Introduction
Cholangiocarcinoma (CCA) is the second most frequent
primitive liver malignant tumor originating from the biliary tract
epithelium. According to their anatomical location, CCAs are
currently classified as intrahepatic (iCCA), perihilar (pCCA) and
distal CCA (dCCA) (1). CCA is
associated not only with a lack of susceptibility to
chemotherapeutic drugs, but also with the development of drug
resistance in chemotherapy. Hence, at present, surgical resection
is the only treatment strategy for this disease. However, due to
the specific anatomic position of this type of tumor, the majority
of patients with CCA are already at the advanced stages of the
disease at the time of diagnosis, and are thus ineligible for
surgery (2). However, even after
radical resection, the post-operative recurrence rates of patients
with CCA range between 67 and 75% (3). The 5-year survival rate for patients
with CCA remains <5% (4).
It is well known that CA199 serves as a
tumor-associated antigen and may be useful for the diagnosis of
various types of cancer (5,6). Nevertheless, the use of CA199 in the
diagnosis of CCA is difficult due to the lack of specificity.
Therefore, the identification of novel effective tumor biomarkers
and therapeutic strategies with which to improve the diagnosis,
therapy and prognosis of patients with CCA is urgently
required.
As the development of CCA occurs at the biliary
epithelium and tumor-related proteins are secreted or shed into the
bile, the identification of markers in bile is a straightforward
approach for the accurate detection of CCA (7). Endoscopic retrograde
cholangiopancreatography (ERCP) is a well-known technique, which
provides information on stricture site, length and the presence of
mucosal irregularity or shouldering during therapeutic procedures.
Importantly, ERCP is beneficial for the diagnosis of CCA, and
provides the option of tumor sampling for cytology (8). However, the variable composition of
secretory proteins in bile poses tremendous technical challenges
for the identification of relevant biomarkers for CCA.
The technique of isobaric tags for relative and
absolute quantitation (iTRAQ) combined with liquid
chromatography-tandem mass spectrometric (LC-MS/MS) is emerging as
one of the most important proteomics approaches, and has become a
novel tool for the detection of protein expression in cancer cell
lines and tumor-related tissues (9,10).
iTRAQ-based proteomics is applied to protein samples with a more
robust labeling and can detect 8 samples in parallel (11); it can also decrease the potential
variation in multiple mass spectrometry detection. More
importantly, iTRAQ has the advantage of providing detailed protein
expression profiles and high resolution, while reducing
experimental error (12). Hence, it
has been widely used for the identification of candidate biomarkers
for multiple tumors.
In this study, quantitative proteomics analysis
using the iTRAQ in combination with LC-MS/MS was performed to
identify CCA cell-enriched secretory proteins compared with those
in a normal biliary epithelial cell line. In total, 778 proteins
were identified as differentially expressed proteins (DEPs) and
were classified according to biological process, cellular component
and as molecular function using bioinformatics analyses. Finally,
we confirmed tumor protein D52 (TPD52) and DnaJ heat shock protein
family (Hsp40) member B1 (DNAJB1) as biomarkers, which were
upregulated in CCA cells, culture supernatants, tissues and bile
samples. In addition, the overexpression of TPD52 and DNAJB1 was
found to be associated with the clinical parameters and prognosis
of patients with CCA.
Materials and methods
Reagents
RPMI-1640 medium, fetal bovine serum (FBS) and
penicillin were purchased from Invitrogen; Thermo Fisher
Scientific. Antibodies against TPD52, transforming acidic
coiled-coil-containing protein 3 (TACC3), Ephrin A1(EFNA1),
transferrin (TRF) and β-actin were obtained from Abcam. DNAJB1,
LDH, trypsin and goat anti-rabbit/mouse secondary antibodies
conjugated to horseradish peroxidase were from Sigma-Aldrich. The
Total RNA extraction kit, SYBR-Green detection system and the
Bradford protein assay kit were purchased from Tiangen Biotech. The
ultrafiltration device, ECL reagent and polyvinylidene difluoride
(PVDF) were from Millipore. The DAB detection kit was from Maixin
Biotech. The iTRAQ labeling Reagents kit was from Applied
Biosystems.
Patients and tissue samples
A tissue microarray (TMA) of CCA was obtained from
National SOBC Biobank, which included 127 CCA specimens and 9
samples of normal biliary epithelial tissues. Among the 127 CCA
specimens, 61 patients were ≤60 years old and 66 patients were
>60 years old. In total, 80 patients were males and 47 were
females. Of these patient samples, 78 were characterized as low
differentiation (I, I–II and II) and 49 as high differentiation
(II–III, III and III–IV). In total, 100 patients had iCCA and 27
patients had extrahepatic CCA. As regards the tumor sizes, 59
patients had tumors ≤5 cm, 58 had tumors >5 cm and in 10
patients, tumor size was unknown. Vessel invasion was found in 100
patients. Information regarding TNM stage and clinical stage was
obtained for the patients. Fresh bile samples of 29 patients with
CCA and 8 healthy individuals were collected at the First
Affiliated Hospital of Xiamen University between January 2015 and
December 2017. This study was approved by the Ethics Committee of
the First Affiliated Hospital of Xiamen University. Informed
consent was obtained from all individual participants included in
the study.
Cells and cell culture
Normal biliary epithelial [human intrahepatic
biliary epithelial cells (HiBECs)] cells were obtained from
PriCells and were cultured in PriCells medium. The human cancer
cell lines (cancers of the hepatobiliary system), QBC939 (CCA),
SK-ChA-1 (CCA), MZ-ChA-1 (gallbladder cancer, for comparison), TFK1
(CCA) and HuCCT1 (CCA), were routinely cultured in RPMI-1640 medium
containing 10% FBS and 100 U/ml penicillin. The cells were
maintained at 37°C in a humidified atmosphere under 5%
CO2. The human CCA cell line QBC939 was kindly provided
by Professor Shu-Guang Wang from Southwest Hospital, the Third
Military Medical University, Chongqing, China. The cell lines
HuCCT1, Sk-ChA-1, MZ-ChA-1 and TFK1 were kindly provided by
Chun-Dong Yu, laboratory of Xiamen University, Xiamen, China.
HiBECs were purchased from Cell Bank of the Type Culture Collection
of Chinese Academy of Sciences (HUM-CELL-0035).
CCA cell culture supernatant
collection
The same number cells, including the 4 CCA cell
lines, a gallbladder cancer cell line (MZ-ChA-1) and a biliary
epithelial cell line (HiBECs) were seeded in 6-well plates
overnight and then cultured in serum-free cell culture medium for
24 h at 37°C. The cell culture supernatants containing secretory
proteins were collected and centrifuged to remove cells. Following
centrifugation at 4,000 × g for 30 min at 4°C to concentrate using
ultrafiltration with 3-kDa filters, the cell culture supernatants
were transferred to new tubes and then stored at −70°C. The CCA
cell culture supernatants were referred to as TRF. LDH was used as
a quality control.
Bile secretory protein collection
The bile samples were centrifugated at 12,000 × g
for 30 min at 4°C to remove impurities, and the supernatants
containing secretory proteins were then transferred to a clean
tube. If the samples have more impurities following the
above-mentioned strategy, repeated centrifugation is then
necessary. The protein concentration was determined using the
Bradford protein assay kit according to the instructions of the
manufacturer. The CCA bile samples were referred to TRF.
Western blot analysis
Cell lysates containing equal amounts of proteins
underwent 8% SDS-polyacrylamide gel electrophoresis followed by
transfer onto PVDF membranes. After blocking non-specific binding
sites with 5% dried skimmed milk solution for 1 h at room
temperature, the membranes were incubated overnight at 4°C with
primary antibodies (TPD52, 1:10,000 dilution, ab155296, Abcam;
DNAJB1, 1:500 dilution, SAB2100604, Sigma-Aldrich; TACC3, 1:500
dilution, ab138262, Abcam; Ephrin A1, 1:500 dilution, ab199697,
Abcam; β-actin, 1:1,000 dilution, ab227387, Abcam). The membranes
were then incubated with secondary antibodies conjugated to
horseradish peroxidase (against mouse, 1:10,000 dilution, A9917,
Sigma-Aldrich; against rabbit, 1:5,000 dilution, A9169,
Sigma-Aldrich) for 1 h at room temperature and the signals were
finally detected with ECL system. β-actin was used as a loading
control, and the data were quantitatively measured using ImageJ
software (version 1.48; National Institutes of Health).
Protein digestion and iTRAQ
labeling
Secretory proteins of CCA cells were extracted as
described above. iTRAQ labeling was performed according to the
manufacturer's protocol. Briefly, 100 µg proteins of each sample
were reduced and alkylated, and subsequently digested with trypsin
overnight at 37°C. Sequentially, 3 iTRAQ regents (117, 118 and 119)
were used to label TFK1, HiBECs and HuCCT1, respectively. Finally,
the samples were lyophilized.
LC-MS/MS analysis
The iTRAQ samples were analyzed using LC-MS/MS.
Briefly, labeled samples were pooled and fractionated by strong
cation exchange (SCX) chromatography. The mixed peptides were
reconstituted with buffer A (10 mM KH2PO4 in 25% acetonitrile, pH
3.0). Separation was carried out using a linear binary gradient of
0–100% buffer B (buffer B was buffer A containing 2 M KCl) at a
rate of l m/min for 10–20 min. The absorbance at 214 and 280 nm was
monitored, and a total of 15 fractions were collected. Following
the desalination process, the SCX fractions were dried and
dissolved in buffer C (0.1% formic acid). They were then separated
on a reverse-phase column (75×10 cm, 5 µm, 300 Ǻ, Agela
Technologies) using the Nano-LC system. Subsequently, the elution
flow rate was maintained at 400 nl/min with 5–80% buffer D
(acetonitrile with 0.1% formic acid) in buffer C for 100 min. A
survey scan was acquired from m/z 350–2000 at a mass resolution of
17,500 full width at half maximum (FWHM) for MS/MS. The experiment
was conducted in 3 technical replicates.
Data analysis
All of the obtained mass spectrometry data were
processed with Proteome Discoverer 1.3 (Thermo Fisher Scientific)
and subsequently searched using MASCOT engine (Matrix Science;
version 2.3.01). The parameters were set as follows: Trypsin as
digestion enzyme, cysteine carbamidomethylation as a fixed
modification, Oxidation (M), Gln→Pyro-Glu (N-term Q), iTRAQ 8 plex
(K), iTRAQ 8 plex (Y) and iTRAQ 8 plex (N-term) as the variable
modification. If the iTRAQ ratio in the CCA cell-enriched secretory
proteins compared with the normal biliary epithelial cell line was
>1.2 and the computed P-value <0.05, the protein was
considered to be differentially expressed.
Bioinformatics analysis
The Gene Ontology (GO, http://www.geneontology.org/) database was used for
the functional annotation among the obtained proteins. The DEPs in
GO were divided into 3 categories, including biological process,
molecular function and cellular component. The Kyoto Encyclopedia
of Genes and Genomes (KEGG, http://www.kegg.jp/) database was used to analyze the
cancer pathways of the DEPs.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted and reverse transcribed from
the cultured cells using RNA simple total RNA extraction kit
(Tiangen Biotech) according to the instructions of the manufacturer
as previously described (13). qPCR
was performed using the SYBR-Green detection system (Tiangen
Biotech). The primers for qPCR reactions are listed in Table SI. The relative expression levels of
the target genes were according to GAPDH. The qPCR thermocycling
conditions were as follows: 95°C for 30 sec, 45 cycles of 90°C for
30 sec, 60°C for 20 sec, 72°C for 25 sec, and 72°C for 15 min. The
relative gene expression levels were analyzed using the
2−∆∆Cq method (14). All
experiments were performed in triplicate.
Immunohistochemistry
(IHC)/immunocytochemistry (ICC)
After dewaxing in xylene and brought to water
through graded alcohols as previously described (13), the 3-µm-thick tissue microarray
sections were incubated with antibodies against TPD52 (1:400
dilution) and DNAJB1 (1:200 dilution) overnight at 4°C. Appropriate
negative controls were performed by omitting the primary antibody.
For ICC, the cells planted on the glass slides were cultured in
RPMI-1640 medium. After fixing with 4% paraformaldehyde, the slides
were permeabilized with 0.5% Triton X-100. Subsequent steps were
performed as for IHC.
Statistical analysis
All statistical analyses were performed using SPSS
16.0 software (SPSS, Inc.). Data were expressed as the means ± SEM
from at least 3 independent experiments. The Student's t-test was
used to analyze two sets of data, and one-way analysis of variance
(one-way ANOVA) with Dunnett's post hoc test was performed to
compare >2 datasets. Pearson's χ2 test was applied to
evaluate the association between the patient clinicopathological
parameters and the expression protein levels of TPD52 and DNAJB1.
Survival curves were evaluated by the Kaplan-Meier method. The
log-rank test was used to compare the significant difference
between groups. Differences were determined to be statistically
significant when the P-value was <0.05.
Results
Quantitative proteomics analysis of
secretory proteins by iTRAQ
To identify dysregulated secretory proteins in CCA,
iTRAQ quantitative proteomics analysis was performed to compare the
CCA cell lines (TFK1 and HuCCT1) with a normal biliary epithelial
cell line (HiBECs). The strategy of iTRAQ analysis between the 2
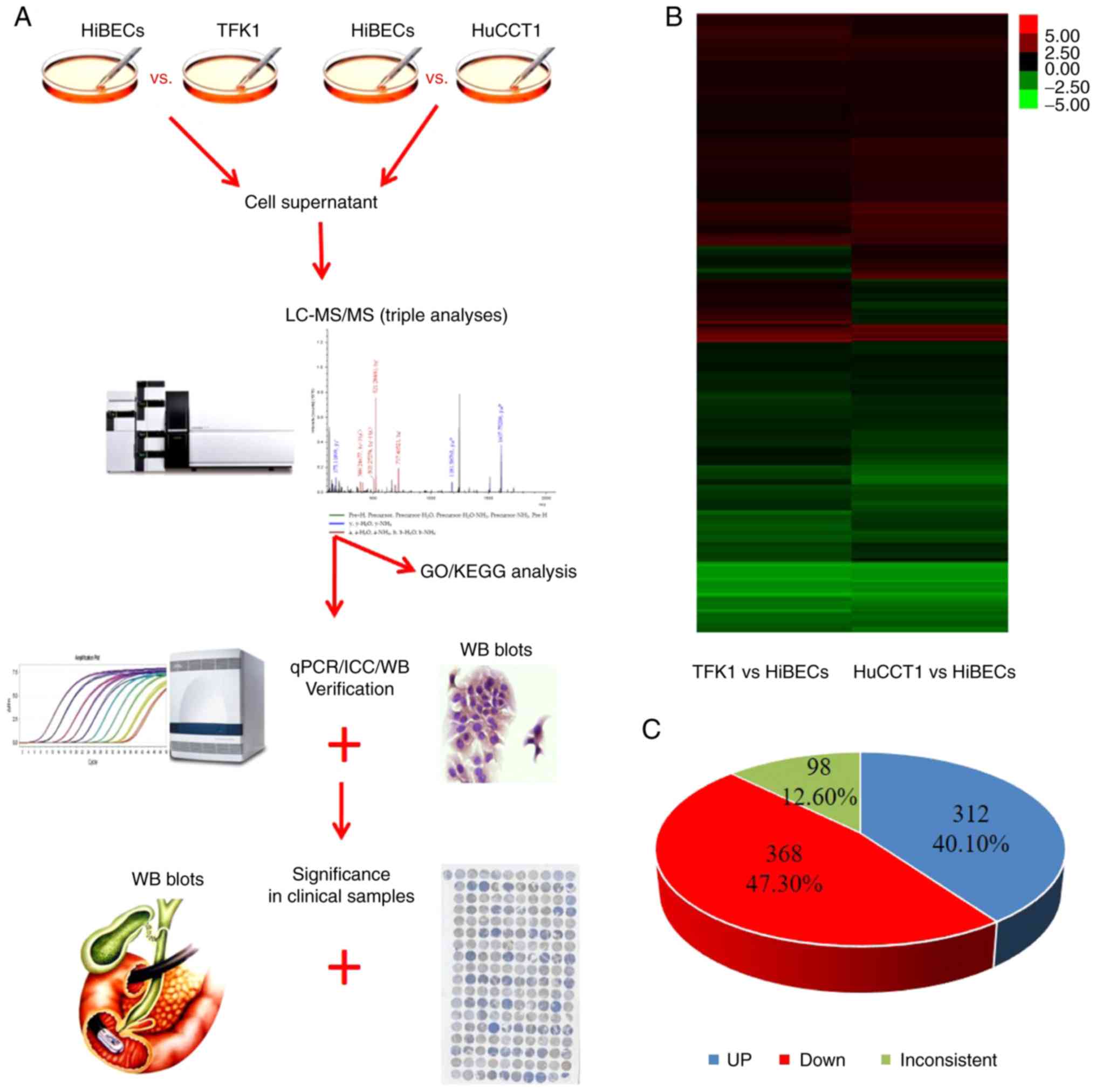
CCA cells (TFK1 and HuCCT1) and the HiBECs is presented in Fig. 1A. When the expression levels of
proteins exhibited differences corresponding to >1.2- or
<1.2-fold changes in expression in the TFK1 or HuCCT1 cells
compared to the HiBECs, these proteins were regarded as DEPs. As
shown in Fig. 1B, a total of 778 DEPs
were detected. Briefly, 680 proteins were significantly altered in
2 CCA cell lines (TFK1 and HuCCT1) compared with the HiBECs. Among
these, 312 proteins (40.10%) displayed a higher expression, whereas
368 proteins (47.30%) exhibited a lower expression. Furthermore, 98
proteins (12.60%) presented a differential expression (either
upregulation or downregulation in the 2 CCA cell lines compared to
the HiBECs (Fig. 1C).
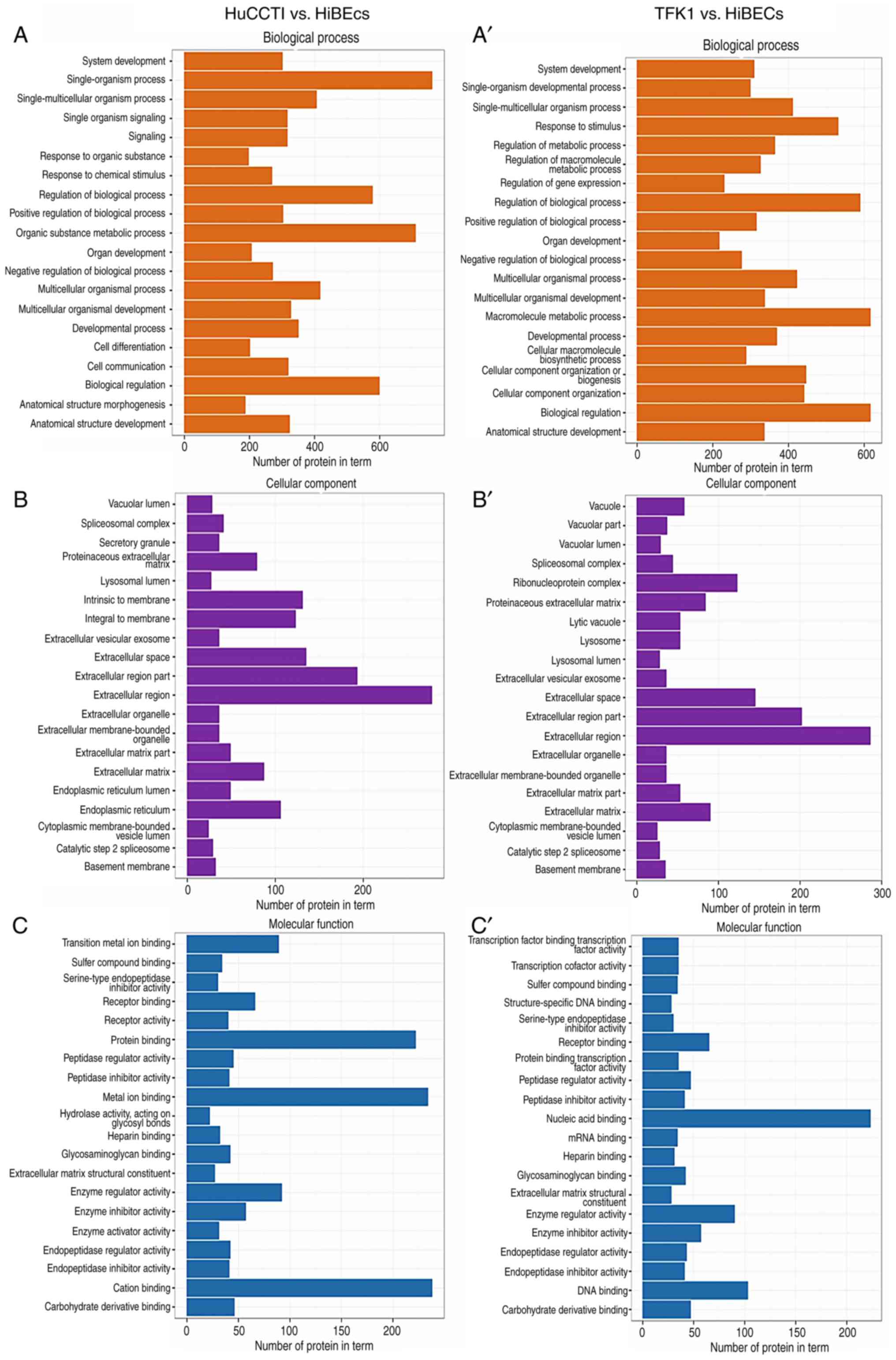
To reveal the biological functions of the
above-mentioned DEPs, we categorized these DEPs between the 2 CCA
cell lines and the HiBECs into 3 large groups, including molecular
functions, biological processes and cellular components, as well as
60 subgroups depending on their functional annotation in the GO
database. As shown in Fig. 2, the
results of GO analysis revealed that the DEPs were mainly
distributed in the extracellular region. In terms of biological
processes, the DEPs mainly participated in macromolecule metabolic
process and biological regulation. As regards molecular function,
the results indicated that the DEPs were mainly involved in nucleic
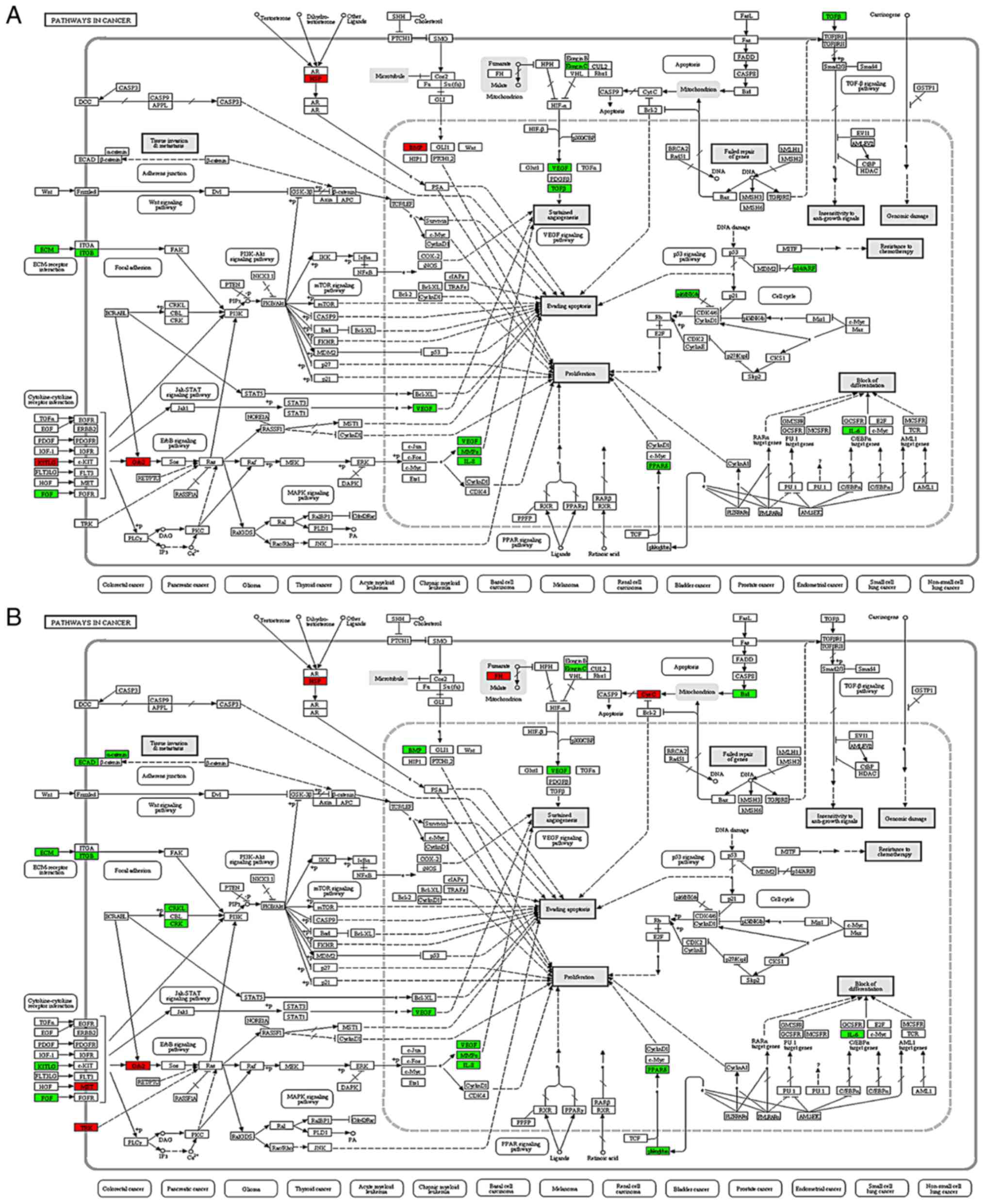
acid binding and protein binding. In addition, KEGG pathway
analysis revealed that the DEPs were associated with processes
involved in the development and progression of cancers, such as
proliferation, the evasion of apoptosis, differentiation, tissue
invasion and resistance to chemotherapy (Fig. 3). Among these, 22 proteins were found
to be connected to carcinogenesis. Most importantly, the expression
levels of TPD52, DNAJB1, TACC3 and Ephrin A1 (EFNA1) were
upregulated in the CCA cells compared with the HiBECs (Figs. S1–S4).
Finally, the above-mentioned 4 candidate DEPs were considered for
further analysis.
Validation of TPD52 and DNAJB1
upregulation in CCA cell lines
To confirm the dysregulated expression of 4 DEPs
(TPD52, DNAJB1, TACC3 and EFNA1), RT-qPCR and western blot analysis
were performed to detect the mRNA and protein expression levels of
the 4 markers in 4 CCA cell lines, a gallbladder cancer cell line
(MZ-ChA-1) and a biliary epithelial cell line (HiBECs). The results
revealed that the mRNA expression levels of the above-mentioned 4
makers in the 4 CCA cell lines and MZ-ChA-1 were all significantly
higher than those in the HiBECs cell line (Fig. 4A); however, only the protein
expression levels of TPD52 and DNAJB1 were in line with the mRNA
results (Fig. 4B). In addition, ICC
was performed to observe the cellular localization, which revealed
that TPD52 and DNAJB1 were strongly stained and predominantly
presented in the cell cytoplasm of the 4 CCA cell lines and
MZ-ChA-1, but weakly or not stained in the HiBECs cell line
(Fig. 4C). Hence, TPD52 and DNAJB1
were selected for further analysis.
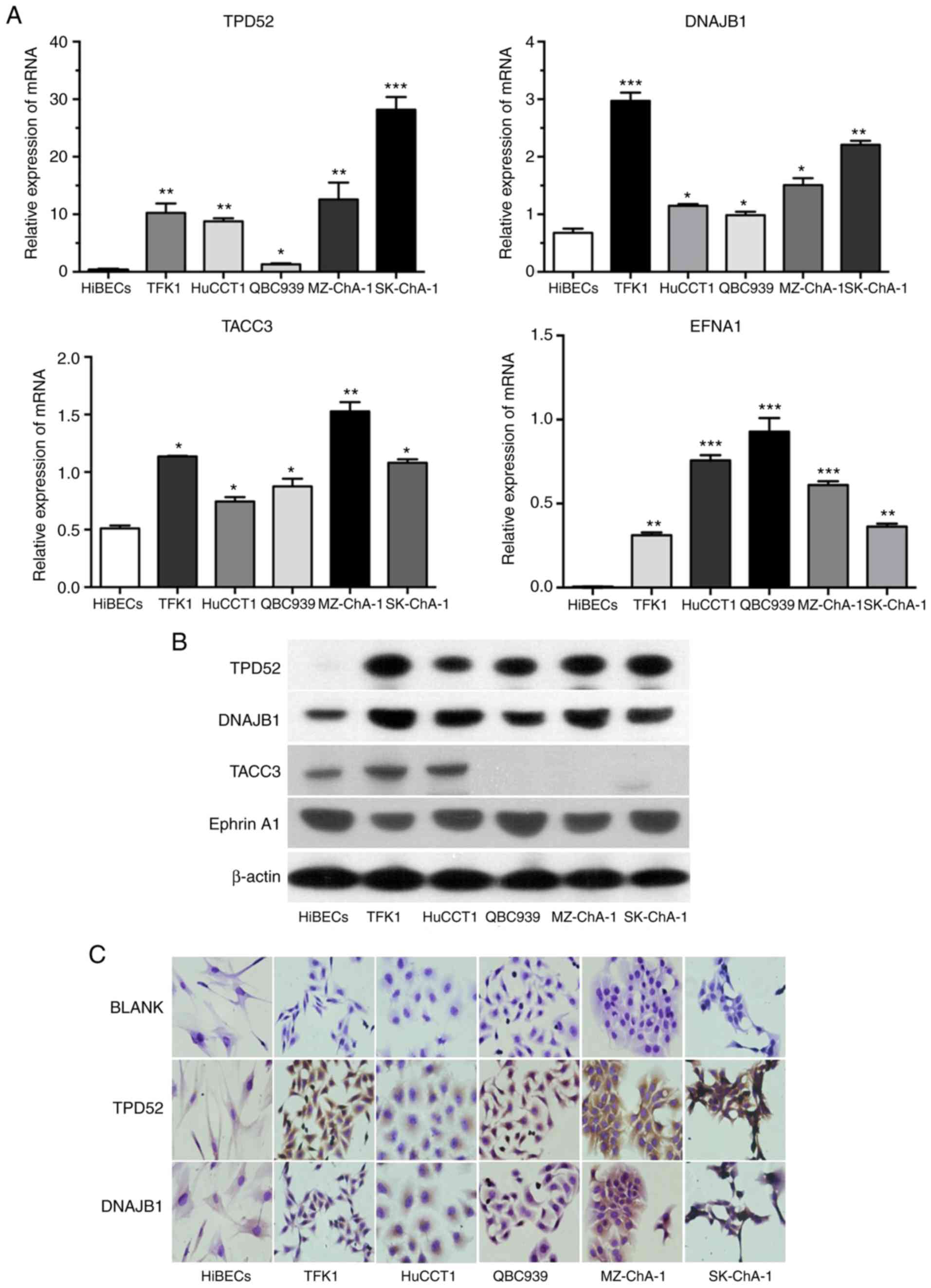 | Figure 4.Initial validation of 2 candidate
biomarkers. (A and B) mRNA and protein expression levels of TPD52,
DNAJB1, TACC3 and Ephrin A1 (EFNA1) in 4 CCA cell lines, a
gallbladder cancer cell line (MZ-ChA-1) and a normal biliary
epithelial cell line (HiBECs) were respectively analyzed by RT-qPCR
and western blot analysis. (C) Representative ICC staining for
TPD52 and DNAJB1 in 4 CCA cell lines, MZ-ChA-1 and HiBECs.
Magnification, ×400. *P<0.05, **P<0.01 and ***P<0.001.
CCA, cholangiocarcinoma; HiBECs, human intrahepatic biliary
epithelial cells; TPD52, tumor protein D52; DNAJB1, DnaJ heat shock
protein family (Hsp40) member B1; TACC3, transforming acidic
coiled-coil-containing protein 3. |
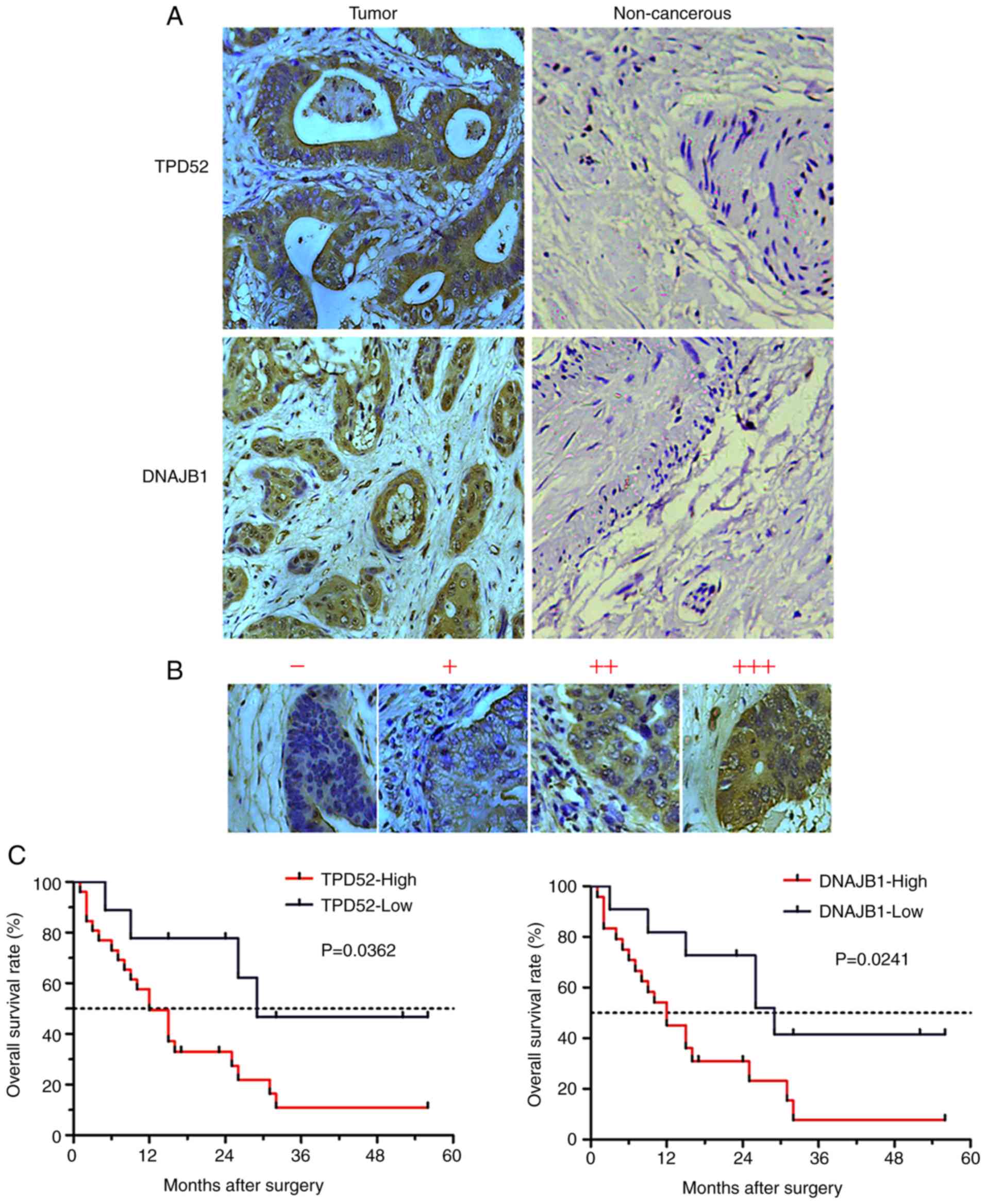
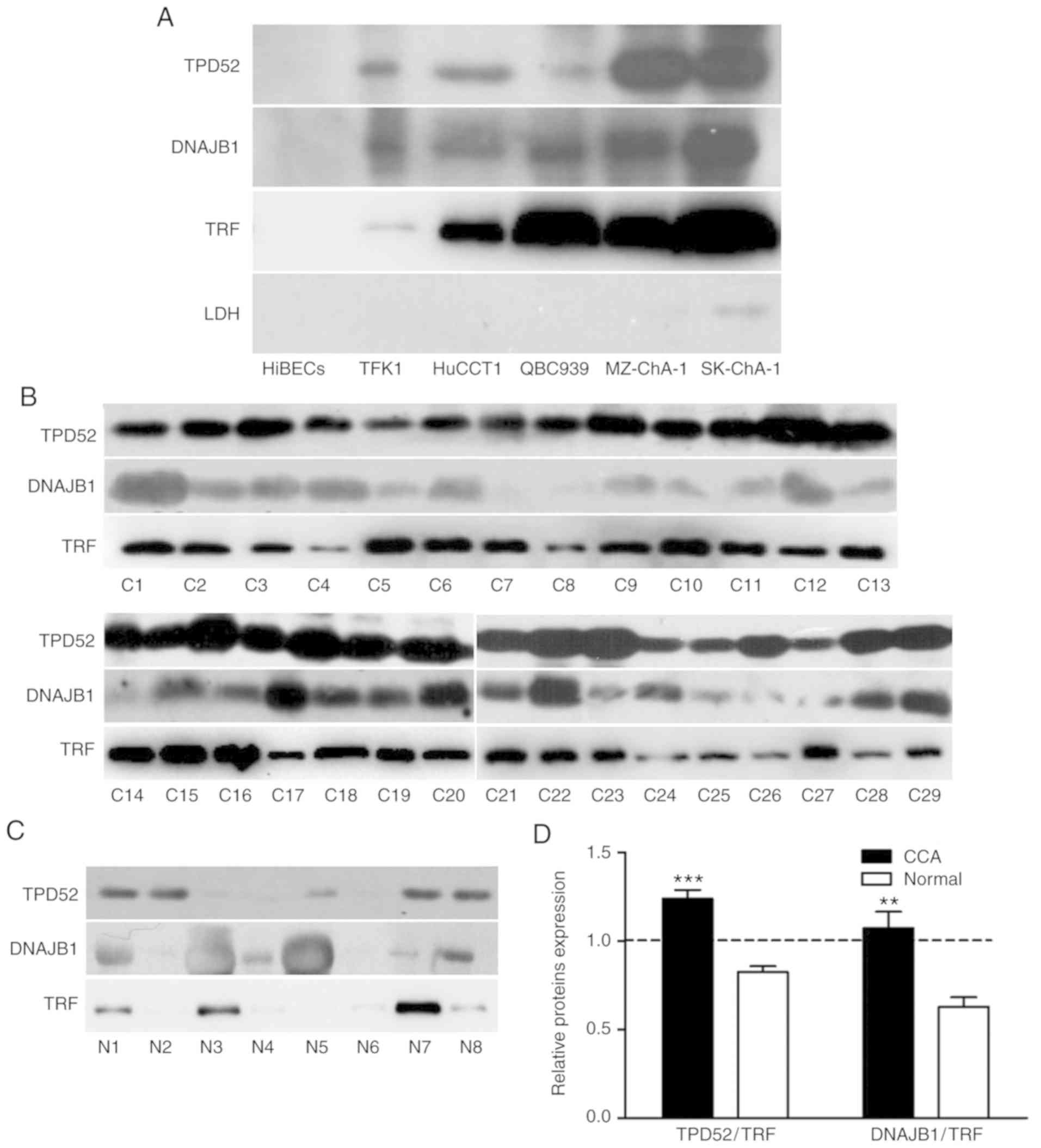
TPD52 and DNAJB1 protein levels are
significantly elevated in tissues of patients with CCA
To further evaluate the TPD52 and DNAJB1 expression
levels in clinical CCA tissues, IHC was performed to analyze the
expression levels of the 2 markers using a commercial TMA
containing an independent cohort of CCA (127 cases) samples. The
results revealed that 2 markers, predominantly presenting in the
cell cytoplasm, were all strongly stained in the CCA tissues, but
weakly or not stained in the normal biliary epithelial tissues
(Fig. 5A). The staining intensity of
the 2 markers in the CCA tissues were categorized as low (− to +)
or high (2+ to 3+) (Fig. 5B). The
high expression rate of TPD52 protein in the CCA tissues was 80.3%
(120/127) and that of DNAJB was 78.7% (100/127); the levels of both
these proteins were much higher in the cancerous samples than in
the normal biliary epithelial tissues (0%) (P<0. 0001; Table I). These data demonstrated that TPD52
and DNAJB1were markedly overexpressed in CCA tissues.
 | Table I.Expression of two tumor markers in
CCA and non-cancerous tissues. |
Table I.
Expression of two tumor markers in
CCA and non-cancerous tissues.
|
|
| TPD52 |
| DNAJB1 |
|
|---|
|
|
|
|
|
|
|
|---|
| Tissues | No. of samples | L | H | P-value | L | H | P-value |
|---|
| Tumor | 127 | 25 | 102 | 0.0001a | 27 | 100 | 0.0001a |
| Non-cancerous | 9 | 9 | 0 |
| 9 | 0 |
|
Associations of TPD52 and DNAJB1
expression with the clinical parameters of patients with CCA
We analyzed the associations between the expression
of TPD52 or DNAJB1 and the clinicopathological parameters of
patients with CCA using the χ2 text. The results of
statistical analysis are summarized in Table II. We observed that the TPD52
expression level was closely associated with pathological
differentiation, T stage and clinical stage (P<0.05); however,
it was not associated with age, sex, localization, tumor size,
vessel invasion, lymph node metastasis and distant metastasis
(P>0.05) in the patients with CCA. The DNAJB1 expression level
was closely associated with pathological differentiation, vessel
invasion, T stage, lymph node metastasis and clinical stage
(P<0.05); however, it was not associated with age, sex,
localization, tumor size and distant metastasis (P>0.05) in the
patients with CCA (Table II). These
results indicated that high expression levels of the 2 proteins may
be associated with the development of CCA.
 | Table II.Associations between the 2 tumor
markers and clinical features of patients with CCA. |
Table II.
Associations between the 2 tumor
markers and clinical features of patients with CCA.
|
| TPD52 |
| DNAJB1 |
|
|---|
|
|
|
|
|
|
|---|
| Features | N | L | H | P-value | L | H | P-value |
|---|
| Age (years) |
|
|
| 0.3836 |
|
| 0.6742 |
|
≤60 | 61 | 14 | 47 |
| 12 | 49 |
|
|
>60 | 66 | 11 | 55 |
| 15 | 51 |
|
| Sex |
|
|
| 0.4191 |
|
| 0.9972 |
|
Male | 80 | 14 | 66 |
| 17 | 63 |
|
|
Female | 47 | 11 | 36 |
| 10 | 37 |
|
|
Differentiation |
|
|
| 0.0332a |
|
| 0.0158a |
| I,
I–II, II | 78 | 20 | 58 |
| 22 | 56 |
|
| II–III,
III, III–IV | 49 | 5 | 44 |
| 5 | 44 |
|
| Localization |
|
|
| 0.7087 |
|
| 0.5042 |
|
Intrahepatic | 100 | 19 | 81 |
| 20 | 80 |
|
|
Extrahepatic | 27 | 6 | 21 |
| 7 | 20 |
|
| Tumor size
(cm) |
|
|
| 0.6811 |
|
| 0.6811 |
| ≤5 | 59 | 13 | 46 |
| 13 | 46 |
|
|
>5 | 58 | 11 | 47 |
| 11 | 47 |
|
|
Unknown | 10 | 1 | 9 |
| 3 | 7 |
|
| Vessel
invasion |
|
|
| 0.3580 |
|
| 0.0474a |
|
Negative | 100 | 18 | 82 |
| 25 | 75 |
|
|
Positive | 27 | 7 | 20 |
| 2 | 25 |
|
| T stage |
|
|
| 0.0255a |
|
| 0.0002a |
| T1 | 15 | 9 | 6 |
| 11 | 4 |
|
| T2 | 39 | 11 | 28 |
| 6 | 33 |
|
| T3 | 17 | 3 | 14 |
| 5 | 12 |
|
| T4 | 4 | 0 | 4 |
| 0 | 4 |
|
|
Unknown | 52 | 2 | 50 |
| 5 | 47 |
|
| N stage |
|
|
| 0.0594 |
|
| 0.0021a |
| N0 | 58 | 20 | 38 |
| 24 | 34 |
|
| N1 | 25 | 3 | 22 |
| 2 | 23 |
|
|
Unknown | 44 | 2 | 42 |
| 1 | 43 |
|
| M stage |
|
|
| 0.1370 |
|
| 0.3436 |
| M0 | 120 | 22 | 98 |
| 25 | 95 |
|
| M1 | 7 | 3 | 4 |
| 0 | 7 |
|
| Clinical stage |
|
|
| 0.0042a |
|
| 0.0174a |
| 1 | 16 | 11 | 5 |
| 9 | 7 |
|
| 2 | 37 | 9 | 28 |
| 10 | 27 |
|
| 4 | 17 | 4 | 13 |
| 2 | 15 |
|
|
Unknown | 57 | 1 | 56 |
| 6 | 51 |
|
Overexpression of TPD52 or DNAJB1 is
associated with the prognosis of patients with CCA
To address the prognostic significance of TPD52 and
DNAJB1 in CCA, Kaplan-Meier analysis was performed to evaluate the
association between the survival time of patients with CCA and the
expression levels of TPD52 or DNAJB1. Of note, the post-operative
overall survival rate of patients with a high TPD52 expression
(median survival time, 12 months) was lower than that of patients
with a low TPD52 expression (median survival time, 29 months)
(P<0.05). In addition, post-operative overall survival rate of
patients with a high DNAJB1 expression (median survival time, 12
months) was also lower than that of patients with a low DNAJB1
expression (median survival time, 29 months) (P<0.05) (Fig. 5C). These results suggest that the
overexpression of TPD52 or DNAJB1 may predict a poor prognosis of
patients with CCA.
A high level of TPD52 or DNAJB1 in
bile is a potential bile marker for CCA
In order to determine whether the 2 proteins can be
used as biliary tumor markers for CCA, we first detected the
existence and the level of the 2 proteins in the cell culture
supernatant of CCA cells compared with the HiBECs. The results from
western blot analysis indicated that TPD52, DNAJB1, as well as TRF,
were not only secreted into the CCA cell supernatant, and that the
levels were higher than those of the HiBECs (Fig. 6A). Furthermore, we then analyzed the
bile concentration of TPD52 and DNAJB1 proteins in 29 patients with
CCA and 8 healthy individuals by western blot analysis. Of note, we
found that the expression levels of TPD52 and DNAJB1 in the bile of
patients with CCA were all markedly higher than those of the
healthy individuals (Fig. 6B and C).
As shown in Fig. 6D, TPD52 expression
was higher in the patients with CCA and lower in the healthy
individuals compared with TRF. These results indicated that TPD52
and DNAJB1 may be more sensitive biliary biomarkers for the
clinical diagnosis of patients with CCA.
Discussion
The investigation of the proteome involved in the
development of cancer has significance in the discovery of
biomarkers for tumors. However, the identification of secretory
proteins related to CCA remains limited. In the present study,
iTRAQ labeling combined with LC-MS/MS was used to detect CCA
cell-enriched secretory proteins compared with the normal biliary
epithelial cell line. A total of 778 DEPs were identified and 4
upregulated proteins associated with cancer were further
identified, including TPD52, DNAJB1, TACC3 and Ephrin A1. In
addition, the GO database revealed cellular component, molecular
functions and biological process of 778 DEPs. The signaling
pathways of these proteins were analyzed using the KEGG database.
Finally, following validation by RT-qPCR and western blot analysis,
TPD52 and DNAJB1 were selected for further analysis.
With the progress of ‘Omics’ technologies, a number
of technologies have been applied to CCA biomarker discovery
(15). Although there are some
studies available on the development of CCA biomarkers using
proteomic-based approaches (16,17), the
identification of secretory proteins in CCA remains unknown. Thus,
this study aimed to detect CCA cell-enriched secretory proteins
using iTRAQ labeling combined with LC-MS/MS, and to further
validate the biomarkers in CCA cell lines, tissue and bile.
Due to the limited number of bile samples, we could
not use β-actin or GAPDH as a control. There is an evidence to
indicate that TRF may serve as a good standard in the secreted
protein (18). The findings of this
study indicated that TPD52 and DNAJB1 were highly expressed tissues
from patients with CCA compared with those healthy individuals
compared with TRF, suggesting that TPD52 and DNAJB1 may be
sensitive biliary biomarkers for the clinical diagnosis of CCA.
TPD52 is a composing member of the TPD52-like
protein family, which are small coiled-coil motif bearing proteins
and conserved from lower organisms to humans (19). TPD52 was first identified through its
overexpression in human breast cancer (20). In recent years, a variety of studies
have demonstrated that TPD52 expression is increased at both the
mRNA and protein level in a vast number of malignancies, such as
lung squamous cell carcinoma (21),
colorectal cancer (22), testicular
germ cell tumor (23), prostate
cancer (24) and ovarian carcinoma
(25). In addition, TPD52 is
associated with cellular transformation, proliferation and
metastasis. It has been reported that TPD52 serves as an oncogene
and plays a role in the development of several types of cancer. For
example, the overexpression of TPD52 has been shown to promote cell
migration and invasion by inducing epithelial-mesenchymal
transition and activating focal adhesion kinase-mediated integrin
signaling and PI3K⁄Akt signaling in colorectal cells (22). In prostate cancer, the silencing of
the TPD52 gene has been shown to significantly inhibit cancer cell
migration and invasion (26). In the
present study, we found that TPD52 was upregulated in CCA cells,
tissues and bile samples. Moreover, the expression level of TPD52
was closely associated with pathological differentiation, T stage
and clinical stage in patients with CCA. Furthermore, the
overexpression of TPD52 was associated with a poor prognosis of
patients with CCA.
DNAJB1, a member of the heat shock 40 protein
family, has been shown to be associated with a variety of cellular
processes, including the proteasome pathway (27), endoplasmic reticulum stress (28) and viral infection (29). Recently, more attention has been paid
to the function of DNAJB1 in the progression of cancer. There is
evidence to indicate that the DNAJB1-PRKACA chimeric transcript is
elevated in 100% of fibrolamellar hepatocellular carcinoma cases,
and this suggests that alterations in the expression of
DNAJB1-PRKACA may contribute to tumor pathogenesis (30). In lung cancer, DNAJB1 has been shown
to suppress MIG6 stabilization to enhance lung cancer cell
proliferation (31). In the meantime,
DNAJB1 also serves as an autophagy-associated protein involved in
the development of tumors. DNAJB1 can target PDCD5 to inhibit
p53-mediated apoptosis and enhance the proliferation of cancer
cells (32). In the current study,
DNAJB1 expression was found to be enhanced in CCA cells, tissues
and bile samples. Furthermore, the expression level of DNAJB1was
closely associated with pathological differentiation, vessel
invasion, T stage, lymph node metastasis and clinical stage in
patients with CCA. In addition, the upregulation of DNAJB1 was
associated with a poor prognosis of patients with CCA.
The diagnosis of CCA from tissues poses difficulties
due to its location, size and desmoplastic characteristics. Bile, a
potential source of diagnostic biomarkers, can be obtained with
ERCP. There is evidence to indicate that some secretory proteins
can be detected in the bile of patients with biliary cancer
(33). Hence, TPD52 and DNAJB1 in
bile samples may be used for differential diagnoses among CCA, bile
duct stones and inflammation.
Although there are interesting discoveries in this
study, our research also has some limitations. First, we only
detected the expression of TPD52 and DNAJB1. The pathological
mechanisms of action of TPD52 and DNAJB1 in CCA warrant further
investigation. Second, our results were obtained using CCA and
normal biliary epithelial tissues; however, our results need to be
validated using a larger sample size, including cholangitis. Third,
whether TPD52 and DNAJB1 can be used as independent prognostic
markers for CCA remains to be determined. Multivariate analysis may
need to be done in the future. Fourth, another project with lager
sample sizes will need to be performed in order to verify our
findings. Finally, further studies are required to validate the
pathways of DEPs in the development and progression of cancers
using KEGG analysis.
In conclusion, in this study, using the iTRAQ
technique, we analyzed the secretory proteins of CCA cells and a
normal biliary epithelial cell line. We clearly identified TPD52
and DNAJB1 as the DEPs in CCA cells, tissues and bile samples, and
these may be potential biomarkers for CCA. Furthermore, the
elevated expression levels of TPD52 and DNAJB1 were strongly
associated with the prognosis of patients with CCA. The findings of
this study may provide new insight into the diagnosis and therapy
of CCA.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This study was supported by the National Natural
Science Foundation of China (grant nos. 81572394 and 81201892),
Xiamen Huimin Project of Science and Technology (grant no.
3502Z20174072), the Youth Nursery Foundation of the Affiliated
Southeast Hospital of Xiamen University, Zhangzhou, Fujian, China
(grant no. 16Y019) and the project managed by Fujian University of
Traditional Chinese Medicine (grant no. XB2018127).
Availability of data and materials
All data generated or analyzed during this study are
from the corresponding author on reasonable request.
Authors' contributions
DS and BC designed the study. HR and ML performed
the experiments. JC, YZhou, XL and YZhan analyzed the data and
processed the figures. HR and BC wrote the manuscript, and DS
revised this manuscript. All authors were also involved in the
discussion and revision of the manuscript.
Ethics approval and consent to
participate
All procedures performed involving human
participants were carried out in accordance with the ethical
standards of the institutional and/or national research committee
and with the 1964 Helsinki declaration and its later amendments or
comparable ethical standards. This study was approved by the Ethics
Committee of the First Affiliated Hospital of Xiamen University.
Informed consent was obtained from all individual participants
included in the study.
Patient consent for publication
Not applicable.
Conflict of interest
The authors declare that they have no competing
interests.
References
|
1
|
Banales JM, Cardinale V, Carpino G,
Marzioni M, Andersen JB, Invernizzi P, Lind GE, Folseraas T, Forbes
SJ, Fouassier L, et al: Expert consensus document:
Cholangiocarcinoma: Current knowledge and future perspectives
consensus statement from the european network for the study of
cholangiocarcinoma (ENS-CCA). Nat Rev Gastroenterol Hepatol.
13:261–280. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Razumilava N and Gores GJ:
Cholangiocarcinoma. Lancet. 383:2168–2179. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Qian Y, Yao W, Yang T, Yang Y, Liu Y, Shen
Q, Zhang J, Qi W and Wang J: aPKC-ι/P-Sp1/Snail signaling induces
epithelial-mesenchymal transition and immunosuppression in
cholangiocarcinoma. Hepatology. 66:1165–1182. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhang W and Yan LN: Perihilar
cholangiocarcinoma: Current therapy. World J Gastrointest
Pathophysiol. 5:344–354. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Khan SA, Davidson BR, Goldin RD, Heaton N,
Karani J, Pereira SP, Rosenberg WM, Tait P, Taylor-Robinson SD,
Thillainayagam AV, et al: Guidelines for the diagnosis and
treatment of cholangiocarcinoma: An update. Gut. 61:1657–1669.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Coelho R, Silva M, Rodrigues-Pinto E,
Cardoso H, Lopes S, Pereira P, Vilas-Boas F, Santos-Antunes J,
Costa-Maia J and Macedo G: CA 19-9 as a marker of survival and a
predictor of metastization in cholangiocarcinoma. GE Port J
Gastroenterol. 24:114–121. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Voigtländer T, Metzger J, Schönemeier B,
Jäger M, Mischak H, Manns M and Lankisch TO: A combined bile and
urine proteomic test for cholangiocarcinoma diagnosis in patients
with biliary strictures of unknown origin. United European
Gastroenterol J. 5:668–676. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Navaneethan U, Njei B, Lourdusamy V,
Konjeti R, Vargo JJ and Parsi MA: Comparative effectiveness of
biliary brush cytology and intraductal biopsy for detection of
malignant biliary strictures: A systematic review and
meta-analysis. Gastrointest Endosc. 81:168–176. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Melle C, Ernst G, Schimmel B, Bleul A,
Koscielny S, Wiesner A, Bogumil R, Möller U, Osterloh D, Halbhuber
KJ and von Eggeling F: A technical triade for proteomic
identification and characterization of cancer biomarkers. Cancer
Res. 64:4099–4104. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ghosh D, Li Z, Tan XF, Lim TK, Mao Y and
Lin Q: iTRAQ based quantitative proteomics approach validated the
role of calcyclin binding protein (CacyBP) in promoting colorectal
cancer metastasis. Mol Cell Proteomics. 12:1865–1880. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Qiao J, Fang CY, Chen SX, Wang XQ, Cui SJ,
Liu XH, Jiang YH, Wang J, Zhang Y, Yang PY and Liu F: Stroma
derived COL6A3 is a potential prognosis marker of colorectal
carcinoma revealed by quantitative proteomics. Oncotarget.
6:29929–29946. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ross PL, Huang YN, Marchese JN, Williamson
B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, et
al: Multiplexed protein quantitation in saccharomyces cerevisiae
using amine-reactive isobaric tagging reagents. Mol Cell
Proteomics. 3:1154–1169. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Huang GL, Luo Q, Rui G, Zhang W, Zhang QY,
Chen QX and Shen DY: Oncogenic activity of retinoic acid receptor γ
is exhibited through activation of the Akt/NF-κB and Wnt/β-catenin
pathways in cholangiocarcinoma. Mol Cell Biol. 33:3416–3425. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Silsirivanit A, Sawanyawisuth K, Riggins
GJ and Wongkham C: Cancer biomarker discovery for
cholangiocarcinoma: The high-throughput approaches. J Hepatobiliary
Pancreat Sci. 21:388–396. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Phoomak C, Park D, Silsirivanit A,
Sawanyawisuth K, Vaeteewoottacharn K, Detarya M, Wongkham C,
Lebrilla CB and Wongkham S: O-GlcNAc-induced nuclear translocation
of hnRNP-K is associated with progression and metastasis of
cholangiocarcinoma. Mol Oncol. 13:338–357. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kristiansen TZ, Harsha HC, Grønborg M,
Maitra A and Pandey A: Differential membrane proteomics using
18O-labeling to identify biomarkers for cholangiocarcinoma. J
Proteome Res. 7:4670–4677. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Perdomo J, Leung HHL, Ahmadi Z, Yan F,
Chong JJH, Passam FH and Chong BH: Neutrophil activation and
NETosis are the major drivers of thrombosis in heparin-induced
thrombocytopenia. Nat Commun. 10:13222019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Boutros R, Fanayan S, Shehata M and Byrne
JA: The tumor protein D52 family: Many pieces, many puzzles.
Biochem Biophys Res Commun. 325:1115–1121. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Byrne JA, Tomasetto C, Garnier JM, Rouyer
N, Mattei MG, Bellocq JP, Rio MC and Basset P: A screening method
to identify genes commonly overexpressed in carcinomas and the
identification of a novel complementary DNA sequence. Cancer Res.
55:2896–2903. 1995.PubMed/NCBI
|
|
21
|
Kumamoto T, Seki N, Mataki H, Mizuno K,
Kamikawaji K, Samukawa T, Koshizuka K, Goto Y and Inoue H:
Regulation of TPD52 by antitumor microRNA-218 suppresses cancer
cell migration and invasion in lung squamous cell carcinoma. Int J
Oncol. 49:1870–1880. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li J, Li Y, Liu H, Liu Y and Cui B: The
four-transmembrane protein MAL2 and tumor protein D52 (TPD52) are
highly expressed in colorectal cancer and correlated with poor
prognosis. PLoS One. 12:e01785152017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Alagaratnam S, Hardy JR, Lothe RA,
Skotheim RI and Byrne JA: TPD52, a candidate gene from genomic
studies, is overexpressed in testicular germ cell tumours. Mol Cell
Endocrinol. 306:75–80. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Rubin MA, Varambally S, Beroukhim R,
Tomlins SA, Rhodes DR, Paris PL, Hofer MD, Storz-Schweizer M,
Kuefer R, Fletcher JA, et al: Overexpression, amplification, and
androgen regulation of TPD52 in prostate cancer. Cancer Res.
64:3814–3822. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Byrne JA, Maleki S, Hardy JR, Gloss BS,
Murali R, Scurry JP, Fanayan S, Emmanuel C, Hacker NF, Sutherland
RL, et al: MAL2 and tumor protein D52 (TPD52) are frequently
overexpressed in ovarian carcinoma, but differentially associated
with histological subtype and patient outcome. BMC Cancer.
10:4972010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Goto Y, Nishikawa R, Kojima S, Chiyomaru
T, Enokida H, Inoguchi S, Kinoshita T, Fuse M, Sakamoto S, Nakagawa
M, et al: Tumour-suppressive microRNA-224 inhibits cancer cell
migration and invasion via targeting oncogenic TPD52 in prostate
cancer. FEBS Lett. 588:1973–1982. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yamazaki S, Uchiumi A and Katagata Y:
Hsp40 regulates the amount of keratin proteins via
ubiquitin-proteasome pathway in cultured human cells. Int J Mol
Med. 29:165–168. 2012.PubMed/NCBI
|
|
28
|
Lenna S, Farina AG, Martyanov V,
Christmann RB, Wood TA, Farber HW, Scorza R, Whitfield ML, Lafyatis
R and Trojanowska M: Increased expression of endoplasmic reticulum
stress and unfolded protein response genes in peripheral blood
mononuclear cells from patients with limited cutaneous systemic
sclerosis and pulmonary arterial hypertension. Arthritis Rheum.
65:1357–1366. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Batra J, Tripathi S, Kumar A, Katz JM, Cox
NJ, Lal RB, Sambhara S and Lal SK: Human heat shock protein 40
(Hsp40/DnaJB1) promotes influenza A virus replication by assisting
nuclear import of viral ribonucleoproteins. Sci Rep. 6:190632016.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Honeyman JN, Simon EP, Robine N,
Chiaroni-Clarke R, Darcy DG, Lim II, Gleason CE, Murphy JM,
Rosenberg BR, Teegan L, et al: Detection of a recurrent
DNAJB1-PRKACA chimeric transcript in fibrolamellar hepatocellular
carcinoma. Science. 343:1010–1014. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Park SY, Choi HK, Seo JS, Yoo JY, Jeong
JW, Choi Y, Choi KC and Yoon HG: DNAJB1 negatively regulates MIG6
to promote epidermal growth factor receptor signaling. Biochim
Biophys Acta. 1853:2722–2730. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Cui X, Choi HK, Choi YS, Park SY, Sung GJ,
Lee YH, Lee J, Jun WJ, Kim K, Choi KC and Yoon HG: DNAJB1
destabilizes PDCD5 to suppress p53-mediated apoptosis. Cancer Lett.
357:307–315. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Rose JB, Correa-Gallego C, Li Y, Nelson J,
Alseidi A, Helton WS, Allen PJ, D'Angelica MI, DeMatteo RP, Fong Y,
et al: The role of biliary carcinoembryonic antigen-related
cellular adhesion molecule 6 (CEACAM6) as a biomarker in
cholangiocarcinoma. PLoS One. 11:e01501952016. View Article : Google Scholar : PubMed/NCBI
|