Introduction
Glioma are the most aggressive brain tumors, arising
from glial cells. Despite considerable advances in treatment,
resistance and relapse of brain tumors remain a significant
clinical challenge. Current treatments for glioma patients include
surgery, radiotherapy and chemotherapy. Although a combined
approach may eradicate most of a glioma mass, the ability of cancer
cells to become concurrently resistant to different drugs remains a
significant setback to successful chemotherapy. A clearer
understanding of the underlying mechanisms of drug resistance is
required to identify novel therapeutic targets and improve
treatment efficacy.
Cancer stem cells (CSCs) likely contribute to
chemotherapy resistance (1). CSCs are
a slow-dividing, small proportion of cancer cells with
self-renewal, differentiation, and propagation abilities. CSCs were
first conceptualized in human myeloid leukemias (2), where CD34+ and
CD38– cells could engraft severe combined
immune-deficient (SCID) mice to produce large numbers of
colony-forming progenitors with characteristics of stem cells
(3). Following this, CSCs were
identified in solid tumors including breast (4), prostate (5), pancreas (6), colon (7),
brain (8), as well as other cancers.
Brain tumor stem cells in vitro are able to form tumor
spheres and express neural stem cell (NSC) markers CD133, nestin,
Sox2, as well as others. In vivo, they can initiate tumors.
Singh et al revealed that a CD133+ cell
subpopulation from human brain tumors exhibited stem cell
properties in vitro and could initiate tumor development in
SCID mouse brain (8,9). Conventional chemotherapy may eradicate
most susceptible cells in a tumor, but leave the CSCs intact,
resulting in development of resistance.
Although CSCs can self-renew, most are generally
quiescent, spending most of their time in the G0 cell cycle phase.
Since chemotherapeutic drugs are designed to target either the cell
cycle or rapidly dividing cells, this contributes to the capacity
of CSCs for drug resistance. A resistance to apoptosis, activation
of detoxification mechanisms, and a capacity for DNA repair are
also contributing factors (10–12).
Glioma CSCs also exhibit active efflux of chemotherapeutic drugs
through the cellular membrane (13),
attributable to drug transportation such as occurs during
overexpression of the adenosine triphosphate-binding cassette (ABC)
superfamily. The ABC superfamily mainly includes P-glycoprotein
[P-gp, also known as multidrug resistance protein 1 (MDR1)],
multidrug resistance associated protein (MRP), and breast cancer
resistance protein (BCRP). These transporters are able to actively
efflux certain dyes out of cells. Due to the characteristics of ABC
transport, flow cytometry is used to detect a side population (SP)
on the basis of the ability of these cells to efflux Hoechst 33342
dye and a combination of surface marker expression. High levels of
MDR1 RNA are often associated with resistance to chemotherapy in
neuroblastoma, suggesting a contribution (14). Furthermore, high expression levels of
ABC drug transporters are a unique feature of stem cells (15). The identification of ABC gene
expression in CSCs has led to attempts to use this to isolate or
characterize them. SP cells in glioma cell lines are able to form
spheres, and have abilities of self-renewal, multi-lineage
differentiation, and tumorigenicity, representing properties of
CSCs (16). Stem cells are
predominantly found in the SP fraction. The SP phenotype then is
exploited to identify stem-like cells (17). Hirshmann-Jax et al first
demonstrated that neuroblastoma SP cells are less sensitive to
mitoxantrone (18). Moreover, the
cancer progenitor-like cells isolated using SP have been revealed
to be increased following treatment with temozolomide (TMZ), which
indicates the existance of drug resistance in progenitor-like cells
(19). CD133+ CSCs from
glioblastoma display significant resistance to conventional
chemotherapeutic agents, which may be correlated with
overexpression of drug resistance genes such as BCRP1 and
DNA-mismatch repair genes such as MGMT, as well as genes related to
inhibition of apoptosis in CD133+ CSCs (20). Although various mechanisms involved in
the drug resistance of glioma cells have been reported, their
precise actions are still not fully understood.
Notably, CD133+ is not the only
characteristic of CSCs. CD133− cells derived from six
different human patients were tumorigenic when implanted into
brains of nude rats and gave rise to CD133+ cells in
rats (21). In addition, when the ABC
transporters ABCG2 (BCRP1), ABCB1 (MDR1), or ABCC1 are knocked out
in mice, the mice remain viable, fertile, and have normal stem cell
compartments (22–24). These results suggest that the SP
phenotype is not necessary for the maintenance of CSCs. Moreover,
not all cells in the SP compartment are stem cells, and non-stem
cells often exhibit high expression of ABC transporters such as
ABCG2 (BCRP1) and ABCB1 (MDR1) (23).
Non-SP cells are able to generate SP cells and have
tumor-initiating capacity as SP cells (25–28).
The rat C6 glioma cell line is a model for studying
cell growth and invasion, and has been intensively studied for
decades (29). However, the results
in C6 cells are contradictory. Zhou et al revealed that only
a small fraction (4.02%) of C6 cells were CSCs that could form
tumor spheres in a simplified serum-free neural stem cell medium
and express CD133 and nestin (30).
In addition, Kondo et al suggested that only 0.4% of C6
cells could be considered CSCs in a complicated serum-free medium
(31). Zheng et al revealed
that most C6 cells were CSCs, although numerous cells were neither
CD133+ nor a side population (32,33). These
conflicting findings underscore the need to improve methods of
identifying CSCs and exploring the precise mechanisms of drug
resistance in glioma cells. Therefore, C6 cells were used as an
experimental model and their involvement in drug resistance was
investigated.
Materials and methods
Cell culture
Rat C6 glioma cells were provided by the Cell Bank
of Type Culture Collection of Chinese Academy of Sciences
(Shanghai, China). Rat C6 glioma cells were cultured in Dulbecco's
modified Eagle's medium (DMEM) containing high glucose and
pyruvate, with 10% fetal bovine serum plus antibiotics penicillin
and streptomycin (serum was obtained from HyClone; GE Healthcare
Life Sciences, while the other reagents were from Invitrogen;
Gibco; Thermo Fisher Scientific, Inc.). Cells were maintained in
T25 tissue culture flasks at 37°C in a humidified 5% CO2
atmosphere. Confluent cells were harvested by washing in
phosphate-buffered saline (PBS) followed by trypsinization (0.25%
in EDTA) for subculture. For the generation of TMZ-resistant C6
subclones, parental C6 cells were treated with 35 µM TMZ for seven
generations. During this period, dead cells were discarded and the
remaining living cells were prepared to passage and the next
exposure to TMZ. Resistant subclones were generated until there
were no more dead cells. Verapamil hydrochloride (Sigma-Aldrich;
Merck KGaA) 250 µM was used to block ABC transporters.
Tumor sphere culture
C6 cells were plated in 25-cm2 tissue
culture flasks at clonal density (2,500-5,000 cells/cm2)
in serum-free medium containing 20 ng/ml epidermal growth factor
(EGF) and basic fibroblast growth factor (FGF) (both from Sigma
Aldrich; Merck KGaA), and B27 supplement (50X; Gibco; Thermo Fisher
Scientific, Inc.). Four days later, floating tumor spheres were
observed and collected.
Invasion assays
Transwell membranes were precoated with 24 mg/ml
Matrigel (R&D Systems, Inc.), and cells were incubated for 8 h.
Cells on the seeded side were removed with a cotton swab, and cells
adhering to the lower surface were fixed with 100% methanol at
−20°C for 10 min, stained with 3% Giemsa solution at room
temperature for 15 min and counted under a microscope in five
randomly selected fields at a magnification of ×200.
Evaluation of C6-PL3 and C6 cell
invasion in rats
A rat glioma model was established to test the in
vivo effect of C6 drug resistance on cell invasion. Six adult
male Sprague-Dawley rats weighing between 250–300 g were used
(three rats/group). Animals were maintained at a constant room
temperature (24°C) and 50% relative humidity with free access to
food and water under a 12-h light/dark cycle. They were
anesthetized with intraperitoneal injection of sodium pentobarbital
(Nembutal; 50 mg/kg) before surgery. Following administration of
sodium pentobarbital, rats became ataxic, lost their righting
reflex, had no responses to pain and eventually remained immobile.
C6-PL3 and C6 cells were orthotopically injected to the right
striatum of Sprague-Dawley rat brains through a pre-settled
stainless-steel tube. Rat health and behaviour were monitored every
2 days. Rats were sacrificed by anesthesia overdose with
intraperitoneal injection of sodium pentobarbital (Nembutal; 200
mg/kg) 20 days after injection and their brain tissues were removed
and embedded immediately in optimal cutting temperature compound as
previously described (34). Brain
tissues were subsequently cut into 7-µm slices using a Leica CM1850
cryostat microtome. Slices were fixed with 95% ethanol and stained
with hematoxylin and eosin (H&E; Sigma-Aldrich; Merck KGaA).
After sealing with neutral mounting medium (Sigma-Aldrich; Merck
KGaA), the slices were observed under an Olympus upright and
inverted microscope (Olympus IX73P1F) at a magnification of
×400.
Ethical statement
Methods involving live animals were carried out in
accordance with the guidelines and regulations enacted and enforced
by the Chinese National Ministry of Science and Technology as well
as the National Ministry of Health. All experimental protocols were
approved by the Institutional Lab Animal Ethics Committee at
Kunming University of Science and Technology (Kunming, China).
Flow cytometry
BCRP1 and MDR1 ABC transporters allow the active
efflux of Hoechst 33342 dye, which is a feature that defines
pluripotential SPs. To determine whether drug resistance in C6-PL3
cells was involved in the SP phenotype, cells were stained with
Hoechst 33342 fluorescent dye. Verapamil, a calcium channel blocker
and non-specific inhibitor of ABC transporters, can inhibit SP
generation. To analyze SP production, harvested C6 cells were
adjusted to 106 cells/ml in pre-warmed DMEM containing
2% fetal bovine serum and 10 mM HEPES buffer, and incubated for 90
min in a 37°C water bath with Hoechst 33342 at 2.5 µg/ml final
concentration prepared from 1 mg/ml stock of the dye dissolved in
water (Invitrogen; Molecular Probes; thermos Fisher Scientific,
Inc.) and with intermittent mixing. Control cells were treated with
verapamil (50 µM final concentration, in 95% ethanol; from
Sigma-Aldrich; Merck KGaA). At the end of incubation, the cells
were centrifuged at 300 × g for 5 min under refrigeration and then
adjusted to 3×106 cells/ml in PBS containing 2% fetal
bovine serum and 10 mM HEPES.
Apoptosis was assessed using an Annexin V Apoptosis
Detection kit (BD Biosciences) according to the manufacturer's
instructions. Fluorescence intensity was measured using a BD
FACSVantage SE flow cytometer. Original data were analyzed using
WinMDI 2.9 software (Windows Multiple Document Interface for Flow
Cytometry) and presented in the form of dot plots, with fluorescein
isothiocyanate (FITC)-conjugated Annexin V as the x-axis and PI
(propidium iodide) as the y-axis.
Western blot analysis
Cells were prepared using a RIPA Lysis Buffer
(product no. P0013B; Beyotime Institute of Biotechnology) in the
presence of a proteinase inhibitor cocktail (cOmplete™, mini; Roche
Diagnostics), then centrifuged at 4°C for 15 min at 13,201 × g.
Protein concentration was assessed using the BCA protein assay kit
(Applygen Technologies, Inc.). Proteins (50 µg) were separated on a
12% SDS polyacrylamide gel and transferred to a 0.20-µm PVDF
membrane (EMD Millipore). Membranes were blocked at room
temperature with 5% BSA for 2 h and incubated with the following
primary antibodies: Nestin (dilution 1:2,000; cat. no. 33475), Sox2
(dilution 1:1,000; 3579), Bmi-1 (dilution 1:2,000; cat. no. 6964),
BRCP1 (dilution 1:2,000; cat. no. 42078) and MDR1 (dilution
1:2,000; cat. no. 13342; all from Cell Signaling Technology, Inc.)
and β-actin (dilution 1:5,000; product no. A5316; Sigma-Aldrich;
Merck KGaA) for overnight at 4°C. Detection was carried out using
HRP-conjugated secondary antibodies (cat. no. 7076, anti-mouse IgG,
1:10,000; and cat. no. 7074, anti-rabbit IgG, 1:10,000; all from
Cell Signaling Technology, Inc.) for 2 h at room temperature and
enhanced chemiluminescence substrate (GE Healthcare).
Immunofluorescence
C6-PL3 and C6 cells were cultured on Lab-Tek chamber
slides (Sigma-Aldrich; Merck KGaA) for 24 h, then fixed with 4%
paraformaldehyde for 15 min and permeabilized with 0.4% Triton
X-100 for 15 min at room temperature. Cells were then blocked with
5% bovine serum albumin (BSA; Sigma-Aldrich; Merck KGaA) for 1.5 h
and incubated with nestin primary antibodies (dilution 1:400, cat.
no. 556309; BD Transduction Laboratories™) at 4°C overnight. They
were subsequently incubated with PE-conjugated secondary antibodies
(cat. no. sc-516191; Santa Cruz Biotechnology, Inc.) at room
temperature for 2 h and labeled with DAPI (Sigma-Aldrich; Merck
KGaA) for 5 min to identify cell nuclei. Fluorescence was detected
using an Olympus IX81S1F-3 laser confocal scanning microscope.
Statistical analysis
All experiments were repeated at least three times
prior to statistical analysis. All experimental data are presented
as the mean ± SEM. Differences between samples were analyzed using
a two-tailed Student's t-test and regarded as statistically
significant at P<0.05.
Results
Acquisition of TMZ resistance affects
cell morphology and enhances cell migration
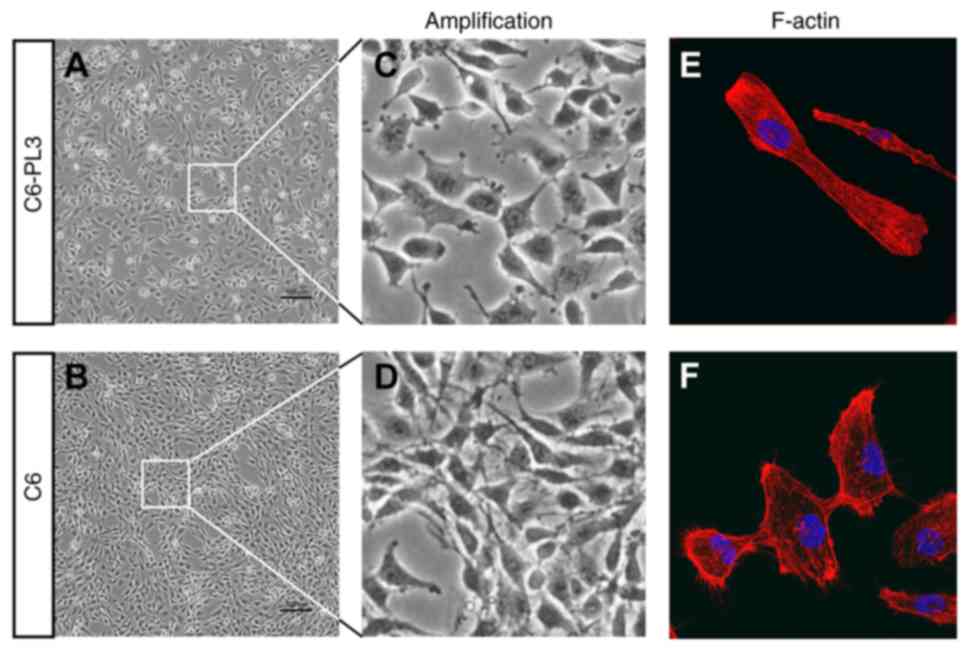
It was revealed that C6-PL3 cells had shorter
extrusions and irregular cell shapes (Fig. 1A-D), as well as reduced cell-to-cell
contact. This suggests that TMZ resistance may interfere with
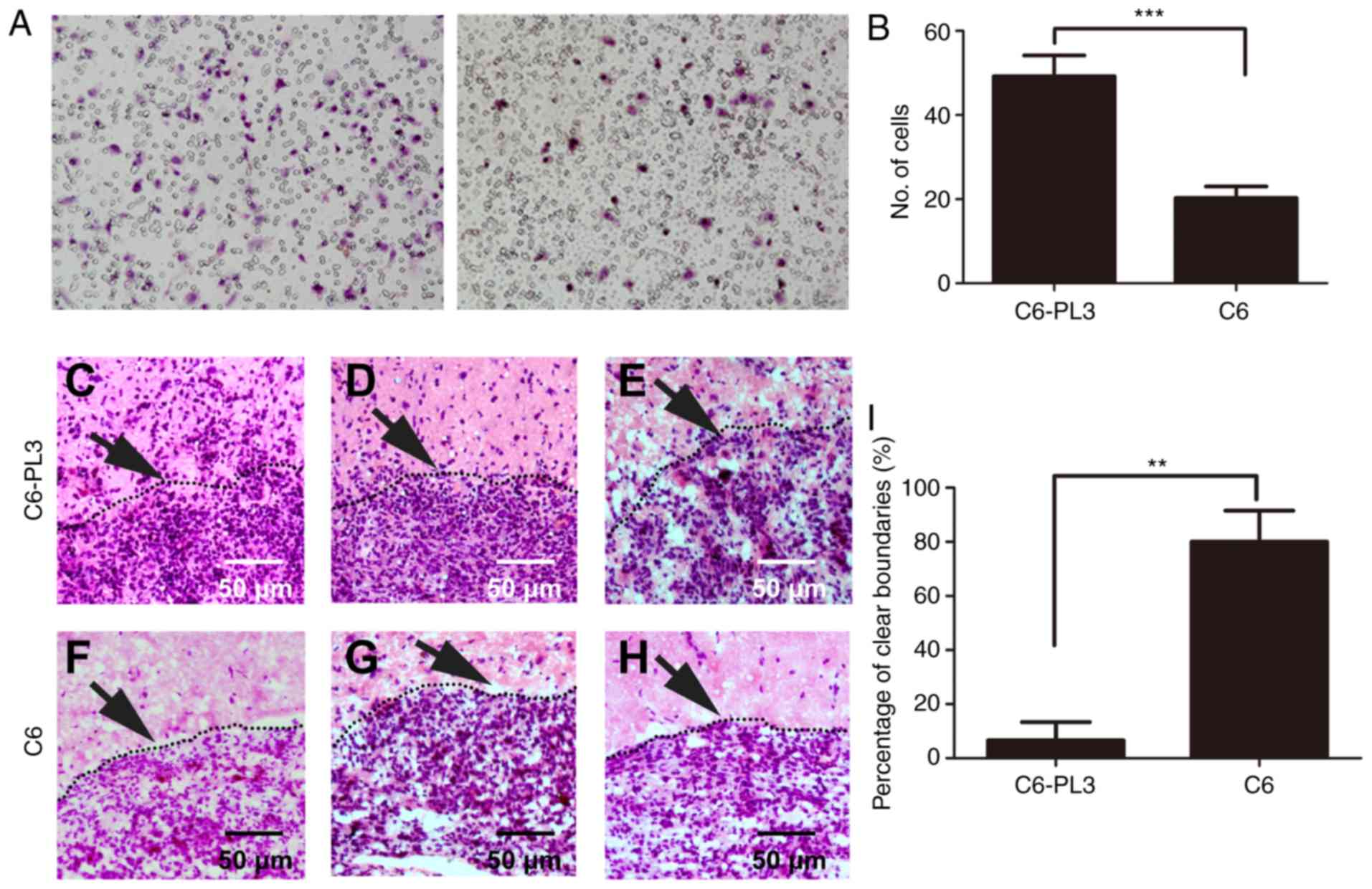
migration and invasion abilities of C6 cells. A Transwell assay
indicated that the invasive ability of C6-PL3 cells was
significantly increased compared to that of C6 cells (Fig. 2A and B). In allografted rats with
C6-PL3 and C6 glioma cells, it was revealed that more C6-PL3 glioma
cells infiltrated normal rat brain tissues than C6 cells (Fig. 2C-H). C6-PL3 glioma cells frequently
infiltrated into normal brain tissues, and <20% of all selected
regions had a clear boundary between tumor and normal brain tissue.
While for C6 cells, >80% of all selected regions had a clear
boundary between tumor and normal brain tissue (Fig. 2I). These results indicated that the
acquisition of TMZ resistance by glioma cells can affect cell
morphology and enhance cell invasion ability, reflecting a more
aggressive phenotype.
Acquisition of TMZ resistance by C6
cells is not associated with CSCs
Previous clonal and population analyses have
indicated that most C6 cells are CSCs (32). Given that CSCs may contribute to
treatment resistance, it was investigated whether the aggressive
phenotype of C6-PL3 was associated with CSC properties of parental
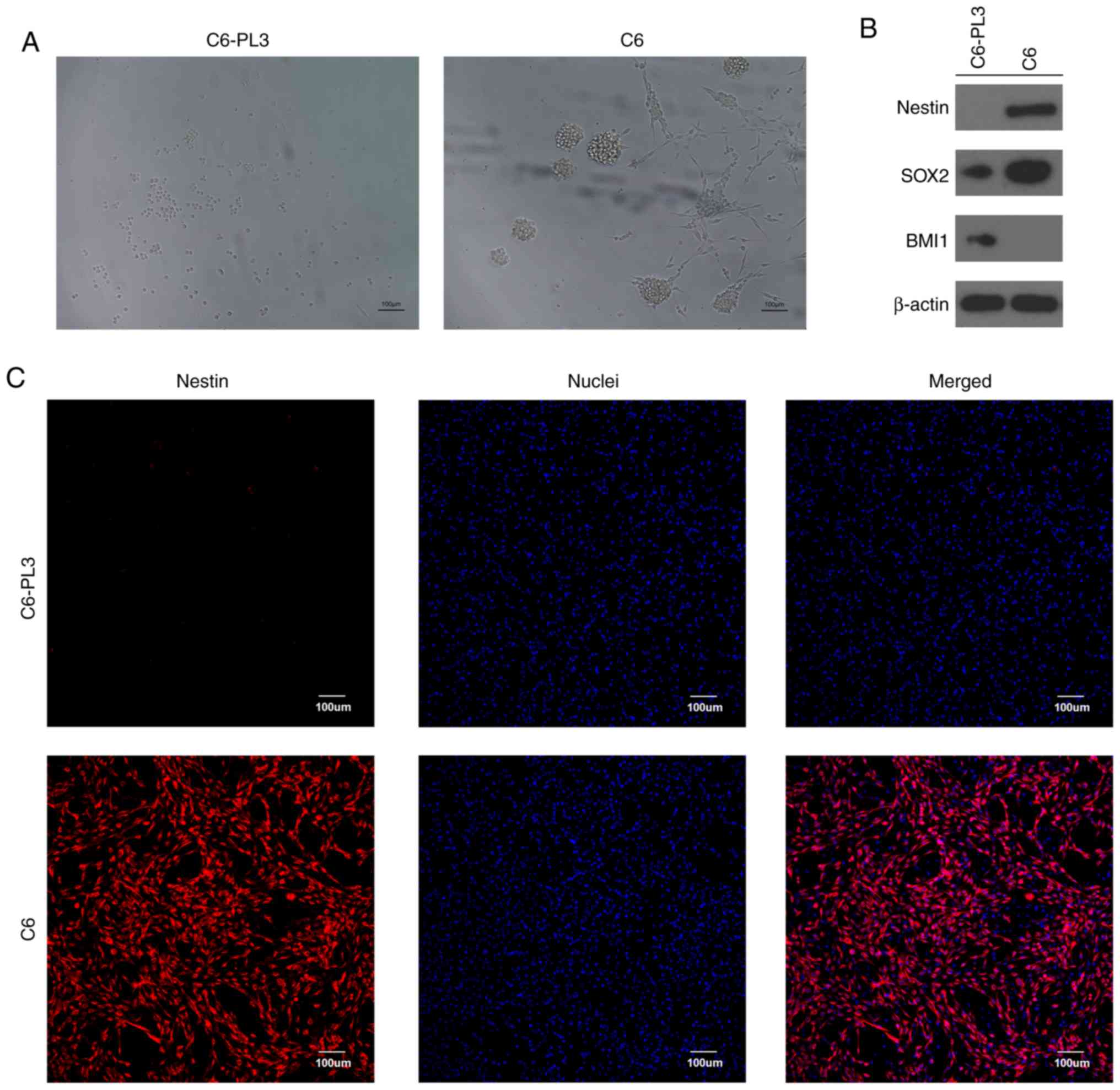
C6 cells. CSCs are able to grow in vitro in aggregates
called tumor spheres and maintain an undifferentiated state
expressing stem cell markers such as nestin (35). Notably, when C6-PL3 and C6 cells were
cultured in serum-free culture medium, most of the cultured C6
cells formed tumor spheres after 10 days, while most of the C6-PL3
cells did not, and grew as flattened cells in a scattered pattern
(Fig. 3A). Western blotting indicated
that C6-PL3 and C6 cells exhibited different expression patterns of
stem cell markers. SOX2 were expressed in both cell lines, while
nestin and BMI1 were expressed only by C6 and C6-PL3 cells,
respectively (Fig. 3B).
Immunofluorescence staining confirmed that nestin was expressed
only in C6 cells (Fig. 3C). These
results indicated that most C6 cells were endowed with CSC
properties, consistent with previous research (32,33).
Conversely, these results also indicated that the acquisition of
TMZ resistance by C6 cells was not associated with CSC.
Acquisition of TMZ resistance by C6
cells is attributable to SP phenotype
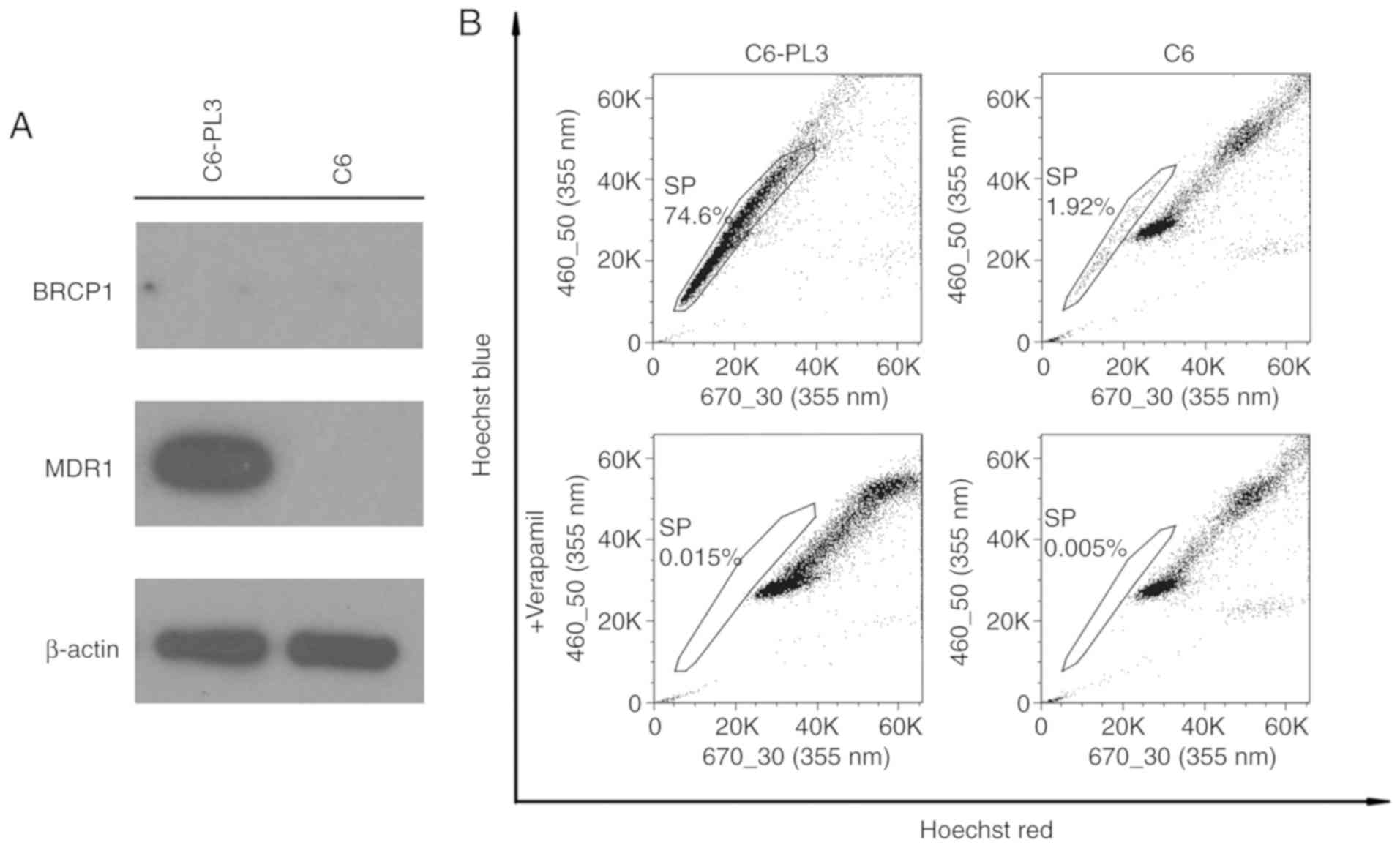
The active efflux of drugs, which requires the
action of the ABC transporters, has been reported to contribute to
drug resistance in glioma. Expression of BCRP1 and MDR1, which are
ABC transporters in C6 cells, indicated that MDR1 expression
occurred only in C6-PL3 cells (Fig.
4A). In addition, neither cell line expressed BCRP1. Flow
cytometric analysis indicated that the C6 glioma cells contained
1.92% SP cells, while 74.6% of C6-PL3 cells were SP cells. Nearly
all SP cells of both lines could be abolished using verapamil,
which indicated that the selected populations were genuine SP cells
(Fig. 4B). This data raises the
possibility that the acquisition of TMZ resistance by C6 cells may
be attributed to the SP phenotype.
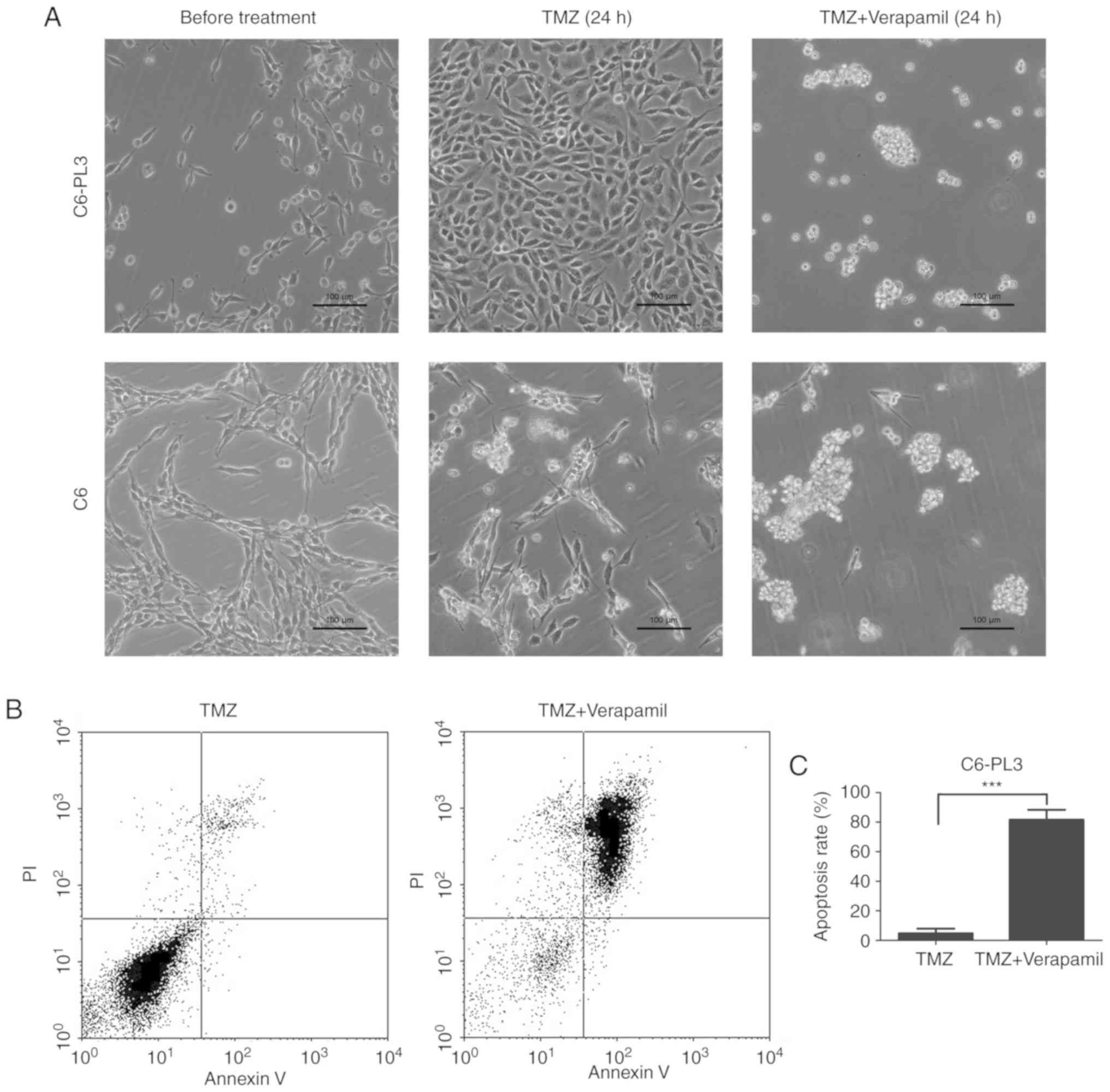
Verapamil increases TMZ-induced tumor
sphere formation and cell apoptosis in C6-PL3 and C6 cells
To further confirm whether SP phenotype was
associated with the drug resistance of C6-PL3 cells, cells were
treated with TMZ for 24 h and verapamil was added for another 24 h.
The results indicated that TMZ could significantly decrease the
number of C6 cells, but not C6-PL3 cells. Verapamil treatment also
restored tumor sphere formation, a CSC trait, in C6-PL3 (Fig. 5A). While C6-PL3 cells remained viable
under TMZ treatment, ABC transporter blockade by verapamil led to a
restored ability of tumor sphere formation. These results indicated
that ABC transporters play a critical role in the TMZ-resistance of
glioma cells, but not CSC. Verapamil blockade of ABC transporters
yielded increased sensitivity to TMZ, as well as TMZ-induced
apoptosis. The average apoptosis rate was 4.79 and 81.61%
respectively (Fig. 5B and C). These
findings indicated that ABC transporters mediated sensitivity to
TMZ in glioma C6 cells.
Discussion
Numerous glioma patients are subject to drug
resistance, relapse, and an overall short survival. Details of the
mechanisms that govern relapse and drug resistance remain
unresolved. Brain CSCs have been revealed to be resistant to
standard-of-care chemotherapy (36).
Exploring the potential role of CSCs in drug resistance in glioma,
and other mechanisms of resistance, may be applicable in
identifying novel therapeutic targets.
The present results revealed that the acquisition of
TMZ resistance permitted C6-PL3 cells to gain a more significant
invasive phenotype, in accordance with a previous demonstration.
Growing evidence indicates a close link between metastatic
potential and therapeutic resistance in gliomas (37,38). Wang
et al revealed that TMZ-resistant glioma cells enhanced cell
migration capacity and acquired epithelial-mesenchymal transition
(EMT)-like changes (37). It was
revealed that the acquisition of TMZ resistance by C6 cells was not
associated with CSCs, but attributable to the SP phenotype and
increased expression of MDR1. This result is in agreement with a
previous finding which identified MDR1 as the most relevant ABC
transporter in drug resistance of rat C6 glioma cells (39). In addition, the BCRP1 gene is a
conserved feature of stem cells from a wide variety of sources, and
serves as a molecular determinant of the SP phenotype (13,40). The
discrepancy between the present results and other studies may be
due to species difference and measurement limitations. In the
present study, rat C6 glioma cells were used, since they are a
well-established in vitro cancer cell line; it is possible
that these differ from primary human glioma cells in genotype and
gene expression pattern. In addition, emerging evidence suggests
that the ABC transporters play a more active role in cell biology,
such as exporting endogenous metabolites as well as signaling
molecules (41). The present results
also indicated that blocking the ABC transporter could increase
sensitivity to TMZ and TMZ-induced apoptosis in C6 cells, which
further supports the role of ABC transporters on drug resistance
development. Further research effort should be devoted to the
effects of ABC transporters and SP phenotype in chemotherapy, which
may lead to new therapeutic targets and better anticancer
strategies.
It was confirmed that acquisition of TMZ resistance
by C6 cells was not due to CSCs. Most C6 cells were capable of
forming tumor spheres, but did not exhibit an SP phenotype, which
is recommended for CSC verification. Based on the present results,
an SP phenotype may not be necessary for CSC development in C6
cells. Further study should clarify whether drug resistance in
cancer is due to CSCs or SPs alone. In addition, current theories
vary as to the characteristics of CSCs. Adding to the controversy
is evidence that terminally differentiated cancer cells can
de-differentiate into pluripotent CSCs (42). Ideally, a perfect enrichment method
for CSCs would be based on a property that defines an essential CSC
feature as their capacity for self-renewal and exact recapitulation
of the original tumor. However, there is currently no such method
in existence for any cell type. Numerous transcription factors or
structural proteins essential for normal NSPC function also mark
glioma CSCs, including SOX2 (43),
OLIG2 (44), Myc (45), BMI1 (43), and nestin (46). Various potential cell surface markers
such as CD133 (43) and CD44
(20) have also been suggested for
identifying CSCs. However, it is not likely that any marker would
be uniformly informative for CSCs, possibly due to the presence of
multiple populations of stem cells in most tissue types due to the
inherent adaptability of cancer cells. In addition to marker
expression, growth as neurospheres in serum-free medium (47) or efflux of fluorescent dyes (31) have also been widely used to
characterize glioma CSCs. Normally, neurosphere culture is carried
out in conditioned medium without added serum, EGF receptor, or FGF
receptor (48). However, this
selection process fails to recapitulate the heterogeneity of the
original tumor. In addition, it has been reported that the majority
of spheres originate from progenitor cells with limited
self-renewal potential, rather than from stem cells (48). Another approach to enrich CSCs is the
use of flow cytometry to identify SPs which contain CSCs, but not
all SPs isolated from different cell lines are self-renewing,
reflecting species-specific challenges of this enrichment method
(49–51). At present, the most reliable method in
validating a putative CSC fraction is their transplantation into
immunodeficient mice. An animal experiment of longer duration or
injecting cells along with Matrigel could significantly increase
the frequency of existing tumor-initiating cells (52,53).
Collectively, these findings indicated further study of additional
criteria to validate CSCs in C6 cells, as stringent standards are
urgently required to help interpret various investigations into
CSCs.
The present findings revealed that the acquisition
of drug resistance by C6 cells conferred increased migration
ability. Additionally, acquisition of drug resistance was not
mediated by the properties of C6 CSCs, but by and increase in SP
phenotype. Blocking the ABC transporter could increase tumor
sensitivity to temozolomide and temozolomide-induced apoptosis in
C6 cells. In summary, drug resistance of C6 cells was revealed to
be dependent on ABC transporters rather than CSCs. These findings
support a potential therapeutic application of ABC transporters in
glioma, and suggest a more accurate choice of experimental
model.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China (no. 81760503 to XY), the
National Natural Science Foundation of China (no. 81760660 to JL),
the Yunnan Applied Basic Research Project [grant no.
2018FE001(−318)], the Yunnan Applied Basic Research Project [grant
no. 2018FE001(−123)] and the Yunnan Health Science and Technology
Plan Projects (grant no. 2016NS207).
Availability of data and materials
All data generated or analyzed during the present
study are included in this published article.
Authors' contributions
XY and JL designed the experiments. XY, YX, SM, YS
performed the experiments. XY, YX and JL analyzed the data and
wrote the manuscript. All authors read and approved the manuscript
and agree to be accountable for all aspects of the research in
ensuring that the accuracy or integrity of any part of the work are
appropriately investigated and resolved.
Ethics approval and consent to
participate
Methods involving live animals were carried out in
accordance with the guidelines and regulations enacted and enforced
by the Chinese National Ministry of Science and Technology as well
as the National Ministry of Health. All experimental protocols were
approved by the Institutional Lab Animal Ethics Committee at
Kunming University of Science and Technology (Kunming, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Auffinger B, Tobias AL, Han Y, Lee G, Guo
D, Dey M, Lesniak MS and Ahmed AU: Conversion of differentiated
cancer cells into cancer stem-like cells in a glioblastoma model
after primary chemotherapy. Cell Death Differ. 21:1119–1131. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bonnet D and Dick JE: Human acute myeloid
leukemia is organized as a hierarchy that originates from a
primitive hematopoietic cell. Nat Med. 3:730–737. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lapidot T, Sirard C, Vormoor J, Murdoch B,
Hoang T, Caceres-Cortes J, Minden M, Paterson B, Caligiuri MA and
Dick JE: A cell initiating human acute myeloid leukaemia after
transplantation into SCID mice. Nature. 367:645–648. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Al-Hajj M, Wicha MS, Benito-Hernandez A,
Morrison SJ and Clarke MF: Prospective identification of
tumorigenic breast cancer cells. Proc Natl Acad Sci USA.
100:3983–3988. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Collins AT, Berry PA, Hyde C, Stower MJ
and Maitland NJ: Prospective identification of tumorigenic prostate
cancer stem cells. Cancer Res. 65:10946–10951. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Li C, Lee CJ and Simeone DM:
Identification of human pancreatic cancer stem cells. Methods Mol
Biol. 568:161–173. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ricci-Vitiani L, Lombardi DG, Pilozzi E,
Biffoni M, Todaro M, Peschle C and De Maria R: Identification and
expansion of human colon-cancer-initiating cells. Nature.
445:111–115. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Singh SK, Hawkins C, Clarke ID, Squire JA,
Bayani J, Hide T, Henkelman RM, Cusimano MD and Dirks PB:
Identification of human brain tumour initiating cells. Nature.
432:396–401. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Singh SK, Clarke ID, Terasaki M, Bonn VE,
Hawkins C, Squire J and Dirks PB: Identification of a cancer stem
cell in human brain tumors. Cancer Res. 63:5821–5828.
2003.PubMed/NCBI
|
|
10
|
Bao S, Wu Q, McLendon RE, Hao Y, Shi Q,
Hjelmeland AB, Dewhirst MW, Bigner DD and Rich JN: Glioma stem
cells promote radioresistance by preferential activation of the DNA
damage response. Nature. 444:756–760. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Peitzsch C, Kurth I, Kunz-Schughart L,
Baumann M and Dubrovska A: Discovery of the cancer stem cell
related determinants of radioresistance. Radiother Oncol.
108:378–387. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Meacham CE and Morrison SJ: Tumour
heterogeneity and cancer cell plasticity. Nature. 501:328–337.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhou S, Schuetz JD, Bunting KD, Colapietro
AM, Sampath J, Morris JJ, Lagutina I, Grosveld GC, Osawa M,
Nakauchi H and Sorrentino BP: The ABC transporter Bcrp1/ABCG2 is
expressed in a wide variety of stem cells and is a molecular
determinant of the side-population phenotype. Nat Med. 7:1028–1034.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Goldstein LJ, Fojo AT, Ueda K, Crist W,
Green A, Brodeur G, Pastan I and Gottesman MM: Expression of the
multidrug resistance, MDR1, gene in neuroblastomas. J Clin Oncol.
8:128–136. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Dean M, Fojo T and Bates S: Tumour stem
cells and drug resistance. Nat Rev Cancer. 5:275–284. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Fukaya R, Ohta S, Yamaguchi M, Fujii H,
Kawakami Y, Kawase T and Toda M: Isolation of cancer stem-like
cells from a side population of a human glioblastoma cell line,
SK-MG-1. Cancer Lett. 291:150–157. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hadnagy A, Gaboury L, Beaulieu R and
Balicki D: SP analysis may be used to identify cancer stem cell
populations. Exp Cell Res. 312:3701–3710. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hirschmann-Jax C, Foster AE, Wulf GG,
Nuchtern JG, Jax TW, Gobel U, Goodell MA and Brenner MK: A distinct
‘side population’ of cells with high drug efflux capacity in human
tumor cells. Proc Natl Acad Sci USA. 101:14228–14233. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chua C, Zaiden N, Chong KH, See SJ, Wong
MC, Ang BT and Tang C: Characterization of a side population of
astrocytoma cells in response to temozolomide. J Neurosurg.
109:856–866. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liu G, Yuan X, Zeng Z, Tunici P, Ng H,
Abdulkadir IR, Lu L, Irvin D, Black KL and Yu JS: Analysis of gene
expression and chemoresistance of CD133+ cancer stem cells in
glioblastoma. Mol Cancer. 5:672006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang J, Sakariassen PO, Tsinkalovsky O,
Immervoll H, Boe SO, Svendsen A, Prestegarden L, Rosland G, Thorsen
F, Stuhr L, et al: CD133 negative glioma cells form tumors in nude
rats and give rise to CD133 positive cells. Int J Cancer.
122:761–768. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Schinkel AH, Smit JJ, van Tellingen O,
Beijnen JH, Wagenaar E, van Deemter L, Mol CA, van der Valk MA,
Robanus-Maandag EC, te Riele HP, et al: Disruption of the mouse
mdr1a P-glycoprotein gene leads to a deficiency in the blood-brain
barrier and to increased sensitivity to drugs. Cell. 77:491–502.
1994. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhou S, Morris JJ, Barnes Y, Lan L,
Schuetz JD and Sorrentino BP: Bcrp1 gene expression is required for
normal numbers of side population stem cells in mice, and confers
relative protection to mitoxantrone in hematopoietic cells in vivo.
Proc Natl Acad Sci USA. 99:12339–12344. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Jonker JW, Buitelaar M, Wagenaar E, Van
Der Valk MA, Scheffer GL, Scheper RJ, Plosch T, Kuipers F, Elferink
RP, Rosing H, et al: The breast cancer resistance protein protects
against a major chlorophyll-derived dietary phototoxin and
protoporphyria. Proc Natl Acad Sci USA. 99:15649–15654. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Platet N, Mayol JF, Berger F, Herodin F
and Wion D: Fluctuation of the SP/non-SP phenotype in the C6 glioma
cell line. FEBS Lett. 581:1435–1440. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Mitsutake N, Iwao A, Nagai K, Namba H,
Ohtsuru A, Saenko V and Yamashita S: Characterization of side
population in thyroid cancer cell lines: Cancer stem-like cells are
enriched partly but not exclusively. Endocrinology. 148:1797–1803.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Burkert J, Otto WR and Wright NA: Side
populations of gastrointestinal cancers are not enriched in stem
cells. J Pathol. 214:564–573. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lichtenauer UD, Shapiro I, Geiger K,
Quinkler M, Fassnacht M, Nitschke R, Ruckauer KD and Beuschlein F:
Side population does not define stem cell-like cancer cells in the
adrenocortical carcinoma cell line NCI h295R. Endocrinology.
149:1314–1322. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Grobben B, De Deyn PP and Slegers H: Rat
C6 glioma as experimental model system for the study of
glioblastoma growth and invasion. Cell Tissue Res. 310:257–270.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhou XD, Wang XY, Qu FJ, Zhong YH, Lu XD,
Zhao P, Wang DH, Huang QB, Zhang L and Li XG: Detection of cancer
stem cells from the C6 glioma cell line. J Int Med Res. 37:503–510.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kondo T, Setoguchi T and Taga T:
Persistence of a small subpopulation of cancer stem-like cells in
the C6 glioma cell line. Proc Natl Acad Sci USA. 101:781–786. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zheng X, Shen G, Yang X and Liu W: Most C6
cells are cancer stem cells: Evidence from clonal and population
analyses. Cancer Res. 67:3691–3697. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Shen G, Shen F, Shi Z, Liu W, Hu W, Zheng
X, Wen L and Yang X: Identification of cancer stem-like cells in
the C6 glioma cell line and the limitation of current
identification methods. In Vitro Cell Dev Biol Anim. 44:280–289.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Li J, Qu J, Shi Y, Perfetto M, Ping Z,
Christian L, Niu H, Mei S, Zhang Q, Yang X and Wei S: Nicotinic
acid inhibits glioma invasion by facilitating Snail1 degradation.
Sci Rep. 7:431732017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Neradil J and Veselska R: Nestin as a
marker of cancer stem cells. Cancer Sci. 106:803–811. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chen J, Li Y, Yu TS, McKay RM, Burns DK,
Kernie SG and Parada LF: A restricted cell population propagates
glioblastoma growth after chemotherapy. Nature. 488:522–526. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wang J, Zhou F, Li Y, Li Q, Wu Z, Yu L,
Yuan F, Liu J, Tian Y, Cao Y, et al: Cdc20 overexpression is
involved in temozolomide-resistant glioma cells with
epithelial-mesenchymal transition. Cell Cycle. 16:2355–2365. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Yi GZ, Liu YW, Xiang W, Wang H, Chen ZY,
Xie SD and Qi ST: Akt and β-catenin contribute to TMZ resistance
and EMT of MGMT negative malignant glioma cell line. J Neurol Sci.
367:101–106. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Decleves X, Bihorel S, Debray M, Yousif S,
Camenisch G and Scherrmann JM: ABC transporters and the
accumulation of imatinib and its active metabolite CGP74588 in rat
C6 glioma cells. Pharmacol Res. 57:214–222. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wee B, Pietras A, Ozawa T, Bazzoli E,
Podlaha O, Antczak C, Westermark B, Nelander S, Uhrbom L,
Forsberg-Nilsson K, et al: ABCG2 regulates self-renewal and stem
cell marker expression but not tumorigenicity or radiation
resistance of glioma cells. Sci Rep. 6:259562016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Nigam SK: What do drug transporters really
do? Nat Rev Drug Discov. 14:29–44. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Gupta PB, Fillmore CM, Jiang G, Shapira
SD, Tao K, Kuperwasser C and Lander ES: Stochastic state
transitions give rise to phenotypic equilibrium in populations of
cancer cells. Cell. 146:633–644. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Hemmati HD, Nakano I, Lazareff JA,
Masterman-Smith M, Geschwind DH, Bronner-Fraser M and Kornblum HI:
Cancerous stem cells can arise from pediatric brain tumors. Proc
Natl Acad Sci USA. 100:15178–15183. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Ligon KL, Huillard E, Mehta S, Kesari S,
Liu H, Alberta JA, Bachoo RM, Kane M, Louis DN, Depinho RA, et al:
Olig2-regulated lineage-restricted pathway controls replication
competence in neural stem cells and malignant glioma. Neuron.
53:503–157. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Kim J, Woo AJ, Chu J, Snow JW, Fujiwara Y,
Kim CG, Cantor AB and Orkin SH: A Myc network accounts for
similarities between embryonic stem and cancer cell transcription
programs. Cell. 143:313–324. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Tunici P, Bissola L, Lualdi E, Pollo B,
Cajola L, Broggi G, Sozzi G and Finocchiaro G: Genetic alterations
and in vivo tumorigenicity of neurospheres derived from an adult
glioblastoma. Mol Cancer. 3:252004. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Goodell MA, Brose K, Paradis G, Conner AS
and Mulligan RC: Isolation and functional properties of murine
hematopoietic stem cells that are replicating in vivo. J Exp Med.
183:1797–1806. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Pastrana E, Silva-Vargas V and Doetsch F:
Eyes wide open: A critical review of sphere-formation as an assay
for stem cells. Cell Stem Cell. 8:486–498. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Broadley KW, Hunn MK, Farrand KJ, Price
KM, Grasso C, Miller RJ, Hermans IF and McConnell MJ: Side
population is not necessary or sufficient for a cancer stem cell
phenotype in glioblastoma multiforme. Stem Cells. 29:452–461. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Bleau AM, Hambardzumyan D, Ozawa T,
Fomchenko EI, Huse JT, Brennan CW and Holland EC: PTEN/PI3K/Akt
pathway regulates the side population phenotype and ABCG2 activity
in glioma tumor stem-like cells. Cell Stem Cell. 4:226–235. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Golebiewska A, Bougnaud S, Stieber D,
Brons NH, Vallar L, Hertel F, Klink B, Schrock E, Bjerkvig R and
Niclou SP: Side population in human glioblastoma is non-tumorigenic
and characterizes brain endothelial cells. Brain. 136:1462–1475.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Quintana E, Shackleton M, Sabel MS, Fullen
DR, Johnson TM and Morrison SJ: Efficient tumour formation by
single human melanoma cells. Nature. 456:593–598. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Bissell MJ and Labarge MA: Context, tissue
plasticity, and cancer: Are tumor stem cells also regulated by the
microenvironment? Cancer Cell. 7:17–23. 2005. View Article : Google Scholar : PubMed/NCBI
|