Introduction
Despite the tremendous improvements in cancer
diagnosis and treatment, pancreatic cancer remains highly lethal
worldwide (1). It is often diagnosed
at advanced stages which is associated with a very poor patient
prognosis (2). Surgical resection,
chemotherapy and radiation therapy are the main treatments for
pancreatic cancer. Nevertheless, most patients eventually
experience recurrence, metastasis and resistance to chemotherapy
and radiotherapy. Metastasis of pancreatic cancer often occurs in
the liver, lung, bone and peritoneum (3). Peritoneal metastasis induces malignant
peritoneum effusion, one of the most common symptoms of late-stage
pancreatic cancer (4). In the clinic,
chemotherapeutic drugs have been applied to attenuate peritoneum
effusion with limited success (5).
Therefore, a novel treatment option specifically targeting
peritoneum effusion is urgently needed.
β-elemene
(1-methyl-1-vinyl-2,4-diisopropenyl-cyclohexane), a compound
isolated from an extract of the traditional Chinese medicinal herb
Curcuma wenyujin, is a broad spectrum antitumor drug that
has been approved by the state Food and Drug Administration of
China for the treatment of specific solid and malignant tumors
(6–8).
Research has shown that β-elemene inhibits tumor cell proliferation
(9), induces tumor cell apoptosis
(10), suppresses tumor angiogenesis
(11) and circumvents drug resistance
(12,13). Particularly, β-elemene emulsion plays
significant roles in the clinical treatment of malignant effusion,
such as pleural effusion in lung cancer (14). However, there are only a few reports
of β-elemene use in the management of peritoneal effusion (15). Additionally, the mechanism of action
behind this phenomenon remains unclear.
Intratumoral hypoxia is thought to be an essential
characteristic of solid tumors, and results in the activation of
hypoxia-inducible factor 1-α (HIF1A). Previous studies have
revealed that HIF1A plays critically important roles in maintaining
the energy metabolism of tumor cells (16), tumor angiogenesis (17), accelerating tumor proliferation and
metastasis (18). In addition, HIF1A
can contribute to peritoneum effusion in several types of cancer
via the promotion of its downstream target, vascular endothelial
growth factor A (VEGFA) (19–21). Importantly, β-elemene can reduce the
expression of HIF1A in cancer (22–24).
In the present study, network pharmacology was
performed to investigate the molecular mechanism underlying the
suppression of peritoneum effusion in pancreatic cancer by
β-elemene. HIF1A was discovered as a target of both β-elemene and
pancreatic cancer. β-elemene suppressed the proliferation of
pancreatic cancer cells from peritoneum effusion. Furthermore,
β-elemene attenuated the protein expression of HIF1A and its
downstream target, VEGFA. These findings suggest that β-elemene
ameliorates malignant peritoneum effusion in pancreatic cancer by
inhibiting the HIF1A/VEGFA pathway.
Materials and methods
Tumor specimens and cell lines
Pancreatic cancer peritoneum effusion samples were
obtained from two patients at Huadong Hospital Affiliated to Fudan
University (Shanghai, China) for testing the cell viability. Fresh
samples were collected from two patients upon obtaining written
informed consent in Oct. 2018. The detailed information concerning
these two patients are summarized in Table I. Cells were acquired from peritoneum
effusion by centrifugation at 1,500 × g for 5 min. This study was
approved by the Internal Review and Ethics Boards of Huadong
Hospital. The pancreatic cancer cell lines PANC-1 and BxPC3 were
purchased from the American Type Culture Collection (ATCC;
Manassas, VA, USA). Cells were cultured in RPMI-1640 medium,
supplemented with 10% fetal bovine serum at 37°C with 5%
CO2.
 | Table I.Clinical characteristics of the
pancreatic cancer patients. |
Table I.
Clinical characteristics of the
pancreatic cancer patients.
| Patient ID | Age (years) | Sex | Histology | Stage | Tumor size
(cm) | Vascular
invasion | Metastasis | CA199 (kU/l) | CEA (ng/ml) |
|---|
| PC1 | 67 | Male | Adenocarcinoma | IV | 3.9×3.7 | Yes | Liver | 79.3 | 8.09 |
| PC2 | 78 | Male | Adenocarcinoma | IV | 3.2×2.4 | Yes | Liver | 289.08 | 5.85 |
Identification of β-elemene-associated
genes
The human genes associated with β-elemene were
acquired from the BATMAN-TCM (http://bionet.ncpsb.org/batman-tcm/) database.
BATMAN-TCM is an online bioinformatics analysis tool specially
designed for studying the molecular mechanisms of traditional
Chinese medicine, and is based on traditional Chinese medicine
ingredients' target prediction (25).
The keyword ‘6918391’, which is the PubChem CID (https://pubchem.ncbi.nlm.nih.gov/compound/6918391) of
β-elemene, was input into the BATMAN-TCM database. To make the
results more credible, the cutoff score was set at 5.
Identification of pancreatic
cancer-associated genes
The human genes associated with pancreatic cancer
were acquired from the DigSee (http://210.107.182.61/geneSearch/) database. DigSee is
a text mining search engine that provides evidence sentences
describing which ‘genes’ are involved in the development of
‘disease’ through ‘biological events’ (26,27). The
keyword ‘pancreatic neoplasms’ was input into the DigSee database.
To make the results more credible, the required number of academic
papers was set to 5.
Network construction
The Search Tool for the Retrieval of Interacting
Genes (STRING, https://string-db.org/, version 10.5)
database, which provides information regarding the predicted and
experimental interactions of proteins, was used to obtain
protein-protein interaction (PPI) data (28). Networks of β-elemene-target genes and
pancreatic cancer-target genes were constructed using PPI analysis
based on β-elemene-target genes and pancreatic cancer-target genes,
respectively. The Kyoto Encyclopedia of Genes and Genomes (KEGG)
pathway analysis of target genes in common between β-elemene and
pancreatic cancer were performed using the Database for Annotation,
Visualization and Integrated Discovery (DAVID, http://david.abcc.ncifcrf.gov/, version 6.8)
database (29,30). A network of the common target genes of
β-elemene and pancreatic cancer was constructed by linking PPI
analysis and KEGG pathway analysis based on the common target
genes. All networks were visualized utilizing Cytoscape software
(31) (https://cytoscape.org/, version 3.4.0). In addition,
the Cytoscape plugin Molecular Complex Detection (MCODE) was
applied to analyze clustering modules in the network of target
genes in common between β-elemene and pancreatic cancer.
Molecular docking simulation
The molecular docking simulation was conducted with
AutoDockTools (32) (http://mgltools.scripps.edu/, version 1.5.6), which is
an automated docking software designed to predict how small
molecules bind to a receptor of known three dimensional (3D)
structure, to find the preferred binding conformation of HIF1A and
β-elemene. The human HIF1A structure was retrieved from the PDB
database (PDB code: 1H2M). PubChem was referenced for the 3D
structures of β-elemene. The preparation work included affixing
hydrogen atoms and removing co-crystallized ligands as well as
water molecules from the 1H2M. The detailed docking operation
process strictly followed the software instruction manual. To cover
all residues with grid map, each grid point in the x, y, and z axes
was 126×126×126 with X=21.796, Y=28.466, Z=30.538 co-ordinates. In
every docking test, 10 runs were executed with the population size
set at 150 and a maximum number of evaluation of 2,500,000. The
remaining parameters were set to default. To visualize the 3D
structure of the docking result, PyMOL software (33) (https://pymol.org/2/, version 2.2) was used.
Co-expression analysis and overall
survival analysis
The Cancer Genome Atlas (TCGA) is a collaboration
between the National Cancer Institute and the National Human Genome
Research Institute that has generated comprehensive,
multi-dimensional maps of the key genomic changes in 33 types of
cancer (34). The dataset of
pancreatic adenocarcinoma (N=177, tumor samples with mRNA data)
from TCGA was selected and a z-score threshold was set at ±2.0.
Genetic alteration and co-expression of HIF1A and VEGFA were
analyzed using cBioPortal (35,36)
(http://www.cbioportal.org/). Analysis of
overall survival (OS) was performed using Kaplan-Meier Plotter
(37) (http://kmplot.com/analysis/).
Cell viability assay
PANC-1, BxPC3, and cells from two peritoneum
effusion samples were seeded in 96-well plates at 5,000 cells/well
in RPMI-1640 medium, supplemented with 10% fetal bovine serum at
37°C with 5% CO2 and cultured overnight. Serial
concentrations of β-elemene [CSPC Pharmaceutical Group Company Ltd.
(YUANDA), Dalian, China] were added to the cells: 0, 0.5, 1, 2, 4,
8 and 16 µM. The stocking solution used here was prepared with an
evenly-distributed aqueous solution of β-elemene, not the
conventional emulsion dosage form of β-elemene. After 72 h of
culture, the cells were incubated with 100 µl of 0.5 mg/ml sterile
MTT (3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium
bromide; Sigma-Aldrich; Merck KGaA) for 4 h at 37°C. Then, the
culture medium was removed and 150 µl of DMSO (Sigma-Aldrich; Merck
KGaA) was added. The absorbance values were measured at 490 nm. The
assay was performed in three replicates, and three parallel
experiments were performed for each sample.
Western blotting
Total protein was extracted from PANC-1 and BxPC3
cells with radioimmunoprecipitation assay lysis buffer (Beijing
Solarbio Science & Technology Co., Ltd., Beijing, China) and
quantified using a Bicinchoninic Acid Protein Assay Kit (Pierce;
Thermo Fisher Scientific, Inc.). A total of 10 µg protein was
denatured and separated via sodium dodecyl sulfate-polyacrylamide
gel electrophoresis (SDS-PAGE). The percentage of stacking gel and
separation gel used in this study was 5 and 10%, respectively. The
proteins were transferred to polyvinylidene difluoride membranes
(EMD Millipore), followed by blocking with 5% bovine serum albumin
and incubation with primary antibodies (dilution 1:1,000) against
GAPDH (cat. no. 2118; Cell Signaling Technology, Inc.), HIF1A (cat.
no. 14179; Cell Signaling Technology, Inc.), and VEGFA (cat. no.
ab52917; Abcam) overnight at 4°C. The membranes were then incubated
with secondary antibody (dilution 1:2,000) (cat. no. 7074; Cell
Signaling Technology, Inc.) for 1 h at 37°C. Finally, the membranes
were imaged using the ChemiDOC™ XRS system (Bio-Rad Laboratories,
Inc.) following detection with an enhanced chemiluminescence kit
(Beijing Solarbio Science & Technology Co., Ltd.). The protein
expression levels were calculated based on the greyscale values of
the blots in ImageJ (version 1.52; NIH) and normalized to that of
GAPDH.
RT-qPCR assay
Total RNA was extracted using the RNeasy mini kit
(Qiagen) according to the manual without further modifications. RNA
purity and quantity were assessed with NanoDrop 100
spectrophotometer (Thermo Scientific, Inc.). cDNA was synthesized
with the Verso cDNA Synthesis kit (Thermo Scientific, Inc.)
following the protocol described by the manufacturer. For qPCR,
GoTap® qPCR Master Mix (Promega) was utilized to set up
the reactions as suggested by the manufacturer. The sequences of
the primers are: GAPDH (upstream GCACCGTCAAGGCTGAGAAC and
downstream TGGTGAAGACGCCAGTGGA); HIF1A (upstream
TCAAAGTCGGACAGCCTCAC and downstream ATCCATTGATTGCCCCAGCA); VEGFA
(upstream TTGCAGATGTGACAAGCCGA and downstream
GGCCGCGGTGTGTCTA).
Statistical analysis
SPSS statistical software (version 22.0; IBM Corp.)
was used for statistical analysis. One-way analysis of variance
followed by Tukey's multiple comparison post-hoc test was performed
and a Student's t-test was used when only two groups were compared.
Data are presented as the mean ± standard deviation from three
independent biological replicates. P<0.05 was considered to be
statistically significant.
Results
Network of β-elemene-target genes
To acquire a list of human genes associated with
β-elemene, the BATMAN-TCM database was employed. The PubChem CID of
β-elemene is 6918391. After entering the PubChem CID of β-elemene
(6918391) as the keyword into the BATMAN-TCM database with a cutoff
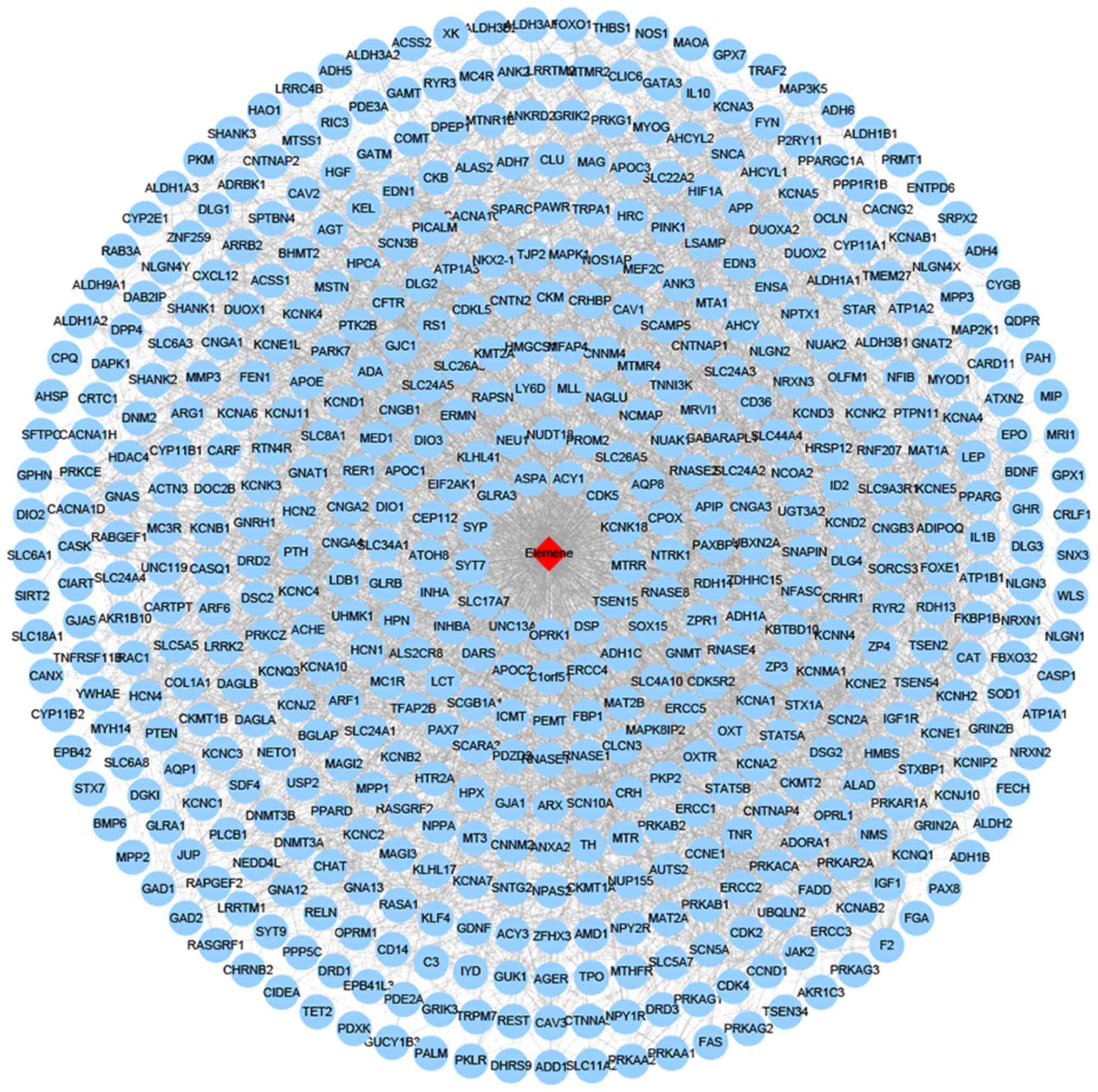
score ≥5, 522 β-elemene-target genes were obtained. A
protein-protein interaction (PPI) network was built using the
String database and Cytoscape software to uncover the relationships
between these β-elemene-target genes. In total, 519 nodes and 3,672
edges were present in the network of β-elemene-target genes
(Fig. 1).
Network of pancreatic cancer-target
genes
The DigSee database was used to search for human
genes relevant to pancreatic cancer. The required number of
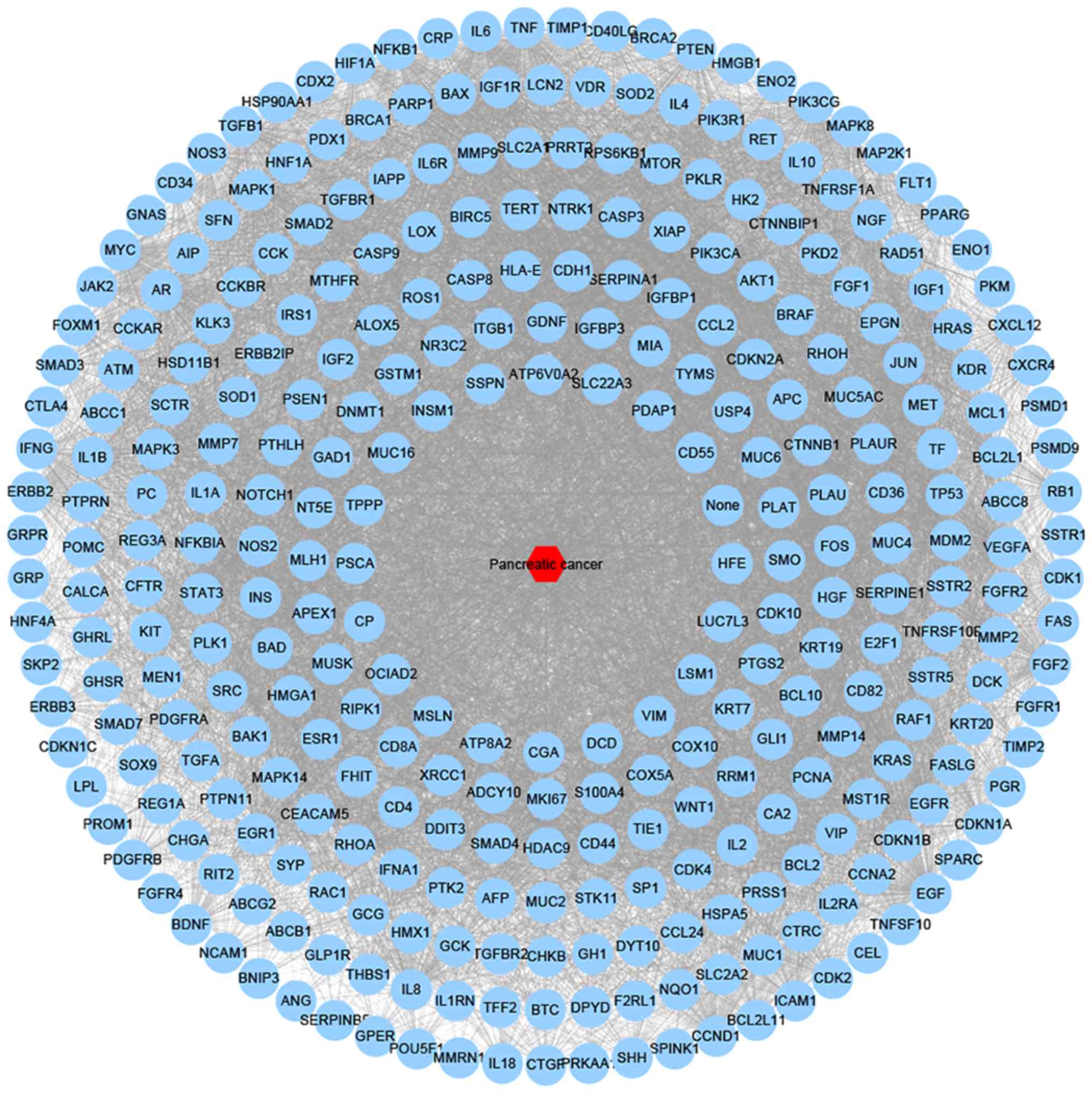
academic papers was set to be >5 on the website and 319
pancreatic cancer-target genes were obtained. Similarly, a PPI
network was constructed to establish the relationship between these
pancreatic cancer-target genes. A total of 318 nodes and 8,733
edges made up the network of the pancreatic cancer-target genes
(Fig. 2).
Network of target genes for β-elemene
against pancreatic cancer
To further uncover the potential pharmacological
mechanisms of β-elemene activity against pancreatic cancer, target
genes common to both β-elemene and pancreatic cancer were selected
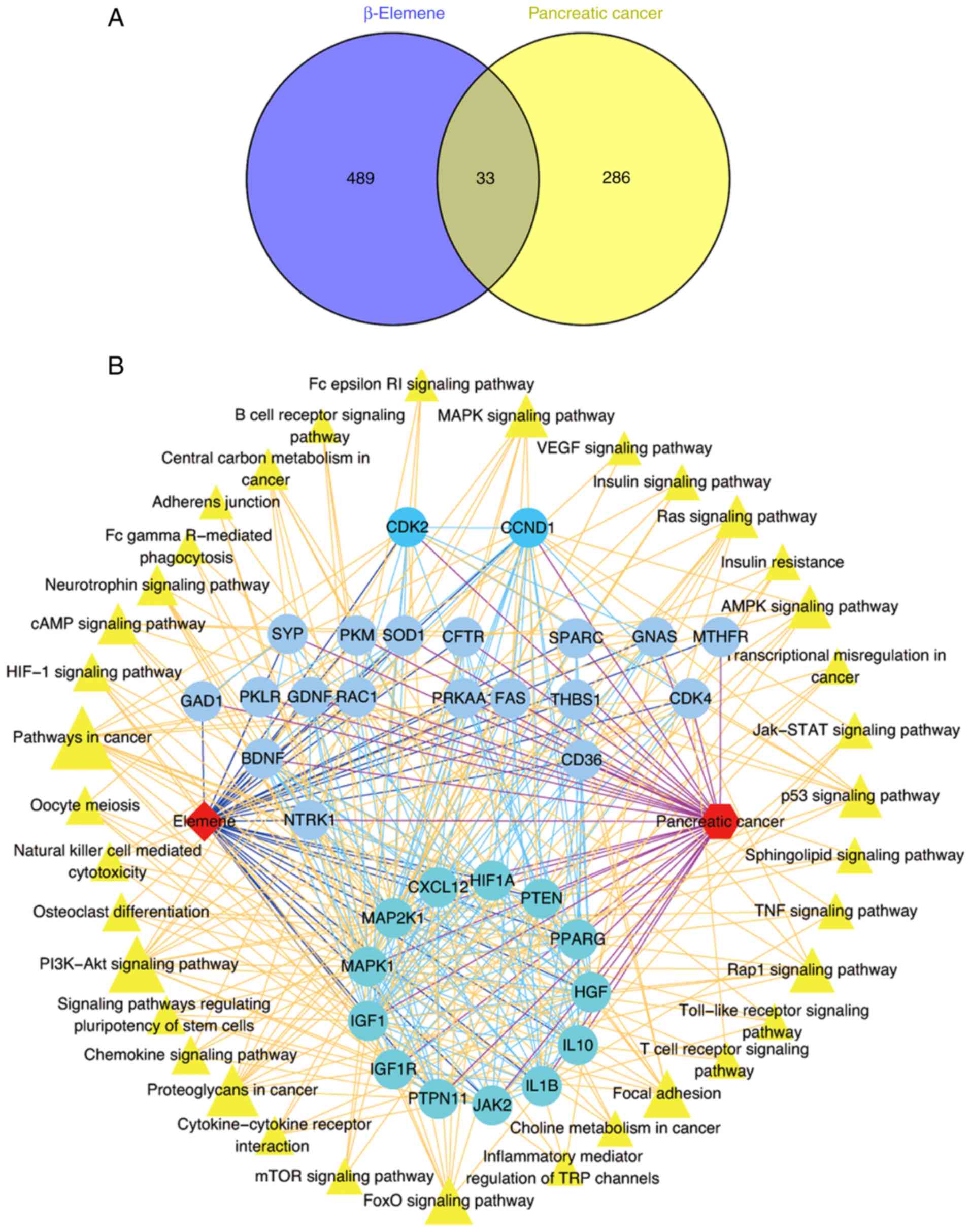
in both networks. A total of 33 genes belonging to both the
β-elemene-target gene and pancreatic cancer-target gene networks
were screened via Venn analysis (Fig.
3A). Additionally, KEGG pathway analysis based on the DAVID
database was performed to determine the potential biological roles
of these 33 genes. Subsequently, a network of target genes for
β-elemene against pancreatic cancer was built using the String
database and Cytoscape software (Fig.
3B). Two modules were identified by MCODE. Module 1 consisted
of HIF1A, PTEN, PPARG, HGF, IL10, IL1B, JAK2, PTPN11, IGF1R, IGF1,
MAPL1, MAP2K1 and CXCL12, and was highly relevant to proteoglycans
in cancer, focal adhesions, and the HIF-1, PI3K-Akt, FoxO, Rap1,
and Ras signaling pathways, among others. Module 2, containing CDK2
and CCND1, was of great importance in the PI3K-Akt, FoxO and p53
signaling pathways.
Molecular docking simulation of
β-elemene to HIF1A
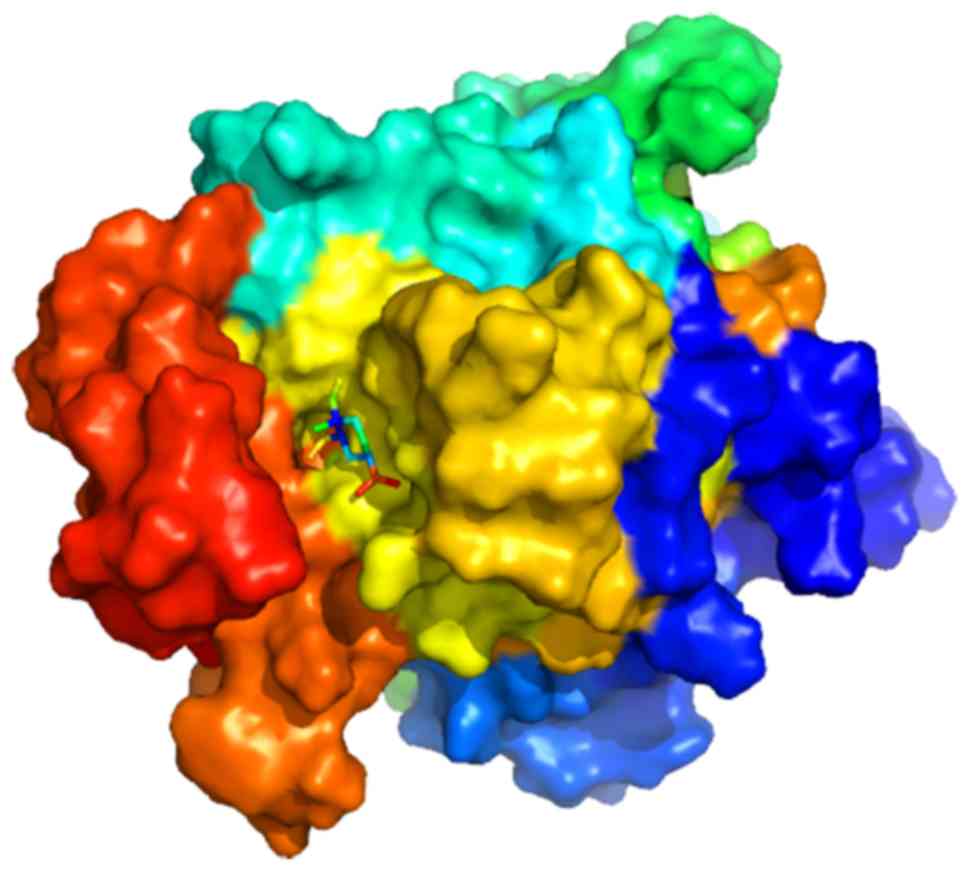
Molecular docking mimics the binding between
molecules and their corresponding receptors. To provide deeper
insight into the binding interactions between β-elemene and HIF1A,
a molecular docking simulation was performed using AutoDockTools
(http://mgltools.scripps.edu/, version
1.5.6) yielding a binding energy of −5.3 kcal/mol, a ligand
efficiency of −0.35 and an inhibitor constant of 129.58 µM.
Collectively, these results suggest the potential of direct binding
between β-elemene and HIF1A (Fig.
4).
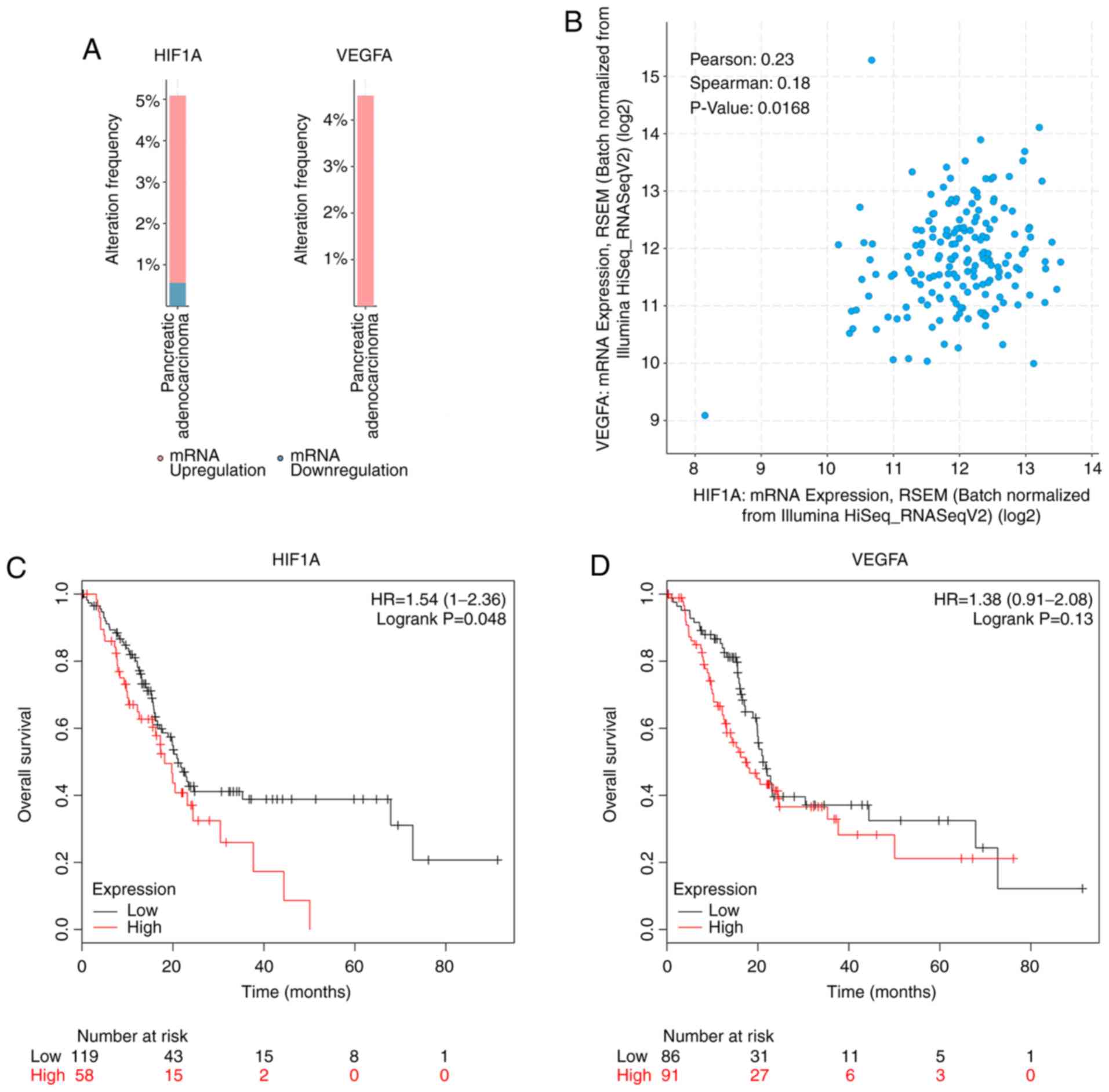
Co-expression of HIF1A and VEGFA
The expression profiles of HIF1A and VEGFA were not
separate from each other. The dataset of pancreatic adenocarcinoma
(N=177, tumor samples with mRNA data) from TCGA was selected and
analyzed with cBioPortal. HIF1A and VEGFA were both upregulated in
pancreatic cancer (Fig. 5A).
Furthermore, there was a trend of co-expression between HIF1A and
VEGFA, verified by bioinformatic analysis. (Fig. 5B). Additionally, elevated expression
of HIF1A and VEGFA corelated with poor overall survival, as
evaluated using Kaplan-Meier Plotter (Fig. 5C and D).
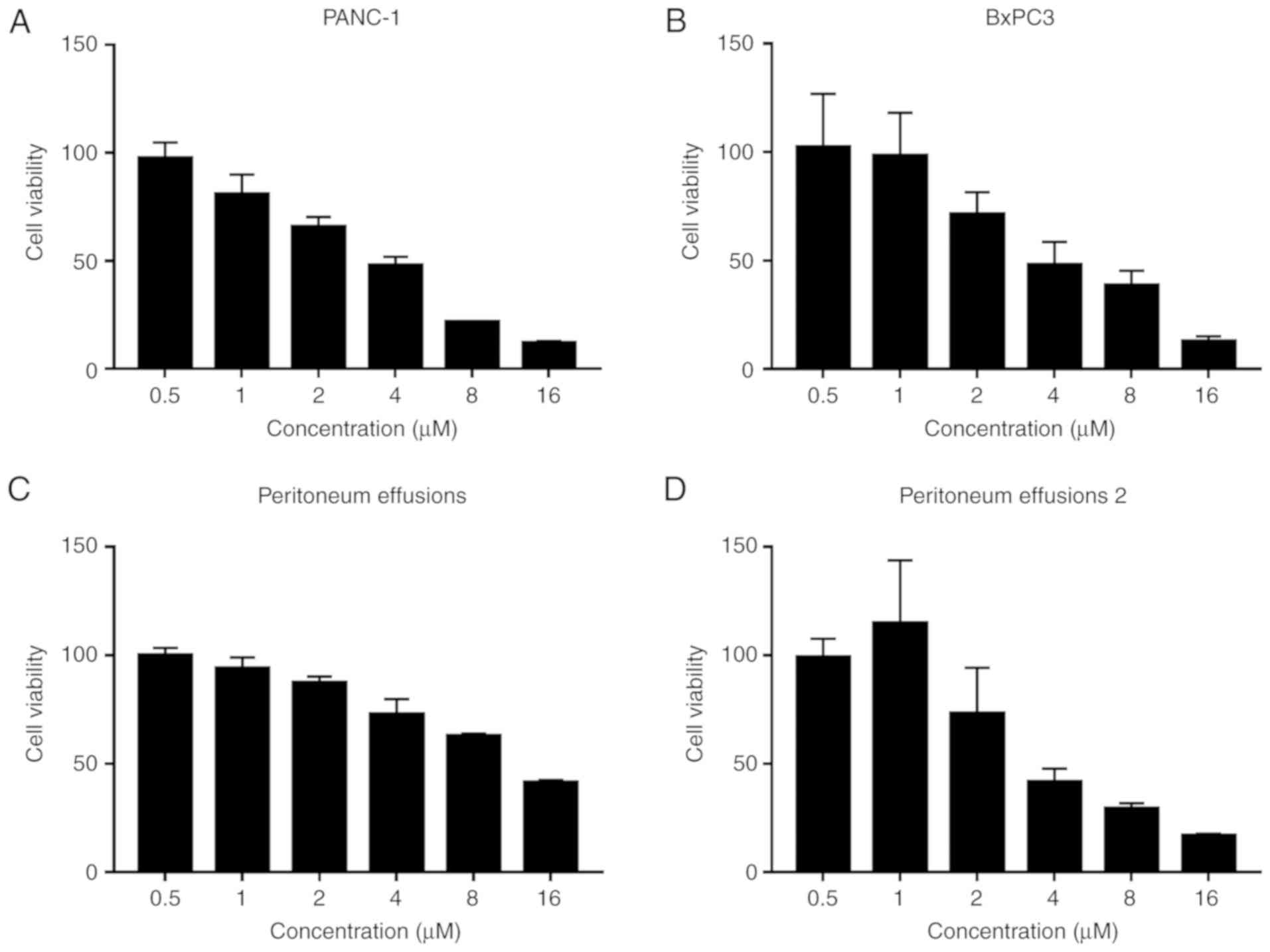
Antitumor effect of β-elemene in
pancreatic cancer
To investigate the antitumor effect of β-elemene in
pancreatic cancer, an MTT assay was performed using PANC-1, BxPC3
cells and cells from the peritoneum effusion of two pancreatic
cancer patients. The cells were treated with serial concentrations
of β-elemene. Analysis of the PANC-1 and BxPC3 cells revealed that
treatment with β-elemene reduced cell viability (Fig. 6A and B). In addition, cells from the
peritoneum effusion of two pancreatic cancer patients demonstrated
decreased cell viability when treated with β-elemene (Fig. 6C and D). The IC50 values of
β-elemene were 6.94±0.86, 17.36±1.25, 15.80±0.63, 14.86±0.69 µM in
the PANC-1 and BxPC3 cell lines, and in the two peritoneum effusion
samples, respectively. Collectively, β-elemene showed
dose-dependent antitumor activity in pancreatic cancer.
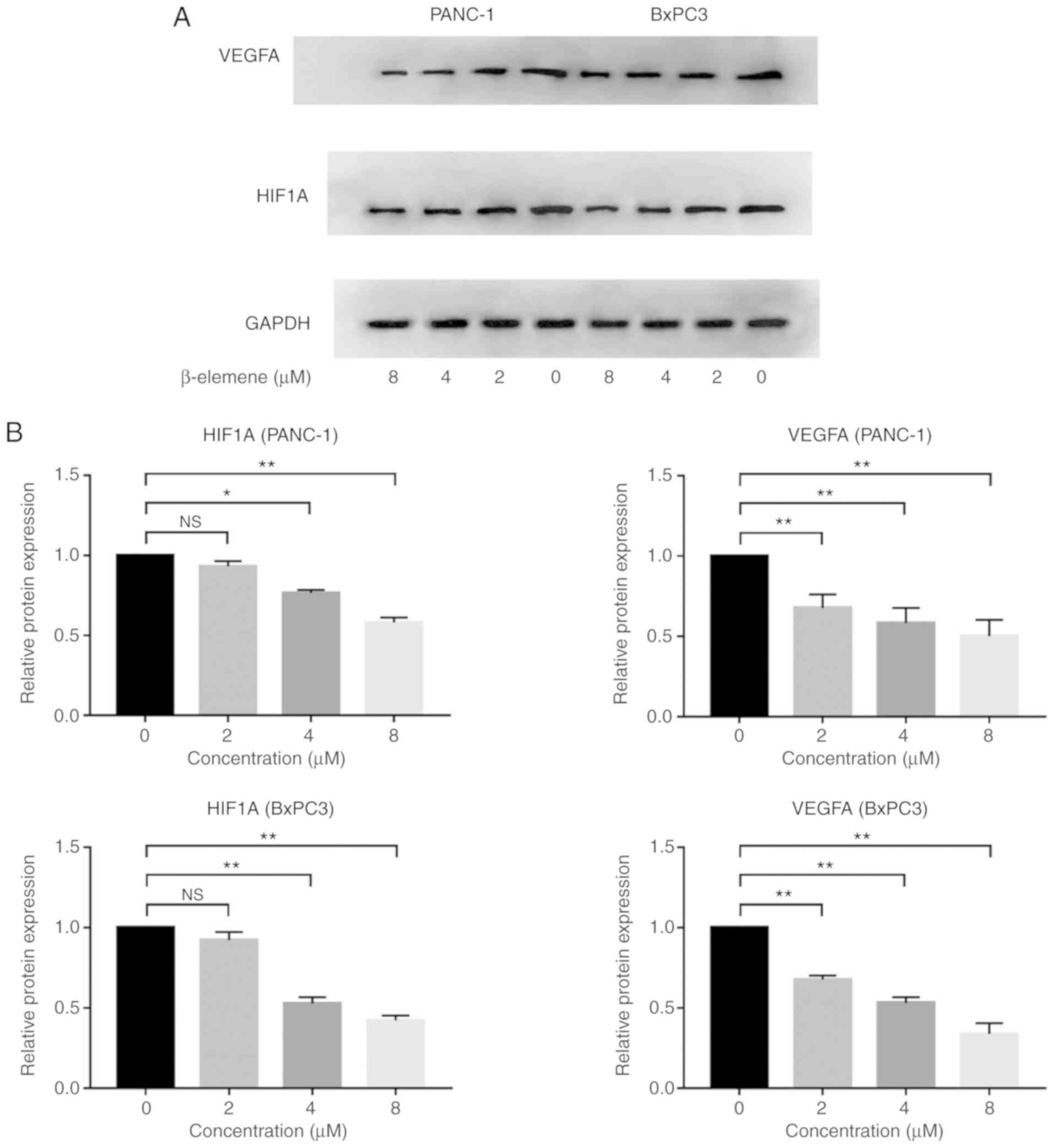
Validation of HIF1A-VEGFA pathway
inhibition by β-elemene in pancreatic cancer
Based on our network pharmacology analysis,
β-elemene is expected to ameliorate malignant peritoneum effusion
in pancreatic cancer by targeting the HIF1A/VEGFA pathway. Thus, an
in vitro experiment was conducted to further explore the
molecular mechanism of β-elemene against peritoneum effusion in
pancreatic cancer. The HIF1A protein levels and those of its
downstream target, VEGFA, were evaluated via western blotting assay
in both PANC-1 and BxPC3. Cells were treated with β-elemene at
concentrations of 0, 2, 4, and 8 µM. As the concentration of
β-elemene increased, the expression of both HIF1A and VEGFA
decreased, especially at the highest dosage (Fig. 7A and B), implying that β-elemene
attenuates the expression of HIF1A, thereby inhibiting the
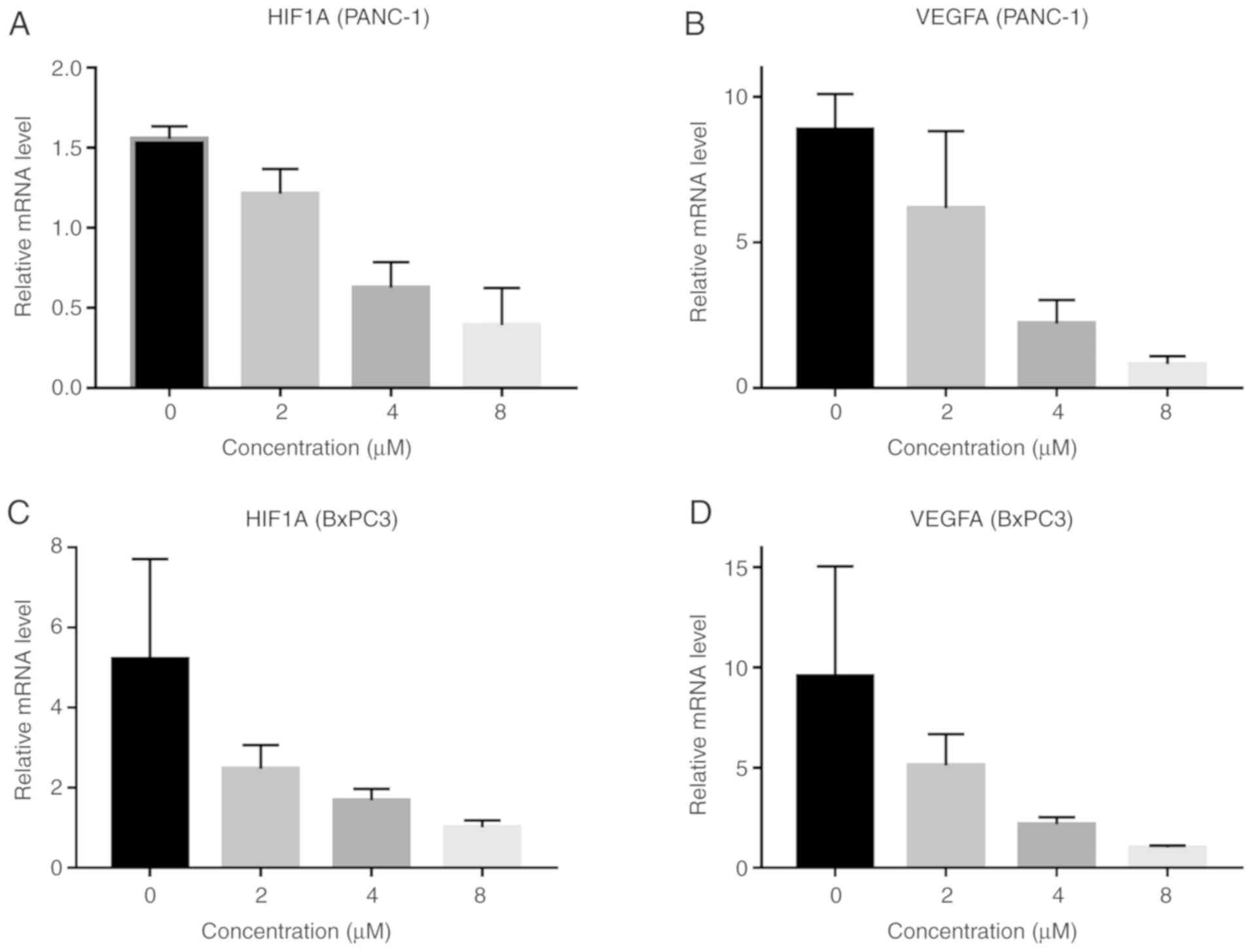
expression of its downstream target, VEGFA. Apart from the
evaluation of the protein expression, RT-qPCR was also conducted to
assess expression of these genes at the RNA level. Similarly, the
mRNA levels of HIF1A and VEGFA were decreased as the concentration
of β-elemene increased in the PANC-1 and BxPC3 cell lines (Fig. 8), which was in concordance with the
western blot assay. Taken together, all these results demonstrate
that β-elemene functions to inhibit the HIF1A/VEGFA pathway,
thereby conferring suppression of malignant peritoneum effusion in
pancreatic cancer.
Discussion
β-elemene is a bioactive compound isolated from the
traditional Chinese medicinal herb Curcuma wenyujin
(38). It exerts a wide range of
antitumor activities and has been approved in China to treat a wide
spectrum of cancers including brain (9), breast (39), ovarian (40), gastric (41), hepatic (42) and lung cancers (43). A meta-analysis, including 46 clinical
controlled trials with 2,992 patients, evaluated the efficacy and
safety of β-elemene in treating malignant pleural effusion, and
showed that β-elemene significantly improved the overall response
rate in controlling malignant pleural effusion [risk ratio (RR) =
1.16; 95% CI: 1.08–1.23; P < 0.05) (14). Results of the study described herein
revealed that β-elemene reduced the viability of PANC-1 and BxPC3
cell lines and cells from the peritoneum effusion of pancreatic
cancer patients, rendering it a promising candidate for the
management of peritoneum effusion in pancreatic cancer patients.
However, only preliminary in vitro studies were conducted to
elucidate the potential of controlling the progression of
peritoneum effusion in pancreatic cancer. More details should be
addressed and the specific process by which β-elemene impacts the
HIF1A/VEGFA pathway requires further exploration.
The advent of the big data era, the continuous
accumulation of omics data, and the progress of bioinformatics
methods provide strong support for network pharmacology development
(44). As a core concept in network
pharmacology, network targets have changed the current ‘single
target’ research approach and provided a potential research
strategy for analyzing the biological basis of traditional Chinese
medicine from the perspective of networks and guiding the discovery
of new active ingredients in traditional Chinese medicine (45,46).
Several studies have applied the network pharmacology approach to
predict the potential targets of traditional Chinese medicine
compounds. HIF1A was discovered as one of the 31 target proteins
that might be regulated by active ingredients in the Fuzheng Huayu
formula in inhibiting hepatic stellate cell viability based on
network pharmacology analysis (47).
In addition, a network pharmacology approach determined that HIF1A
is one of the most important potential protein targets of
tetramethylpyrazine hydrochloride and paeoniflorin in the
Chuanxiong-Chishao herb-pair for promoting angiogenic activity
(48). In our study, HIF1A was
identified as a target of β-elemene using the BATMAN-TCM database,
as well as a target of pancreatic cancer using the DigSee database.
Particularly, experimental validation by MTT assay and western blot
analysis showed that the protein expression of HIF1A was hindered
along with decreased cell viability following β-elemene treatment
in pancreatic cancer cells. Taken together, network pharmacology
analysis provided a theoretical direction for the exploration of a
molecular mechanism of β-elemene against peritoneum effusion in
pancreatic cancer, and established a foundation for subsequent
studies.
In other studies, β-elemene was also shown to have a
negative impact on HIF1A expression. In one study, the expression
level of HIF1A was decreased in A549 cells treated with β-elemene
and radiation compared to the corresponding groups receiving only
radiation treatment (22–24). In addition, knockdown of
bcl-2-associated transcription factor 1 in hepatocellular carcinoma
cell lines significantly reduced the expression of HIF1A, leading
to decreased transcription of VEGFA, which in turn suppressed
proliferation of hepatocellular carcinoma cells (17). Similarly, rhaponticin exhibits potent
anti-metastatic and anti-angiogenic activities due to the
inhibition of VEGFA as a consequence of HIF1A suppression (49). Previously, several studies have
demonstrated that the HIF1A/VEGFA pathway is related to peritoneum
effusion generation (19–21). For instance, peritoneum effusion is
enhanced with elevated VEGFA expression and HIF-related hypoxic
response (19). Furthermore,
lysophosphatidic acid contributes to the generation of peritoneum
effusion in ovarian cancer patients by stimulating HIF1A and VEGFA
expression through activation of the c-Myc and Sp-1 transcription
factors (21). Taken together, our
results reveal a possible molecular mechanism of β-elemene activity
against peritoneum effusion, and further highlight the importance
of targeting the HIF1A-VEGFA pathway as a therapeutic approach to
treat peritoneum effusion in pancreatic cancer patients. However,
there exists several limitations in this study. In the first place,
the complicated mediation of HIF1A-VEGFA involves multiple pathways
regulating the entire process. We only proved the β-elemene could
affect the expression of HIF1A and VEGFA at the protein and mRNA
levels. In addition, β-elemene could induce cytotoxicity via other
mechanisms, including apoptosis. All of these diverse aspects of
β-elemene warrant further exploration in future research.
In conclusion, we identified the HIF1A/VEGFA
signaling pathway as a promising target relevant to the mechanism
of action of β-elemene in ameliorating malignant peritoneum
effusion in pancreatic cancer via a network pharmacology-based
strategy. Docking simulation demonstrated the direct binding
potential between HIF1A and β-elemene. Preliminary in vitro
studies have been conducted to prove the antitumor effect of
β-elemene and its potential to lower the expression levels of HIF1A
and VEGFA, indicating that β-elemene could mediate the expression
of HIF1A and affect its downstream response. Our study provides the
theoretical foundation for the application of β-elemene to treat
malignant peritoneum effusion in the clinic. Further research is
required for deeper exploration of the antitumor mechanism of
β-elemene moving forward.
Acknowledgements
We sincerely express our gratitude to Polaris
Biology (Shanghai, China) and Dr Zhi Yang for their heartful and
prompt assistance in this study.
Funding
Not applicable.
Availability of data and materials
The datasets and materials used and/or analyzed in
this study are available from the corresponding author on
reasonable request.
Authors' contributions
JZ and BL contributed equally to this work. Both
designed, conducted, and drafted the major parts of this
manuscript. YJ, LZ and YZ participated in the network pharmacology
analysis, including searching for target genes and constructing
individual networks. HZ proposed the idea for this study and
facilitated and solved the difficulties encountered in this
research. All authors read and approved the final manuscript and
agree to be accountable for all aspects of the research in ensuring
that the accuracy or integrity of any part of the work are
appropriately investigated and resolved.
Ethics approval and consent to
participate
This study was approved by the Internal Review and
Ethics Boards of Huadong Hospital Affiliated to Fudan University
(Shanghai, China). Fresh samples were collected from two patients
upon obtaining written informed consent in October 2018.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kamisawa T, Wood LD, Itoi T and Takaori K:
Pancreatic cancer. Lancet. 388:73–85. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Vivaldi C, Caparello C, Musettini G,
Pasquini G, Catanese S, Fornaro L, Lencioni M, Falcone A and Vasile
E: First-line treatment with FOLFOXIRI for advanced pancreatic
cancer in clinical practice: Patients' outcome and analysis of
prognostic factors. Int J Cancer. 139:938–945. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Takahara N, Isayama H, Nakai Y, Ishigami
H, Satoi S, Mizuno S, Kogure H, Matsubara S, Yamamoto N, Yamaguchi
H, et al: Intravenous and intraperitoneal paclitaxel with S-1 for
treatment of refractory pancreatic cancer with malignant ascites.
Investigational New Drugs. 34:636–642. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Thomassen I, Lemmens VE, Nienhuijs SW,
Luyer MD, Klaver YL and de Hingh IH: Incidence, prognosis, and
possible treatment strategies of peritoneal carcinomatosis of
pancreatic origin: A population-based study. Pancreas. 42:72–75.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Xu HB, Zheng LP, Li L, Xu LZ and Fu J:
Elemene, one ingredient of a Chinese herb, against malignant
tumors: A literature-based meta-analysis. Cancer Invest.
31:156–166. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chang Z, Gao M, Zhang W, Song L, Jia Y and
Qin Y: Beta-elemene treatment is associated with improved outcomes
of patients with esophageal squamous cell carcinoma. Surg Oncol.
26:333–337. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Jiang X, Hidru TH, Zhang Z, Bai Y, Kong L
and Li X: Evidence of elemene injection combined radiotherapy in
lung cancer treatment among patients with brain metastases: A
systematic review and meta-analysis. Medicine (Baltimore).
96:e69632017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Feng HB, Wang J, Jiang HR, Mei X, Zhao YY,
Chen FR, Qu Y, Sai K, Guo CC, Yang QY, et al: β-elemene selectively
inhibits the proliferation of glioma stem-like cells through the
downregulation of Notch1. Stem Cells Transl Med. 6:830–839. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liu Y, Jiang ZY, Zhou YL, Qiu HH, Wang G,
Luo Y, Liu JB, Liu XW, Bu WQ, Song J, et al: β-elemene regulates
endoplasmic reticulum stress to induce the apoptosis of NSCLC cells
through PERK/IRE1α/ATF6 pathway. Biomed Pharmacother. 93:490–497.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sun W, Huang Y, Yin T, Wang J, Du R, Qiu
J, Zhang Y, Wang Y, Chen J and Wang G: Effects of elemene on
inhibiting proliferation of vascular smooth muscle cells and
promoting reendothelialization at the stent implantation site.
Biomater Sci. 5:1144–1155. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Jiang Z, Jacob JA, Loganathachetti DS,
Nainangu P and Chen B: β-elemene: Mechanistic studies on cancer
cell interaction and its chemosensitization effect. Front
Pharmacol. 8:1052017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li X, Lin Z, Zhang B, Guo L, Liu S, Li H,
Zhang J and Ye Q: β-elemene sensitizes hepatocellular carcinoma
cells to oxaliplatin by preventing oxaliplatin-induced degradation
of copper transporter 1. Sci Rep. 6:210102016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang QT, Zhang ZL, Xiong H, Zhou DS, Li J,
Liang J and Wang YF: Evaluation of the efficacy and safety of
elemene in treating malignant pleural effusion caused by tumors: A
PRISMA guided meta-analysis. Medicine (Baltimore). 97:e125422018.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang J, Zhang H and Sun Y: Phase III
clinical trial of elemenum emulsion in the management of malignant
pleural and peritoneal effusions. Zhonghua Zhong Liu Za Zhi.
18:464–467. 1996.(In Chinese). PubMed/NCBI
|
|
16
|
Hu LP, Zhang XX, Jiang SH, Tao LY, Li Q,
Zhu LL, Yang MW, Huo YM, Jiang YS, Tian GA, et al: Targeting
purinergic receptor P2Y2 prevents the growth of pancreatic ductal
adenocarcinoma by inhibiting cancer cell glycolysis. Clin Cancer
Res. 25:1318–1330. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wen Y, Zhou X, Lu M, He M, Tian Y, Liu L,
Wang M, Tan W, Deng Y, Yang X, et al: Bclaf1 promotes angiogenesis
by regulating HIF-1α transcription in hepatocellular carcinoma.
Oncogene. 38:1845–1859. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li AG, Murphy EC, Culhane AC, Powell E,
Wang H, Bronson RT, Von T, Giobbie-Hurder A, Gelman RS, Briggs KJ,
et al: BRCA1-IRIS promotes human tumor progression through PTEN
blockade and HIF-1α activation. Proc Natl Acad Sci USA.
115:E96002018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Pérez S, Pereda J, Sabater L and Sastre J:
Pancreatic ascites hemoglobin contributes to the systemic response
in acute pancreatitis. Free Radic Biol Med. 81:145–155. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Pasquet M, Golzio M, Mery E, Rafii A,
Benabbou N, Mirshahi P, Hennebelle I, Bourin P, Allal B, Teissie J,
et al: Hospicells (ascites-derived stromal cells. promote
tumorigenicity and angiogenesis. Int J Cancer. 126:2090–2101.
2010.PubMed/NCBI
|
|
21
|
Song Y, Wu J, Oyesanya RA, Lee Z,
Mukherjee A and Fang X: Sp-1 and c-Myc mediate lysophosphatidic
acid-induced expression of vascular endothelial growth factor in
ovarian cancer cells via a hypoxia-inducible factor-1-independent
mechanism. Clin Cancer Res. 15:492–501. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zou K, Tong E, Xu Y, Deng X and Zou L:
Down regulation of mammalian target of rapamycin decreases HIF-1α
and survivin expression in anoxic lung adenocarcinoma A549 cell to
elemene and/or irradiation. Tumor Biol. 35:9735–9741. 2014.
View Article : Google Scholar
|
|
23
|
Tong E, Xu Y, Li G, Zou K and Zou L: The
effects of β-elemene on the expression of mTOR, HIF-1A, survivin in
lung adenocarcinoma A549 cell. Afr J Tradit Complement Altern Med.
10:18–23. 2013.PubMed/NCBI
|
|
24
|
Li G, Xie B, Li X, Chen Y, Wang Q, Xu Y,
Xu-Welliver M and Zou L: Down-regulation of survivin and
hypoxia-inducible factor-1 α by β-elemene enhances the
radiosensitivity of lung adenocarcinoma xenograft. Cancer Biother
Radiopharm. 27:56–64. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu Z, Guo F, Wang Y, Li C, Zhang X, Li H,
Diao L, Gu J, Wang W, Li D, et al: BATMAN-TCM: A bioinformatics
analysis tool for molecular mechANism of traditional Chinese
medicine. Sci Rep. 6:211462016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kim J, So S, Lee HJ, Park JC, Kim JJ and
Lee H: DigSee: Disease gene search engine with evidence sentences
(version cancer). Nucleic Acids Res. 41:W510–W517. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kim J, Kim JJ and Lee H: An analysis of
disease-gene relationship from Medline abstracts by DigSee. Sci
Rep. 7:40154. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Szklarczyk D, Morris JH, Cook H, Kuhn M,
Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, et al:
The STRING database in 2017: Quality-controlled protein-protein
association networks, made broadly accessible. Nucleic Acids Res.
45:D362–D368. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nature Protoc. 4:44–57. 2008. View Article : Google Scholar
|
|
30
|
Huang da W, Sherman BT and Lempicki RA:
Bioinformatics enrichment tools: Paths toward the comprehensive
functional analysis of large gene lists. Nucleic Acids Res.
37:1–13. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Goodsell DS and Olson AJ: Automated
docking of substrates to proteins by simulated annealing. Proteins.
8:195–202. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Schrödinger LLC: The PyMOL molecular
graphics system, version 1.8. 2015.
|
|
34
|
Tomczak K, Czerwińska P and Wiznerowicz M:
The Cancer Genome Atlas (TCGA): An immeasurable source of
knowledge. Contemp Oncol (Pozn). 19:A68–A77. 2015.PubMed/NCBI
|
|
35
|
Gao J, Aksoy BA, Dogrusoz U, Dresdner G,
Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al:
Integrative analysis of complex cancer genomics and clinical
profiles using the cBioPortal. Sci Signal. 6:pl12013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Cerami E, Gao J, Dogrusoz U, Gross BE,
Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et
al: The cBio cancer genomics portal: An open platform for exploring
multidimensional cancer genomics data. Cancer Discov. 2:401–404.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lánczky A, Nagy Á, Bottai G, Munkácsy G,
Szabó A, Santarpia L and Győrffy B: miRpower: A web-tool to
validate survival-associated miRNAs utilizing expression data from
2178 breast cancer patients. Breast Cancer Res Treat. 160:439–446.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
An YW, Hu G, Yin GP, Zhu JJ, Zhang QW,
Wang ZM, Peng J and Fan B: Quantitative analysis and discrimination
of steamed and non-steamed rhizomes of Curcuma wenyujin by
GC-MS and HPLC. J Chromatogr Sci. 52:961–970. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Tang CY, Zhu LX, Yu JD, Chen Z, Gu MC, Mu
CF, Liu Q and Xiong Y: Effect of β-elemene on the kinetics of
intracellular transport of d-luciferin potassium salt (ABC
substrate) in doxorubicin-resistant breast cancer cells and the
associated molecular mechanism. Eur J Pharm Sci. 120:20–29. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Li X, Wang G, Zhao J, Ding H, Cunningham
C, Chen F, Flynn DC, Reed E and Li QQ: Antiproliferative effect of
beta-elemene in chemoresistant ovarian carcinoma cells is mediated
through arrest of the cell cycle at the G2-M phase. Cell Mol Life
Sci. 62:894–904. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Liu JS, Che XM, Chang S, Qiu GL, He SC,
Fan L, Zhao W, Zhang ZL and Wang SF: β-elemene enhances the
radiosensitivity of gastric cancer cells by inhibiting Pak1
activation. World J Gastroenterol. 21:9945–9956. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Dai ZJ, Tang W, Lu WF, Gao J, Kang HF, Ma
XB, Min WL, Wang XJ and Wu WY: Antiproliferative and apoptotic
effects of β-elemene on human hepatoma HepG2 cells. Cancer Cell
Int. 13:272013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Cheng H, Ge X, Zhuo S, Gao Y, Zhu B, Zhang
J, Shang W, Xu D, Ge W and Shi L: β-elemene synergizes with
gefitinib to inhibit stem-like phenotypes and progression of lung
cancer via down-regulating EZH2. Front Pharmacol. 9:14132018.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Hopkins AL: Network pharmacology: The next
paradigm in drug discovery. Nat Chem Biol. 4:682–690. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zheng J, Wu M, Wang H and Li S, Wang X, Li
Y, Wang D and Li S: Network pharmacology to unveil the biological
basis of health-strengthening herbal medicine in cancer treatment.
Cancers (Basel). 10:E4612018. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Wang N, Yang B, Zhang X, Wang S, Zheng Y,
Li X, Liu S, Pan H, Li Y, Huang Z, et al: Network
pharmacology-based validation of caveolin-1 as a key mediator of Ai
Du Qing inhibition of drug resistance in breast cancer. Front
Pharmacol. 9:11062018. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Xing X, Chen S, Li L, Cao Y, Chen L, Wang
X and Zhu Z: The active components of Fuzheng Huayu formula and
their potential mechanism of action in inhibiting the hepatic
stellate cells viability-a network pharmacology and transcriptomics
approach. Front Pharmacol. 9:5252018. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Wang Y, Guo G, Yang BR, Xin QQ, Liao QW,
Lee SM, Hu YJ, Chen KJ and Cong WH: Synergistic effects of
Chuanxiong-Chishao herb-pair on promoting angiogenesis at network
pharmacological and pharmacodynamic levels. Chin J Integr Med.
23:654–662. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Kim A and Ma JY: Rhaponticin decreases the
metastatic and angiogenic abilities of cancer cells via suppression
of the HIF-1α pathway. Int J Oncol. 53:1160–1170. 2018.PubMed/NCBI
|