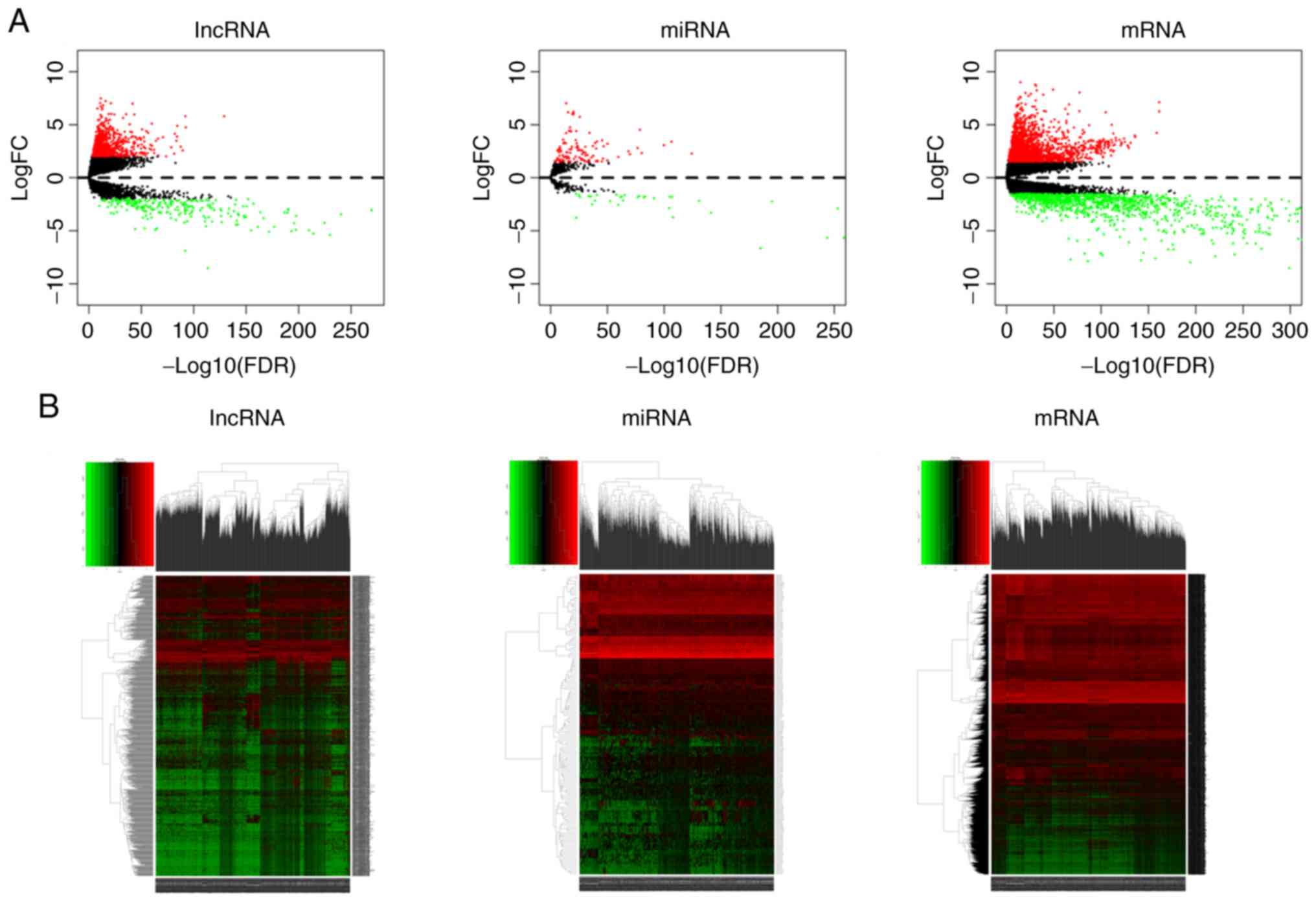
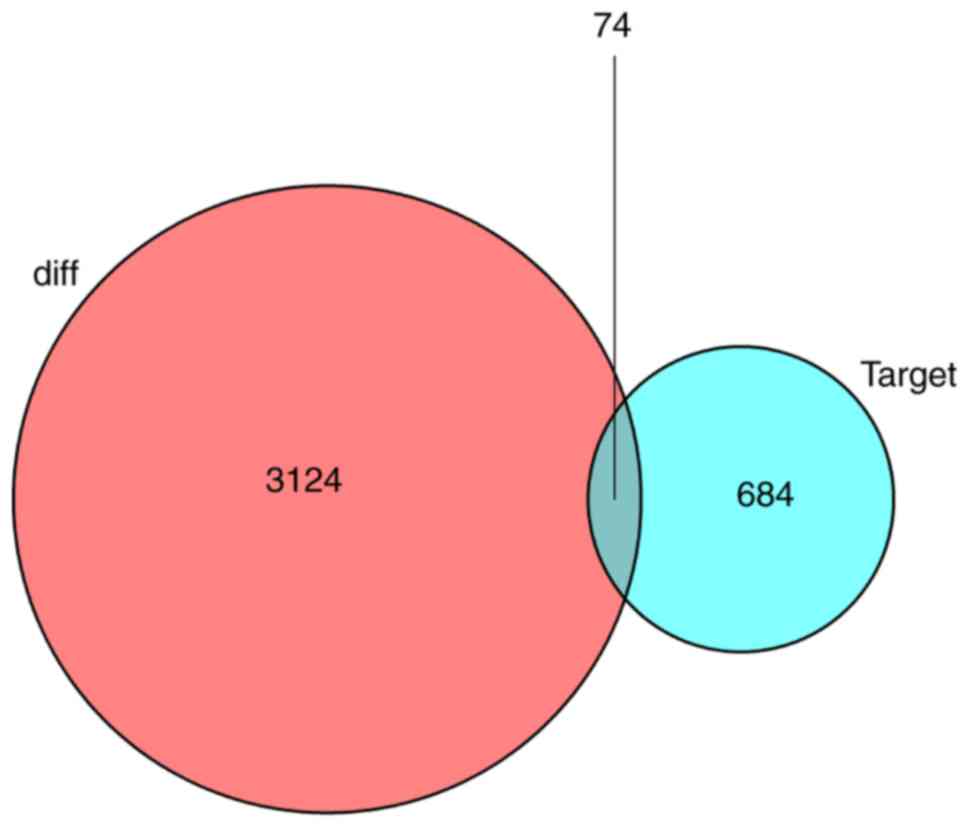
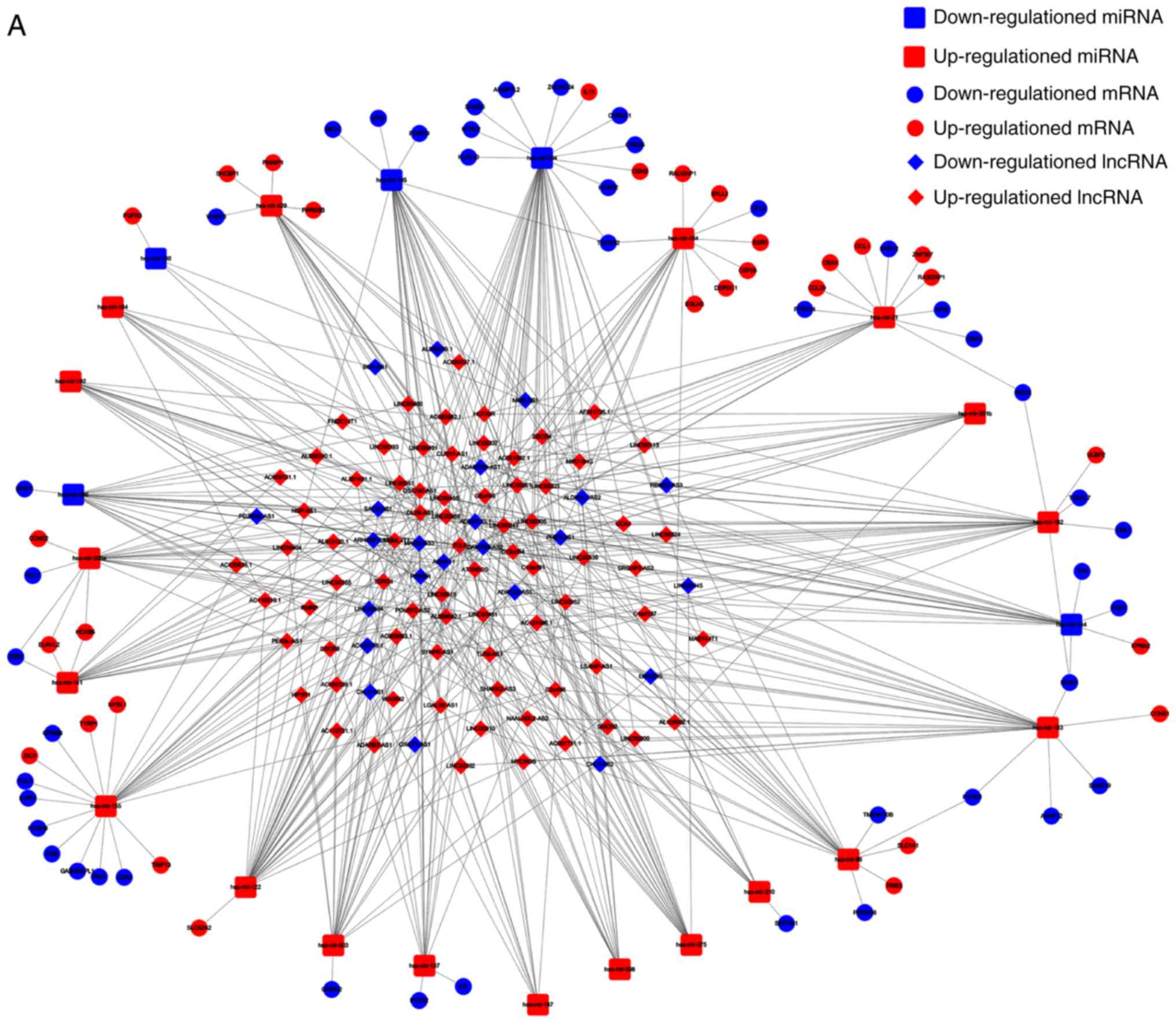
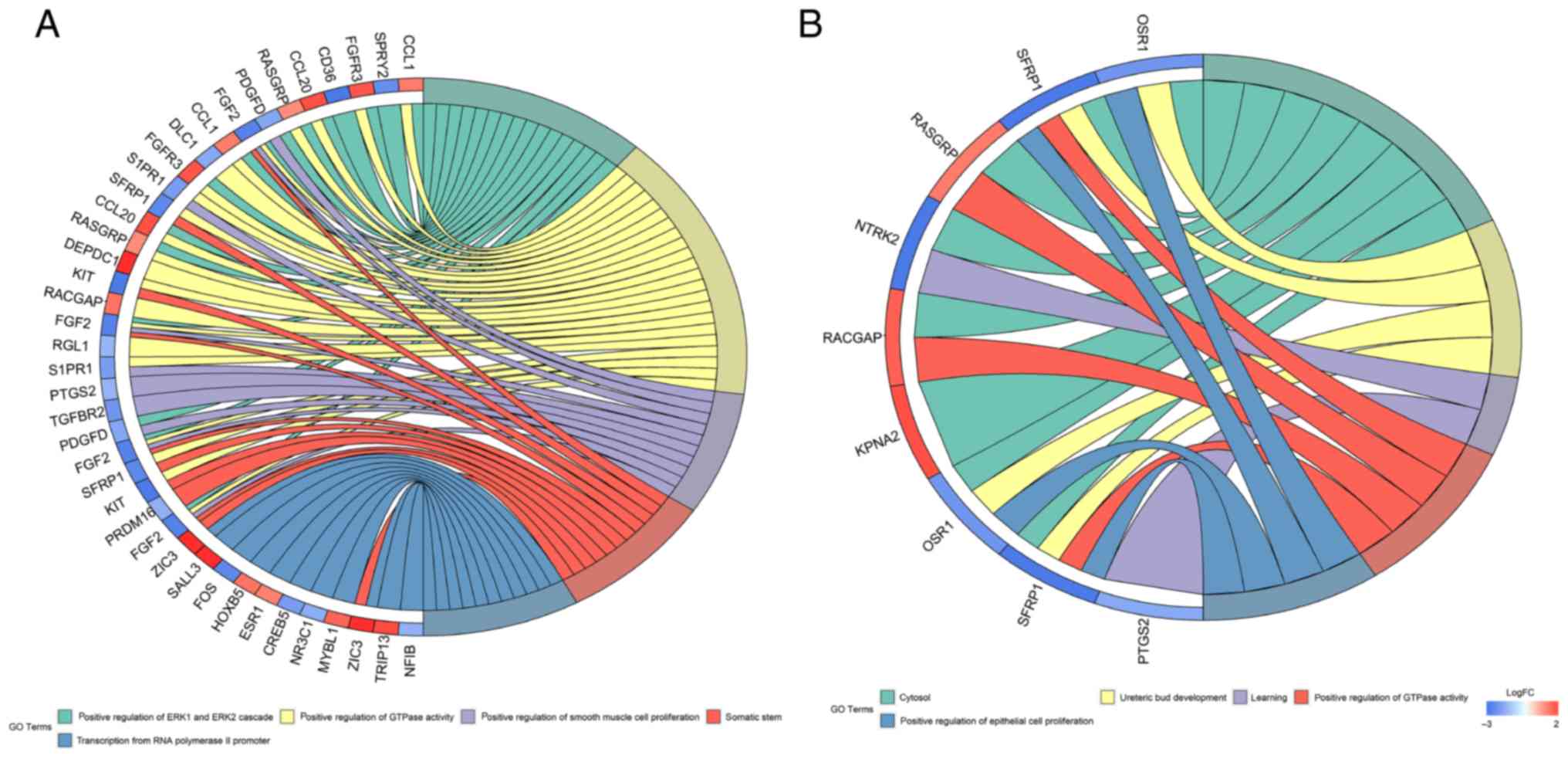
|
1
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
statistics, 2014. CA Cancer J Clin. 64:9–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wang Y, Zhou Y, Yang Z, Chen B, Huang W,
Liu Y and Zhang Y: MiR-204/ZEB2 axis functions as key mediator for
MALAT1-induced epithelial-mesenchymal transition in breast cancer.
Tumour Biol. 39:10104283176909982017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kroenke CH, Michael YL, Poole EM, Kwan ML,
Nechuta S, Leas E, Caan BJ, Pierce J, Shu XO, Zheng Y and Chen WY:
Postdiagnosis social networks and breast cancer mortality in the
after breast cancer pooling project. Cancer. 123:1228–1237. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Luschin G and Habersack M: Oral
information about side effects of endocrine therapy for early
breast cancer patients at initial consultation and first follow-up
visit: An online survey. Health Commun. 29:421–426. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Cheng Y, Tao L, Xu J, Li Q, Yu J, Jin Y,
Chen Q, Xu Z, Zou Q and Liu X: CD44/cellular prion protein interact
in multidrug resistant breast cancer cells and correlate with
responses to neoadjuvant chemotherapy in breast cancer patients.
Mol Carcinog. 53:686–697. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bao L, Messer K, Schwab R, Harismendy O,
Pu M, Crain B, Yost S, Frazer KA, Rana B, Hasteh F, et al:
Mutational profiling can establish clonal or independent origin in
synchronous bilateral breast and other tumors. PLoS One.
10:e01424872015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: The Rosetta Stone of a hidden RNA
language? Cell. 146:353–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rapicavoli NA, Qu K, Zhang J, Mikhail M,
Laberge RM and Chang HY: A mammalian pseudogene lncRNA at the
interface of inflammation and anti-inflammatory therapeutics.
Elife. 2:e007622013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: Insights into functions. Nat Rev Genet.
10:155–159. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Anamaria N, Soumillon M, Warnefors M,
Liechti A, Daish T, Zeller U, Baker JC, Grützner F and Kaessmann H:
The evolution of lncRNA repertoires and expression patterns in
tetrapods. Nature. 505:635–640. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xue B and He L: An expanding universe of
the non-coding genome in cancer biology. Carcinogenesis.
35:1209–1216. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Xie C, Yuan J, Li H, Li M, Zhao G, Bu D,
Zhu W, Wu W, Chen R and Zhao Y: NONCODEv4: Exploring the world of
long non-coding RNA genes. Nucleic Acids Res. 42((Database Issue)):
D98–D103. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kumar M.S..Armenteros-Monterroso E, East
P, Chakravorty P, Matthews N, Winslow MM and Downward J: HMGA2
functions as a competing endogenous RNA to promote lung cancer
progression. Nature. 505:212–217. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang C, Chen L, Yang Y, Zhang M and Wong
G: Identification of bladder cancer prognostic biomarkers using an
ageing gene-related competitive endogenous RNA network. Oncotarget.
8:111742–111753. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhou M, Wang X, Shi H, Cheng L, Wang Z,
Zhao H, Yang L and Sun J: Characterization of long non-coding
RNA-associated ceRNA network to reveal potential prognostic lncRNA
biomarkers in human ovarian cancer. Oncotarget. 7:12598–12611.
2016.PubMed/NCBI
|
|
17
|
Chiu YC, Hsiao TH, Chen Y and Chuang EY:
Parameter optimization for constructing competing endogenous RNA
regulatory network in glioblastoma multiforme and other cancers.
BMC Genomics. 16 (Suppl 4):S12015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wu Q, Xiang S, Ma J, Hui P, Wang T, Meng
W, Shi M and Wang Y: Long non-coding RNA CASC15 regulates gastric
cancer cell proliferation, migration and epithelial mesenchymal
transition by targeting CDKN1A and ZEB1. Mol Oncol. 12:799–813.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yao K, Wang Q, Jia J and Zhao H: A
competing endogenous RNA network identifies novel mRNA, miRNA and
lncRNA markers for the prognosis of diabetic pancreatic cancer.
Tumour Biol. 39:10104283177078822017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Shan Y, Ma J, Pan Y, Hu J, Liu B and Jia
L: LncRNA SNHG7 sponges miR-216b to promote proliferation and liver
metastasis of colorectal cancer through upregulating GALNT1. Cell
Death Dis. 9:7222018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Paci P, Colombo T and Farina L:
Computational analysis identifies a sponge interaction network
between long non-coding RNAs and messenger RNAs in human breast
cancer. BMC Syst Biol. 8:832014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Berger AC, Korkut A, Kanchi RS, Hegde AM,
Lenoir W, Liu W, Liu Y, Fan H, Shen H, Ravikumar V, et al: A
comprehensive pan-cancer molecular study of gynecologic and breast
cancers. Cancer Cell. 33:690–705.e9. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhao W, Luo J and Jiao S: Comprehensive
characterization of cancer subtype associated long non-coding RNAs
and their clinical implications. Sci Rep. 4:65912014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Jeggari A, Marks DS and Larsson E:
miRcode: A map of putative microRNA target sites in the long
non-coding transcriptome. Bioinformatics. 28:2062–2063. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li JH, Liu S, Zhou H, Qu LH and Yang JH:
StarBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA
interaction networks from large-scale CLIP-Seq data. Nucleic Acids
Res. 42((Database Issue)): D92–D97. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wong N and Wang X: miRDB: An online
resource for microRNA target prediction and functional annotations.
Nucleic Acids Res. 43((Database Issue)): D146–D152. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chou CH, Shrestha S, Yang CD, Chang NW,
Lin YL, Liao KW, Huang WC, Sun TH, Tu SJ, Lee WH, et al: MiRTarBase
update 2018: A resource for experimentally validated
microRNA-target interactions. Nucleic Acids Res. 46:D296–D302.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
Elife. 4:2015.doi: 10.7554/eLife.05005. View Article : Google Scholar
|
|
29
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Dennis G Jr, Sherman BT, Hosack DA, Yang
J, Gao W, Lane HC and Lempicki RA: DAVID: Database for Annotation,
visualization, and Integrated Discovery. Genome Biol. 4:P32003.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Aiex RM, Resende MGC and Ribeiro CC: TTT
plots: A perl program to create time-to-target plots. Optimization
Lett. 1:355–366. 2007. View Article : Google Scholar
|
|
32
|
Jia P, Liu Y and Zhao Z: Integrative
pathway analysis of genome-wide association studies and gene
expression data in prostate cancer. BMC Syst Biol. 6 (Suppl
3):S132012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Coomans de Brachène A and Demoulin JB:
FOXO transcription factors in cancer development and therapy. Cell
Mol Life Sci. 73:1159–1172. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Stagl JM, Bouchard LC, Lechner SC,
Blomberg BB, Gudenkauf LM, Jutagir DR, Glück S, Derhagopian RP,
Carver CS and Antoni MH: Long-term psychological benefits of
cognitive-behavioral stress management for women with breast
cancer: 11-year follow-up of a randomized controlled trial. Cancer.
121:1873–1881. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Haecker I and Renne R: HITS-CLIP and
PAR-CLIP advance viral miRNA targetome analysis. Crit Rev Eukaryot
Gene Expr. 24:101–116. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wang K, Guo WX, Li N, Gao CF, Shi J, Tang
YF, Shen F, Wu MC, Liu SR and Cheng SQ: Serum LncRNAs profiles
serve as novel potential biomarkers for the diagnosis of
HBV-positive hepatocellular carcinoma. PLoS One. 10:e01449342015.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Denzler R, Agarwal V, Stefano J, Bartel DP
and Stoffel M: Assessing the ceRNA hypothesis with quantitative
measurements of miRNA and target abundance. Mol Cell. 54:766–776.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Xia T, Liao Q, Jiang X, Shao Y, Xiao B, Xi
Y and Guo J: Long noncoding RNA associated-competing endogenous
RNAs in gastric cancer. Sci Rep. 4:60882014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Liu C, Liu R, Zhang D, Deng Q, Liu B, Chao
HP, Rycaj K, Takata Y, Lin K, Lu Y, et al: MicroRNA-141 suppresses
prostate cancer stem cells and metastasis by targeting a cohort of
pro-metastasis genes. Nat Commun. 8:142702017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Mikhaylova O, Stratton Y, Hall D, Kellner
E, Ehmer B, Drew AF, Gallo CA, Plas DR, Biesiada J, Meller J and
Czyzyk-Krzeska MF: VHL-regulated MiR-204 suppresses tumor growth
through inhibition of LC3B-mediated autophagy in renal clear cell
carcinoma. Cancer Cell. 21:532–546. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Hall DP, Cost NG, Hegde S, Kellner E,
Mikhaylova O, Stratton Y, Ehmer B, Abplanalp WA, Pandey R, Biesiada
J, et al: TRPM3 and miR-204 establish a regulatory circuit that
controls oncogenic autophagy in clear cell renal cell carcinoma.
Cancer Cell. 26:738–753. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Ying Z, Li Y, Wu J, Zhu X, Yang Y, Tian H,
Li W, Hu B, Cheng SY and Li M: Loss of miR-204 expression enhances
glioma migration and stem cell-like phenotype. Cancer Res.
73:990–999. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Shen SQ, Huang LS, Xiao XL, Zhu XF, Xiong
DD, Cao XM, Wei KL, Chen G and Feng ZB: miR-204 regulates the
biological behavior of breast cancer MCF-7 cells by directly
targeting FOXA1. Oncol Rep. 38:368–376. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Fort RS, Mathó C, Oliveira-Rizzo C, Garat
B, Sotelo-Silveira JR and Duhagon MA: An integrated view of the
role of miR-130b/301b miRNA cluster in prostate cancer. Exp Hematol
Oncol. 7:102018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Yuan SX, Tao QF, Wang J, Yang F, Liu L,
Wang LL, Zhang J, Yang Y, Liu H, Wang F, et al: Antisense long
non-coding RNA PCNA-AS1 promotes tumor growth by regulating
proliferating cell nuclear antigen in hepatocellular carcinoma.
Cancer Lett. 349:87–94. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zhu N, Hou J, Wu Y, Liu J, Li G, Zhao W,
Ma G, Chen B and Song Y: Integrated analysis of a competing
endogenous RNA network reveals key lncRNAs as potential prognostic
biomarkers for human bladder cancer. Medicine (Baltimore).
97:e118872018. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Xia Y, Liu Z, Yu W, Zhou S, Shao L, Song W
and Liu M: The prognostic significance of long noncoding RNAs in
bladder cancer: A meta-analysis. PLoS One. 13:e01986022018.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Xing Y, Zhao Z, Zhu Y, Zhao L, Zhu A and
Piao D: Comprehensive analysis of differential expression profiles
of mRNAs and lncRNAs and identification of a 14-lncRNA prognostic
signature for patients with colon adenocarcinoma. Oncol Rep.
39:2365–2375. 2018.PubMed/NCBI
|
|
49
|
Li Z, Yao Q, Zhao S, Wang Y, Li Y and Wang
Z: Comprehensive analysis of differential co-expression patterns
reveal transcriptional dysregulation mechanism and identify novel
prognostic lncRNAs in esophageal squamous cell carcinoma. Onco
Targets Ther. 10:3095–3105. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Wang H, Fu Z, Dai C, Cao J, Liu X, Xu J,
Lv M, Gu Y, Zhang J, Hua X, et al: LncRNAs expression profiling in
normal ovary, benign ovarian cyst and malignant epithelial ovarian
cancer. Sci Rep. 6:389832016. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Sonohara F, Inokawa Y, Hayashi M, Kodera Y
and Nomoto S: Epigenetic modulation associated with carcinogenesis
and prognosis of human gastric cancer. Oncol Lett. 13:3363–3368.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Shi L, Zhang B, Sun X, Lu S, Liu Z, Liu Y,
Li H, Wang L, Wang X and Zhao C: MiR-204 inhibits human NSCLC
metastasis through suppression of NUAK1. Br J Cancer.
111:2316–2327. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Alhawaj R: Heme biosynthesis and
metabolism are important contributors to the pathophysiology of
chronic hypoxia-induced pulmonary hypertension. Dissertations &
Theses-Gradworks, 2014. http://xueshu.baidu.com/usercenter/paper/show?paperid=c770828df9516fbf2613928c9fbecb09&site=xueshu_se&hitarticle=1
|
|
54
|
Liu J and Li Y: Trichostatin A and
Tamoxifen inhibit breast cancer cell growth by miR-204 and ERα
reducing AKT/mTOR pathway. Biochem Biophys Res Commun. 467:242–247.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Vivacqua A, De Marco P, Santolla MF,
Cirillo F, Pellegrino M, Panno ML, Abonante S and Maggiolini M:
Estrogenic gper signaling regulates mir144 expression in cancer
cells and cancer-associated fibroblasts (cafs). Oncotarget.
6:16573–16587. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Liu SY, Li XY, Chen WQ, Hu H, Luo B, Shi
YX, Wu TW, Li Y, Kong QZ, Lu HD and Lu ZX: Demethylation of the
MIR145 promoter suppresses migration and invasion in breast cancer.
Oncotarget. 8:61731–61741. 2017.PubMed/NCBI
|
|
57
|
Muti P, Sacconi A, Hossain A, Donzelli S,
Ben Moshe NB, Ganci F, Sieri S, Krogh V, Berrino F, Biagioni F, et
al: Downregulation of microRNAs 145-3p and 145-5p is a long-term
predictor of postmenopausal breast cancer risk: The ORDET
prospective study. Cancer Epidemiol Biomarkers Prev. 23:2471–2481.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Faraoni I, Antonetti FR, Cardone J and
Bonmassar E: miR-155 gene: A typical multifunctional microRNA.
Biochim Biophys Acta. 1792:497–505. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Volinia S, Galasso M, Costinean S,
Tagliavini L, Gamberoni G, Drusco A, Marchesini J, Mascellani N,
Sana ME, Abu Jarour R, et al: Reprogramming of miRNA networks in
cancer and leukemia. Genome Res. 20:589–599. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Chuang MK, Chiu YC, Chou WC, Hou HA,
Chuang EY and Tien HF: A 3-microRNA scoring system for
prognostication in de novo acute myeloid leukemia patients.
Leukemia. 29:1051–1059. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Braun R, Finney R, Yan C, Chen QR, Hu Y,
Edmonson M, Meerzaman D and Buetow K: Discovery analysis of TCGA
data reveals association between germline genotype and survival in
ovarian cancer patients. PLoS One. 8:e550372013. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Yasuda S, Stevens RL, Terada T, Takeda M,
Hashimoto T, Fukae J, Horita T, Kataoka H, Atsumi T and Koike T:
Defective expression of Ras guanyl nucleotide-releasing protein 1
in a subset of patients with systemic lupus erythematosus. J
Immunol. 179:4890–4900. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Horinouchi M, Yagi M, Imanishi H, Mori T,
Yanai T, Hayakawa A, Takeshima Y, Hijioka M, Okamura N, Sakaeda T,
et al: Association of genetic polymorphisms with hepatotoxicity in
patients with childhood acute lymphoblastic leukemia or lymphoma.
Pediatr Hematol Oncol. 27:344–354. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Depeille P, Henricks LM, van de Ven RA,
Lemmens E, Wang CY, Matli M, Werb Z, Haigis KM, Donner D, Warren R
and Roose JP: RasGRP1 opposes proliferative EGFR-SOS1-Ras signals
and restricts intestinal epithelial cell growth. Nat Cell Biol.
17:804–815. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Sharma A, Fonseca LL, Rajani C, Yanagida
JK, Endo Y, Cline JM, Stone JC, Ji J, Ramos JW and Lorenzo PS:
Targeted deletion of RasGRP1 impairs skin tumorigenesis.
Carcinogenesis. 35:1084–1091. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Zhang X, Zhuang H, Han F, Shao X, Liu Y,
Ma X, Wang Z, Qiang Z and Li Y: Sp1-regulated transcription of
RasGRP1 promotes HCC proliferation. Liver Int. 38:2006–2017. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Wang S, Beeghly-Fadiel A, Cai Q, Cai H,
Guo X, Shi L, Wu J, Ye F, Qiu Q, Zheng Y, et al: Gene expression in
triple-negative breast cancer in relation to survival. Breast
Cancer Res Treat. 171:199–207. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Lauchle JO, Kim D, Le DT, Akagi K, Crone
M, Krisman K, Warner K, Bonifas JM, Li Q, Coakley KM, et al:
Response and resistance to MEK inhibition in leukaemias initiated
by hyperactive Ras. Nature. 461:411–414. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Ding H, Peterson KL, Correia C, Koh B,
Schneider PA, Nowakowski GS and Kaufmann SH: Histone deacetylase
inhibitors interrupt HSP90 RASGRP1 and HSP90 CRAF interactions to
upregulate BIM and circumvent drug resistance in lymphoma cells.
Leukemia. 31:1593–1602. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Mortezavi A, Hermanns T, Seifert HH,
Baumgartner MK, Provenzano M, Sulser T, Burger M, Montani M,
Ikenberg K, Hofstädter F, et al: KPNA2 expression is an independent
adverse predictor of biochemical recurrence after radical
prostatectomy. Clin Cancer Res. 17:1111–1121. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Sengel-Turk CT, Hascicek C, Bakar F and
Simsek E: Comparative evaluation of nimesulide-loaded nanoparticles
for anticancer activity against breast cancer cells. Aaps
PharmSciTech. 18:393–403. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Hass HG, Vogel U, Scheurlen M and Jobst J:
Gene-expression Analysis identifies specific patterns of
dysregulated molecular pathways and genetic subgroups of human
hepatocellular carcinoma. Anticancer Res. 36:5087–5095. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Buhmeida A, Merdad A, Al-Maghrabi J,
Al-Thobaiti F, Ata M, Bugis A, Syrjänen K, Abuzenadah A, Chaudhary
A, Gari M, et al: RASSF1A methylation is predictive of poor
prognosis in female breast cancer in a background of overall low
methylation frequency. Anticancer Res. 31:2975–2981.
2011.PubMed/NCBI
|
|
74
|
Tao HC, Wang HX, Dai M, Gu CY, Wang Q, Han
ZG and Cai B: Targeting SHCBP1 inhibits cell proliferation in human
hepatocellular carcinoma cells. Asian Pac J Cancer Prev.
14:5645–5650. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Li WX, He K, Tang L, Dai SX, Li GH, Lv WW,
Guo YC, An SQ, Wu GY, Liu D and Huang JF: Comprehensive
tissue-specific gene set enrichment analysis and transcription
factor analysis of breast cancer by integrating 14 gene expression
datasets. Oncotarget. 8:6775–6786. 2016.
|
|
76
|
Yang XL, Liu KY, Lin FJ, Shi HM and Ou ZL:
CCL28 promotes breast cancer growth and metastasis through
MAPK-mediated cellular anti-apoptosis and pro-metastasis. Oncol
Rep. 38:1393–1401. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Kono M, Fujii T, Lim B, Karuturi MS,
Tripathy D and Ueno NT: Androgen receptor function and androgen
receptor-targeted therapies in breast cancer: A Review. JAMA Oncol.
3:1266–1273. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Katayama K, Yoshioka S, Tsukahara S,
Mitsuhashi J and Sugimoto Y: Inhibition of the mitogen-activated
protein kinase pathway results in the down-regulation of
P-glycoprotein. Mol Cancer Ther. 6:2092–2102. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Jia Y, Zhou J, Luo X, Chen M, Chen Y, Wang
J, Xiong H, Ying X, Hu W, Zhao W, et al: KLF4 overcomes tamoxifen
resistance by suppressing MAPK signaling pathway and predicts good
prognosis in breast cancer. Cell Signal. 42:165–175. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Serini S and Calviello G: Modulation of
Ras/ERK and phosphoinositide signaling by long-chain n-3 PUFA in
breast cancer and their potential complementary role in combination
with targeted drugs. Nutrients. 9:E1852017. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Wright KL, Adams JR, Liu JC, Loch AJ, Wong
RG, Jo CE, Beck LA, Santhanam DR, Weiss L, Mei X, et al: Ras
signaling is a key determinant for metastatic dissemination and
poor survival of luminal breast cancer patients. Cancer Res.
75:4960–4972. 2015. View Article : Google Scholar : PubMed/NCBI
|