Introduction
Rapid growth is one of the main characteristics of
non-small cell lung cancer (NSCLC) (1). Patients burdened with large size
tumors usually have poor clinical prognosis. Even though new drugs
targeting various critical oncogenes significantly reduce tumor
size before surgery, side-effects seriously restrict the effective
clinical application of these drugs (2). Thus, the identification of new
therapeutic targets remains vital for effective NSCLC
treatment.
Long non-coding RNAs (lncRNAs) are a group of
evolutionarily conserved RNAs that are >200 nucleotides in
length with no protein-coding capacity (3). Recent studies have shown that lncRNAs
play critical roles in various biological processes including
proliferation, invasion and angiogenesis. lncRNA-XIST was found to
be upregulated in lung cancer cells and accelerated lung cancer
cell growth through targeting miR-140 (4). lncRNA-PVT1 directly interacted with
miR-200a and miR-200b to facilitate NSCLC cell invasion by
upregulating MMP9 expression (5).
Inhibition of lincRNA-p21 was found to induce global downregulation
of the expression of angiogenesis-related genes including VEGFA in
human NSCLC (6).
Recently, lncRNA-CCAT1 (simply CCAT1) was
demonstrated to promoted tumor growth and metastasis in human
cancers (7). In osteosarcoma, CCAT1
was found to be upregulated in osteosarcoma tissues and cell lines
(8). High levels of CCAT1 promoted
cell proliferation and migration by inhibiting miR-148. CCAT1 was
also found to be upregulated in renal cell carcinoma (RCC)
(9). CCAT1 knockdown inhibited cell
viability and increased apoptosis of RCC cells in vitro.
Overexpression of CCAT1 could bind miR155-5p and let7b-5p to
account for the poor prognosis of human oral squamous cell
carcinoma (10). However, the
expression and biological functions of CCAT1 in NSCLC are still
unclear.
In the present study, we determined the expression
of CAAT1 in NSCLC tissues and cell lines. Upregulation of CAAT1 was
associated with large tumor size and poor prognosis in NSCLC
patients. CCAT1 knockdown inhibited NSCLC tumor growth in
vitro and in vivo by rescuing the expression of miR-218
and simultaneously inhibiting the expression of BMI-1, which is a
downstream target of miR-218.
Materials and methods
Clinical samples and cell lines
Fifty paired NSCLC and matched tumor-adjacent
tissues were collected from Linyi Central Hospital (Linyi, China)
and North China University of Science and Technology Affiliated
Hospital (North China University of Science and Technology, China).
These patients included 31 males and 19 females who received
surgical resection between March 2011 and January 2013. The age
range was between 41 and 73 years. The use of clinical data was
approved by the Biomedical Ethics Committee of Linyi Central
Hospital. All patients enrolled in this study signed an informed
consent. Normal lung epithelial BEAS-2B cells and 4 NSCLC cell
lines (A549, H1299, H1975 and HCC827) were purchased from the
American Type Culture Collection (ATCC; Manassas, VA, USA) and
cultured in Dulbecco's modified Eagle's medium (DMEM; Invitrogen;
Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with
10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific,
Inc.), 1% penicillin and 1% streptomycin.
Real-time quantitative reverse
transcription-PCR (qRT-PCR)
Total RNA from tissues and cells was isolated using
TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.).
One-Step Perfect Real-Time RT-qPCR (SYBR-Green I) kit (Takara
Biotechnology Co., Ltd., Dalian, China) was used to detected the
expression levels of lncRNA-CCAT1, miR-218 and BMI-1 mRNA. miR-218
and RNU6 Bulge-Loop™ primers were purchased from RiboBio Co., Ltd.
(Guangzhou, China). lncRNA-CCAT1, BMI-1 and GAPDH primers were
synthetized by Sangon Co., Ltd. (Shanghai, China). Data were
analyzed using the 2−ΔΔCq method (11). The sequences of primers used are
shown as follows: lncRNA-CCAT1 (sense, 5′-AGAAACACTATCACCTACGC-3′
and antisense, 5′-CTTAACAGGGCATTGCTAATCT-3′); BMI-1 (sense,
5′-GTGCTTTGTGGAGGGTACTTCAT-3′ and antisense,
5′-TTGGACATCACAAATAGGACAATACTT-3′); GAPDH (sense,
5′-CCAGGGCTGCTTTTAACTCT-3′ and antisense,
5′-GGACTCCACGACGTACTCA-3′). The thermocycling conditions were:
holding stage, 50°C for 2 min, 95°C for 10 min; PCR stage (40
times), 95°C for 15 sec, 60°C for 1 min.
Cell infection and transfection
The control short hairpin RNA (shRNA) (sh-ctrl)
plasmids and CCAT1-specific shRNA (sh-CCAT1) plasmids were
purchased from GeneCopoeia (Guangzhou, China) and cloned into
lentivirus vectors. Lentiviruses were packaged and amplified in
293T cells (ATCC) according to themanufacturers instructions.
Lentivirus supernatant was used to infect the tumor cells. The
effect of infection was determined by qRT-PCR. The negative control
inhibitor (anti-miR-ctrl) and miR-218 inhibitor (anti-miR-218) were
purchased from Riobio Co., Ltd. (Guangzhou, China). These
inhibitors were transfected into tumor cells using Lipofectamine
3000 (Invitrogen; Thermo Fisher Scientific, Inc.) reagent. Cells
were used for further experiments after 24-h transfection.
Cell Counting Kit-8 (CCK-8) cell
proliferation assay
For cell proliferation detection, we used CCK-8
assay according to the manufacturer's recommendations (Sangon
Biotech Co., Ltd., Shanghai, China). Infected or transfected cells
were cultured in a 96-well plate at a density of 2,000 cells/well
in quintuplicate. CCK-8 solution (10 µl) was added to each well
plate and was incubated for 4 h in an incubator. Cells were
incubated at 37°C for 4 h. The absorbance at 450 nm was measured by
a microplate reader.
Cell apoptosis assay
To evaluate the apoptosis of NSCLC cells, flow
cytometric assay was performed to evaluate the percentage of
apoptotic cells. In brief, cells were harvested and re-suspended in
phosphate-buffered saline (PBS) solution, and then stained with
Annexin V-FITC/PI detection kit (BD Biosciences, San Jose, CA, USA)
and subjected to FACS analysis (FlowJo, v7.60; FlowJo LLC, Ashland,
OR, USA).
In vivo experiments
Ten BALB/cA, 4-week-old, 16–18 g, female nude mice
were obtained from the Animal Center of Jining Medical College. The
mice were housed in sterilized cages under appropriate
environmental conditions (25°C, 45% humidity), were fed a regular
chow diet with water ad libitum, and were handled according
to the requirements of the National Institutes of Health guidelines
for care and use of laboratory animals. All animal protocols were
approved by the Biomedical Ethics Committee of Linyi Central
Hospital. Mice were randomly divided into 2 groups. Recombinant
A549 and H1975 cells were subcutaneously injected into the nude
mice to investigate the functions of CCAT1 for tumor growth. The
mice were sacrificed after 4 weeks by CO2 euthanasia
(the flow rate of CO2 was 20% displacement/min). Final
tumor volumes were calculated based on the following formula:
(length × width2)/2.
Ki-67 immunohistochemical
staining
Subcutaneous tumor tissues were made into
paraformaldehyde-fixed paraffin sections. Sections were processed
with xylene, alcohol and PBS, sequentially. Sections were incubated
with Ki-67 primary antibodies [cat. no. 9027; Cell Signaling
Technology (CST), Inc., Danvers, MA, USA] (1:100) at 4°C overnight.
Biotinylated secondary antibodies were used to combine Ki-67
antibodies, detected by HRP-streptavidin conjugates and visualized
by DAB. The Ki-67 index was calculated according to a previous
study (12).
Western blotting
Total cell proteins were extracted with RIPA buffer,
separated on 10% SDS-PAGE gel and transferred onto a nitrocellulose
membrane (Invitrogen; Thermo Fisher Scientific, Inc.). The
membranes were incubated with the following primary rabbit
anti-human antibodies purchased from (CST) at 4°C overnight: BMI-1
(cat. no 6964; dilution at 1:1,000) and GAPDH (cat. no 5174;
dilution at 1:1,000). Then, the membranes were incubated with
HRP-linked goat anti-rabbit antibodies (cat. no 7074; dilution at
1:5,000; CST), and the signals were detected using the Bio-Rad Gel
imaging system (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Statistical analysis
All quantitative data are expressed as mean ± SD.
The Statistical Product and Service Solutions v21.0 software (SPSS
21.0; IBM Corp., Armonk, NY, USA) was used to perform statistical
analysis. The Pearson's Chi-square test was used to analyze the
relationship between CCAT1 expression and clinical features.
Student's t-test was performed for data in 2 groups. ANOVA was used
to analyze the data in 3 or more groups and Bonferroni's method was
used as a post hoc test. Overall survival and progression-free
survival analysis were performed using the Kaplan-Meier method for
plotting and the log-rank test for comparison. P<0.05 indicated
a statistically significant difference.
Results
CCAT1 is upregulated in NSCLC tissues
and is associated with poor patient prognosis
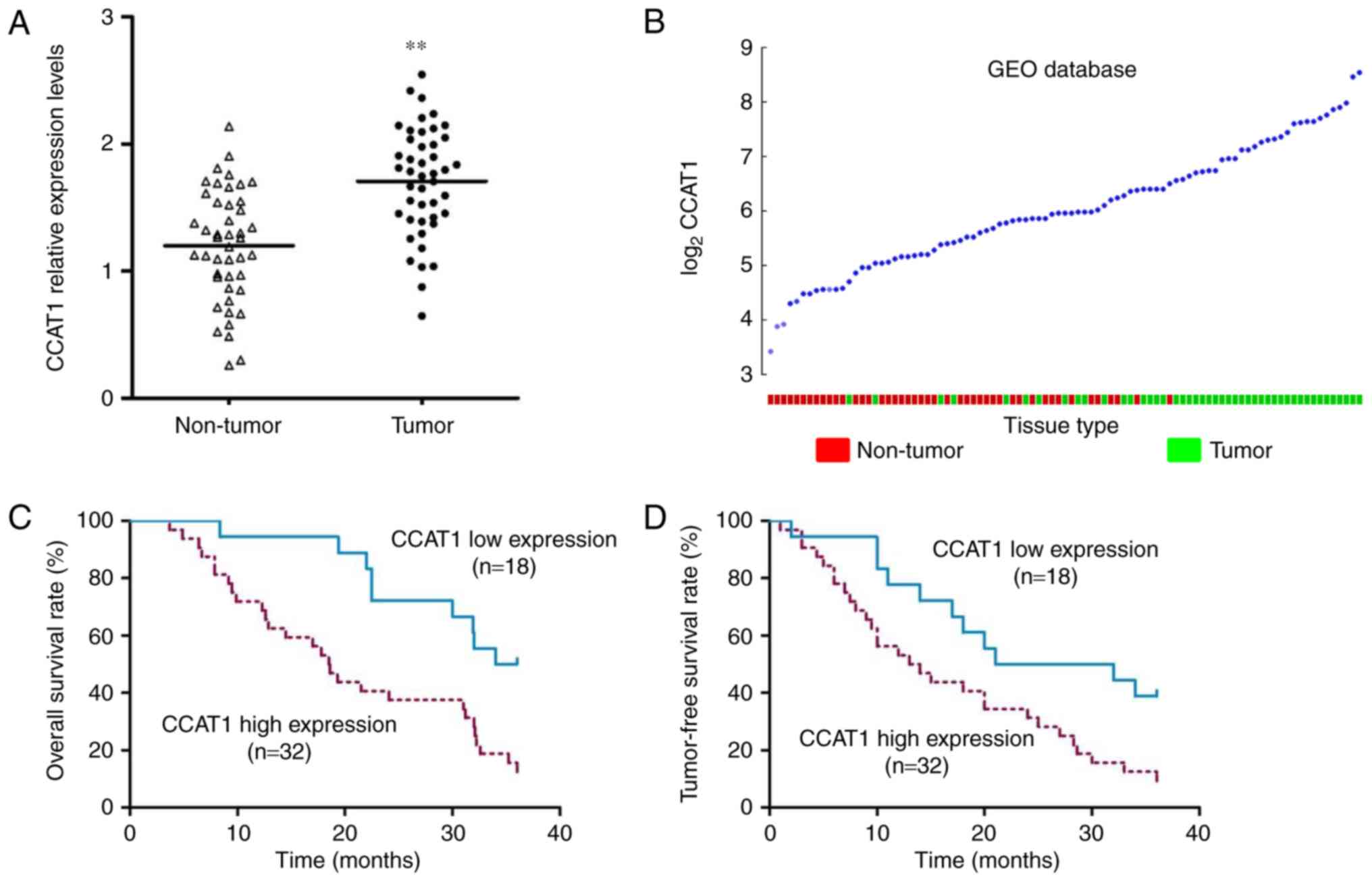
As shown in Fig. 1A,
the expression of CCAT1 was elevated in NSCLC tissues (P<0.001).
Furthermore, consistent with our results, the expression of CCAT1
was also increased in 91 NSCLC tissues available in Gene Expression
Omnibus (GEO) database (GSE18842) (13) (Fig.
1B). To further investigate the clinical significance of CCAT1,
we used the mean expression value of CCAT1 (=1.804) to divide these
50 patients into 2 subgroups: CCAT1 high expression group (n=32)
and CCAT1 low expression group (n=18). As shown in Table I, high expression of CCAT1 was
associated with large tumor size (P=0.018). Moreover, patients in
the high CAAT1 expression group had poor 3-year overall survival
rate (Fig. 1C, P=0.003, HR=2.736)
and tumor-free survival rate (Fig.
1D, P=0.014, HR=2.224).
 | Table I.Correlation between CCAT1 and clinical
features in NSCLC (n=50). |
Table I.
Correlation between CCAT1 and clinical
features in NSCLC (n=50).
|
|
| CCAT1 expression |
|
|
|---|
|
|
|
|
|
|
|---|
| Clinical
characteristics | Total | High n=32 | Low n=18 | χ2 | P-value |
|---|
| Sex |
| Male | 31 | 22 | 9 | 1.719 | 0.190 |
|
Female | 19 | 10 | 9 |
|
|
| Age (years) |
|
<50 | 21 | 11 | 10 | 2.122 | 0.145 |
| ≥50 | 29 | 21 | 8 |
|
|
| Smoking status |
| Yes | 26 | 19 | 7 | 1.937 | 0.164 |
| No | 24 | 13 | 11 |
|
|
| Tumor size (cm) |
|
>3 | 25 | 20 | 5 | 5.556 | 0.018a |
| ≤3 | 25 | 12 | 13 |
|
|
| Lymphatic
metastasis |
|
Present | 30 | 22 | 8 | 2.836 | 0.092 |
|
Absent | 20 | 10 | 10 |
|
|
| TNM stage |
|
I+II | 26 | 14 | 12 | 2.424 | 0.119 |
|
III+IV | 24 | 18 | 6 |
|
|
CCAT1 knockdown inhibits proliferation
and induces apoptosis in NSCLC cells
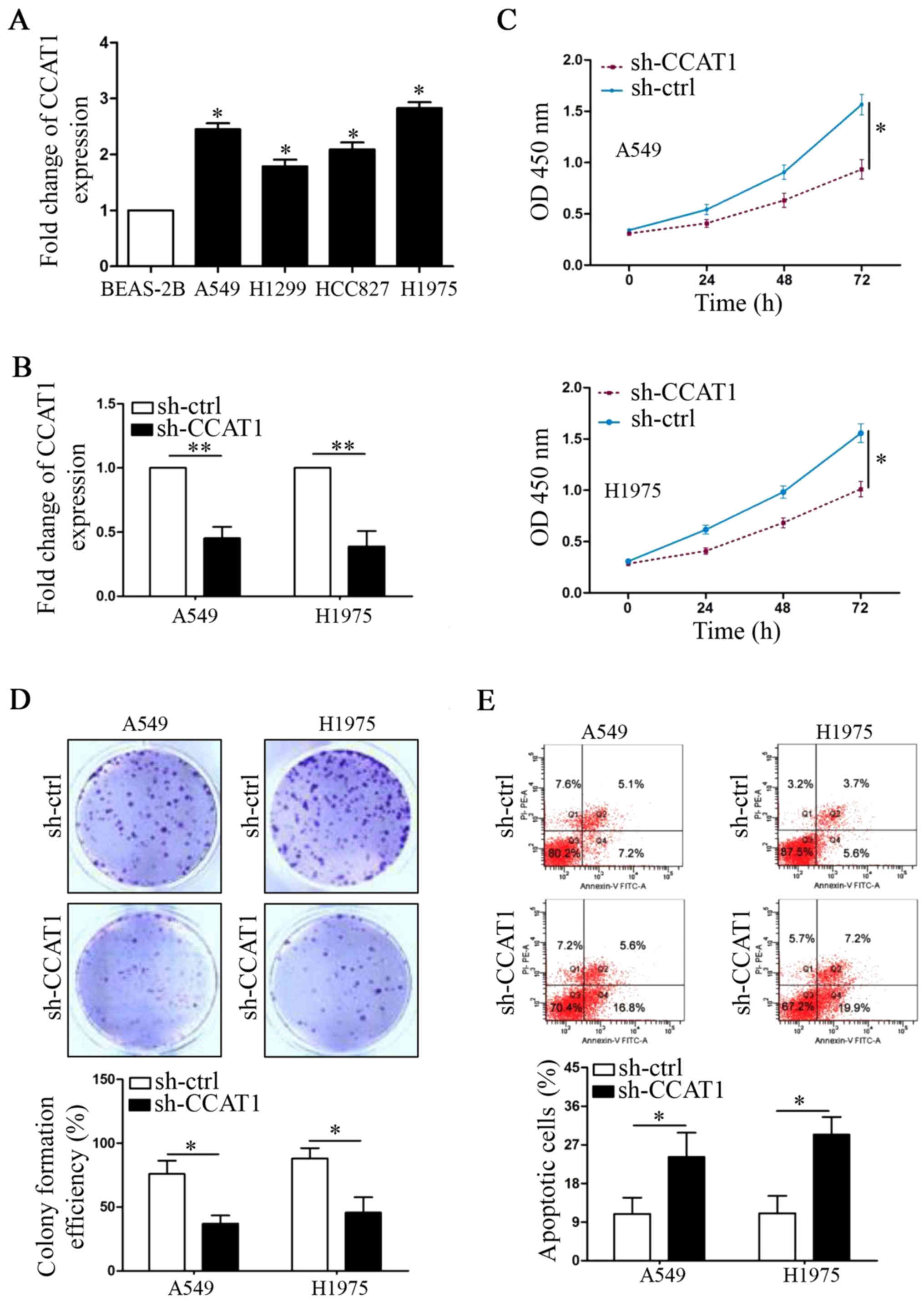
Next, compared with BEAS-2B cells, we also found
that the expression levels of CCAT1 were upregulated in all NSCLC
cell lines (Fig. 2A, P<0.05,
respectively). We used shRNA to knock down CCAT1 expression in A549
and H1975 cells, which were two cell lines that expressed the
highest CCAT1 levels (Fig. 2B,
P<0.01, respectively). CCK-8 assays showed that CCAT1 knockdown
significantly decreased cell viability in the two cell lines
(Fig. 2C, P<0.05, respectively).
Furthermore, CCAT1 knockdown also decreased colony formation number
of A549 and H1975 cells (Fig. 2D,
P<0.05, respectively). On the contrary, flow cytometry (FCW)
assays demonstrated that CCAT1 knockdown increased the percentage
of apoptotic NSCLC cells (Fig. 2E,
P<0.05, respectively).
CCAT1 knockdown inhibits tumor growth
in vivo
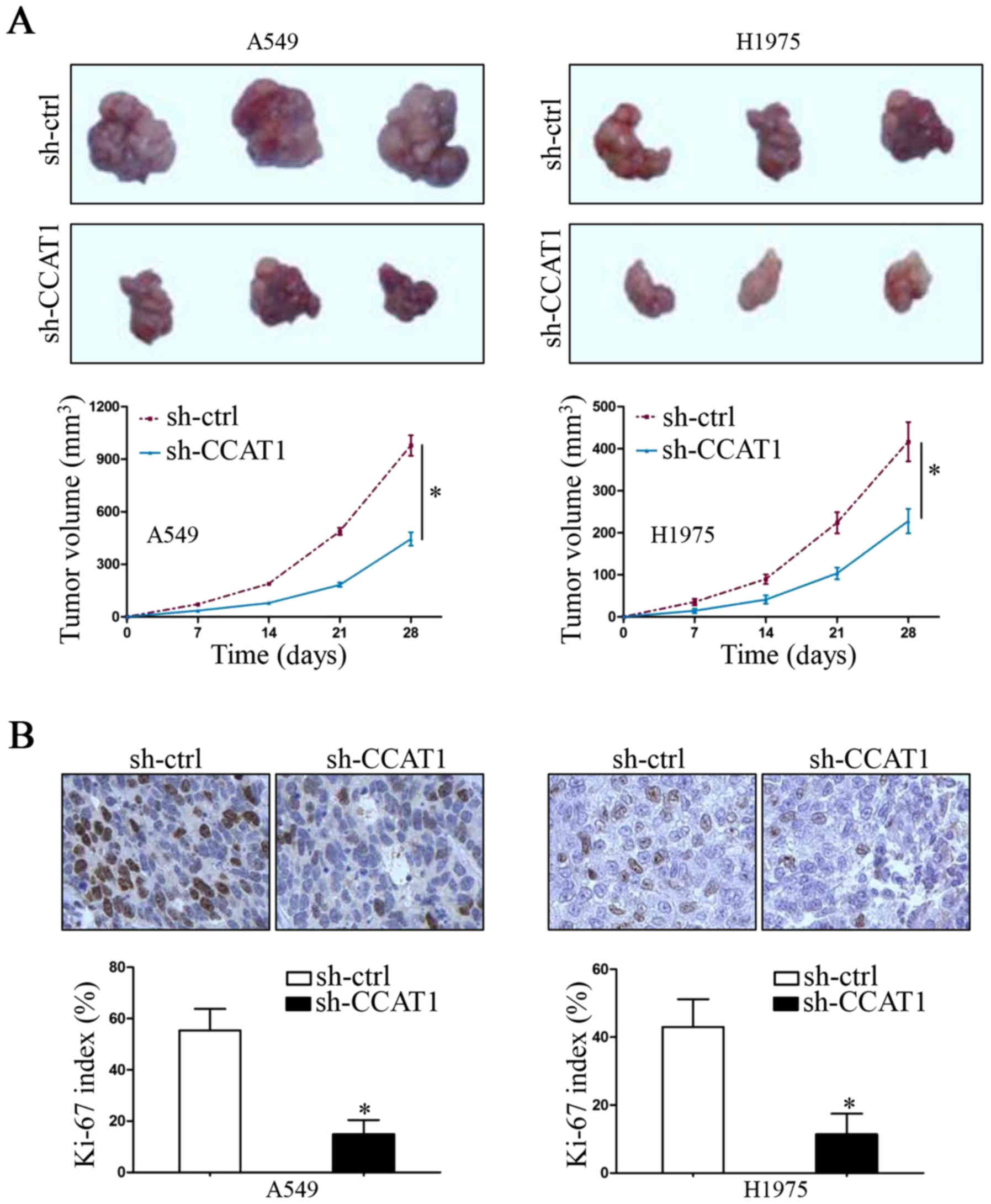
When we demonstrated that CCAT1 knockdown inhibited
NSCLC cell proliferation in vitro, we also utilized a
subcutaneous xenograft growth model to investigate the growth
promoting functions of CCAT1 in vivo. As shown in Fig. 3A, downregulation of CCAT1 inhibited
the tumor growth in nude mice (P<0.05, respectively). This
effect was further confirmed by Ki-67 immunohistochemical staining
in mouse tumor tissue sections (Fig.
3B, P<0.05, respectively). These data suggest that
lncRNA-CCAT1 promotes tumor growth of NSCLC in vitro and
in vivo.
CCAT1 exerts its function by targeting
the miR-218/BMI-1 axis in NSCLC cells
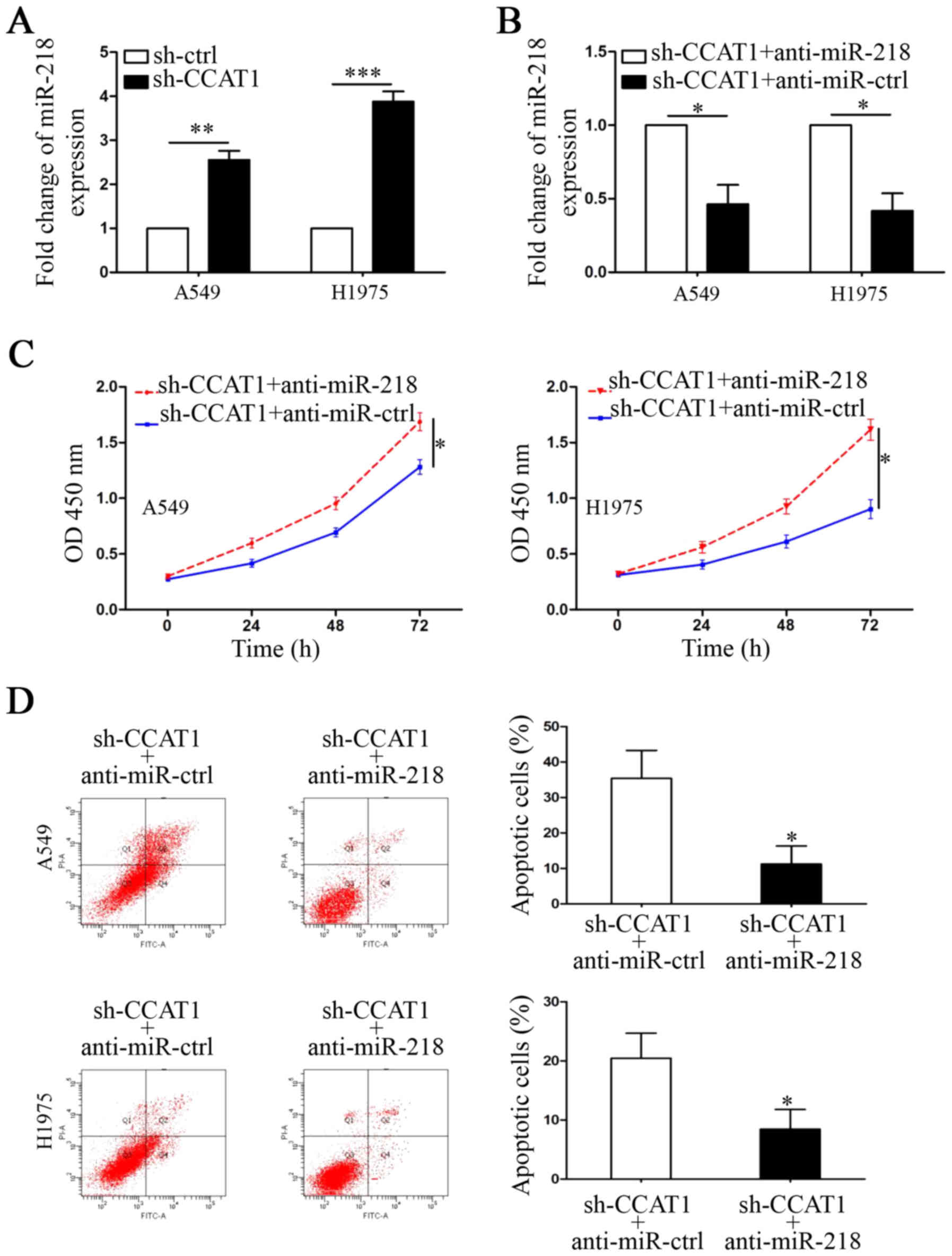
miR-218 was predicted as a potential interactive
target of CCAT1 by informatic analysis (MiRanda: http://www.microrna.org/). The following evidence was
used to demonstrate that CCAT1 exerted its function by
downregulating miR-218 in NSCLC cells. First, qRT-PCR results
showed that the expression of miR-218 was upregulated in
CCAT1-knockdown A549 and H1975 cells (Fig. 4A, P<0.01 and P<0.001,
respectively). Second, miR-218 inhibitors were used to repress its
expression in CCAT1-knockdown cells (Fig. 4B, P<0.05, respectively). CCK-8
and FCW assays were performed to show that the CCAT1
knockdown-induced anti-growth effect was abrogated by
downregulation of miR-218 (Fig. 4C and
D, P<0.05, respectively). BMI-1 was reported as a downstream
target of miR-218 in NSCLC. Finally, we detected the mRNA and
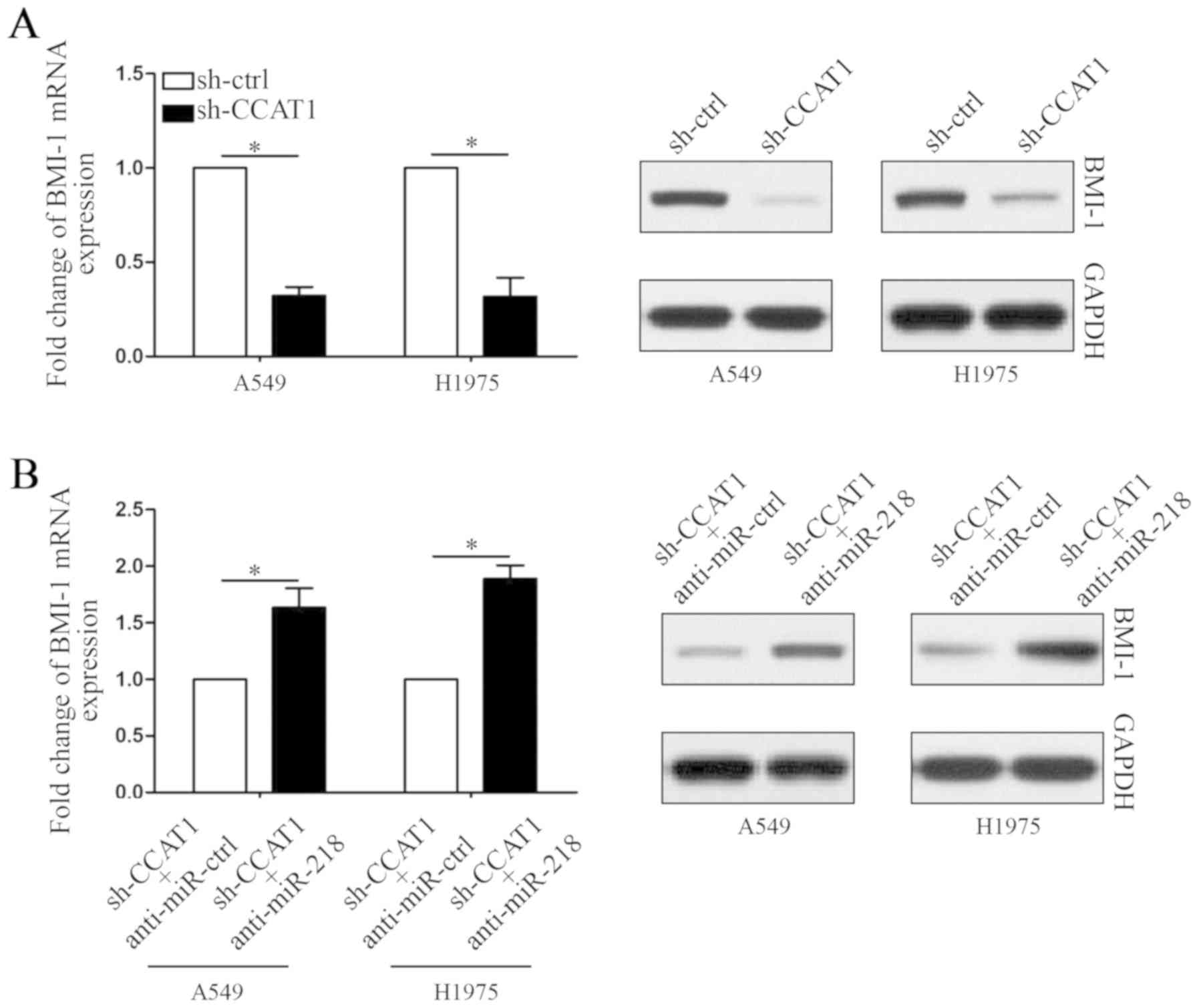
protein levels of BMI-1 in NSCLC cells. As shown in Fig. 5A, both mRNA and protein levels of
BMI-1 were decreased by CCAT1 knockdown (P<0.05, respectively).
Moreover, the expression of BMI-1 was rescued in CCAT1-knockdown
cells by inhibition of miR-218 (Fig.
5B, P<0.05, respectively). Thus, these results may partly
explain the anti-growth mechanisms of lncRNA-CCAT1 knockdown in
NSCLC.
Discussion
Tumor growth is an important risk factor for the
poor prognosis of NSCLC patients. Numerous studies have
demonstrated that long non-coding RNAs (lncRNAs) function as tumor
growth promotors or inhibitors in NSCLC (14,15).
Long non-coding RNA MEG3 was found to inhibit NSCLC cell
proliferation, arrest the cell cycle and promote apoptosis by
inducing p53 expression (16).
Moreover, decreased expression of MEG3 was also associated with
advanced pathologic stage and tumor size in NSCLC patients
(17). However, long non-coding RNA
ANRIL was found to be upregulated in NSCLC tissues and cell lines
(18). ANRIL promoted cell
proliferation and suppressed apoptosis by silencing KLF2 and P21
expression. In the present study, our results showed that
lncRNA-CCAT1 is overexpressed in NSCLC tissues and cell lines.
Importantly, elevated expression of CCAT1 was also confirmed in a
GEO database including 91 NSCLC patients. Moreover, high expression
of CCAT1 was related to large tumor size and short survival time
after surgery. This prognostic prediction value also exists in
breast cancer (19) and gastric
carcinoma (20). Taken together,
these data indicated that lncRNA-CCAT1 may play a critical role in
the progression of NSCLC.
Most studies have shown that CCAT1 promotes cell
migration and invasion in human cancers, but few studies have
investigated its function in tumor growth (21). In the present study, we silenced
CCAT1 expression in NSCLC cells. In vitro analysis confirmed
that CCAT1 knockdown inhibited cell viability and proliferation,
and induced apoptosis in NSCLC cells. This tumor growth promotion
effect of CCAT1 was also blocked by shRNAs in a nude mouse
subcutaneous xenograft model. These results may partly explain why
high expression of CCAT1 is associated with large tumor size in
NSCLC patients.
lncRNAs usually exert their functions through
sponging various microRNAs (22).
lncRNA-CCAT1 has been reported to sponge let-7 in hepatocellular
carcinoma (23), miR-410 in glioma
(24) and miR-181a in nasopharynx
cancer (25). In the present study,
we used an online database to select a new potential target miRNA
of CCAT1 named microRNA-218 (miR-218). Previous research has
established the inhibitory role of miR-218 in tumor growth
(26). In our CCAT1-knockdown NSCLC
cells, upregulation of miR-218 was noted. The cell proliferation
was increased whereas apoptosis was inhibited when we used the
miR-218 inhibitor to block its expression. That indicates that
lncRNA-CCAT1 increases NSCLC tumor growth by sponging miR-218.
B lymphoma Mo-MLV insertion region 1 homolog (BMI-1)
is a component of polycomb repressive complex 1 (PRC1) (27). In NSCLC, BMI-1 has been demonstrated
as an oncogene due to its functions in tumor proliferation,
invasion and chemoresistance (28,29).
In the present study, it was found that the expression of BMI-1 was
downregulated by CCAT1 silencing in NSCLC cells. On the contrary,
the expression levels of BMI-1 were reversed in the CCAT1-knockdown
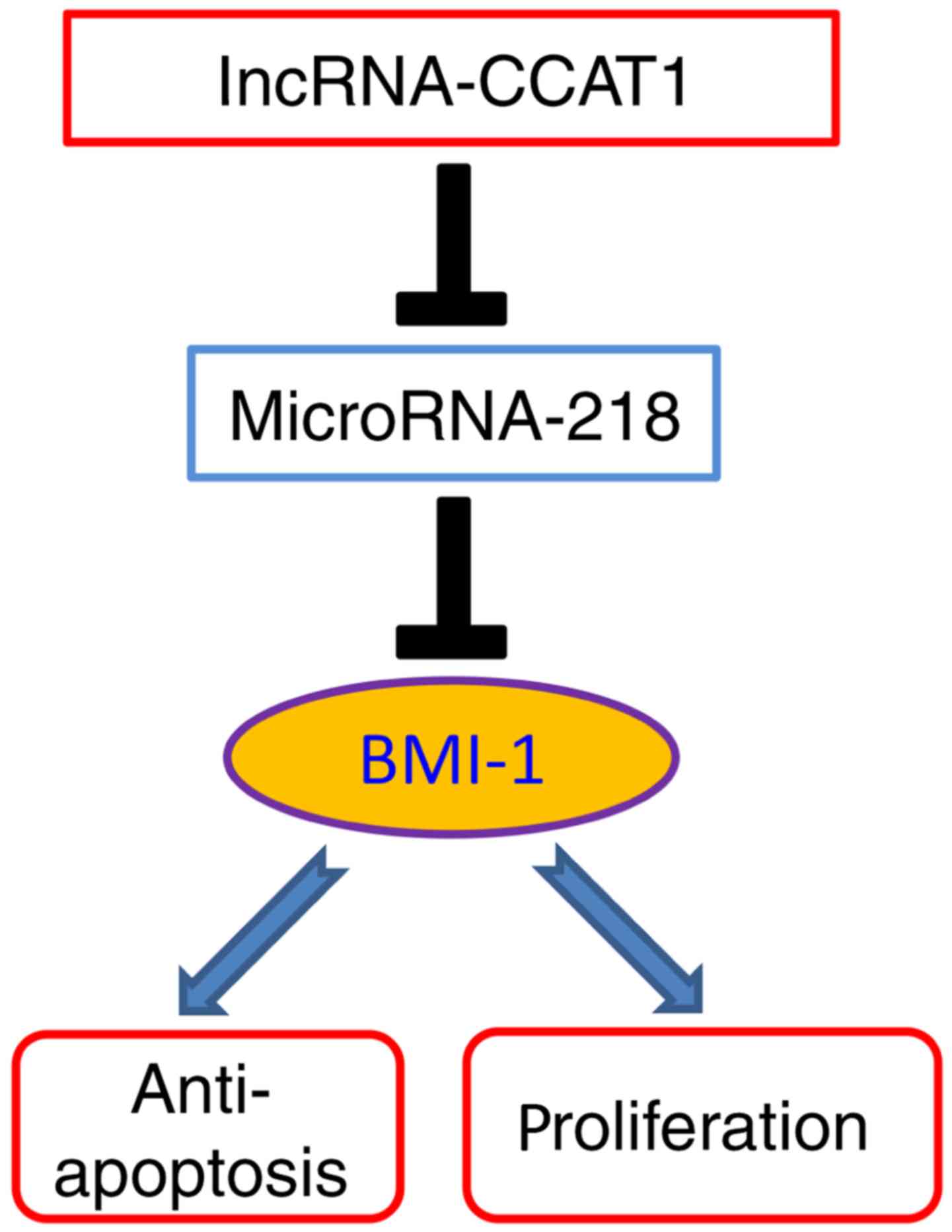
NSCLC cells by inhibiting miR-218 expression. Thus, we conclude
that CCAT1 promotes NSCLC growth by increasing BMI-1 expression
through decreasing the expression levels of miR-218 (Fig. 6).
In conclusion, we demonstrated that long non-coding
RNA CCAT1 is upregulated in NSCLC tissues and cell lines, and its
elevated expression is associated with malignant clinical features.
lncRNA-CCAT1 promotes NSCLC growth behaviors by targeting the
miR-218/BMI-1 axis. These results suggest that lncRNA-CCAT1 is a
potential therapeutic target for NSCLC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
LZ and PM conceived and designed the study. LZ, LW
and YW performed the experiments. LZ wrote the paper. LZ, LW, YW
and PM reviewed and edited the manuscript. All authors read and
approved the manuscript and agree to be accountable for all aspects
of the research in ensuring that the accuracy or integrity of any
part of the work are appropriately investigated and resolved.
Ethics approval and consent to
participate
The use of clinical data was approved by the
Biomedical Ethics Committee of Linyi Central Hospital (Linyi,
China). Patients enrolled in this study signed an informed consent.
All animal protocols were approved by the Biomedical Ethics
Committee of Linyi Central Hospital.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Yu Y, Guan H, Xing LG and Xiang YB: Role
of gross tumor volume in the prognosis of non-small cell lung
cancer treated with 3D conformal radiotherapy: a meta-analysis.
Clin Ther. 37:2256–2266. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Daga A, Ansari A, Patel S, Mirza S, Rawal
R and Umrania V: Current drugs and drug targets in non-small cell
lung cancer: limitations and opportunities. Asian Pac J Cancer
Prev. 16:4147–4156. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Schmitt AM and Chang HY: Long non-coding
RNAs in cancer pathways. Cancer Cell. 29:452–463. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tang Y, He R, An J, Deng P, Huang L and
Yang W: lncRNA XIST interacts with miR-140 to modulate lung cancer
growth by targeting iASPP. Oncol Rep. 38:941–948. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chen W, Zhu H, Yin L, Wang T, Wu J, Xu J,
Tao H, Liu J and He X: lncRNA-PVT1 facilitates invasion through
upregulation of MMP9 in nonsmall cell lung cancer cell. DNA Cell
Biol. 36:787–793. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Castellano JJ, Navarro A, Viñolas N,
Marrades RM, Moises J, Cordeiro A, Saco A, Muñoz C, Fuster D,
Molins L, et al: LincRNA-p21 impacts prognosis in resected
non-small cell lung cancer patients through angiogenesis
regulation. J Thorac Oncol. 11:2173–2182. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Guo X and Hua Y: CCAT1: an oncogenic long
non-coding RNA in human cancers. J Cancer Res Clin Oncol.
143:555–562. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhao J and Cheng L: Long non-coding RNA
CCAT1/miR-148a axis promotes osteosarcoma proliferation and
migration through regulating PIK3IP1. Acta Biochim Biophys Sin.
49:503–512. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chen S, Ma P, Li B, Zhu D, Chen X, Xiang
Y, Wang T, Ren X, Liu C and Jin X: LncRNA CCAT1 inhibits cell
apoptosis of renal cell carcinoma through up-regulation of Livin
protein. Mol Cell Biochem. May 3–2017.(Epub ahead of print). doi:
10.1007/s11010-017-3043-8. View Article : Google Scholar
|
|
10
|
Arunkumar G, Murugan AK, Prasanna
Srinivasa Rao H, Subbiah S, Rajaraman R and Munirajan AK: Long
non-coding RNA CCAT1 is overexpressed in oral squamous cell
carcinomas and predicts poor prognosis. Biomed Rep. 6:455–462.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li C, Miao R, Liu S, Wan Y, Zhang S, Deng
Y, Bi J, Qu K, Zhang J and Liu C: Down-regulation of miR-146b-5p by
long non-coding RNA MALAT1 in hepatocellular carcinoma promotes
cancer growth and metastasis. Oncotarget. 8:28683–28695.
2017.PubMed/NCBI
|
|
13
|
Sanchez-Palencia A, Gomez-Morales M,
Gomez-Capilla JA, Pedraza V, Boyero L, Rosell R and Fárez-Vidal ME:
Gene expression profiling reveals novel biomarkers in nonsmall cell
lung cancer. Int J Cancer. 129:355–364. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Feng J, Sun Y, Zhang EB, Lu XY, Jin SD and
Guo RH: A novel long non-coding RNA IRAIN regulates cell
proliferation in non small cell lung cancer. Int J Clin Exp Pathol.
8:12268–12275. 2015.PubMed/NCBI
|
|
15
|
Cheng N, Li X, Zhao C, Ren S, Chen X, Cai
W, Zhao M, Zhang Y, Li J, Wang Q, et al: Microarray expression
profile of long non-coding RNAs in EGFR-TKIs resistance of human
non-small cell lung cancer. Oncol Rep. 33:833–839. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lu KH, Li W, Liu XH, Sun M, Zhang ML, Wu
WQ, Xie WP and Hou YY: Long non-coding RNA MEG3 inhibits NSCLC
cells proliferation and induces apoptosis by affecting p53
expression. BMC Cancer. 13:4612013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Su L, Han D, Wu J and Huo X: Skp2
regulates non-small cell lung cancer cell growth by Meg3 and
miR-3163. Tumour Biol. 37:3925–3931. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Nie FQ, Sun M, Yang JS, Xie M, Xu TP, Xia
R, Liu YW, Liu XH, Zhang EB, Lu KH, et al: Long non-coding RNA
ANRIL promotes non-small cell lung cancer cell proliferation and
inhibits apoptosis by silencing KLF2 and P21 expression. Mol Cancer
Ther. 14:268–277. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang XF, Liu T, Li Y and Li S:
Overexpression of long non-coding RNA CCAT1 is a novel biomarker of
poor prognosis in patients with breast cancer. Int J Clin Exp
Pathol. 8:9440–9445. 2015.PubMed/NCBI
|
|
20
|
Yang F, Xue X, Bi J, Zheng L, Zhi K, Gu Y
and Fang G: Long non-coding RNA CCAT1, which could be activated by
c-Myc, promotes the progression of gastric carcinoma. J Cancer Res
Clin Oncol. 139:437–445. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liu SP, Yang JX, Cao DY and Shen K:
Identification of differentially expressed long non-coding RNAs in
human ovarian cancer cells with different metastatic potentials.
Cancer Biol Med. 10:138–141. 2013.PubMed/NCBI
|
|
22
|
Bischof O and Martínez-Zamudio RI:
MicroRNAs and lncRNAs in senescence: A re-view. IUBMB Life.
67:255–267. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
23
|
Deng L, Yang SB, Xu FF and Zhang JH: Long
non-coding RNA CCAT1 promotes hepatocellular carcinoma progression
by functioning as let-7 sponge. J Exp Clin Cancer Res. 34:182015.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang ZH, Guo XQ, Zhang QS, Zhang JL, Duan
YL, Li GF and Zheng DL: Long non-coding RNA CCAT1 promotes glioma
cell proliferation via inhibiting microRNA-410. Biochem Biophys Res
Commun. 480:715–720. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang Q, Zhang W and Hao S: LncRNA CCAT1
modulates the sensitivity of paclitaxel in nasopharynx cancers
cells via miR-181a/CPEB2 axis. Cell Cycle. 16:795–801. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tu K, Li C, Zheng X, Yang W, Yao Y and Liu
Q: Prognostic significance of miR-218 in human hepatocellular
carcinoma and its role in cell growth. Oncol Rep. 32:1571–1577.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Cao L, Bombard J, Cintron K, Sheedy J,
Weetall ML and Davis TW: BMI1 as a novel target for drug discovery
in cancer. J Cell Biochem. 112:2729–2741. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Xiong D, Ye Y, Fu Y, Wang J, Kuang B, Wang
H, Wang X, Zu L, Xiao G, Hao M, et al: Bmi-1 expression modulates
non-small cell lung cancer progression. Cancer Biol Ther.
16:756–763. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhang X, Tian T, Sun W, Liu C and Fang X:
Bmi-1 overexpression as an efficient prognostic marker in patients
with nonsmall cell lung cancer. Medicine. 96:e73462017. View Article : Google Scholar : PubMed/NCBI
|