Introduction
The CACNA1A gene, located on chromosome
19p13, encodes the pore-forming α-1A subunit of human neuronal
voltage-gated Cav2.1 (P/Q-type) calcium channels (1,2).
Mutations in the CACNA1A gene cause several
autosomal-dominant neurological disorders and clinical
symptoms/phenotypes such as migraines, spinocerebellar ataxia,
familial hemiplegic migraine (FHM), and episodic ataxia (EA)
(3–6).
In our previous study, using whole-exome sequencing,
a homozygous variant of Cav2.1 was identified in a three-generation
consanguineous Chinese Han family with progressive myoclonic
epilepsy (PME), which is a heterogeneous neurodegenerative disorder
(7). The mutant Cav2.1 was revealed
in two siblings but not in fifty normal donors, indicating the
mutation gene spectrum of CACNA1A can be used as a valuable
predictor for PME.
A repeated polymorphic cytosine-adenine-guanine
(CAG) mutation was also identified in the PME patient samples that
encodes an elongated tract of glutamine residues in the C-terminus
of Cav2.1. It has been suggested that trinucleotide repeat
expansion in the CAG in protein coding regions in the genome may
cause polyglutamine (polyQ) disease, which induces a set of
dominantly inherited neurodegenerative disorders including
Huntington's disease (HD), spinocerebellar ataxia, type 1/2/3/6/7
(SCA-1/2/3/6/7), dentatorubropallidoluysian atrophy (DRPLA),
Machado-Joseph disease (MJD) and spinobulbar muscularatrophy
(8,9).
Although polyQ diseases are well known clinically,
their molecular mechanism and cell biological behavior are still
unclear, especially in the context of PME. To address this issue,
in the present study, a wild-type (wt) human Cav2.1 (Cav2.1 wt)
with 13 repeats of CAG as well as a mutant-type (mt) Cav2.1 (Cav2.1
mt) with 26 repeats of CAG in the C-terminus of this protein were
constructed and delivered into cells of the SH-SY5Y neuroblastoma
cell line, which is always used as the model cell line for
investigations of human nervous system diseases (10–12).
The results revealed that the forced expression of Cav2.1 mt
significantly inhibited SH-SY5Y cell proliferation by inducing
apoptosis. By constructing a series of truncated Cav2.1 mt
molecules and a Cav2.1 mt-GFP fusion molecule, a marked nuclear
translocation phenomenon was revealed. Furthermore, it was observed
that Cav2.1 mt may activate apoptosis-relevant factors, namely,
Bcl-2/Bax, caspase and PARP, and thus induces cell apoptosis. The
present study clearly revealed the apoptosis-inducing effects of
polyQ mutations in human nerve cells, a finding that may provide a
new insight useful for interpreting the pathogenesis of and
developing new therapeutic targets for PME.
Materials and methods
Cell culture
Human SH-SY5Y neuroblastoma cells were kindly
provided by the Stem Cell Bank, Chinese Academy of Sciences (cat.
no. SCSP-5014) and were authenticated by STR profiling (Shanghai
Genechem Co., Ltd.). The cells were routinely maintained in
Dulbecco's modified Eagle's medium (DMEM; GE Healthcare Life
Sciences) supplemented with 10% fetal calf serum (FCS; GE
Healthcare Life Sciences), 100 µg/ml streptomycin and 100 U/ml
penicillin (HyClone; GE Healthcare Life Sciences) under standard
conditions (37°C, 5% CO2). The cell culture medium was
changed every two days. Upon reaching 80% confluence, the cells
were digested with 0.25% trypsin (GE Healthcare Life Sciences) for
1 min at 37°C.
Plasmid construction
The DNA molecules expressing Cav2.1 wt (Q13, 13 CAG
repeats) and Cav2.1 mt (Q26, 26 CAG repeats) were synthesized by
Sangon Biotech Co., Ltd. The first CAG starts from the 6955 site of
CACNA1A cDNA (NM_023035.2). DNA molecules were cloned into
pcDNA3.1.neo vector with BamHI and XbaI restriction
enzyme sites. The DNA expressing a series of CACNA1A
deletion mutants (Cav2.1dm) were synthesized by PCR reaction and
cloned into pcDNA3.1.neo vector with BamHI and XbaI.
The primer sequences of the Cav2.1dm were as follows: Cav2.1N
forward, 5′-CAAGGGATCCGCCACCATGGCCCGCTTCGGAGAC-3′ and reverse,
5′-GCCCCTCTAGATTAGGCCTTACGGATCACAGG-3′; Cav2.1dm1 forward,
5′-CAAGGGATCCGCCACCATGCAGGTGTTTGGTAACATTG-3′ and reverse,
5′-GCCCCTCTAGATTAGCACCAATCATCGTCAC-3′; Cav2.1dm2 forward,
5′-CAAGGGATCCGCCACCATGAAGCGTTCAGCCTCCG-3′ and reverse,
5′-GCCCCTCTAGATTAGCACCAATCATCGTCAC-3′; Cav2.1C forward,
5′-CAAGGGATCCGCCACCATGGCGGTGGCCAGGCCGGGC-3′ and reverse,
5′-GCCCCTCTAGATTAGCACCAATCATCGTCAC-3′. DNA sequences of Cav2.1dm3,
Cav2.1dm4 and Cav2.1dm5 were synthesized by Sangon Biotech Co.,
Ltd. and cloned into the same vector via BamHI and
XbaI restriction enzyme sites. To ascertain the cellular
distribution of Cav2.1dm, GFP was fused to the C-terminus of the
Cav2.1dm3 molecule. The plasmid expressing Cav2.1dm3G fusion
protein was constructed.
Polypeptide structure prediction
The 3D structure of polypeptide was built using an
online tool (http://zhanglab.ccmb.med.umich.edu/).
Transfection
According to the manufacturer's instructions, 2 µg
of each of the aforementioned plasmids and the empty vector (EV,
control) were transfected into SH-SY5Y cells using Lipofectamine
3000 (Invitrogen; Thermo Fisher Scientific, Inc.). All the cellular
experiments were carried out at or after 48 h
post-transfection.
RNA extraction, reverse
transcriptional reaction and real-time quantitative PCR (qPCR)
Total RNA was isolated using EasyPure RNA
Purification Kit (cat. no. ER701; Beijing TransGen Biotech Co.,
Ltd.). RNA (1,000 ng) was reverse-transcribed with TransScript
All-in-One First-Strand cDNA Synthesis SuperMix for qPCR (One-Step
gDNA Removal) (cat. no. AT341; Beijing TransGen Biotech Co., Ltd.).
qPCR was conducted based on the instructions of RealStar Green Fast
Mixture (GenStar). Primers for qPCR are listed as follows: Forward
primer for Cav2.1 wt, Cav2.1 mt, Cav2.1N, and Cav2.1dm4 is
5′-AAGGATCGGAAGCATCGACAG-3′ and reverse primer is
5′-GTGCTGGTACCAGATGTTGAG-3′; forward primer for Cav2.1dm1,
Cav2.1dm2, Cav2.1dm3, Cav2.1C and Cav2.1dm5 is
5′-GACTCCCCAACGGCTACTAC-3′ and reverse primer is
5′-CACCAATCATCGTCACTCTCG-3′. qPCR was performed on ABI 7500
(Applied Biosystems; Thermo Fisher Scientific, Inc.) and the PCR
program consisted of 10 sec at 95°C, 40 cycles at 95°C for 5 sec
and 60°C for 35 sec. The relative expression of the aforementioned
target gene was calculated based on the 2−ΔΔCq method
(13).
Cell growth assay
At 48 h post-transfection, SH-SY5Y cells were
trypsinized and seeded into a 96-well plate at a density of
5×103 cells/well and cultured for another 48 h. Cell
proliferation was determined using WST-1 cell proliferation reagent
(Roche Applied Science). According to the manufacturer's
instructions, 20 µl WST-1 was added to 200 µl cell culture medium
and incubated at 37°C in the dark for 2.5 h. The absorbances at 450
and 630 nm were assessed by a microplate reader (BioTek
Instruments). The final OD (optical density) was designated as
OD450-OD630-ODblank.
Cell cycle and apoptosis assay
At 72 h post-transfection, at least 2×106
cells were harvested and washed with phosphate-buffered saline
(PBS) three times. For cell cycle analysis, cell pellets were
resuspended in 200 µl PBS containing 10 µg/ml propidium iodide (PI;
Sigma-Aldrich; Merck KGaA) and incubated for 30 min in the dark at
room temperature. For cell apoptosis, the transfected cells were
stained with an Annexin V-FITC/PI Apoptosis Detection kit (BD
Biosciences) for 15 min in the dark and analyzed using a flow
cytometer (FACScan; BD Biosciences) with FlowJo analysis software
(version 10.0; FlowJo LLC). The cells in the quadrants represented
the different cell states as follows: Late-apoptotic cells were
presented in the upper right quadrant, viable cells were presented
in the lower left quadrant, and early apoptotic cells were
presented in the lower right quadrant.
Protein extraction
Protein was extracted from the whole cell, cell
membrane, or cell nucleus of the SH-SY5Y cells, respectively. The
whole cell protein was extracted with a Protein Extraction kit
(cat. no. KGP2100; Nanjing KeyGen Biotech. Co., Ltd.). Cell
membrane protein and nucleoprotein were extracted with a Cell
Membrane Protein/Cytoplasmic Protein/Nucleoprotein Extraction kit
(cat. no. KGBSP002; Nanjing KeyGen Biotech. Co., Ltd.).
Western blotting
Equal amounts of 25 µg protein from each sample were
loaded to 10% SDS polyacrylamide gel and separated by
electrophoresis. Then, proteins were transferred to PVDF membranes
(EMD Millipore). The membranes were incubated in 5% BSA (cat. no.
E661003; Sangon Biotech Co., Ltd.)/TBST solution at 37°C for 1 h
and then washed in TBST 3 times for 5 min. The membranes were then
incubated with primary antibodies at 4°C overnight. The primary
antibodies were: Anti-Cav2.1 N-terminus (product code ab191140;
recognizes Human CACNA1A aa 430–447; 1:1,500); anti-Cav2.1
C-terminus (product code ab181371; recognizes Human CACNA1A aa
2050–2150; 1:1,000); anti-Bcl-2 rabbit polyclonal antibody (product
code ab59348; 1:15,00); and anti-Bax (product code ab32503;
1:1,500; all from Abcam); anti-cleaved caspase-3 (product no. 9664;
1:1,000; Cell Signaling Technology, Inc.); anti-cleaved poly
ADP-ribose polymerase (PARP) (product code ab32064; 1:1,500);
anti-GFP (product code ab6556, 1:3,000); and anti-β-actin (product
code ab8226, 1:3,000; all from Abcam). After washing in TBST 3
times for 5 min, the membranes were incubated with HRP-coupled goat
anti-rabbit or goat anti-mouse secondary antibody (cat. no. A0208
and cat. no. A0216, respectively; 1:2,000; Beyotime Institute of
Biotechnology) at room temperature for 45 min. The
chemiluminescence signals of the target proteins were visualized
using ChemiDoc XRS+ system (Bio-Rad Laboratories, Inc.).
Densitometric values of the blots were analyzed using Quantity One
software (version 4.6; Bio-Rad Laboratories, Inc.).
Immunofluorescence staining and
nuclear counterstain
Cav2.1dm3G transfectants (5×105) in a
six-well plate were washed with 2 ml cold PBS 2 times for 2 min and
fixed with 3.7% formaldehyde for 15 min at room temperature (RT)
(Sigma-Aldrich; Merck KGaA) in PBS. After the fixation buffer was
removed, the cells were washed with PBS another 2 times for 2 min.
Then, the cells were incubated with 0.5% Triton X-100 (Sigma; Merck
KGaA) in PBS for 10 min at room temperature and washed with PBS 3
times for 2 min. Cells were blocked with 5% powdered milk in TBST
buffer (Sangon Biotech Co., Ltd.) plus 0.02% sodium azide
(Sigma-Aldrich; Merck KGaA) for 45 min at RT. After the blocking
buffer was removed, the cells were incubated with TBST buffer
containing anti-Cav2.1 antibody (1:800; cat. no. HPA064258;
Sigma-Aldrich; Merck KGaA) for 1 h at RT. After washing 3 times for
5 min, the cells were incubated with TBST/0.5% BSA diluted Alexa
Fluor 555 conjugate goat anti-rabbit IgG secondary antibody (cat.
no. A-21428; Thermo Fisher Scientific, Inc.) at a concentration of
4 µg/ml (according to the manufacturer's instructions) for 45 min
at RT.
To visualize the intracellular location of
Cav2.1dm3G, cell nuclei were stained with DAPI. According to the
manufacturer's instructions, SH-SY5Y cells expressing Cav2.1dm3G
were washed with PBS 2 times for 2 min and 1.5 ml DAPI (Beyotime
Institute of Biotechnology) was added to the wells. Then the cells
were maintained at room temperature for 5 min.
An Olympus IX83 fluorescence microscope (Olympus
Corporation) was used to observe the cellular morphology and
fluorescence.
Statistical analysis
Statistical analysis was conducted with SPSS 17.0
software (SPSS, Inc.). All data were expressed as the mean ±
standard deviation. All of the experiments were performed in
triplicate and repeated at least three times. Significance was
determined using Student's t-test or one-way analysis of variance
(ANOVA) followed by LSD or Tukey's post hoc test. P<0.05 and
P<0.01 were considered to indicate statistically significant
differences (*P<0.05 and **P<0.01 as indicated in the images
and figure legends).
Results
Cav2.1 mt with 26 polyQ repeats
suppresses the proliferation of the SH-SY5Y cells
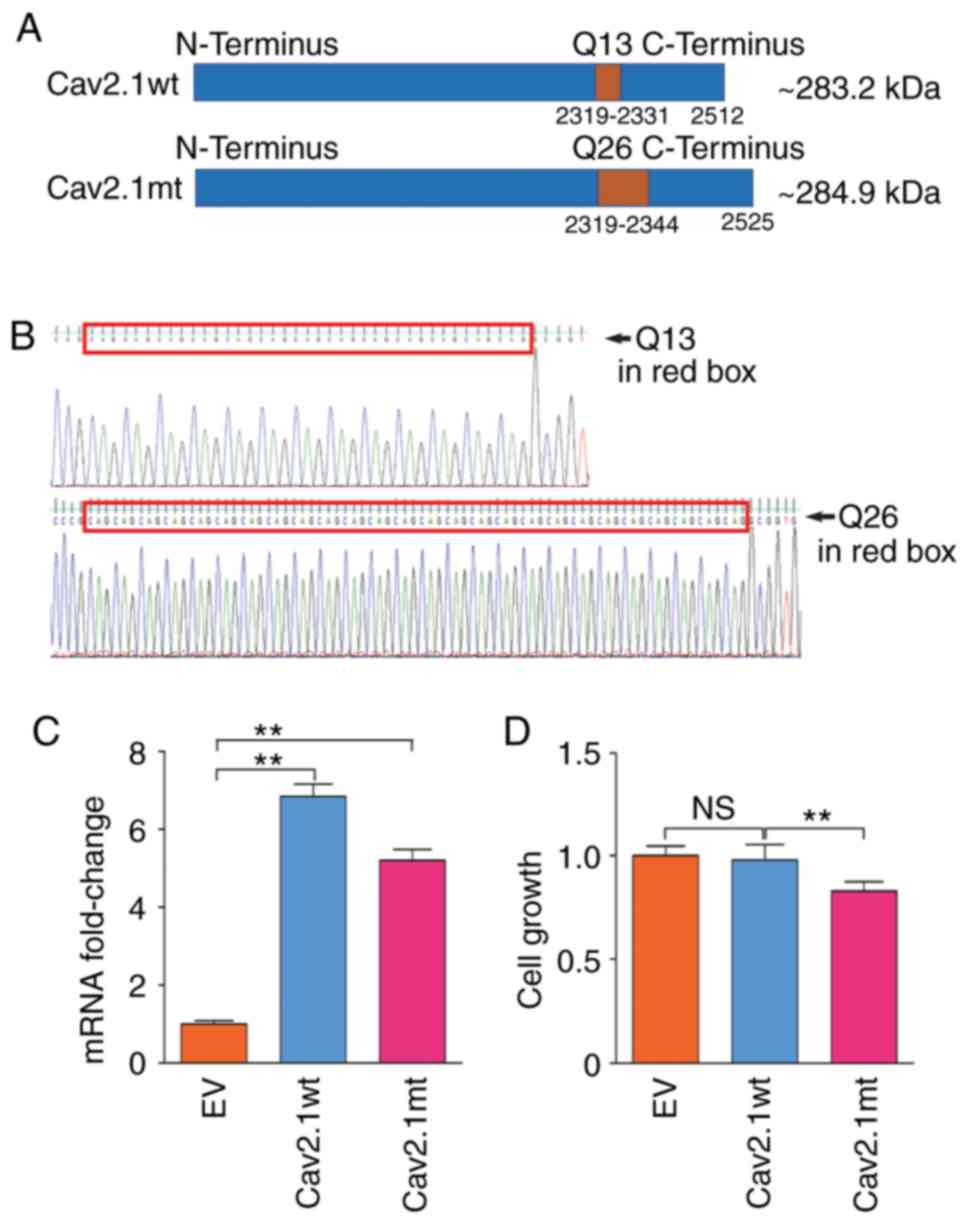
The DNA molecules expressing Cav2.1 wt and Cav2.1 mt
were synthesized and confirmed by Sanger sequencing (Fig. 1A and B). After the two expressing
plasmids, pcDNA3.1-Cav2.1 wt and pcDNA3.1-Cav2.1 mt, were delivered
into SH-SY5Y cells and confirmed by qPCR (Fig. 1C) cell proliferation was analyzed.
As revealed in Fig. 1D, compared
with the Cav2.1 wt-Q13 and the EV transfectants, the forced
expression of Cav2.1 mt-Q26 significantly decreased the cell
proliferation of the SH-SY5Y cells (P<0.01).
Biological function of the truncated
Cav2.1 mt molecules
It has been reported that Cav2.1 truncating mutation
is associated with neurological disorders (14). Herein, to address whether the
truncated Cav2.1 mt molecules affect the biological functions of
the SH-SY5Y cells, a series of CACNA1A gene deletion
mutations were prepared and the corresponding expressing plasmids
that are schematically represented in Fig. 2A, were constructed. Cav2.1N is the
aa1 to aa2312 on the N-terminus of Cav2.1 containing no polyQ;
Cav2.1dm1 is the aa1716 to aa2525 of Cav2.1 mt containing 26 polyQ
repeats; Cav2.1dm2 is the aa2127 to aa2525 of Cav2.1 mt containing
26 polyQ repeats; Cav2.1dm3 is the aa2319 to aa2525 of Cav2.1 mt,
starting from the fist Q and containing 26 polyQ repeats; Cav2.1C
is the aa2345 to aa2525 on the C-terminus of Cav2.1 containing no
polyQ. The approximate molecular weight (kDa) of each deletion
mutation was calculated by an online tool (http://www.detaibio.com/sms2/protein_mw.html).
SH-SY5Y cells were then transfected with all the aforementioned
plasmids and the transfection efficacy was confirmed by qPCR
(Fig. 2B). The cell proliferation
of all the transfected cells were detected by WST-1 and the result
indicated that Cav2.1N and Cav2.1C, containing no polyQ repeats,
did not affect the cell proliferation of the SH-SY5Y cells.
Conversely, the forced expression of Cav2.1dm1, Cav2.1dm2 and
Cav2.1dm3, containing 26 polyQ repeats, significantly impaired cell
proliferation (Fig. 2C, P<0.01).
Notably, the Cav2.1dm3 transfectants had the lowest proliferation
(Fig. 2C).
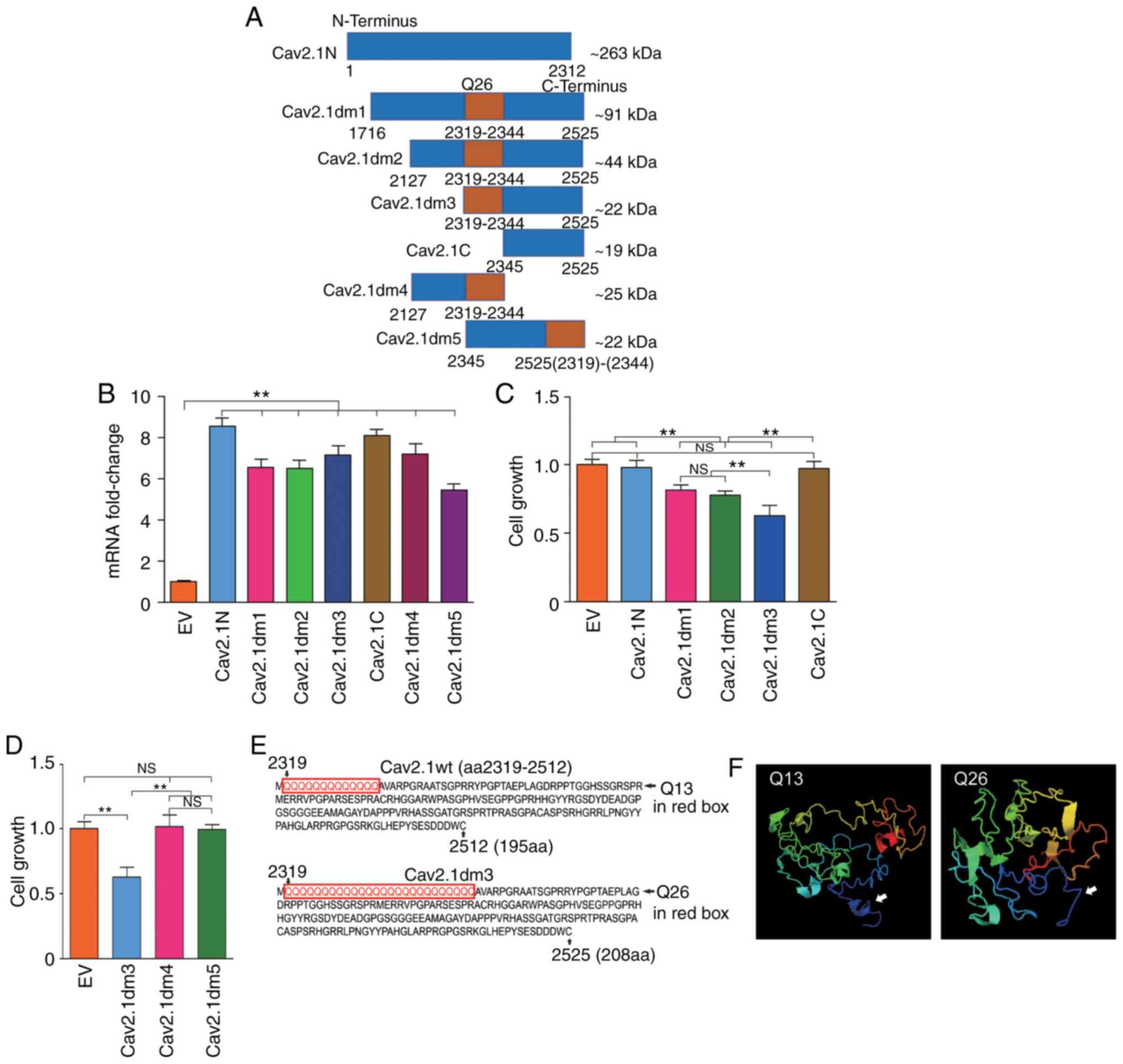 | Figure 2.Construction of a series of truncated
Cav2.1 mt molecules and addressing their effects on cell
proliferation. (A) Diagram of the structures of seven truncated
Cav2.1 mt molecules, an ~263 kDa N-terminus of Cav2.1 without polyQ
repeat (Cav2.1N), a ~91 kDa C-terminus of Cav2.1 mt with 26 polyQ
repeats (Cav2.1dm1), an ~44 kDa C-terminus of Cav2.1 mt with 26
polyQ repeats (Cav2.1dm2), an ~22 kDa C-terminus of Cav2.1 mt
beginning with Q26 (Cav2.1dm3), an ~19 kDa C-terminus of Cav2.1 mt
without polyQ repeat (Cav2.1C), an ~25 kDa C-terminus of Cav2.1 mt
ending with Q26 (Cav2.1dm4) and a reversely assembled Cav2.1dm3
ending with Q26 (Cav2.1dm5). (B) mRNA expression levels of Cav2.1N,
Cav2.1dm1, Cav2.1dm2, Cav2.1dm3, Cav2.1C, Cav2.1dm4 and Cav2.1dm5
in the transfected cells were confirmed by qPCR (t-test). (C)
Comparison of proliferation of the SH-SY5Y cells transfected with a
series of truncated Cav2.1 mt molecules. The WST-1 result indicated
that Cav2.1N and Cav2.1C, containing no polyQ repeat did not affect
the cell proliferation of the SH-SY5Y cells. Conversely, the forced
expression of Cav2.1dm1, Cav2.1dm2 and Cav2.1dm3, containing 26
polyQ repeats significantly impaired cell proliferation.
**P<0.01, a significant difference in cell proliferation was
observed between EV/Cav2.1N/Cav2.1C and
Cav2.1dm1/Cav2.1dm2/Cav2.1dm3, and Cav2.1dm1/Cav2.1dm2 and
Cav2.1dm3. NS, no difference of cell proliferation was observed
among EV, Cav2.1N and Cav2.1C, and Cav.1dm1 and Cav2.1dm2 (Tukey's
test). (D) Comparison of the proliferation of the SH-SY5Y cells
transfected with Cav2.1dm3 and its two variations. The result
indicated that only those molecules composed with integrity Q26 and
C-terminus of Cav2.1 harbored cytotoxicity (Tukey's test). (E)
Partial amino acid sequences of Cav2.1 wt-Q13 and Cav2.1dm3. (F)
Comparison of the 3D structure of Cav2.1 wt and Cav2.1dm3. White
arrows indicate the differences on the secondary structure between
Q13 and Q26. **P<0.01. Cav2.1mt, mutant-type Cav2.1; Cav2.1 wt,
wild-type Cav2.1; EV, empty vector; NS, no significance. |
Since the Cav2.1dm3 molecule is composed by Q26 and
Cav2.1C, and Cav2.1C itself does not affect cell proliferation,
therefore it was speculated that Q26 plays a critical role in the
Cav2.1-induced cell growth inhibition. To ascertain our hypothesis,
another two plasmids were further constructed. Cav2.1dm4, aa2127 to
aa2344 of Cav2.1 mt, contains 26 polyQ repeats and ends at the last
Q, with no Cav2.1 C-terminus. Cav2.1dm5 is the reversed arrangement
of Q26 (aa2319-2344) and Cav2.1C (aa2345-2525). After transfection,
it was revealed that neither Cav2.1dm4 nor Cav2.1dm5 affected the
cell growth of the SH-SY5Y cells (Fig.
2D, P>0.05). These data indicated that: i) The expanded
mutation of the polymorphic Q repeat tract, e.g., from 13 repeats
to 26 repeats, has cytotoxicity and impairs the cell growth of the
SH-SY5Y cells; and ii) the Q26 tract must be linked with the Cav2.1
C-terminus in a cis-arrangement to exert its biological
function.
It was surmised that the structure of the
glutamine-expanded Cav2.1 is instable, and the mutant protein
cleaves and generates truncated molecules. Therefore the 3D
structure of the truncated Cav2.1 beginning with Q13 and Q26
(Fig. 2E) was compared using an
online tool (http://zhanglab.ccmb.med.umich.edu/). Because the
website has set a limit on the amino acid quantity, the 3D
structures of the two molecules with 195aa and 208aa are only
presented. As revealed in Fig. 2F,
the structures of the molecules with Q13 and Q26 at the N-terminus
of the truncated Cav2.1 were quite different.
Truncated Cav2.1 does not affect the
cell cycle distribution of the SH-SY5Y cells
To elucidate the anti-proliferation mechanisms of
the truncated Cav2.1, a cell cycle assay was performed using flow
cytometry. However, no variation of cell cycle distribution was
observed among the EV, Cav2.1 wt, Cav2.1 mt and Cav2.1dm3
transfectants (Fig. 3,
P>0.05).
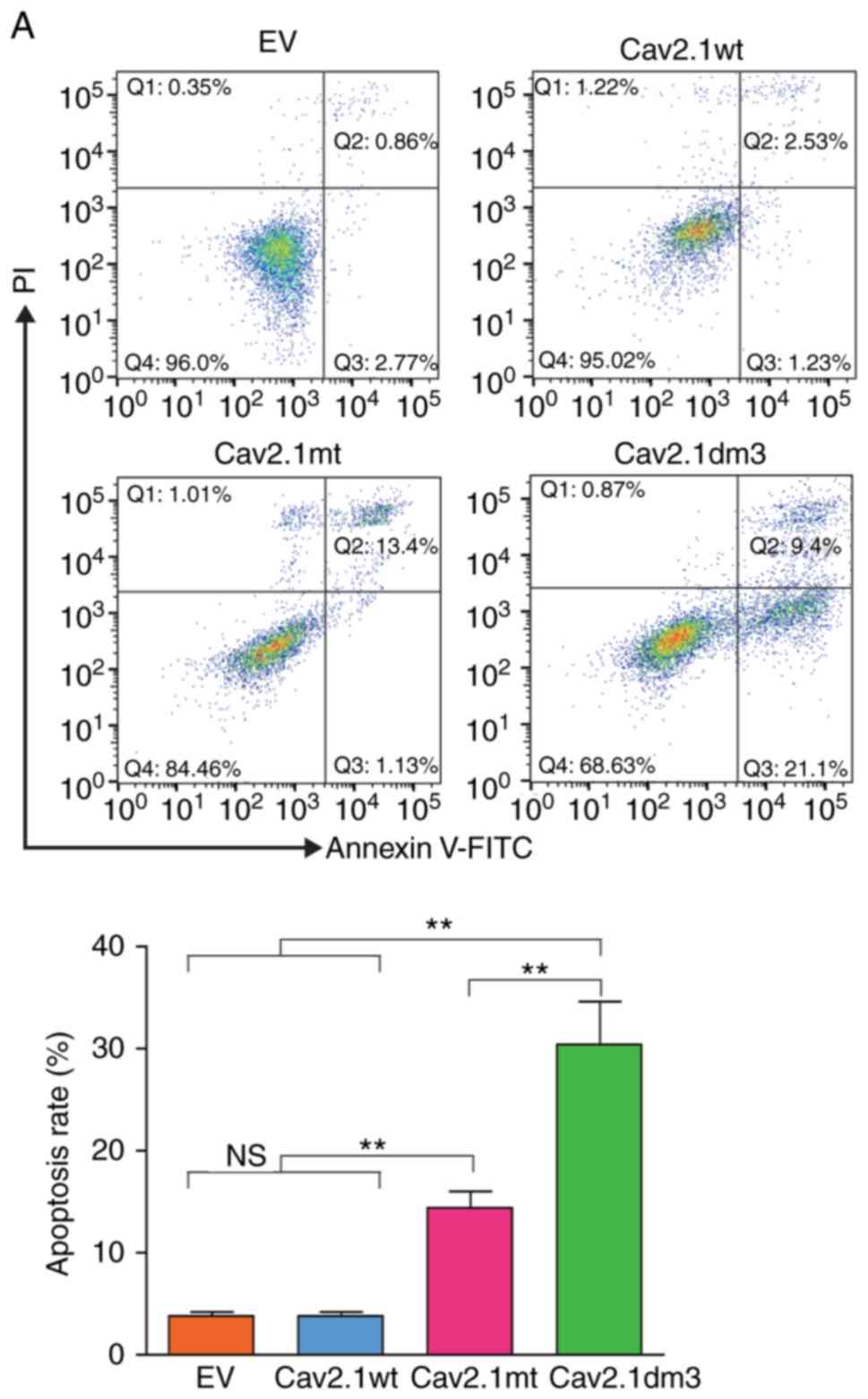
Truncated Cav2.1 promotes apoptosis of
the SH-SY5Y cells
Subsequently, a cell apoptosis assay was performed
using flow cytometry. As revealed in Fig. 4, compared to the Cav2.1 wt and EV
transfection groups, the forced expression of Cav2.1 mt and
Cav2.1dm3 in SH-SY5Y cells significantly induced apoptosis. The
apoptotic rate of the Cav2.1dm3 group was 30.5%, significantly
higher than that of 14.53% in the Cav2.1 mt group (Fig. 4A, P<0.01). The cell morphology
also indicated the increased apoptosis in the Cav2.1dm3
transfectants (Fig. 4B).
Truncated Cav2.1 molecules are
translocated to the nucleus of the SH-SY5Y cells
To gain deeper insight into the role of the
truncated Cav2.1 mt molecules in cell apoptosis, a Cav2.1dm3 and
GFP fusion molecule was constructed. To avoid impairing the Q26
bioactivity, GFP was fused to the C-terminus of Cav2.1dm3, and
named Cav2.1dm3G (Fig. 5A). After
the plasmid was delivered into SH-SY5Y cells, the expression of
Cav2.1dm3G was detected by western blotting. As controls, the
expression of Cav2.1 wt and Cav2.1 mt in the transfectants were
also detected by western blotting, concurrently. As revealed in
Fig. 5B, the western blot results
revealed that the Cav2.1 wt molecules were mainly located on the
cell membrane (87.0%, the upper right panel, P<0.01), Cav2.1 mt
molecules were located both on the cell membrane and in the cell
nucleus (46.7% vs. 53.3%, the left lower panel, P<0.05), whereas
most Cav2.1dm3G molecules were located in the cell nucleus (85.9%,
the right lower panel, P<0.01).
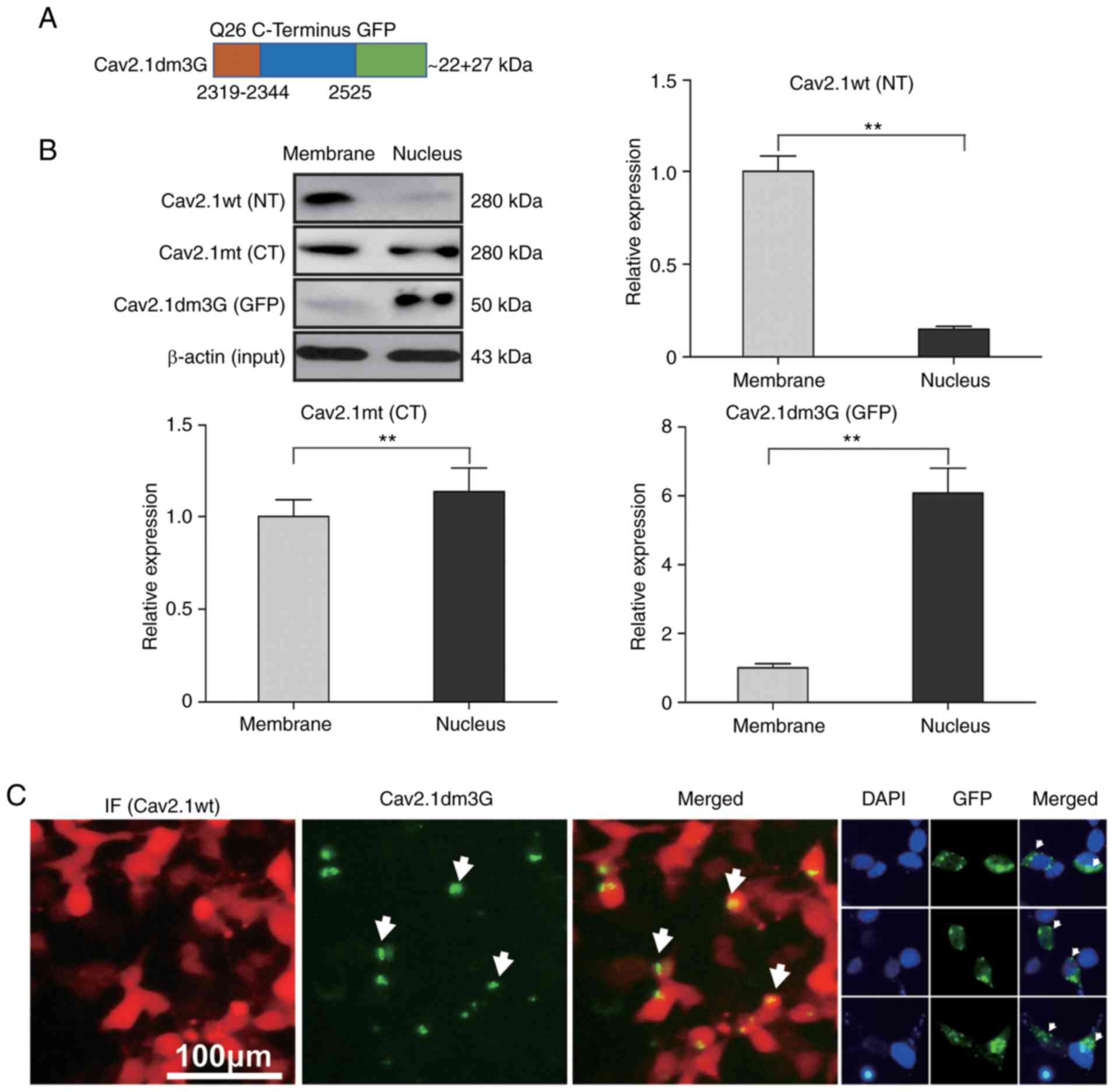 | Figure 5.Cav2.1 mutant molecules are
translocated to the nucleus of the SH-SY5Y cells. Plasmids
expressing Cav2.1 wt, Cav2.1 mt and Cav2.1dm3G were transfected
into SH-SY5Y cells, and proteins were collected from cell membranes
or the nucleus and were detected by western blotting. (A) Diagram
of the Cav2.1dm3-GFP fusion molecule. (B) Western blotting
indicated that Cav2.1 wt molecules were located on the cell
membrane, Cav2.1 mt molecules were located on both the cell
membrane and in the cell nucleus, and Cav2.1dm3G molecules were
translocated to the cell nucleus. β-actin was used to illustrate
that the membrane proteins and nucleoproteins were extracted from
the same quantity of transfected cells. Upper left panel, bands on
the blotting membrane; the other panels, relative band density
(t-test). (C) Immunofluorescence staining revealed the nuclear
translocation phenomenon of Cav2.1 mt molecules. Cav2.1 mt,
mutant-type Cav2.1; Cav.1 wt, wild-type Cav2.1; NT, N terminus; CT,
C terminus. **P<0.01. |
A cell immunofluorescence staining experiment was
also performed and it was observed that the Cav2.1 wt molecules
were distributed on the cell membrane, whereas the GFP fluorescence
of Cav2.1dm3G molecules was mainly distributed inside the cell
nucleus (Fig. 5C). These data
indicated that the truncated Cav2.1 mt molecules were translocated
to nucleus, increasing apoptosis of the SH-SY5Y cells.
Truncated Cav2.1 mt induces cell
apoptosis via the Bcl-2/Bax/caspase-3/PARP pathway
It has been reported that polyQ interacts with the
mitochondrial apoptotic pathway and caspase apoptotic proteins to
exert cytotoxicity (15).
Therefore, the expression of Bcl-2, Bax, cleaved caspase-3 and
cleaved PARP were detected. Western blot results revealed that the
Bcl-2/Bax ratio was markedly decreased in Cav2.1 mt and Cav2.1dm3
transfectants (Fig. 6, upper left
and upper right panel, P<0.01). The caspase-3 and PARP cell
apoptotic pathways were also activated (Fig. 6 upper left panel, lower left/right
panel, P<0.01).
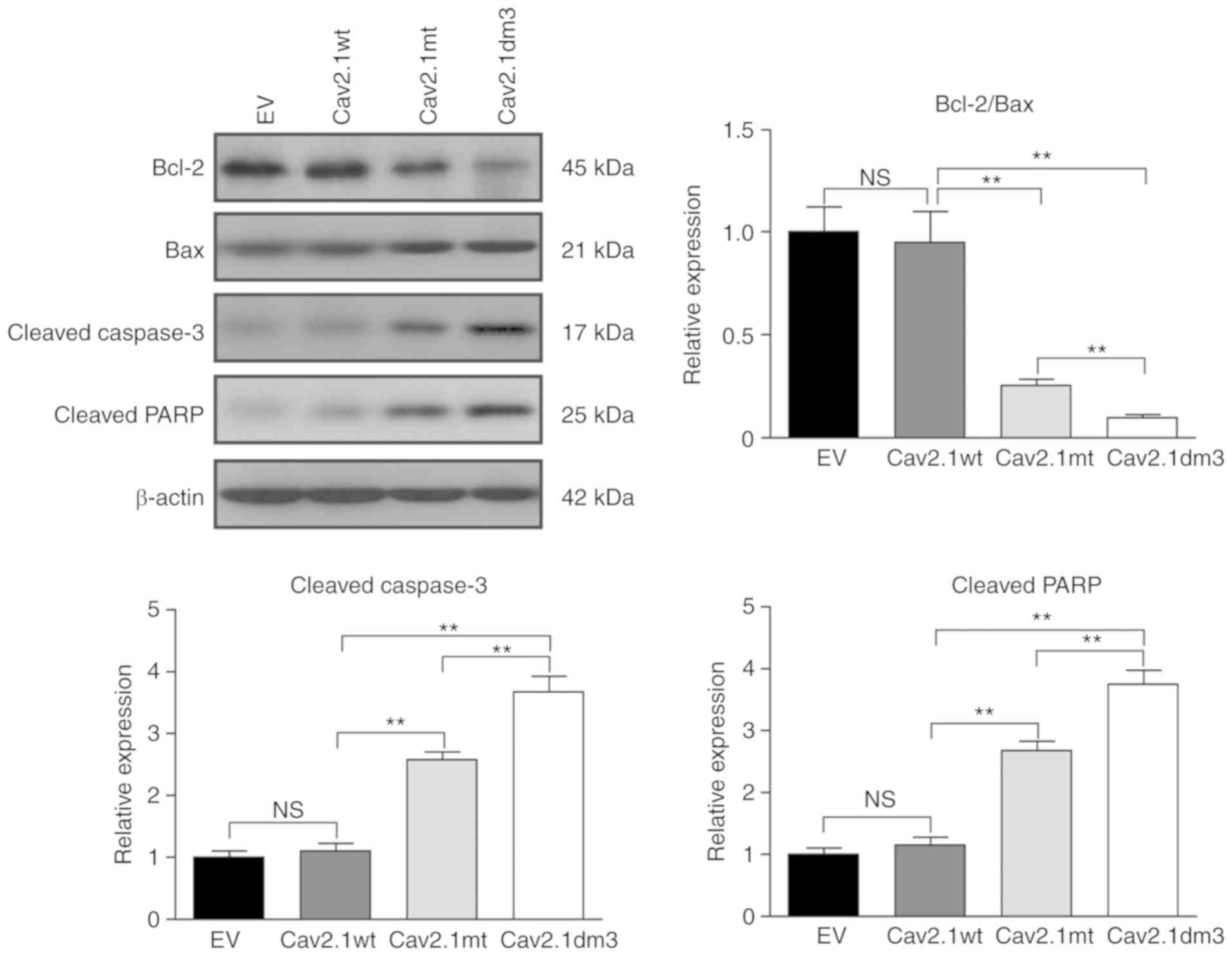 | Figure 6.Cav2.1 mt molecules induce cell
apoptosis via the Bcl-2/Bax, caspase-3 and PARP pathways. SH-SY5Y
cells were transfected with empty vector, Cav2.1 wt, Cav2.1 mt and
Cav2.1dm3 plasmid and the levels of Bcl-2, Bax, cleaved caspase-3
and cleaved PARP were detected by western blotting. Upper left
panel, western blot result indicated that Cav2.1 mt molecules
activated the Bcl-2/Bax, caspase-3 and PARP apoptotic pathways. The
other panels, relative band density (Tukey's test). **P<0.01.
Cav2.1 mt, mutant-type Cav2.1; Cav.1 wt, wild-type Cav2.1; PARP,
poly ADP-ribose polymerase; EV, empty vector; NS, no
significance. |
Discussion
PME comprises a group of rare neurodegenerative
disorders characterized by ataxia, myoclonus, seizures, and
progressive cognitive impairment (16). The occurrence and development of PME
is always accompanied by certain types of gene mutations. Tonin
et al identified a newly discovered synonymous c.363A>G
(Gly82Gly) mutation in the β-glucosidase (GBA) gene in a
type 3 Gaucher disease patient that causes the loss of an exon
splicing enhancer (17). He et
al revealed a novel c.995-1G>A homozygous splicing mutation
in the SCARB2 gene in two PME patients that leads to loss of
function of the SCARB2 protein (18).
In recent years, an increasing number of mutations
in the CACNA1A gene have been reported as causative factors
for several types of neurological disorders. Using next-generation
sequencing, Grieco et al identified an R2157G missense
variant in a Moroccan hemiplegic migraine patient and an I1512T
missense variation in patients with FHM (6). Using whole-exome sequencing,
Khaiboullina et al identified an R583Q missense mutation in
patients with FHM (5). Algahtani
et al summarized that there are more than 170
disease-causing variants of CACNA1A, including mutations
with small deletions, gross deletions, missense variants, nonsense
variants, splice variants, small insertions, complex rearrangements
and repeat variations (19).
In a previous study, using whole-exome sequencing,
we identified a homozygous variant in PME patients: a repeated CAG
mutation in the CACNA1A gene, which encodes an elongated
tract of glutamine residues in the C-terminus of the Cav2.1
protein. Although the intranuclear accumulation of mutant proteins
with an expanded polyQ tract has been well established, the
relationship between neurotoxicity and glutamine aggregation
remains to be investigated. In the present study, it was revealed
that the Cav2.1 protein with Q26 was translocated to the nucleus of
the SH-SY5Y cells. By constructing a series of truncated molecules,
it was revealed that overexpression of the Cav2.1 protein with a
Q26 mutation in the C-terminus induced cell apoptosis. Notably, the
shorter the truncated molecule was, the more obvious the cell
toxicity was found to be. Gerhardstein et al reported that
Cav1.2, an L-type voltage-gated calcium channel, was
proteolytically cleaved in neurons and cardiac myocytes, yielding a
truncated channel and a cytoplasmic C-terminal fragment (20). Gomez-Ospina et al reported
that a high level of the C-terminal Cav1.2 fragment was localized
in the nucleus of brain cells, whereas the N-terminal portion of
the channel was localized in the membrane and cytoplasmic fractions
(21). Park et al reported a
frameshift mutation (c.1642del, p.H548Tfs*24) in the CACNA1A
gene leading to a prematurely truncated Cav2.1 protein (22). Therefore, it was concluded that: i)
Cav2.1 and similar voltage-gated proteins tend to be cleaved in the
C-terminus due to the unstable protein structure caused by the
polyQ fragment; ii) the cleavage yields two (perhaps more)
truncated molecules, and the C-terminal fragment tends to
translocate to the nucleus; and iii) the prematurely truncated
molecules are generated from either proteolysis or gene
mutation.
The truncated voltage-gated proteins may exert their
biological function in two ways. Through one mechanism, the
truncated fragment binds to gene regulatory elements, e.g.,
promoters. For instance, Gomez-Ospina et al revealed that
the C-terminal fragment of Cav1.2 in the nucleus interacted with
multiple transcriptional regulators and affected the transcription
of a wide variety of endogenous genes important for neuronal
signaling and excitability (21).
The other mechanism depends on the polyQ structure, which is
manifested in a so-called polyQ disease. For example, Huntington's
disease is caused by polyQ (more than 35 repeats) in the Huntington
protein that results in neuronal cell death (23). In addition, aberrant expression of
superoxide dismutase and accumulation of oxidative products were
also observed in a CAG-repeat disease (24).
Cell apoptosis is mediated by caspases, which are
activated by signals from the plasma membrane and mitochondria.
Bcl-2/Bax is the key mitochondrial apoptotic pathway controlling
cell apoptosis. Sanchez et al reported that the
polyglutamine repeat (Q79)/caspase 8 axis-induced cell death was
critical in Huntington's disease (25). Tien et al demonstrated that
the polyglutamine repeat (Q26 and Q78) may decrease the mRNA level
of Bcl-2 in Machado-Joseph disease cells (26). In the present study, it was revealed
that the truncated Cav2.1 mutant molecule induced cell apoptosis
through the activation of the Bcl-2/Bax, caspase-3 and PARP
pathways. The production of PAR chains by PARPs is one of the first
steps in the process of DNA damage repair (27). In addition, the increased level of
cleaved caspase-3 can lead to cleaved PARP, which is an inactive
form, thereby disrupting DNA repair and resulting in apoptosis
(28). Considering the relationship
among Bcl-2/Bax, caspase-3 and PARP, the present results are
reasonable. However, since few investigations have focused on the
correlation between polyQ and PARP, to support our conclusion,
further experiments are required.
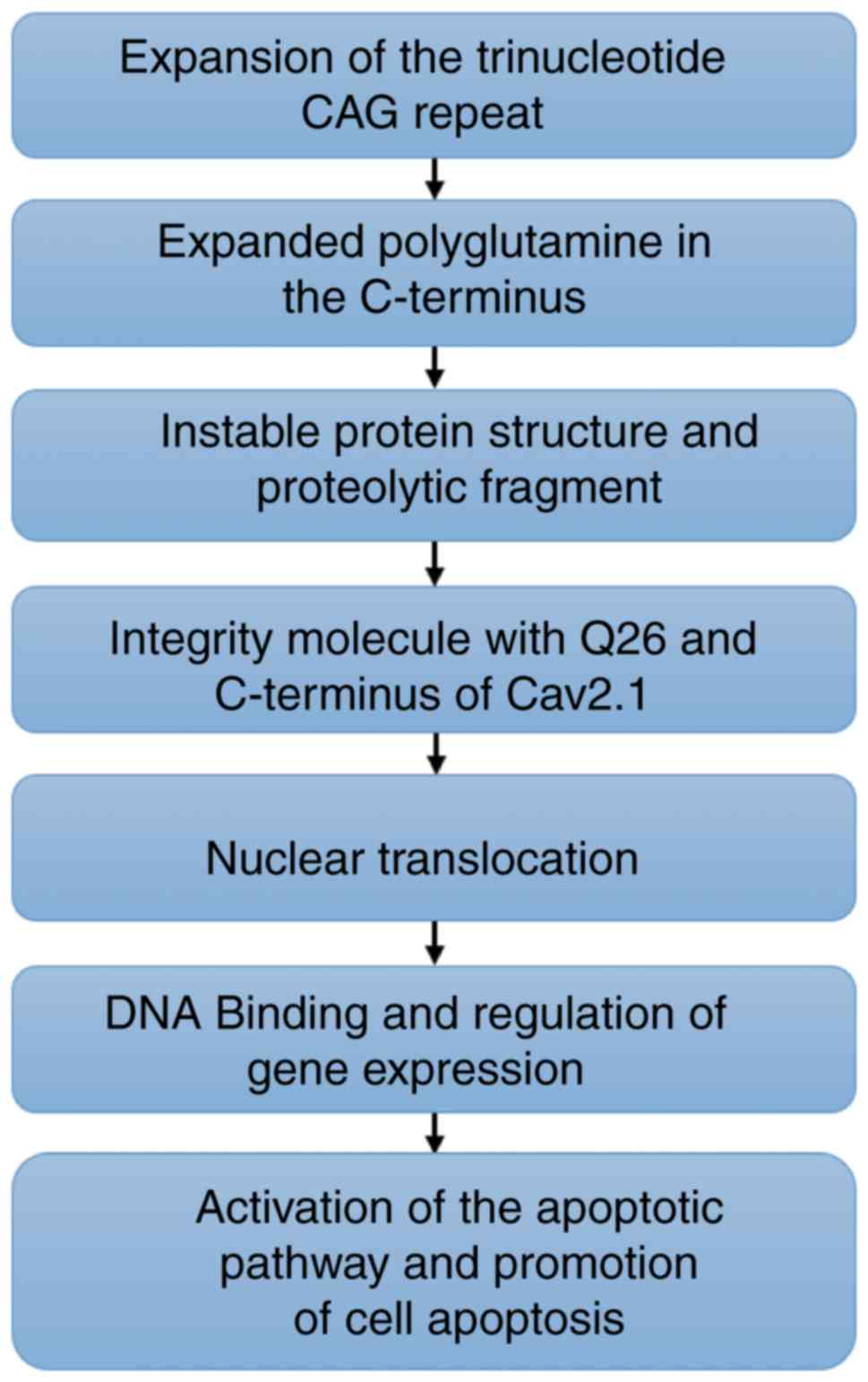
In summary, in the present study, it was
demonstrated that an aberrant elongated tract of polyQ in the
Cav2.1 protein may proteolytically generate a short fragment that
is translocated to the nucleus, inducing cell apoptosis in human
SH-SY5Y cells (Fig. 7). The present
study may provide new insights for the interpretation of the
pathogenesis of PME and for the relationship among polyQ,
CACNA1A gene mutation and PME.
Acknowledgements
We would like to thank Professor Dehai Yu at The
Laboratory of Cancer Precision Medicine of the First Hospital of
Jilin University for revising this article critically for important
intellectual content.
Funding
The present study was supported in part by grants
from the Translational-and-Clinical Collaborative Research Project
from the First Hospital of Jilin University (grant no. 2018-ZL-12
to YL); the National Natural Science Foundation of China (grant no.
81701270 to YL); the Transformation Fund of the First Hospital of
Jilin University (grant no. JDYYZH-1902033 to YL); the Health
Special Fund from Jilin Province Department of Finance (grant no.
2018SCZWSZX-048 to XS).
Availability of data and materials
The datasets used and analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
DM and YL contributed to the conception and design
of the study. JS and XS contributed to the acquisition, analysis
and interpretation of data. JS drafted the manuscript. XS and ZL
analyzed the data and was involved in performing the experiments.
All authors have read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ophoff RA, Terwindt GM, Vergouwe MN, van
Eijk R, Oefner PJ, Hoffman SM, Lamerdin JE, Mohrenweiser HW, Bulman
DE, Ferrari M, et al: Familial hemiplegic migraine and episodic
ataxia type-2 are caused by mutations in the Ca2+ channel gene
CACNL1A4. Cell. 87:543–552. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lea RA, Curtain RP, Hutchins C, Brimage PJ
and Griffiths LR: Investigation of the CACNA1A gene as a candidate
for typical migraine susceptibility. Am J Med Genet. 105:707–712.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Petrovicova A, Brozman M, Kurca E, Gobo T,
Dluha J, Kalmarova K, Nosal V, Hikkelova M, Krajciova A,
Burjanivova T and Sivak S: Novel missense variant of CACNA1A gene
in a Slovak family with episodic ataxia type 2. Biomed Pap Med Fac
Univ Palacky Olomouc Czech Repub. 161:107–110. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bavassano C, Eigentler A, Stanika R,
Obermair GJ, Boesch S, Dechant G and Nat R: Bicistronic CACNA1A
gene expression in neurons derived from spinocerebellar ataxia type
6 patient-induced pluripotent stem cells. Stem Cells Dev.
26:1612–1625. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Khaiboullina SF, Mendelevich EG, Shigapova
LH, Shagimardanova E, Gazizova G, Nikitin A, Martynova E, Davidyuk
YN, Bogdanov EI, Gusev O, et al: Cerebellar atrophy and changes in
cytokines associated with the CACNA1A R583Q mutation in a russian
familial hemiplegic migraine type 1 family. Front Cell Neurosci.
11:2632017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Grieco GS, Gagliardi S, Ricca I, Pansarasa
O, Neri M, Gualandi F, Nappi G, Ferlini A and Cereda C: New CACNA1A
deletions are associated to migraine phenotypes. J Headache Pain.
19:752018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lv Y, Wang Z, Liu C and Cui L:
Identification of a novel CACNA1A mutation in a Chinese family with
autosomal recessive progressive myoclonic epilepsy. Neuropsychiatr
Dis Treat. 13:2631–2636. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhang Q, Chen ZS, An Y, Liu H, Hou Y, Li
W, Lau KF, Koon AC, Ngo JCK and Chan HYE: A peptidylic inhibitor
for neutralizing expanded CAG RNA-induced nucleolar stress in
polyglutamine diseases. RNA. 24:486–498. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Roshan R, Choudhary A, Bhambri A, Bakshi
B, Ghosh T and Pillai B: MicroRNA dysregulation in polyglutamine
toxicity of TATA-box binding protein is mediated through STAT1 in
mouse neuronal cells. J Neuroinflammation. 14:1552017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
von Niederhausern N, Ducray A, Zielinski
J, Murbach M and Mevissen M: Effects of radiofrequency
electromagnetic field exposure on neuronal differentiation and
mitochondrial function in SH-SY5Y cells. Toxicol In Vitro.
61:1046092019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Karikari TK, Nagel DA, Grainger A,
Clarke-Bland C, Crowe J, Hill EJ and Moffat KG: Distinct
conformations, aggregation and cellular internalization of
different tau strains. Front Cell Neurosci. 13:2962019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liu M, Bai X, Yu S, Zhao W, Qiao J, Liu Y,
Zhao D, Wang J and Wang S: Ginsenoside Re inhibits ROS/ASK-1
dependent mitochondrial apoptosis pathway and activation of
Nrf2-Antioxidant response in beta-amyloid-challenged SH-SY5Y cells.
Molecules. 24:E26872019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kordasiewicz HB, Thompson RM, Clark HB and
Gomez CM: C-termini of P/Q-type Ca2+ channel alpha1A subunits
translocate to nuclei and promote polyglutamine-mediated toxicity.
Hum Mol Genet. 15:1587–1599. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tsoi H, Lau TC, Tsang SY, Lau KF and Chan
HY: CAG expansion induces nucleolar stress in polyglutamine
diseases. Proc Natl Acad Sci USA. 109:13428–13433. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mizuguchi T, Suzuki T, Abe C, Umemura A,
Tokunaga K, Kawai Y, Nakamura M, Nagasaki M, Kinoshita K, Okamura
Y, et al: A 12-kb structural variation in progressive myoclonic
epilepsy was newly identified by long-read whole-genome sequencing.
J Hum Genet. 64:359–368. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tonin R, Catarzi S, Caciotti A, Procopio
E, Marini C, Guerrini R and Morrone A: Progressive myoclonus
epilepsy in gaucher disease due to a new Gly-Gly mutation causing
loss of an exonic splicing enhancer. J Neurol. 266:92–101. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
He J, Lin H, Li JJ, Su HZ, Wang DN, Lin Y,
Wang N and Chen WJ: Identification of a novel homozygous
splice-site mutation in SCARB2 that causes progressive myoclonus
epilepsy with or without renal failure. Chin Med J (Engl).
131:1575–1583. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Algahtani H, Shirah B, Algahtani R,
Al-Qahtani MH, Abdulkareem AA and Naseer MI: A novel mutation in
CACNA1A gene in a Saudi female with episodic ataxia type 2 with no
response to acetazolamide or 4-aminopyridine. Intractable Rare Dis
Res. 8:67–71. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gerhardstein BL, Gao T, Bunemann M, Puri
TS, Adair A, Ma H and Hosey MM: Proteolytic processing of the C
terminus of the alpha(1C) subunit of L-type calcium channels and
the role of a proline-rich domain in membrane tethering of
proteolytic fragments. J Biol Chem. 275:8556–8563. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gomez-Ospina N, Tsuruta F, Barreto-Chang
O, Hu L and Dolmetsch R: The C terminus of the L-type voltage-gated
calcium channel Ca(V)1.2 encodes a transcription factor. Cell.
127:591–606. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Park D, Kim SH, Lee YJ, Song GJ and Park
JS: A novel CACNA1A mutation associated with episodic ataxia 2
presenting with periodic paralysis. Acta Neurol Belg. 118:137–139.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Walker FO: Huntington's disease. Lancet.
369:218–228. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Miyata R, Hayashi M, Tanuma N, Shioda K,
Fukatsu R and Mizutani S: Oxidative stress in neurodegeneration in
dentatorubral-pallidoluysian atrophy. J Neurol Sci. 264:133–139.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Sanchez I, Xu CJ, Juo P, Kakizaka A,
Blenis J and Yuan J: Caspase-8 is required for cell death induced
by expanded polyglutamine repeats. Neuron. 22:623–633. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tien CL, Wen FC and Hsieh M: The
polyglutamine-expanded protein ataxin-3 decreases bcl-2 mRNA
stability. Biochem Biophys Res Commun. 365:232–238. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Maiuri T, Suart CE, Hung CLK, Graham KJ,
Barba Bazan CA and Truant R: DNA damage repair in huntington's
disease and other neurodegenerative diseases. Neurotherapeutics.
16:948–956. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hwang JY, Park JH, Kim MJ, Kim WJ, Ha KT,
Choi BT, Lee SY and Shin HK: Isolinderalactone regulates the
BCL-2/caspase-3/PARP pathway and suppresses tumor growth in a human
glioblastoma multiforme xenograft mouse model. Cancer Lett.
443:25–33. 2019. View Article : Google Scholar : PubMed/NCBI
|