breast cancer deaths
are caused by local invasion and distant metastasis of tumor cells
(3). The metastatic cascade
represents a complex multistage process which includes local tumor
cell invasion, entry into the blood or lymphatic system followed by
the exit of cancer cells from the circulation and formation of
metastases at the distal sites (4).
Dissemination of cancer cells from the primary tumor is an
essential step for cancer invasion and metastasis (5). Thus, identifying novel therapeutic
targets to prevent these processes is of great importance for the
treatment of breast cancer.
MicroRNAs (miRNAs) are a family of endogenous,
evolutionarily conserved, non-coding RNAs that can
post-transcriptionally affect gene expression by negatively
regulating protein-coding mRNAs through base-pair matching within
the target mRNA 3′ untranslated region (UTR) (6). Abundant evidence has demonstrated that
miRNAs play important regulatory roles in diverse physiological and
pathological processes through regulating the expressions of target
genes (7–9). Accumulated evidence indicates aberrant
miRNA expression may profoundly influence cancer-related signaling
pathways and play critical roles in almost all aspects of cancer
biology such as cell differentiation, proliferation and apoptosis,
migration, metabolism and stem cell renewal (10,11).
Increasing evidence has suggested that the aberrant expression of
miRNAs contributes to the initiation and progression of cancer by
regulating the expression of their downstream target genes. In
addition, it has been reported that the pattern of miRNA expression
can be correlated with cancer type, stage, and other clinical
variables, which suggests that miRNAs can be used as a potential
novel biomarker for cancer detection and prognosis, as well as a
promising target in cancer therapy (12).
Previous studies have shown that dysregulation of
miRNA expression was implicated in the progression of breast cancer
(13) and varies with lymph node
metastasis and other clinicopathological features (14). These miRNAs can be classified as
oncogenes (oncomirs) or tumor-suppressor genes according to their
potential roles played in the pathogenesis and development of
breast cancer (15). Generally,
oncomirs are upregulated in cancer cells and exert tumor-promoting
activity by targeting tumor-suppressor genes. Examples of breast
cancer oncomirs are miR-10b, miR-21, miR-155, miR-373, and miR-520c
(16). Tumor-suppressor miRNAs
exhibit a lower expression in cancer cells and exert antitumor
effect by suppressing oncogene expression. Emerging evidence
suggests that miR-139-3p may serve as a tumor suppressor with
several types of cancer. It was reported that miR-139-3p level in
colorectal cancer patient serum and cancer tissues was
significantly lower than in control subjects, which suggests that
miR-139-3p may be used as a biomarker for colorectal cancer
diagnosis (17). In addition, lower
levels of miR-139-3p were significantly associated with poor
overall survival in colon cancer patients (18). Furthermore, miR-139-3p was
significantly downregulated in bladder cancer tissues, and reduced
miR-139-3p expression is associated with enhanced migration and
invasion abilities of bladder cancer cells (19). By analyzing breast cancer tissue
specimens, we found that miR-139-3p was downregulated in breast
cancer tissues. However, the role and underlying mechanism of
action of miR-139-3p in the progression of breast cancer remains
unknown.
In the present study, we aimed to detect the
expression of miR-139-3p in breast cancer, and analyzed the effect
of miR-139-3p on the migration and invasion abilities of breast
cancer cells, and further explored the underlying mechanism. We
found that miR-139-3p was downregulated in breast cancer tissues,
and decreased miR-139-3p expression was associated with a poor
prognosis in breast cancer patients. In vitro experiments
indicated that overexpression of miR-139-3p inhibited cell
proliferation, migration and invasion of breast cancer. Further
study revealed that RAB1A is a potential target of miR-139-3p.
These findings suggest that miR-139-3p may serve as a tumor
suppressor gene in breast cancer by targeting RAB1A.
Materials and methods
Ethics statement
This study was approved by the Academic Committee of
the Ethical Committee of the First Affiliated Hospital of Medical
College, Xi'an Jiaotong University, Xi'an, China, and conducted
according to the principles expressed in the Declaration of
Helsinki. Specimens were obtained with informed consent from all
patients.
Clinical tissues and cell culture
From January 2017 to December 2017, 86 pairs of
breast cancer tissues and corresponding adjacent non-cancerous
tissues were obtained from patients (ranging in age from 20 to 79
years with median age 52.0 years; including 11 intraductal
carcinoma, 35 infiltrative ductal carcinoma, 33 infiltrative
lobular carcinoma, and 7 other infiltrative carcinoma) who had been
diagnosed with primary breast cancer and underwent surgical
treatment at the Department of Breast Surgery in the First
Affiliated Hospital of Xi'an Jiaotong University. Tissues and
information of patients were gained with written informed consent.
No chemotherapy or radiotherapy was applied before surgery in any
of the included patients. After surgical removal, the tissues were
collected and immediately frozen in liquid nitrogen, and stored at
−80°C or embedded in paraffin. Safety margin is defined as 2 cm
from the margin of cancerous tissues. The non-cancerous tissues
were confirmed by two independent experienced pathologists after
H&E staining. The breast cancer cell lines MDA-MB-231, MCF-7,
MDA-MB-468, MDA-MB-453, SK-BR-3 and T-47D were obtained from the
Chinese Academy of Sciences Cell Bank of Type Culture Collection
(CBTCCCAS, Shanghai, China). All cells were cultured in recommended
medium containing 10% fetal bovine serum (FBS; BioWest, Nuaillé,
France) and 1% penicillin/streptomycin (Sigma, St. Louis, MO, USA)
in a humidified incubator containing 5% CO2 at 37°C.
RT-qPCR
Total RNA was isolated from breast cancer cells or
tissues using TRIzol reagent (Invitrogen, Carlsbad, CA, USA)
according to the manufacturer's instructions. cDNA was synthesized
from 2 µg of total RNA using a reverse transcription kit (Takara
Biotechnology, Dalian, China) according to the manufacturer's
instructions. RT-qPCR products were amplified using a miScript
SYBR-Green PCR Kit (Qiagen, Hilden, Germany) following the
manufacturer's instructions. Amplicons were detected by an iQ5
Multicolor Real-Time PCR Detection System (Bio-Rad, Hercules, CA,
USA). The following PCR conditions were used: One cycle at 94°C for
5 min, followed by 35 cycles of denaturation at 95°C for 5 sec and
annealing and extension at 60°C for 30 sec. Gene expression levels
were calculated using the ΔΔ Cq method (20). GAPDH was selected as the reference
gene. The PCR primer sequences used were: miR-139-3p,
5′-GGAGACGCGGCCCTGTTGGAGT-3′; RAB1A, 5′-GGAGCCCATGGCATCATA-3′
(forward) and 5′-TTGAAGGACTCCTGATCTGTCA-3′ (reverse); GAPDH:
5′-CTCTGATTTGGTCGTATTGGG-3′ (forward) and
5′-TGGAAGATGGTGATGGGATT-3′ (reverse). All primers used in this
study were synthesized by DingGuo ChangSheng Biotechnology Co.,
Ltd. (Beijing, China). Expression of miR-139-3p was analyzed by
relative quantity (RQ) using the equation RQ=2−ΔΔCq
(Cq=quantification cycle to detect fluorescence) according to a
previous study (21). The mean RQ was
2.32±1.19, and 65 (75.6%) patients were classified as low
miR-139-3p (≤2.32) and 21 (24.4%) as high miR-139-3p
(>2.32).
MTT assay
Cell proliferation was measured with the
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
method. After the cells had been transfected for 24 h, they were
seeded in 96-well microtiter plates (5,000 cells per well) and
cultured for the indicated times. Then 20 µl MTT (5 mg/ml diluted
in 1X PBS) was added to each well, and incubated for an additional
4 h at 37°C. After incubation, 150 µl of dimethyl sulfoxide (DMSO)
was added to dissolve the crystals. Finally, the absorbance of each
well was measured at 490 nm using a microplate reader (Bio-Rad,
Hercules, CA, USA). Tests were performed in triplicate.
Cell migration and invasion
assays
Matrigel uncoated and coated Transwell inserts (8 µm
pore size; Millipore, Billerica, MA, USA) were used to examine the
migration and invasion abilities of breast cancer cell in
vitro. Briefly, a total of 5×104 transfected cells
were suspended in 150 µl serum-free DMEM and added to the upper
chamber of the Transwell chambers, and 600 µl of 20% FBS containing
DMEM was loaded in the lower chamber. After 48 h, the non-migratory
cells on the upper side were removed using a cotton swab, cells
located on the lower surface of the chamber were fixed in 4%
paraformaldehyde for 20 min and then stained with 0.1% crystal
violet for 15 min. The mean number of migrated or invaded cells was
counted by averaging the numbers of cells in 10 fields in both
inserts under a microscope (Olympus, Tokyo, Japan). All experiments
were performed in duplicate and repeated at least three times.
Western blot analysis
Total proteins were extracted from human breast
cancer cells with RIPA lysis buffer and quantified with a BCA
Protein Assay Kit (Qiagen, Valencia, CA, USA). Protein samples (30
µg) were run on 10% SDS-PAGE gels, and then were transferred to
PVDF membranes (Millipore, Billerica, MA, USA). After being blocked
with 5% skimmed milk at room temperature for 2 h, the membranes
were incubated with primary antibodies: Against E-cadherin (Cell
Signaling Technology, Beverly, MA, USA; rabbit monoclonal Ab no.
3195, 1:1,000 dilution)/N-cadherin (Cell Signaling Technology;
rabbit monoclonal Ab no. 13116, 1:1,000 dilution)/MMP-2 (Cell
Signaling Technology; rabbit monoclonal Ab no. 40994, 1:800
dilution)/MMP-9 (Cell Signaling Technology; rabbit monoclonal Ab
no. 13667, 1:750 dilution) and GAPDH (Abcam, Cambridge, MA, USA;
rabbit monoclonal EPR16891, 1:750 dilution) overnight at 4°C. After
washing, the membranes were incubated with HRP-conjugated secondary
antibodies (Abcam, Cambridge, MA, USA; goat anti-rabbit, ab7090) at
room temperature for 2 h. After washing, the immunoreactive
proteins were visualized using an enhancing chemiluminescence kit
(Amersham, Little Chalfont, UK). GAPDH was used as a protein
loading control.
Colony formation assay
For colony formation, 1,000 transfected cells were
seeded into 35-mm petri dish and cultured for 2 weeks. At indicated
time points (24, 48 and 72 h), colonies were fixed in 4%
paraformaldehyde for 20 min and then stained with 0.1% crystal
violet for 15 min. Images were captured under a microscope
(Olympus, Tokyo, Japan) at a magnification of ×400.
Immunohistochemical (IHC)
analysis
The tissue sections were deparaffinized in xylene
and rehydrated with ethanol. Endogenous peroxidase activity was
blocked via incubating the sections with hydrogen peroxide (0.3% in
methanol) at room temperature for 10 min. After blocking with 10%
goat serum, the slides were incubated overnight at 4°C with primary
monoclonal antibodies for RAB1A (1:200, Cell Signaling Technology).
After several rinses in phosphate-buffered saline, the slides were
incubated in the biotinylated secondary antibodies, and antibodies
were fixed using the streptavidin-peroxidase (SP)
immunohistochemical method (22). The
semi-quantitative results were analyzed according to the staining
intensity and percentage of positively labeled cells.
Cell transfection
miR-139-3p mimics/inhibitors and corresponding
control vectors were obtained from GenePharma (Shanghai, China).
The RAB1A overexpression plasmids (pcDNA3.1-RAB1A) were synthesized
by Bioworld Biotech Co., Ltd. (Shanghai, China). Breast cancer
cells were seeded in 24-well plates, and once cells had grown to
70% confluence, transfection with the above vectors was conducted
using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according
to the manufacturer's instructions. Cells were harvested after 48 h
for subsequent analysis.
Luciferase reporter assay
The 3′-UTR sequence of RAB1A is predicted to
interact with miR-139-3p by binding to the corresponding target
sequence or a mutated sequence within the predicted target site.
The normal and mutated 3′-UTRs were therefore inserted into the
pGL3 reporter luciferase vector (GeneChem). The breast cancer
MDA-MB-231 and MCF-7 cells were seeded in 24-well plates. Aliquots
of 50 nM of miR-139-3p mimic or control miRNA were co-transfected
with 0.1 mg of the pGL3-30UTR wild or mutant RAB1A plasmid DNA
using the Lipofectamine 2000 reagent (Invitrogen). After
transfection for 48 h, luciferase activity was measured using the
dual luciferase reporter assay system (Promega, Madison, WI, USA)
according to the manufacturer's instructions. The relative
luciferase activity was normalized to Renilla luciferase
activity. The experiment was performed in triplicate.
Bioinformatics prediction
TargetScan Human 7.0 (http://www.targetscan.org/) was used to predict the
potential targets of miR-139-3p.
The Kaplan-Meier plotter
The prognostic value of miR-139-3p expression was
evaluated using an online database, Kaplan-Meier Plotter
(http://kmplot.com/analysis/), according
to a previous study (23). To analyze
the overall survival (OS) of patients with breast cancer, patient
samples were split into two groups by median expression (high vs.
low expression) and assessed by a Kaplan-Meier survival plot. The
hazard ratio (HR), 95% confidence intervals (CI), and log-rank
P-values were calculated and displayed.
Statistical analysis
All statistical analyses were conducted using SPSS
16.0 software (SPSS; Inc., Chicago, IL, USA) and data were
presented as the mean ± standard deviation. Student's t-test was
performed to assess difference between two groups. Differences
between multiple groups were assessed using one-way analysis of
variance (ANOVA) with Dunnett's test for post hoc analysis. The
relationship between miR-139-3p expression level and
clinicopathological features of the patients was analyzed using the
Chi-square test. Each experiment was performed at least for three
times. Differences were defined as significant at P<0.05.
Results
miR-139-3p was down-regulated in
breast cancer and decreased miR-139-3p expression is associated
with a poor prognosis in breast cancer patients
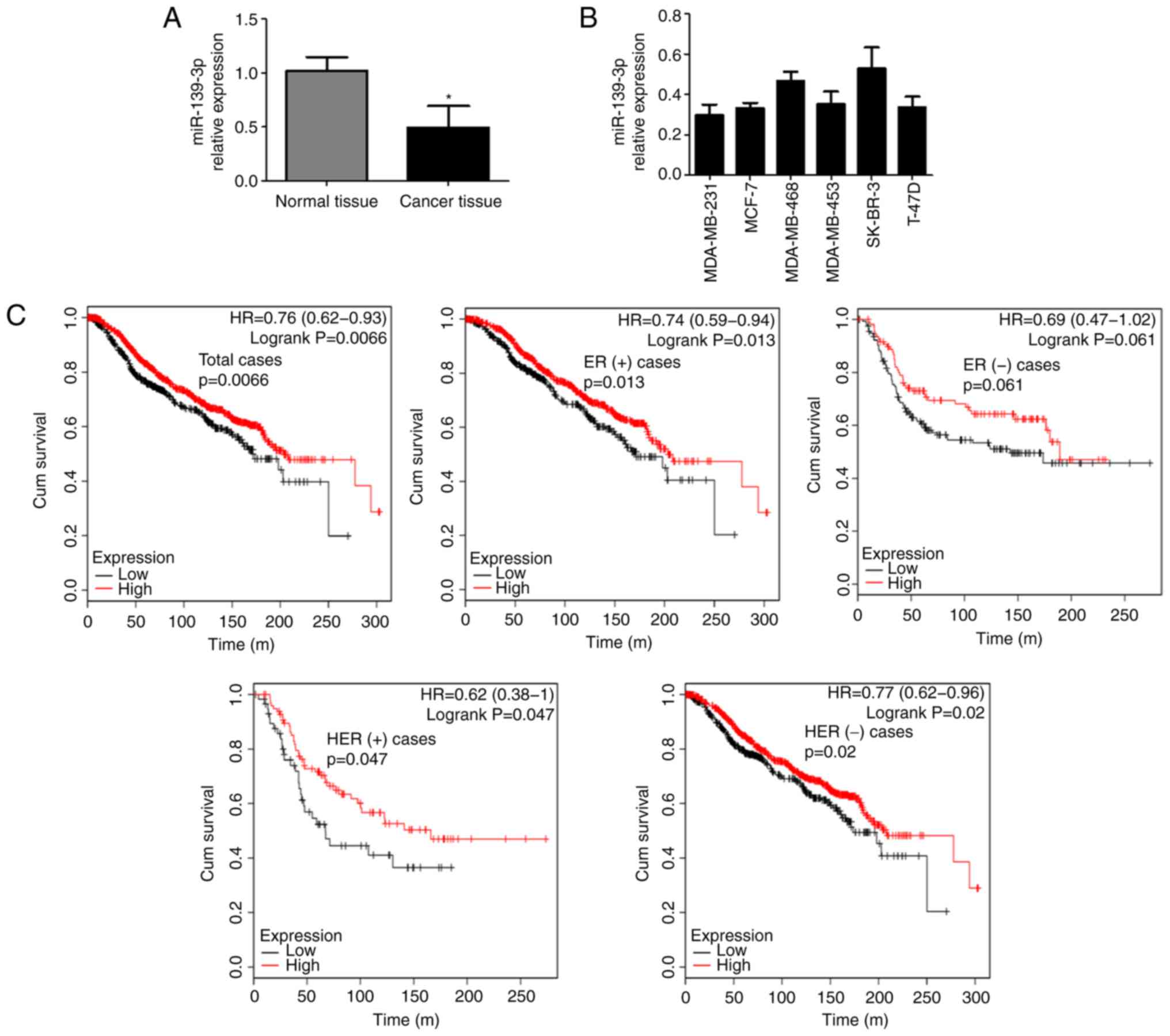
Firstly, we examined the miR-139-3p expression in
breast cancer. In total, 86 pairs of breast cancer tissues and
adjacent non-cancerous tissues were collected and subjected to
RT-qPCR analysis. As shown in Fig.
1A, the expression levels of miR-139-3p in breast cancer
tissues were significantly decreased compared with those in
non-cancerous breast tissues. By analyzing the relationship between
clinicopathological features and miR-139-3p expression, we found
that low miR-139-3p expression was significantly associated with
TNM stage (P=0.02) and lymph node metastasis (P=0.001), but not
with patients' age, tumor size, estrogen receptor (ER) status,
progesterone receptor (PR) status, human epidermal growth factor
receptor (HER) status and distant metastasis (P>0.05, Table I). We then detected miR-139-3p
expression in breast cancer cell lines (MDA-MB-231, MCF-7,
MDA-MB-468, MDA-MB-453, SK-BR-3 and T-47D). The results showed that
miR-139-3p was widely expressed in all tested breast cancer cell
lines (Fig. 1B). We further evaluated
the relationships of miR-139-3p expression with OS of breast cancer
patients using an online database (http://kmplot.com/analysis/). The Kaplan-Meier curve
and log-rank test analyses revealed that low miR-139-3p expression
was associated with poorer overall survival (OS) in breast cancer
patients (Fig. 1C). Although there
was no statistical difference in ER (−) population, there was still
a trend that cases with low miR-139-3p expression experienced a
worse prognosis regardless of ER or HER status. These results
suggest that miR-139-3p is decreased in breast cancer and
associated with worse prognosis of breast cancer patients, which
indicated that miR-139-3p may exert anti-tumor effect in breast
cancer.
 | Table I.Relationship between the expression of
miR-139-3p and the clinical pathological characteristics in breast
cancer tissues. |
Table I.
Relationship between the expression of
miR-139-3p and the clinical pathological characteristics in breast
cancer tissues.
|
|
| miR-139-3p
expression |
|
|---|
|
|
|
|
|
|---|
| Parameters | n | High (n=21) | Low (n=65) | P-value |
|---|
| Age |
|
|
| 0.78 |
| ≤50 | 35 | 8 | 27 |
|
|
>50 | 51 | 13 | 38 |
|
| Tumor size |
|
|
| 0.38a |
| ≤2
cm | 15 | 4 | 11 |
|
| >2
cm, <5 cm | 67 | 15 | 52 |
|
| >5
cm | 4 | 2 | 2 |
|
| TNM stage |
|
|
| 0.02 |
| Early
stage (I/II) | 39 | 14 | 25 |
|
| Late
stage (III/IV) | 47 | 7 | 40 |
|
| ER status |
|
|
|
|
|
Negative | 41 | 11 | 30 | 0.62 |
|
Positive | 45 | 10 | 35 |
|
| PR status |
|
|
| 0.45 |
|
Negative | 43 | 9 | 34 |
|
|
Positive | 43 | 12 | 31 |
|
| HER status |
|
|
| 0.72 |
|
Negative | 64 | 15 | 49 |
|
|
Positive | 22 | 6 | 16 |
|
| Lymph node
metastasis |
|
|
| 0.001 |
|
Negative | 35 | 15 | 20 |
|
|
Positive | 51 | 6 | 45 |
|
| Distant
metastasis |
|
|
|
|
| No | 73 | 20 | 53 | 0.17a |
|
Yes | 13 | 1 | 12 |
|
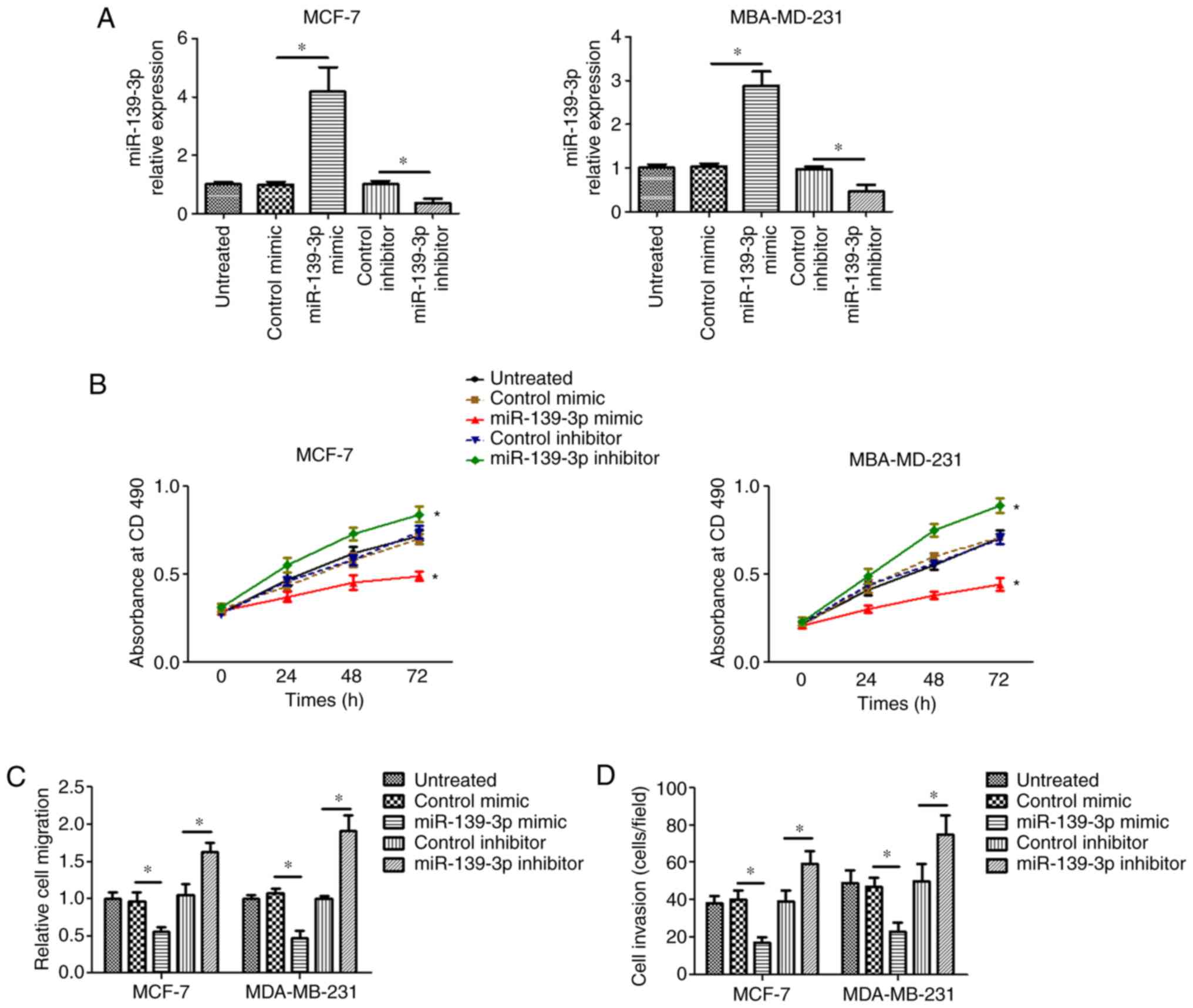
Overexpression of miR-139-3p inhibited
cell proliferation, migration and invasion of breast cancer
Next, we investigated the effect of miR-139-3p on
the proliferation, migration and invasion of breast cancer cells.
miR-139-3p mimic or inhibitor was transfected into two breast
cancer cell lines with different ER status [MCF-7, ER (+);
MBA-MD-231, ER (−)] to increase or decrease miR-139-3p expression
in these two cell lines separately (Fig.
2A). The MTT method was used to investigate the effect of
miR-139-3p on the proliferation of breast cancer cells. As shown in
Fig. 2B, overexpression of miR-139-3p
with miR-139-3p mimic significantly suppressed the proliferative
ability of MCF-7 and MBA-MD-231 cells. In contrast, downregulation
of miR-139-3p with miR-139-3p inhibitor increased the proliferative
ability of MCF-7 and MBA-MD-231 cells. In cell migration
experiments (Fig. 2C), overexpression
of miR-139-3p with miR-139-3p mimic significantly decreased the
migratory ability in MCF-7 and MBA-MD-231 cells. While the
migratory ability of MCF-7 and MBA-MD-231 cells were significantly
increased after silencing miR-139-3p. The Transwell assay (Fig. 2D) showed that the invasive ability of
breast cancer cells was decreased when cells were transfected with
miR-139-3p mimic. By contrast, silencing miR-139-3p expression by
miR-139-3p inhibitor enhanced the invasive ability of breast cancer
cells. These results indicate that miR-139-3p may play a tumor
suppression role in breast cancer progression.
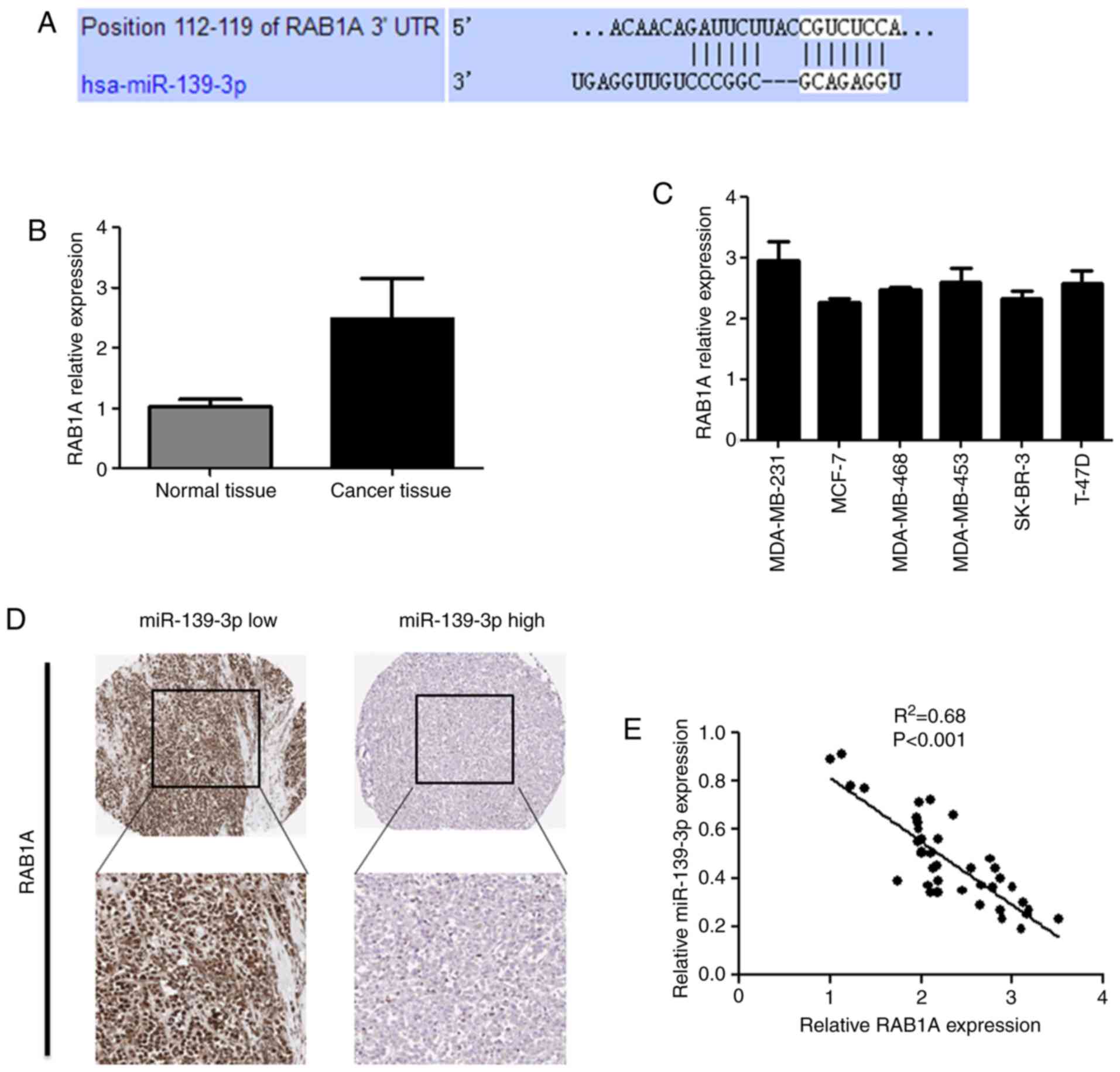
RAB1A is a potential target of
miR-139-3p and is inversely correlated with miR-139-3p expression
in breast cancer tissues
We performed a target search using TargetScan
(TargetScan Human 7.0 software) and predicted that RAB1A is a
direct target of miR-139-3p (Fig.
3A). We next detected the expression of RAB1A in breast cancer
tissues by RT-qPCR. The result showed that the expression level of
RAB1A was significantly increased in breast cancer tissues compared
with the normal breast tissues (Fig.
3B). Similarly, higher expression levels of RAB1A were detected
in breast cancer cell lines (Fig.
3C). Moreover, we could see a negative correlation between
RAB1A expression and miR-139-3p expression from immunohistochemical
staining for RAB1A (Fig. 3D). This
was further confirmed by a linear correlation analysis, which
showed that RAB1A expression was inversely proportional to
miR-139-3p expression (Fig. 3E).
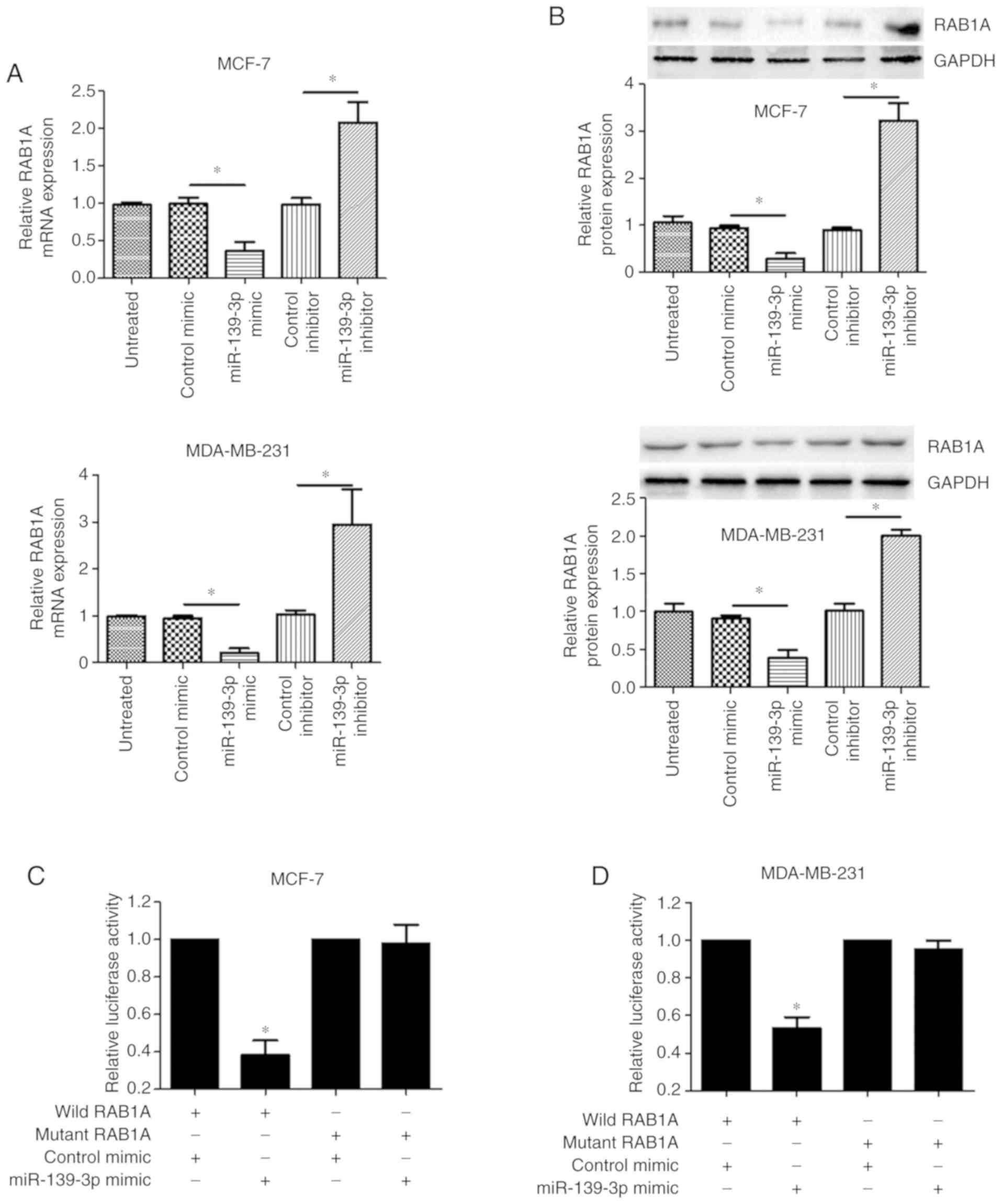
To verify that RAB1A is a potential target of
miR-139-3p, we analyzed the relationship between miR-139-3p and
RAB1A. Both real-time PCR (Fig. 4A)
and western blot analysis (Fig. 4B)
showed that overexpression of miR-139-3p with miR-139-3p mimic led
to a corresponding decrease of RAB1A expression in MCF-7 and
MDA-MB-231 cells. Conversely, RAB1A expression was upregulated in
MCF-7 cells with reduced levels of miR-139-3p by miR-139-3p
inhibitor. To further detect whether RAB1A was indeed directly
regulated by miR-139-3p in breast cancer cells, a dual luciferase
assay was conducted. The result showed that the luciferase activity
in the wild RAB1A-transfected cells, was significantly decreased
compared to the luciferase activity in the mutant cells (Fig. 4C and D). Collectively, these data
suggest that RAB1A is a direct target gene of miR-139- 3p in breast
cancer.
miR-139-3p suppresses cell migration,
invasion and EMT by targeting RAB1A in breast cancer
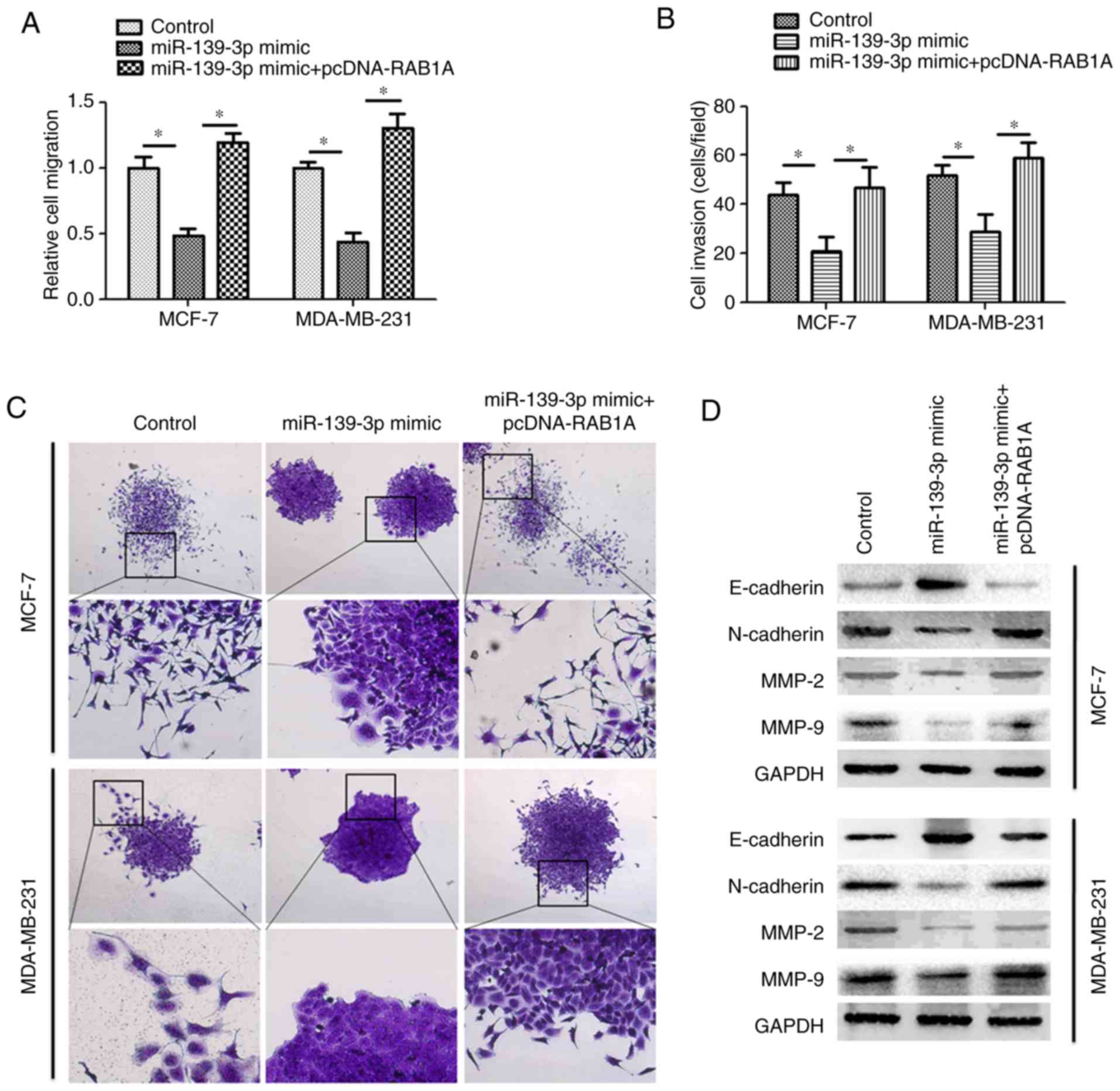
To further investigate whether miR-139-3p suppresses
cell migration and invasion by targeting RAB1A, we overexpressed
miR-139-3p and RAB1A in MCF-7 cells by co-transfecting pcDNA-RAB1A
and miR-139-3p mimic into MCF-7 cells. Consistently, the results
(Fig. 5A and B) showed that MCF-7
cells only transfected with miR-139-3p mimic showed reduced cell
migration and invasion abilities. However, when MCF-7 cells were
co-transfected with miR-139-3p mimic and pcDNA-RAB1A, the migration
and invasion properties were significantly increased as compared
with the cells transfected with miR-139-3p mimic only. When
conducting clone formation experiments, it was found that cancer
cells tended to form tightly connected clones when ectopically
overexpressing miR-139-3p in MCF-7 and MDA-MB-231 cells (Fig. 5C). By contrast, cancer cells
co-transfected with miR-139-3p mimic and pcDNA-RAB1A exhibited
dispersed and highly scattered growth patterns and adopted a
spindle-shaped morphology during clone formation experiments. This
is similar to the processes occurring during cancer invasion and
metastasis termed as epithelial-to-mesenchymal transition (EMT)
(24,25). Western blotting was performed to
detect the expression of MMP-2, MMP-9, E-cadherin and N-cadherin.
As expected, both MCF-7 and MDA-MB-231 cells showed reduced protein
expression of MMP-2 and MMP-9 when transfected with miR-139-3p
mimic. However, this effect was counteracted by co-transfection of
MCF-7 and MDA-MB-231 cells with miR-139-3p mimic and pcDNA-RAB1A.
More importantly, both MCF-7 and MDA-MB-231 cells showed increased
E-cadherin and reduced N-cadherin when transfected with miR-139-3p
mimic, while this cadherin switching was prevented by
overexpressing RAB1A ectopically (Fig.
5D). These data indicate that RAB1A plays an important role in
miR-139-3p-induced cell migration, invasion and EMT in breast
cancer cells.
Discussion
In this study, we investigated the expression,
function, and mechanisms underlying the activities of miR-139-3p in
breast cancer. Through bioinformatics analysis and in vitro
experimental studies, we found that miR-139-3p was downregulated in
breast cancer tissues and cell lines, and decreased miR-139-3p is
associated with a poor prognosis in breast cancer patients.
Overexpression of miR-139-3p significantly inhibits cell migration,
invasion, and EMT by suppressing RAB1A expression. These data
suggested that miR-139-3p might play a tumor suppressive role
during breast cancer progression.
miR-139-3p has been found to be significantly
downregulated in HPV-positive HNC (head and neck cancer) and
cervical cancer cases. Further study revealed that HPV-16 mediated
downregulation of miR-139-3p by promoter methylation created a
favorable environment for viral replication and oncogenesis via
activating pro-oncogenic pathways. In contrast, miR-139-3p
overexpression decreased cell proliferation, migration, and
increased the sensitivity of HPV-16-positive cells to chemotherapy
(26). Moreover, reduced miR-139-3p
expression in cervical cancer tissues and cell lines has been
confirmed by another study (27),
which revealed that overexpression of miR-139-3p suppressed cell
proliferation, migration, invasion and induced apoptosis via
downregulation of NOB1 expression. In the present study, we found
that miR-139-3p was downregulated in breast cancer tissues and cell
lines, and decreased miR-139-3p level is associated with a poor
prognosis in breast cancer patients. In vitro experiments
indicated that miR-139-3p suppresses cell migration, invasion and
EMT by targeting RAB1A in breast cancer. Taken together, the above
evidence supports the tumor suppressive role of miR-139-3p in
various cancer types.
RAB1A is a small GTPase which belongs to the Ras
oncogene family (28). In previous
studies, aberrant expression of RAB1A in human cancer has been
observed (29–32). By using microarray analysis, it was
found that RAB1A was overexpressed in about 98% of tongue squamous
cell carcinomas and 93% of premalignant lesions, which indicated
that RAB1A is a potential biomarker for tongue carcinogenesis
(33). In addition, it was reported
that RAB1A is overexpressed in colorectal cancer, breast and liver
tumors (28). More importantly,
overexpression of RAB1A is correlated with tumor invasion,
progression, poor prognosis, and rapamycin sensitivity in
colorectal cancer. Further in vitro experiment indicated
RAB1A overexpression is sufficient to transform immortalized
fibroblasts and promote malignant growth of established tumor cells
via activation of mTORC1 signaling, whereas, RAB1A knockdown
selectively attenuates oncogenic growth of RAB1A-overexpressing
cancer cells. In addition, another study indicated that RAB1A also
acts as an oncogene in breast cancer, and downregulation of RAB1A
inhibited the growth, migration, invasion and EMT of breast cancer
cells (32). These results suggest
that overexpression of RAB1A is a general phenomenon in human
malignancies and can promote oncogenic transformation and malignant
growth. However, to the best of our knowledge, how RAB1A can be
regulated in cancer has never been investigated. In the present
study, we found that RAB1A is a direct target gene of miR-139-3p,
which was downregulated in breast cancer. Furthermore,
overexpression of RAB1A promotes the migration, invasion, and EMT
of breast cancer. These results confirm the oncogenic role of RAB1A
in cancer and suggest that RAB1A could be a critical therapeutic
target for breast cancer.
In summary, we provide the first data demonstrating
that miR-139-3p plays a tumor suppressive role in breast cancer by
downregulation of RAB1A. Our findings suggest that miR-139-3p may
be a novel prognostic indicator of and potential therapeutic target
for breast cancer.
Acknowledgements
Not applicable.
Funding
This study was supported by the National Key
Research and Development Program of Shaanxi (2018SF_156) and the
Science Foundation of the First Affiliated Hospital of Xi'an
Jiaotong University (2017QN-16).
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
WZ and JH conceived and designed the experiments.
WZ, JX and KW performed the experiments and provided assistance for
data acquisition, data analysis and statistical analysis. WZ and XT
contributed reagents/materials/analysis tools and analyzed the
data. WZ wrote the paper. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
This study was approved by the Academic Committee of
the Ethical Committee of the First Affiliated Hospital of Medical
College, Xi'an Jiaotong University, Xi'an, China, and conducted
according to the principles expressed in the Declaration of
Helsinki. Specimens were obtained with informed consent from all
patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wang Y and Zhou BP: Epithelial-mesenchymal
transition in breast cancer progression and metastasis. Chin J
Cancer. 30:603–611. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
van Zijl F, Krupitza G and Mikulits W:
Initial steps of metastasis: Cell invasion and endothelial
transmigration. Mutat Res. 728:23–34. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chambers AF, Groom AC and MacDonald IC:
Dissemination and growth of cancer cells in metastatic sites. Nat
Rev Cancer. 2:563–572. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Abak A, Amini S, Sakhinia E and Abhari A:
MicroRNA-221: Biogenesis, function and signatures in human cancers.
Eur Rev Med Pharmacol Sci. 22:3094–3117. 2018.PubMed/NCBI
|
|
7
|
Ardekani AM and Naeini MM: The Role of
MicroRNAs in human diseases. Avicenna J Med Biotechnol. 2:161–179.
2010.PubMed/NCBI
|
|
8
|
Ullah S, John P and Bhatti A: MicroRNAs
with a role in gene regulation and in human diseases. Mol Biol Rep.
41:225–232. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tüfekci KU, Oner MG, Meuwissen RL and Genc
S: The role of microRNAs in human diseases. Methods Mol Biol.
1107:33–50. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Waldman SA and Terzic A: Applications of
microRNA in cancer: Exploring the advantages of miRNA. Clin Transl
Sci. 2:248–249. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Reddy KB: MicroRNA (miRNA) in cancer.
Cancer Cell Int. 15:382015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lee YS and Dutta A: MicroRNAs in cancer.
Annu Rev Pathol. 4:199–227. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chen L, Li Y, Fu Y, Peng J, Mo MH,
Stamatakos M, Teal CB, Brem RF, Stojadinovic A, Grinkemeyer M, et
al: Role of deregulated microRNAs in breast cancer progression
using FFPE tissue. PLoS One. 8:e542132013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang B, Li J, Sun M, Sun L and Zhang X:
miRNA expression in breast cancer varies with lymph node metastasis
and other clinicopathologic features. IUBMB Life. 66:371–377. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tang J, Ahmad A and Sarkar FH: The role of
microRNAs in breast cancer migration, invasion and metastasis. Int
J Mol Sci. 13:13414–13437. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
van Schooneveld E, Wildiers H, Vergote I,
Vermeulen PB, Dirix LY and Van Laere SJ: Dysregulation of microRNAs
in breast cancer and their potential role as prognostic and
predictive biomarkers in patient management. Breast Cancer Res.
17:212015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ng L, Wan TM, Man JH, Chow AK, Iyer D,
Chen G, Yau TC, Lo OS, Foo DC, Poon JT, et al: Identification of
serum miR-139-3p as a non-invasive biomarker for colorectal cancer.
Oncotarget. 8:27393–27400. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Liu X, Duan B, Dong Y, He C, Zhou H, Sheng
H, Gao H and Zhang X: MicroRNA-139-3p indicates a poor prognosis of
colon cancer. Int J Clin Exp Pathol. 7:8046–8052. 2014.PubMed/NCBI
|
|
19
|
Yonemori M, Seki N, Yoshino H, Matsushita
R, Miyamoto K, Nakagawa M and Enokida H: Dual tumor-suppressors
miR-139-5p and miR-139-3p targeting matrix metalloprotease 11 in
bladder cancer. Cancer Sci. 107:1233–1242. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative C(T) method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lee JA, Lee HY, Lee ES, Kim I and Bae JW:
Prognostic implications of MicroRNA-21 overexpression in invasive
ductal carcinomas of the breast. J Breast Cancer. 14:269–275. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhai J, Yang X, Zhang Y, Qi Q, Hu J and
Wang Q: Reduced expression levels of the death-associated protein
kinase and E-cadherin are correlated with the development of
esophageal squamous cell carcinoma. Exp Ther Med. 5:972–976. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lánczky A, Nagy Á, Bottai G, Munkácsy G,
Szabó A, Santarpia L and Győrffy B: miRpower: A web-tool to
validate survival-associated miRNAs utilizing expression data from
2178 breast cancer patients. Breast Cancer Res Treat. 160:439–446.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Dongre A, Rashidian M, Reinhardt F,
Bagnato A, Keckesova Z, Ploegh HL and Weinberg RA:
Epithelial-to-mesenchymal transition contributes to
immunosuppression in breast carcinomas. Cancer Res. 77:3982–3989.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Bill R and Christofori G: The relevance of
EMT in breast cancer metastasis: Correlation or causality. FEBS
Lett. 589:1577–1587. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sannigrahi MK, Sharma R, Singh V, Panda
NK, Rattan V and Khullar M: Role of host miRNA Hsa-miR-139-3p in
HPV-16-induced carcinomas. Clin Cancer Res. 23:3884–3895. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Huang P, Xi J and Liu S: miR-139-3p
induces cell apoptosis and inhibits metastasis of cervical cancer
by targeting NOB1. Biomed Pharmacother. 83:850–856. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Jin M, Saucan L, Farquhar MG and Palade
GE: Rab1a and multiple other Rab proteins are associated with the
transcytotic pathway in rat liver. J Biol Chem. 271:30105–30113.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tanaka M, Mun S, Harada A, Ohkawa Y,
Inagaki A, Sano S, Takahashi K, Izumi Y, Osada-Oka M, Wanibuchi H,
et al: Hsc70 contributes to cancer cell survival by preventing
Rab1A degradation under stress conditions. PLoS One. 9:e967852014.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Thomas JD, Zhang YJ, Wei YH, Cho JH,
Morris LE, Wang HY and Zheng XFS: Rab1A is an mTORC1 activator and
a colorectal oncogene. Cancer Cell. 30:181–182. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang X, Liu F, Qin X, Huang T, Huang B,
Zhang Y and Jiang B: Expression of Rab1A is upregulated in human
lung cancer and associated with tumor size and T stage. Aging
(Albany NY). 8:2790–2798. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Xu H, Qian M, Zhao B, Wu C, Maskey N, Song
H, Li D, Song J, Hua K and Fang L: Inhibition of RAB1A suppresses
epithelial-mesenchymal transition and proliferation of
triple-negative breast cancer cells. Oncol Rep. 37:1619–1626. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Shimada K, Uzawa K, Kato M, Endo Y, Shiiba
M, Bukawa H, Yokoe H, Seki N and Tanzawa H: Aberrant expression of
RAB1A in human tongue cancer. Br J Cancer. 92:1915–1921. 2005.
View Article : Google Scholar : PubMed/NCBI
|