Introduction
Colorectal cancer accounts for approximately 10% of
all tumors diagnosed each year and cancer-related deaths (1). It is the fourth most deadly cancer
with approximately 900,000 colorectal cancer related-deaths
annually worldwide (2). The risk
factors of colorectal cancer include an unhealthy diet (especially
excessive consumption of red and processed meats and alcohol),
obesity, tobacco smoking, and lack of physical activity as well as
genetic (familial adenomatous polyposis and hereditary
non-polyposis colon cancer) and pre-cancer conditions such as
inflammatory bowel diseases (Crohn's disease and ulcerative
colitis) (3). Endoscopy is the most
useful clinic tool for early detection and diagnosis of colorectal
cancer while computed tomography (CT), magnetic resonance imaging
(MRI), and positron-emission tomography (PET) are helpful for
detecting colorectal cancer metastasis (3).
Treatment of colorectal cancer, like most other
malignancies, comprises surgical resection of tumor lesions and
adjuvant or neoadjuvant chemoradiotherapy or palliative care. These
treatments depend on the tumor stage (4,5), while
recent immunotherapy and molecularly targeted therapy are also used
to successfully treat colorectal cancer (6,7).
However, the prognosis of colorectal cancer remains unfavorable,
especially at the advanced stages of disease (8–10).
Therefore, a better understanding of colorectal carcinogenesis and
molecular mechanisms could aid in more effectively preventing the
start or progression of colorectal cancer clinically.
Tight junctions are the main structures that
maintain intercellular connections for cell-cell adhesion,
intercellular spaces, and conservation of solutions and other
macromolecules through the paracellular pathway (11). To date, at least 40 different
proteins have been identified that play roles in the tight
junctions. Most of these proteins belong to the three major
transmembrane protein groups: Occludin, claudins, and junction
adhesion molecules (JAM) (12).
Accumulating evidence has revealed that aberrant
expression of the tight junction proteins contributes to the
development and metastasis of numerous epithelial-derived cancers
(13). Tricellulin, also known as
MARVELD2, was the first identified protein to be exclusively
localized at the tri-epithelial junctions and to be revealed to
regulate the junctional tension of epithelial cells (14) and provide a fulcrum for various
epithelial stereo structures (15).
The adhesion between epithelial cells is enhanced when tricellulin
expression increases, whereas the tight junction function of these
cells is lost when tricellulin is deleted (16). A previous study reported that
nuclear tricellulin expression was associated with poorly
differentiated pancreatic cancer and invasion of pancreatic cancer
cells (17). To date, few studies
have investigated the role of tricellulin in human cancers.
In the present study, first, tricellulin expression
was analyzed in colorectal cancer tissues to determine its
association with clinicopathological features of colorectal cancer
patients and then the underlying molecular events were investigated
via quantitative proteomic analysis and in vitro
experiments. The aim of the present study was to provide novel and
valuable information regarding the detection of tricellulin as a
biomarker for colorectal cancer.
Materials and methods
Patients and tissue samples
The present study was approved by the Ethics
Committee of the First Affiliated Hospital of Guangxi Medical
University (Nanning, China) with the approval no. BBMCEC2012063 and
was conducted in accordance with the guidelines of the Helsinki
Declaration. The patient cohort of 98 colorectal cancer tissues and
15 normal tissues were collected from September 2013 and September
2014 at The First Affiliated Hospital of Guangxi Medical University
where colorectal cancer was histologically diagnosed and staged
according to the National Comprehensive Cancer Network (NCCN)
colorectal cancer classifications (18). These 98 patients included 60 (61.2%)
males and 38 (38.8%) females and their ages ranged from 25 to 83
years with a mean age of 56 years. There were 36 tumors located in
the left-sided colon and 62 in the right-side colorectum. Four
tumors were mucinous adenocarcinoma and 94 were tubular
adenocarcinoma and there were 10 patients at stage I, 35 at stage
II, 26 at stage III, and 27 at stage IV of the disease.
Immunohistochemistry (IHC)
For IHC the sections (4–5 µm) were deparaffinized in
xylene and rehydrated in a series of ethanol solutions and then
immunostained following the procedures of a previous study
(19) with a primary antibody
against human tricellulin (cat. no. SAB1306444; Sigma-Aldrich;
Merck KGaA) at a dilution of 1:100. The immunoreactivity score
(IRS) system as described in a previous study (20) was used to semi-quantify the
immunostaining data, i.e., the staining intensity (SI) × the
percentage of positive cells.
Cell culture, infection, and
treatment
Human colorectal adenocarcinoma cell lines HT29,
HCT116, RKO, SW620, Caco2 and HCT-8 and a normal human colon
mucosal epithelial cell line NCM460 were obtained from the Cell
Resource Center, Shanghai Institute of Biochemistry and Cell
Biology at the Chinese Academy of Sciences (Shanghai, China). The
cell lines had been validated by performing short tandem repeat
(STR) profiling. The cells were cultured in RPMI-1640 medium
(Corning, Inc.) supplemented with 10% FBS (Biological Industries)
in a humidified incubator containing 5% CO2 at 37°C.
A tricellulin shRNA (targeting sequences of
5′-GATGAGCAGATTGCCACATCA-3′) was cloned into a pcDNA6. 3-EGFP
vector (Invitrogen; Thermo Fisher Scientific, Inc.). The vector or
negative control vector (lenti-EGFP-miR) was used to generate
lentiviruses in 293 cells, and the lentiviruses were used to infect
HCT116 cells and named as tricellulin knocked down cells (TRIC-KD)
or negative control cells (NC). In addition, lentiviral vector
carrying human tricellulin cDNA (lenti-tricellulin-IRES-EGFP) or
vector-only was used to infect HCT116 cells for
tricellulin-overexpression (TRIC-OE) in cells. The efficiency of
tricellulin knockdown or overexpression was confirmed by RT-qPCR
and western blotting. To determine the effects of tricellulin on
regulation of the canonical NF-κΒ pathway, we treated these stable
cells or parental cells (CON) with 50 ng/ml TNF-α (R&D Systems)
or 100 µM pyrrolidinecarbodithioic acid (PDTC; Sigma-Aldrich; Merck
KGaA) for 24 h for subsequent experiments.
Immunofluorescence and phalloidin
staining
Fresh colorectal cancer tissues were cryosectioned
and fixed in 4% paraformaldehyde and then immunostained following
the procedures of a previous study (21) with a primary antibody against
tricellulin (product no. 48-8400; Invitrogen; Thermo Fisher
Scientific, Inc.) at a dilution of 1:200 at 4°C overnight.
Subsequently, the sections were washed and incubated with the
respective secondary antibody (Alexa Fluor® 594 goat
anti-rabbit; product no. 8889S; 1:1,000; Cell Signaling Technology,
Inc.) and 4′,6-diamidino-2-phenylindole (DAPI) (Wuhan Boster
Biological Technology, Ltd.).
To visualize the cell actin cytoskeleton,
FITC-Phalloidin was utilized to stain HCT116 cells (NC, TRIC-KD,
and TRIC-OE). In brief, cells (1×104) were seeded onto
coverslips and grown overnight and then fixed and permeabilized in
the same way as the cryosections. The cells were next incubated
with the Alexa Fluor® 488 Phalloidin working solution
(Beijing Solarbio Science & Technology, Inc.) for 30 min at
room temperature in the dark and then with DAPI for 10 min. Images
were obtained from the stained cells with a fluorescence Olympus
BX53F microscope (magnification, ×600; Olympus Corporation).
RNA isolation and reverse
transcription (RT)-quantitative (q)PCR
Total cellular RNA was isolated from cells using the
Eastep™ Super Total RNA Extraction Kit and reversely transcribed
into cDNA using a Reverse Transcription Kit (both from Promega
Corporation). qPCR was performed using the Power SYBR Green Master
Mix (Promega Corporation) on an 7500 ABI Real-time PCR system
(Applied Biosystems; Thermo Fisher Scientific, Inc.). The primers
used were tricellulin forward, 5′-CCAGCTATAGCGCCAGATCTCAA-3′ and
reverse, 5′-CAGACACCGGCTTATCCCATTC-3′; and GAPDH forward,
5′-GCACCGCAAGGCTGAGAAC-3′ and reverse, 5′-TGGTGAAGACGCCAGTGGA-3′,
which were all obtained from Takara Bio, Inc.. The cycling
parameters were as follows: Denaturing at 95°C for 10 min, 40
cycles of denaturing at 95°C for 15 sec, primer annealing at 60°C
for 1 min and extension temperature at 95°C for 15 sec; final
extension at 60°C for 15 sec and final denaturing at 95°C for 15
sec. Results were quantified by using the 2−ΔΔCq method
(22).
Western blotting
Cells were lysed using the RIPA lysis buffer
(Beyotime Institute of Biotechnology) and quantified by using the
bicinchoninic acid (BCA) protein assay kit (Beyotime Institute of
Biotechnology). Equal amounts (30 µg/lane) of protein samples were
separated in 10 or 12% sodium dodecyl sulfate-polyacrylamide gel
electrophoresis (SDS-PAGE) gels and transferred onto polyvinylidene
fluoride membranes (PVDF; EMD Millipore).
Western blotting was performed according to our
previous study (23); after being
blocked in a 5% skim milk for 1 h at the room temperature, the
membranes were incubated with a primary antibody against GAPDH
(cat. no. 60004-1-Ig; 1:3,000), MMP2 (cat. no. 10373-2-AP; 1:800),
vimentin (cat. no. 10366-1-AP; 1:500), Snail (cat. no. 13099-1-AP;
1:500), caspase-3 (product no. 19677-1-AP; 1:800) (all from
ProteinTech Group, Inc.), tricellulin (cat. no. 48-8400; 1:800;
Invitrogen; Thermo Fisher Scientific, Inc.), p65 (product no.
8242S; 1:1,000), p-p65 Ser536 (product no. 3033; 1:1,000),
E-cadherin (product no. 3195; 1:1,000), survivin (product no.
2808S; 1:1,000), or Bcl-2 (product no. 15071S; 1:1,000) (all from
Cell Signaling Technology, Inc.) overnight at 4°C. Subsequently,
the membranes were incubated with a secondary antibody (1:10,000;
LI-COR IRDye® 680RD Goat anti-Rabbit P/N 926-68071 or
LI-COR IRDye® 680RD Goat anti-Mouse P/N 926-68070;
LI-COR Biosciences) at room temperature for 1 h. The protein bands
were quantified by using Odyssey infrared imaging (LI-COR
Biosciences). The final data were semi-quantification was carried
out using ImageJ software (version 1.50i; National Institutes of
Health).
Transwell migration and invasion
assays
The cells (1×105) were added to the upper
chamber of a Transwell (8-µm pores) with noncoated membranes
(migration assay) or Matrigel-coated Transwell chambers (invasion
assay) with serum-free medium, while a cell culture medium
containing 20% FBS was added to the bottom chambers. After 24 or 48
h culture, the migrated or invasive cells at the bottom of the
membrane were fixed in 4% methanol for 20 min at room temperature,
and stained using 0.1% crystal for 10 min at room temperature, and
counted from five-randomly selected fields under the 100X objective
of an inverted microscope (Nikon Corporation).
Hoechst 33258 staining
Cells were seeded into a 24-well flat bottom plate
at a density of 1×105 cells/well and grown for 24 h.
Next, the cells were washed with phosphate-buffered saline (PBS),
fixed in 4% methanol, and incubated with Hoechst 33258 stain
solution (Beijing Solarbio Science & Technology, Inc.) for 10
min at room temperature. The cell apoptotic morphology was observed
under an Olympus BX53F fluorescence microscope (magnification,
×600).
Tandem mass tag (TMT)-labeling and
liquid chromatography-mass spectrometry (LC-MS/MS)
TMT-LC-MS/MS was performed by R&S Biotechnology
Co., Ltd.. In brief, total protein was extracted from cells and
processed in a 6-plex TMT labeling kit (Thermo Fisher Scientific,
Inc.) according to a procedure of a previous study (24). Fractionation was then carried out by
basic pH reverse-phase liquid chromatography with fraction
combining according to a previous study (25). The separated samples were loaded
onto the Nano-Aquity UPLC system (Waters Corporation) and the
resulting peptide mixture was loaded onto a trap column (2.1×150 mm
X Bridge BEH300; Waters Corporation) attached to an analytical
column (ZORBAX 300SB-C18 column, 5 µm, 300 Å, 0.1×150 mm; Microm
for mass-spectrometer analysis (26). The linear gradient was from 2% D to
80% D in 90 min (solution D: 0.1% formic acid in ACN) and the
triple TOF 5600 MS, switched between MS and serial (MS/MS)
acquisition. The MS data were acquired using a spectral
accumulation time of 250 msec in the mass range of 350-1,300 m/z.
The scanning tandem mass spectrometry was from 100–1250 m/z with
rolling collision energy. The 20 strongest precursors were selected
according to the procedure of a previous study (26).
Identification of differentially
expressed proteins and enrichment analysis in tricellulin
overexpression (TRIC-OE) or NC cells
The data were then analyzed using the free online
platform of Majorbio I-Sanger Cloud Platform (www.i-sanger.com). The ratio cutoff values were set to
1.2 and 0.8 for fold-change and more than two unique peptides were
used for protein identification. A P-value <0.05 was considered
as the threshold to identify significant changes. We then performed
Gene Ontology (GO) (27) and used
the Kyoto Encyclopedia of Genes and Genomes (KEGG) (28) database to identify the functions or
interactions between these differentially expressed proteins (DEPs)
in colorectal cancer cells.
Protein-protein interaction (PPI)
network analysis
A PPI network was constructed using a Search Tool
[STRING (29); http://string.embl.de/]. A confidence score of 0.4 was
set as the cut-off criterion and the DEPs with connection numbers
10 were considered as hub genes. The top 10 hub nodes with higher
degrees were screened using R (version 3.6.0, http://cran.r-project.org/).
Statistical analysis
All data were presented as the means ± SD and
statistically analyzed using the SPSS v17.0 software package (IBM
Corp.) and GraphPad Prism v7.0 (GraphPad Software, Inc.). The
association between tricellulin expression and clinicopathological
characteristics of colorectal cancer patients was analyzed using
the Chi-squared (χ2) test, while the survival rates
stratified by tricellulin expression were calculated by
Kaplan-Meier curves and the log-rank test. Unpaired Student's
t-tests and one-way ANOVA tests were used to determine statistical
significance between two groups or among more than two groups,
respectively. After a one-way ANOVA test was performed, for post
hoc evaluation, Tukey's HSD test was used. A P-value <0.05 was
considered to indicate a statistically significant difference.
Results
Tricellulin upregulation and
association with poor colorectal cancer prognosis
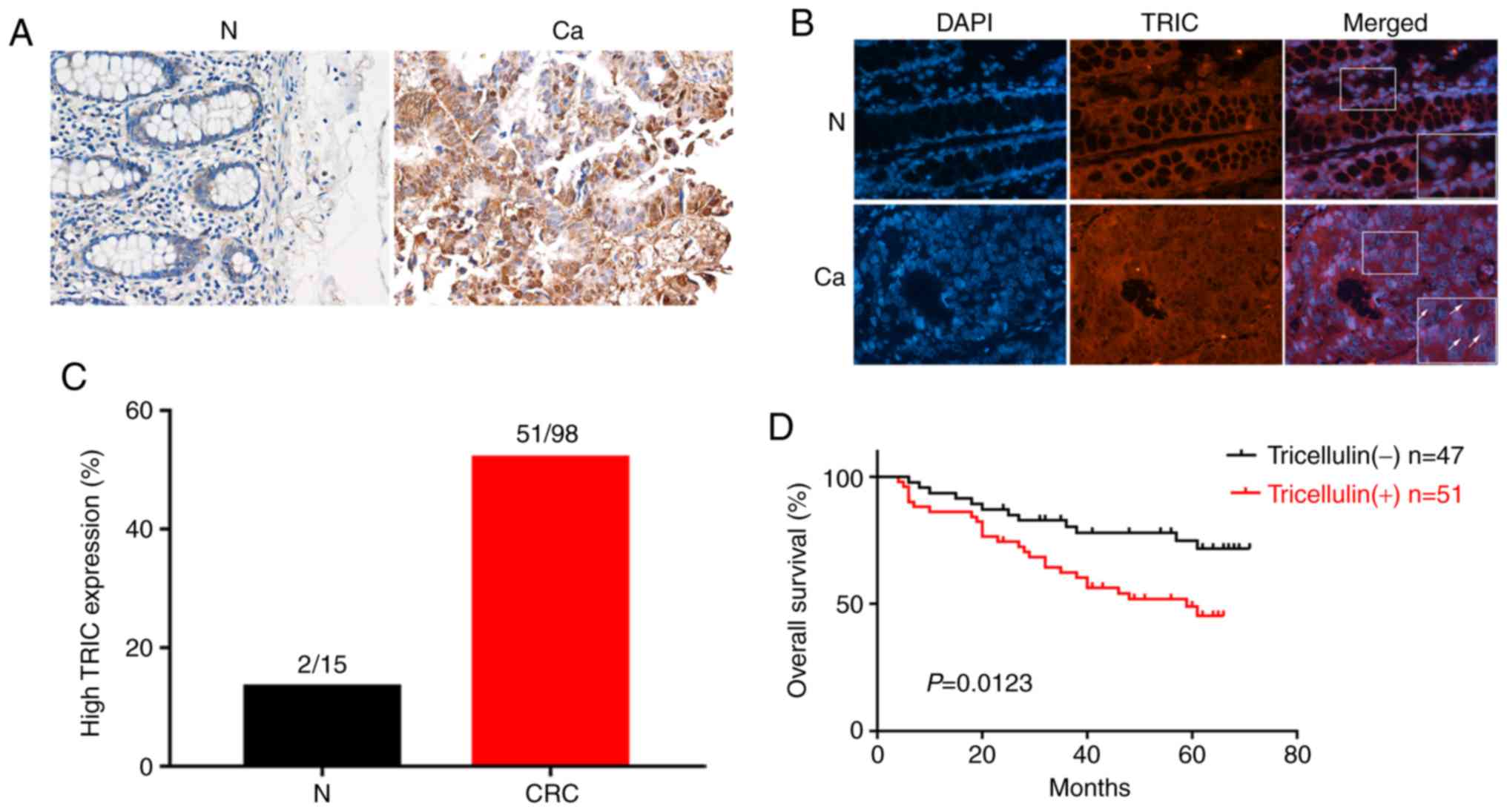
Tricellulin expression was analyzed in 98 cases of
colorectal cancer tissues vs. 15 normal mucosae and the results
revealed that tricellulin expression was significantly upregulated
in colorectal cancer tissues compared to normal mucosae (Fig. 1A). Tricellulin was mainly expressed
in the cytoplasm or nucleus in colorectal cancer tissues, which was
quite different from that of normal mucosae in the cytoplasm
(Fig. 1B). Of these 98 cancer
cases, 51 (52.0%) had tricellulin overexpression vs. 2 (13.3%) with
tricellulin expression among the 15 normal mucosae (Fig. 1C).
Then, the association of tricellulin expression with
clinicopathological characteristics of colorectal cancer patients
was assessed and it was revealed that tricellulin expression
(52.0%; 51/98) was associated with tumor distant metastasis (M
stage) and advanced tumor-lymph node metastasis (TNM stage)
(Table I), but not with the sex and
age, tumor size, and N stage of patients. The Kaplan-Meier curves
and the long rank test revealed that the upregulated tricellulin
expression was associated with shorter overall survival (OS;
P=0.012; Fig. 1D).
 | Table I.Association of tricellulin expression
with clinicopathological features from colorectal patients. |
Table I.
Association of tricellulin expression
with clinicopathological features from colorectal patients.
|
| Tricellulin
expression |
|
|---|
|
|
|
|
|---|
|
Characteristics | Negative | Positive | P-value |
|---|
| Sex |
|
| 0.202 |
|
Male | 33 | 28 |
|
|
Female | 16 | 23 |
|
| Age (years) |
|
| 0.817 |
|
≥65 | 13 | 12 |
|
|
<65 | 37 | 38 |
|
| Treatment |
|
| 0.249 |
|
Surgery | 21 | 17 |
|
|
Comprehensive | 26 | 34 |
|
| N stage |
|
| 0.220 |
| N0 | 30 | 25 |
|
|
N1+N2 | 19 | 26 |
|
| M stage |
|
| 0.025 |
| M0 | 39 | 30 |
|
| M1 | 10 | 21 |
|
| TNM stage |
|
| 0.004 |
|
I+II | 30 | 17 |
|
|
III+IV | 19 | 34 |
|
Effects of tricellulin knockdown and
overexpression on regulation of colorectal cancer cell malignant
phenotypes in vitro
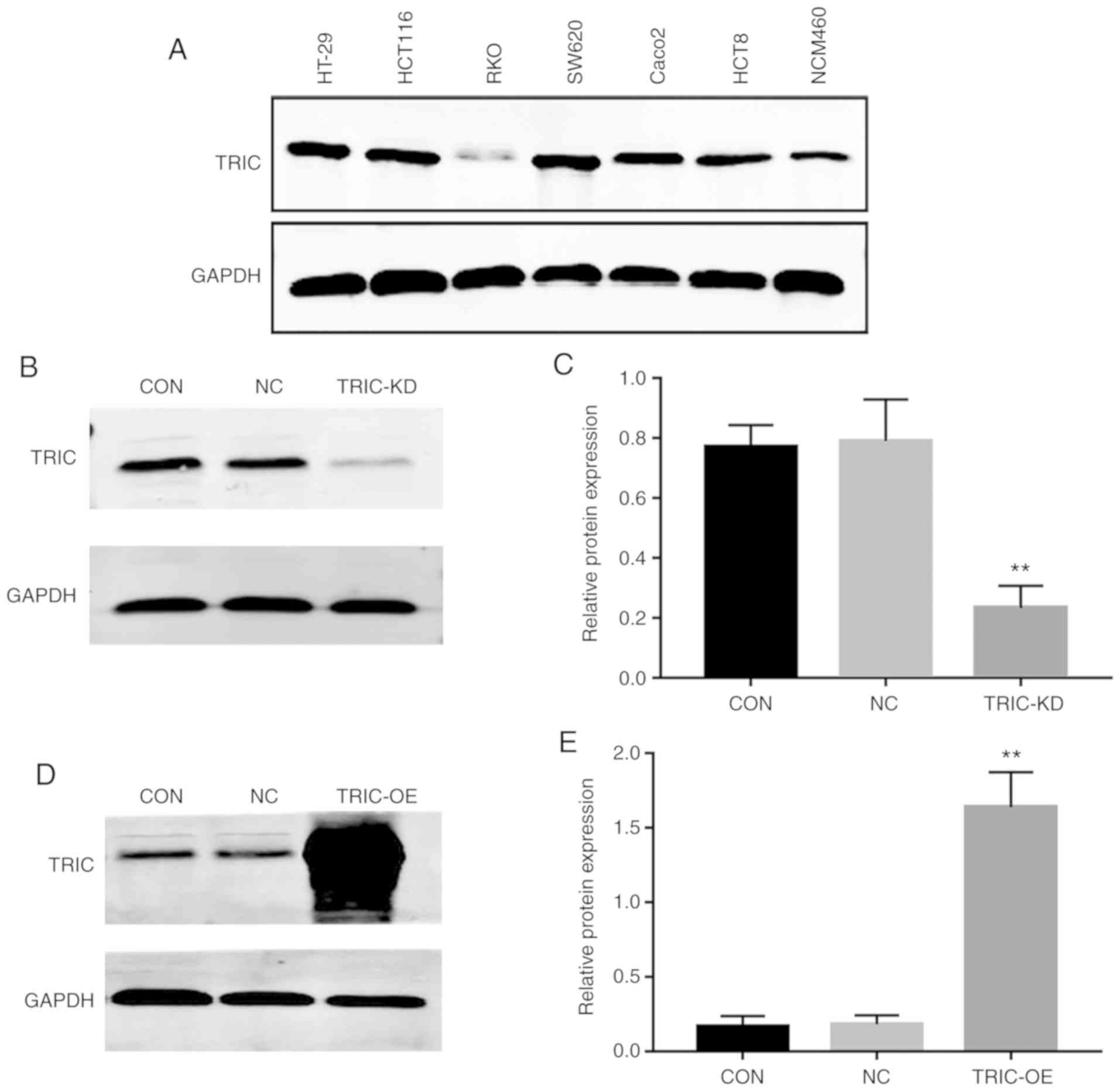
To assess the effects of tricellulin knockdown and
overexpression on regulation of colorectal cancer cell malignant
phenotypes in vitro, tricellulin expression was first
analyzed in human colorectal adenocarcinoma cell lines HT-29,
HCT116, RKO, SW620, Caco2, and HCT8 as well as a normal human colon
mucosal epithelial cell line, NCM460. It was revealed that the
level of tricellulin protein was higher in HT29, HCT116, and SW620
cells than in other all cancerous and normal cells (Fig. 2A). We therefore selected HCT116 with
moderate endogenous expression of tricellulin for stable
tricellulin knockdown or overexpression. Tricellulin knockdown
(TRIC-KD) or overexpression (TRIC-OE) was confirmed by RT-qPCR and
western blotting (Fig. 2B-E).
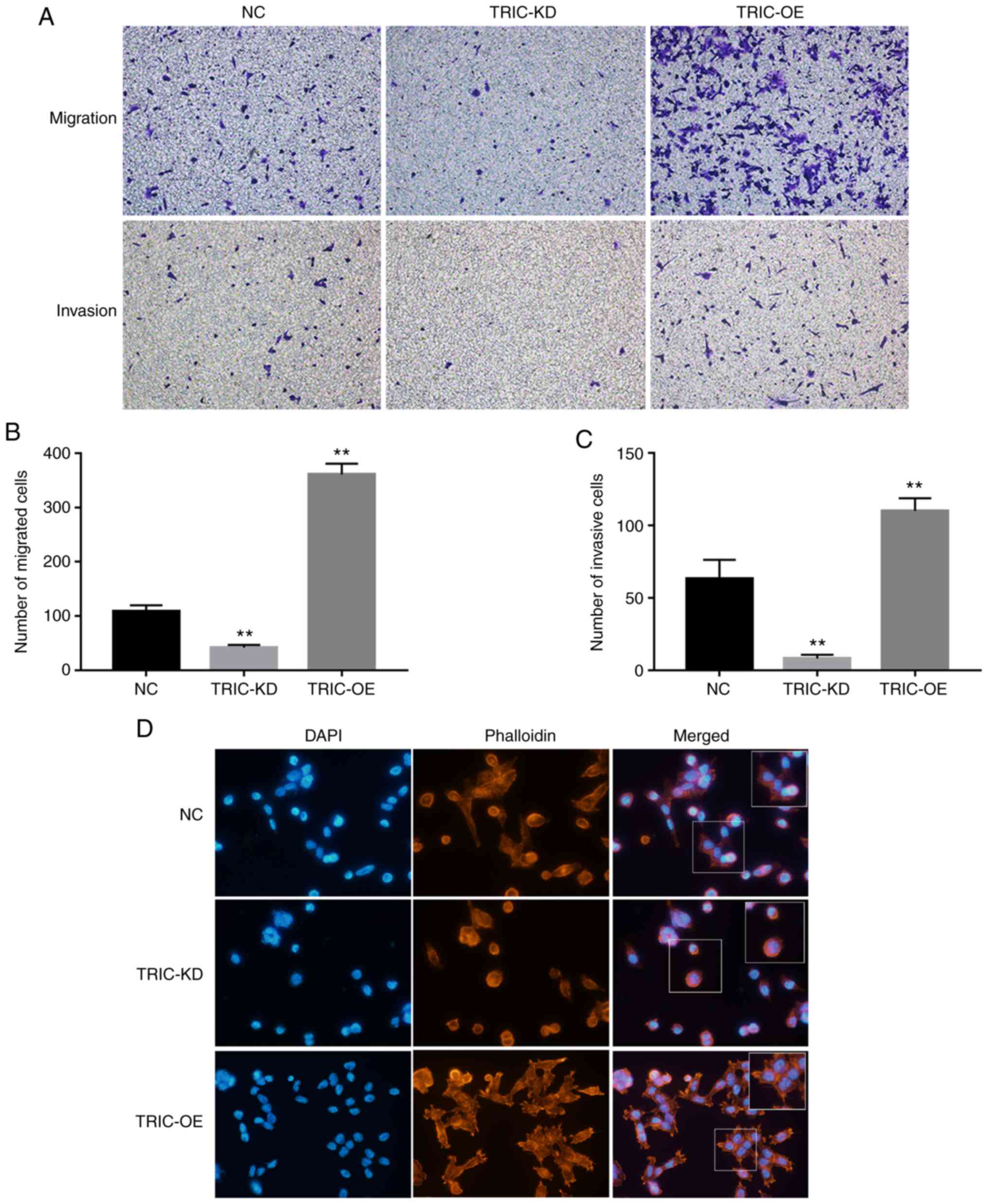
Next, it was determined that TRIC-OE significantly
increased HCT116 cell migration and invasion capacities, whereas
TRIC-KD reduced tumor cell migration and invasion (Fig. 3A-C). The FITC-Phalloidin staining
revealed that tricellulin regulated pseudopodium formation and
cytoskeletal reorganization; downregulation of tricellulin
expression notably inhibited the length and number of pseudopodia
and stress fibers. In contrast, upregulation of tricellulin
expression increased the length and number of pseudopodia and
stress fibers (Fig. 3D).
Tricellulin regulation of
differentially expressed proteins in colorectal cancer cells by
quantitative proteomic analysis
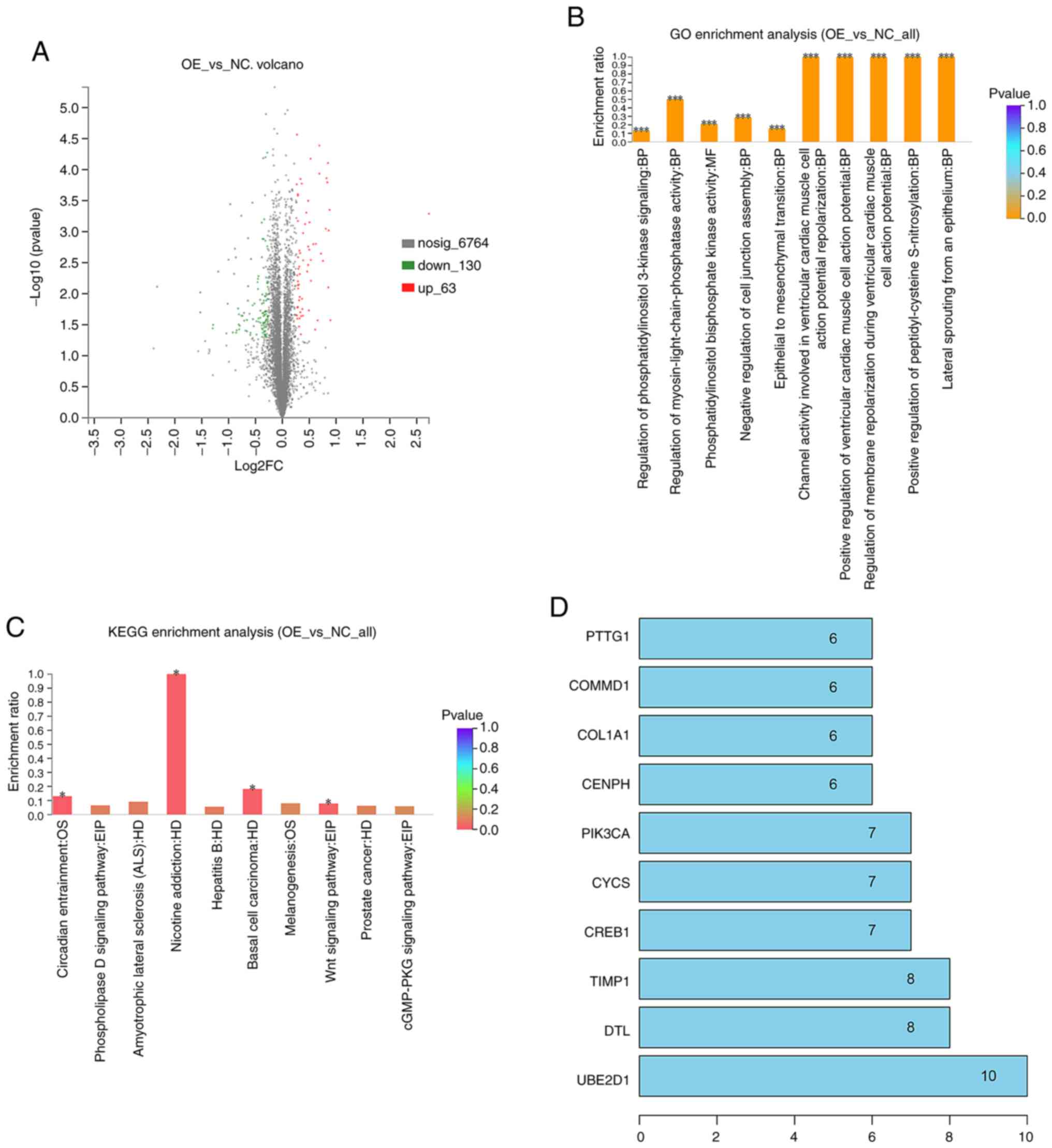
To elucidate the potential molecular mechanisms
underlying tricellulin, quantitative proteomic analysis was
performed to profile DEPs in the TRIC-OE group vs. the NC group. We
identified a total of 6,957 unique proteins in the TRIC-OE group
and 193 were DEPs vs. the NC group, in which 63 were upregulated
and 130 were downregulated. The DEPs volcano plots are presented in
Fig. 4A.
To determine their biological and functional
properties in the TRIC-OE compared to the NC group, GO was
performed and the KEGG database was used. The GO data revealed that
tricellulin could regulate the biological process (BP), molecular
function (MF), and cellular component (CC). These DEPs were
significantly enriched in regulation of the ventricular cardiac
muscle cell action potential, peptidyl-cysteine S-nitrosylation,
lateral sprouting from an epithelium, epithelial-mesenchymal
transition (EMT), and phosphatidylinositol 3-kinase signaling
(Fig. 4B). The KEGG analysis
revealed that DEPs were mainly enriched in nicotine addiction,
basal cell carcinoma, Wnt signaling pathway, circadian entrainment,
and pathways in cancer (Fig.
4C).
The PPI network data with the confidence score set
at 0.4 revealed that the hub genes with higher degrees included
UBE2D1, DTL, TIMP1, CREB1, CYCS, PIK3CA, CENPH, COL1A1, COMMD1, and
PTTG1 (Fig. 4D).
Tricellulin regulation of colorectal
cancer cell EMT via the canonical nuclear
factor-κ-light-chain-enhancer of activated B cells (NF-κB)
EMT, a key developmental regulatory program, plays a
critical role in the promotion of cancer progression and metastasis
in human carcinomas (30). In our
previous study, we demonstrated that the NF-κB signaling was
closely associated with colorectal cancer cell EMT (31). Thus, we further determined the role
of the NF-κB pathway in mediating the tricellulin oncogenic
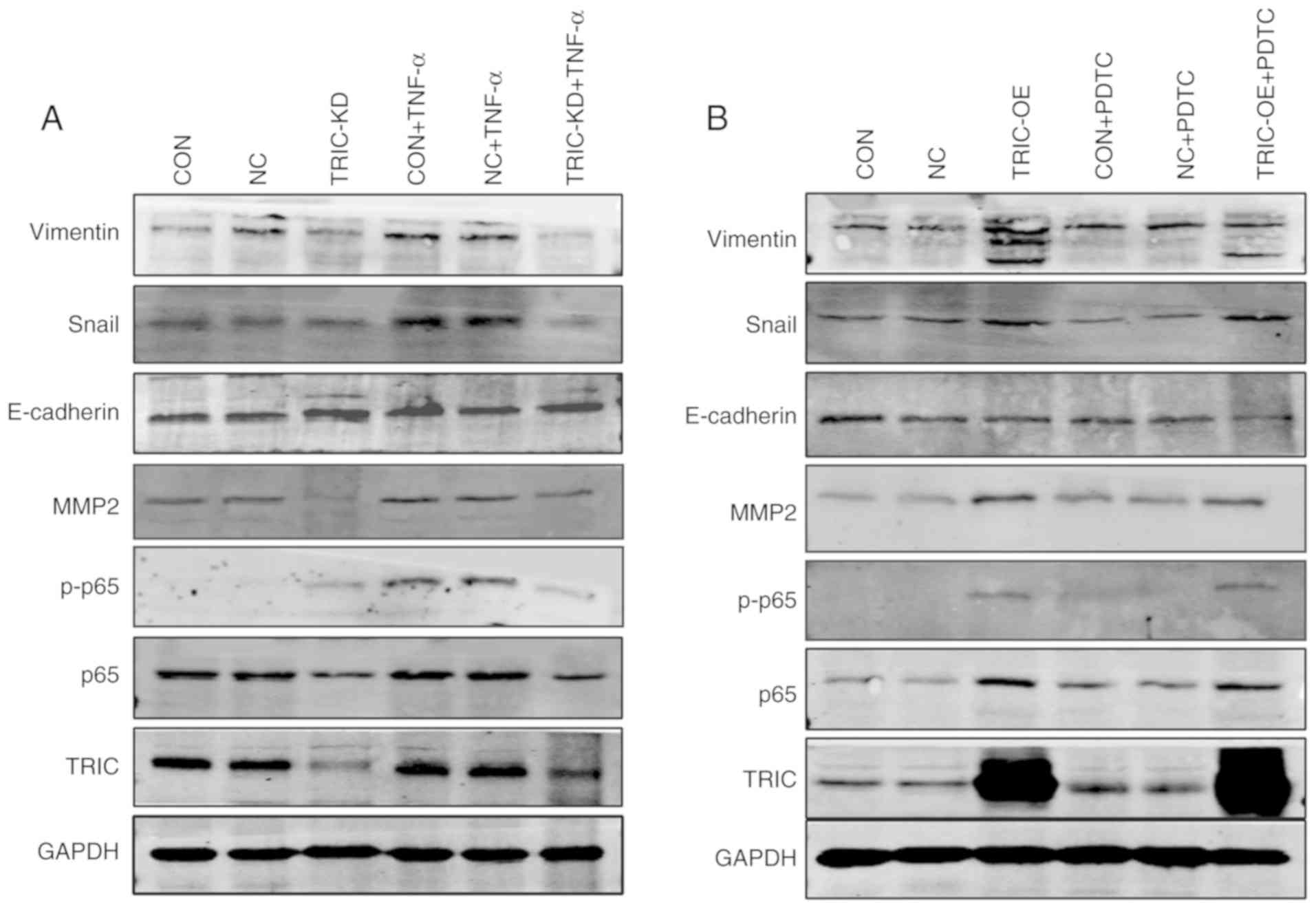
activities in colorectal cancer cells. The expression of p65,
p-p65, mesenchymal markers (N-cadherin, Snail and vimentin), and an
epithelial marker (E-cadherin) was analyzed in HCT116 cells after
tricellulin overexpression of knockdown. The data revealed that
tricellulin overexpression upregulated the levels of vimentin and
Snail but downregulated E-cadherin, which was not completely
inhibited by an NF-κB inhibitor PDTC (Fig. 5A). In contrast, knockdown of
tricellulin expression decreased vimentin and Snail, but increased
E-cadherin, which was also not completely activated by an NF-κB
activator TNF-α (Fig. 5B). These
results indicated that tricellulin may regulate the EMT in
colorectal cancer cell through the canonical NF-κB signaling
pathways.
Tricellulin regulation of HCT116 cell
apoptosis in vitro
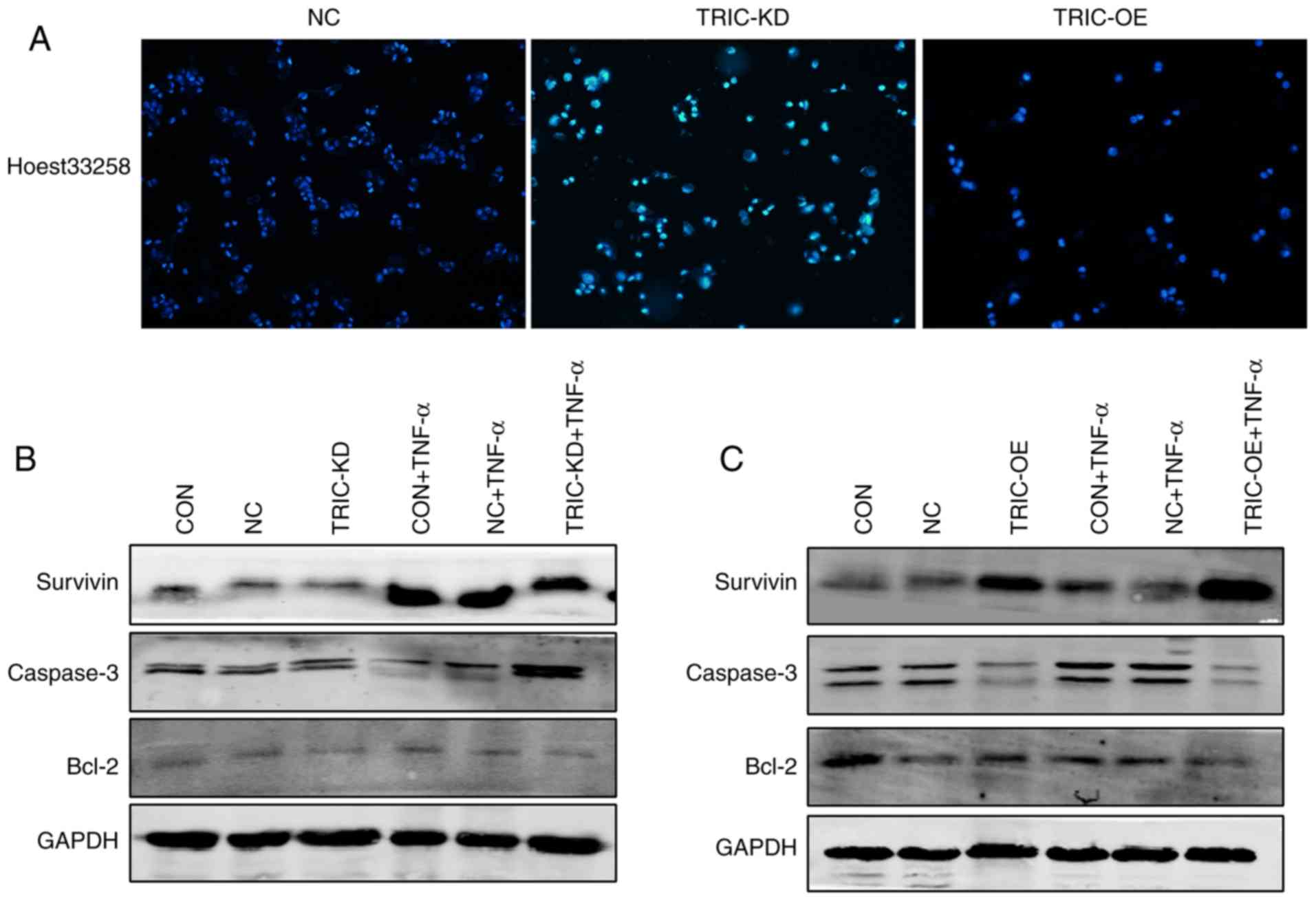
Hoechst 33258 staining and western blotting were
first performed to further assess the role of tricellulin in
apoptosis. The present data revealed that the apoptotic cells had
significant nuclear condensation and morphological changes in
TRIC-KD cells compared to the negative control group (NC).
Conversely, tricellulin overexpression (TRIC-OE) had normal nuclear
morphology (Fig. 6A). Assessment of
the levels of apoptosis-related proteins revealed that tricellulin
overexpression reduced caspase-3 and increased Bcl-2 and survivin
levels, whereas knockdown of tricellulin expression reduced Bcl-2
and survivin levels and enhanced caspase-3 levels (Fig. 6B and C). These data indicated that
tricellulin enhanced tumor cell survival.
Discussion
Aberrant expression and functions of the tight
junction-related proteins contribute to the development of various
human cancers (32). Tricellulin
localizes at tricellular tight junctions as a member of the
tight-junction-associated MARVEL protein (TAMP) family (33). Nuclear or cytoplasmic tricellulin
expression was associated with pancreatic cancer cell invasion and
metastasis as well as a poor prognosis (17).
To date, there is no study revealing the role of
tricellulin in colorectal cancer. Thus, the present study was the
first, to the best of our knowledge, to demonstrate tricellulin
overexpression in colorectal cancer tissues with both cytoplasmic
and nuclear localizations as well as tricellulin expression
associated with colorectal cancer lymph node and distant metastasis
and poor prognosis, which is consistent with those occurring in
pancreatic cancer (17).
Using in vitro functional assays, it was
revealed that knockdown of tricellulin expression inhibited
colorectal cancer cell invasion and migration, whereas tricellulin
overexpression enhanced tumor cell invasion and migration. Our
later experiments also showed that tricellulin expression
upregulated actin and cytoskeletal reorganization and EMT in HCT116
cells. In fact, EMT drives carcinoma progression and plays an
essential role in metastasis (34).
Moreover, multiple signaling pathways are involved in EMT
development (35); for example, the
NF-κB signaling is involved in EMT regulation (31,36).
The present study further confirmed the involvement
of the NF-κB signaling pathway in the regulation of
tricellulin-induced colorectal cancer cell EMT in vitro. In
addition, the GO data revealed that tricellulin could regulate the
BP, MF, CC, and EMT, while the PPI network data revealed that the
hub genes included UBE2D1, DTL, TIMP1, CREB1, CYCS, PIK3CA, CENPH,
COL1A1, COMMD1, and PTTG1. However, further studies are required to
fully reveal the functions and regulations of tricellulin in
colorectal cancer development and progression. In fact, the cell
tight-junction helps to hold cells together and enable cell-cell
communication. It also acts a barrier so that cell polarity is
maintained and it prevents the lateral diffusion of integral
membrane proteins and molecules and loss of ions (3). For example, knockdown of tricellulin
expression using RNAi compromised the epithelial barrier and
tricellular contacts and disorganized the bicellular tight-junction
(37).
It remains unknown how tricellulin overexpression
dysregulated cancer cell invasion and metastasis, including
colorectal cancer. A previous study revealed that nuclear
tricellulin possessed an oncogenic activity in pancreatic cancer
(17). Knockdown of the tricellular
tight junction protein lipolysis-stimulated lipoprotein receptor
(LSR) expression induced tricellulin relocalization from the
tricellular region to the bicellular region at the membrane to
promote endometrial cancer cell proliferation, migration, and
invasion (38). However,
tricellulin expression revealed a significant negative correlation
with differentiation in pancreatic ductal adenocarcinoma (38).
Tricellulin expression was associated with favorable
prognosis in human hepatoblastoma patients (39). In normal gastric mucosa, tricellulin
was localized at the tricellular tight junction, whereas
tricellulin was distributed in the cytoplasm of gastric cancer
cells (40). Transduction of Snail
reduced the level of tricellulin and E-cadherin but increased
vimentin and N-cadherin, thus indicating that suppression of
tricellulin expression mediated Snail-induced EMT in human gastric
cancer cells (40). However,
another previous study revealed tricellulin overexpression and an
association with poor prognosis of hepatocellular carcinoma (HCC),
although higher grades of intrahepatic cholangiocarcinoma revealed
decreases in tricellulin expression, which was correlated with poor
prognosis (41). The present data
are consistent with these latter studies.
Collectively, tricellulin expression and cellular
localization have different roles in different human cancers
(17,38,40).
Furthermore, NF-κB is a protein complex and functions to regulate
gene transcription, cytokine production, and cell survival
(42,43). In human cancers, NF-κB activity is
frequently induced for tumor cell proliferation and survival
(43,44).
In the present study, it was revealed that
tricellulin overexpression upregulated levels of vimentin and Snail
but downregulated E-cadherin, which was not completely inhibited by
an NF-κB inhibitor PDTC. In contrast, knockdown of tricellulin
expression decreased vimentin and Snail, but increased E-cadherin,
which was also not completely activated by an NF-κB activator
TNF-α, thus indicating that tricellulin may regulate the EMT in
colorectal cancer cells through the canonical NF-κB signaling
pathway. In fact, a previous study revealed that upregulation of
TRIC, p-JNK and p-IκB was observed after treatment of human
pancreatic cancer cell lines HPAC with IL-1β, TNF-α and IL-1α for
24 h, whereas change in phospho-IκB was inhibited by JNK and NF-κB
inhibitors. However, after treating hTERT-transfected human
pancreatic duct epithelial cells (hTERT-HPDE cells) with
IL-1β,TNF-α, and IL-1α for 24 h, tricelluin expression was
upregulated (45).
To date, no studies have shown whether tricellulin
regulated EMT, thereby affecting invasion and metastasis of
colorectal cancer via the NF-κB signaling pathway. A previous study
reported that NF-κB induced the expression of anti-apoptotic genes,
inhibited the expression of apoptotic proteins, and regulated
members of the Bcl-2 family of apoptosis regulators, while tumor
cells may also rely on the NF-κB pathway to escape from apoptosis,
which has been demonstrated to be one of the important markers of
cancer (46). Thus, the present
study provided such data, although further studies are required to
further reveal the exact underlying mechanisms.
In summary, the present study demonstrated that
tricellulin expression was associated with colorectal cancer
metastasis and poor prognosis, while tricellulin expression
promoted colorectal cancer cell invasion and EMT in vitro.
The present data support the theory that tricellulin is involved in
colorectal cancer metastasis, and further studies will validate the
detection of tricellulin as a novel marker for prediction of
colorectal cancer prognosis. Mechanistically, tricellulin could
activate the canonical NF-κB signaling pathway for promotion of
colorectal cancer cell EMT and inhibition of tumor cell
apoptosis.
Acknowledgements
Not applicable.
Funding
The present study was supported in part by grants
from the Natural Science Foundation of China (grant no. 81760516),
the 2018 Innovation Project of Guangxi Graduate Education (grant
no. YCBZ2018046), and the Nature Science Foundation of Guangxi
(grant no. 2019GXNSFAA185030).
Availability of data and materials
All data generated or analyzed in the current study
are included in this publication and are available on reasonable
request.
Authors' contributions
JAH, JXZ and MBQ conceived and designed the
experiments. JXZ and ZY wrote the manuscript. ZY, SML, PP, and QS
performed the experiments. LL, LHX, YZ and SQL analyzed the data.
All authors read and approved the manuscript and agree to be
accountable for all aspects of the research in ensuring that the
accuracy or integrity of any part of the work are appropriately
investigated and resolved.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of the First Affiliated Hospital of Guangxi Medical
University (Guangxi, China) with the approval no. BBMCEC2012063 and
performed in accordance with the guidelines of the Declaration of
Helsinki. Written informed consent was obtained from each
patient.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Dekker E, Tanis PJ, Vleugels JL, Kasi PM
and Wallace MB: Colorectal cancer. Lancet. 394:1467–1480. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Fujita T: Colorectal cancer. Lancet.
376:331–332. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Stein A, Atanackovic D and Bokemeyer C:
Current standards and new trends in the primary treatment of
colorectal cancer. Eur J Cancer. 47 (Suppl 3):S312–S314. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Fakih MG: Metastatic colorectal cancer:
Current state and future directions. J Clin Oncol. 33:1809–1824.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Boland PM and Ma WW: Immunotherapy for
colorectal cancer. Cancers (Basel). 9:502017. View Article : Google Scholar
|
|
7
|
Syn NL, Teng MWL, Mok TSK and Soo RA:
De-novo and acquired resistance to immune checkpoint targeting.
Lancet Oncol. 18:e731–e741. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Brenner H and Chen C: The colorectal
cancer epidemic: Challenges and opportunities for primary,
secondary and tertiary prevention. Br J Cancer. 119:785–792. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang W, Kandimalla R, Huang H, Zhu L, Li
Y, Gao F, Goel A and Wang X: Molecular subtyping of colorectal
cancer: Recent progress, new challenges and emerging opportunities.
Semin Cancer Biol. 55:37–52. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zacharakis M, Xynos ID, Lazaris A, Smaro
T, Kosmas C, Dokou A, Felekouras E, Antoniou E, Polyzos A,
Sarantonis J, et al: Predictors of survival in stage IV metastatic
colorectal cancer. Anticancer Res. 30:653–660. 2010.PubMed/NCBI
|
|
11
|
Garcia MA, Nelson WJ and Chavez N:
Cell-cell junctions organize structural and signaling networks.
Cold Spring Harb Perspect Biol. 10:a0291812018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Anderson JM and Van Itallie CM: Physiology
and function of the tight junction. Cold Spring Harb Perspect Biol.
1:a0025842009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Martin TA: The role of tight junctions in
cancer metastasis. Semin Cell Dev Biol. 36:224–231. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Schuetz A, Radusheva V, Krug SM and
Heinemann U: Crystal structure of the tricellulin C-terminal
coiled-coil domain reveals a unique mode of dimerization. Ann NY
Acad Sci. 1405:147–159. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Oda Y, Otani T, Ikenouchi J and Furuse M:
Tricellulin regulates junctional tension of epithelial cells at
tricellular contacts through Cdc42. J Cell Sci. 127:4201–4212.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Morampudi V, Graef FA, Stahl M, Dalwadi U,
Conlin VS, Huang T, Vallance BA, Yu HB and Jacobson K: Tricellular
tight junction protein tricellulin is targeted by the
enteropathogenic escherichia coli effector EspG1, leading to
epithelial barrier disruption. Infect Immun. 85:e00700–16.
2016.PubMed/NCBI
|
|
17
|
Takasawa A, Murata M, Takasawa K, Ono Y,
Osanai M, Tanaka S, Nojima M, Kono T, Hirata K, Kojima T and Sawada
N: Nuclear localization of tricellulin promotes the oncogenic
property of pancreatic cancer. Sci Rep. 6:335822016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Benson AB III, Venook AP, Cederquist L,
Chan E, Chen YJ, Cooper HS, Deming D, Engstrom PF, Enzinger PC,
Fichera A, et al: Colon cancer, version 1.2017, NCCN clinical
practice guidelines in oncology. J Natl Compr Canc Netw.
15:370–398. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yang F, Xu J, Li H, Tan M, Xiong X and Sun
Y: FBXW2 suppresses migration and invasion of lung cancer cells via
promoting β-catenin ubiquitylation and degradation. Nat Commun.
10:13822019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sadeghi MR, Jeddi F, Soozangar N, Somi MH,
Shirmohamadi M, Khaze V and Samadi N: Nrf2/P-glycoprotein axis is
associated with clinicopathological characteristics in colorectal
cancer. Biomed Pharmacother. 104:458–464. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hwang JY, Park JH, Kim MJ, Kim WJ, Ha KT,
Choi BT, Lee SY and Shin HK: Isolinderalactone regulates the
BCL-2/caspase-3/PARP pathway and suppresses tumor growth in a human
glioblastoma multiforme xenograft mouse model. Cancer Lett.
443:25–33. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Qin M, Zhang J, Xu C, Peng P, Tan L, Liu S
and Huang J: Knockdown of NIK and IKKβ-binding protein (NIBP)
reduces colorectal cancer metastasis through down-regulation of the
canonical NF-κΒ signaling pathway and suppression of MAPK signaling
mediated through ERK and JNK. PLoS One. 12:e01705952017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ting L, Rad R, Gygi SP and Haas W: MS3
eliminates ratio distortion in isobaric multiplexed quantitative
proteomics. Nat Methods. 8:937–940. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tolonen AC and Haas W: Quantitative
proteomics using reductive dimethylation for stable isotope
labeling. J Vis Exp. 514162014.
|
|
26
|
Wang K, Shan Z, Duan L, Gong T, Liu F,
Zhang Y, Wang Z, Shen J and Lei L: iTRAQ-based quantitative
proteomic analysis of Yamanaka factors reprogrammed breast cancer
cells. Oncotarget. 8:34330–34339. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The gene
ontology consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kanehisa M, Sato Y, Kawashima M, Furumichi
M and Tanabe M: KEGG as a reference resource for gene and protein
annotation. Nucleic Acids Res. 44:D457–D462. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Szklarczyk D, Franceschini A, Wyder S,
Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos
A, Tsafou KP, et al: STRING v10: Protein-protein interaction
networks, integrated over the tree of life. Nucleic Acids Res.
43:D447–D452. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhao GX, Xu YY, Weng SQ, Zhang S, Chen Y,
Shen XZ, Dong L and Chen S: CAPS1 promotes colorectal cancer
metastasis via Snail mediated epithelial mesenchymal
transformation. Oncogene. 38:4574–4589. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Xu CY, Qin MB, Tan L, Liu SQ and Huang JA:
NIBP impacts on the expression of E-cadherin, CD44 and vimentin in
colon cancer via the NF-κB pathway. Mol Med Rep. 13:5379–5385.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zeisel MB, Dhawan P and Baumert TF: Tight
junction proteins in gastrointestinal and liver disease. Gut.
68:547–561. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
France MM and Turner JR: The mucosal
barrier at a glance. J Cell Sci. 130:307–314. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Pastushenko I and Blanpain C: EMT
transition states during tumor progression and metastasis. Trends
Cell Biol. 29:212–226. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Gonzalez DM and Medici D: Signaling
mechanisms of the epithelial-mesenchymal transition. Sci Signal.
7:re82014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Xu CY, Liu SQ, Qin MB, Zhuge CF, Qin L,
Qin N, Lai MY and Huang JA: SphK1 modulates cell migration and
EMT-related marker expression by regulating the expression of p-FAK
in colorectal cancer cells. Int J Mol Med. 39:1277–1284. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ikenouchi J, Furuse M, Furuse K, Sasaki H
and Tsukita S and Tsukita S: Tricellulin constitutes a novel
barrier at tricellular contacts of epithelial cells. J Cell Biol.
171:939–945. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Shimada H, Satohisa S, Kohno T, Takahashi
S, Hatakeyama T, Konno T, Tsujiwaki M, Saito T and Kojima T: The
roles of tricellular tight junction protein lipolysis-stimulated
lipoprotein receptor in malignancy of human endometrial cancer
cells. Oncotarget. 7:27735–27752. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Schlachter K, Gyugos M, Halasz J, Lendvai
G, Baghy K, Garami M, Gyöngyösi B, Schaff Z and Kiss A: High
tricellulin expression is associated with better survival in human
hepatoblastoma. Histopathology. 65:631–641. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Masuda R, Semba S, Mizuuchi E, Yanagihara
K and Yokozaki H: Negative regulation of the tight junction protein
tricellulin by snail-induced epithelial-mesenchymal transition in
gastric carcinoma cells. Pathobiology. 77:106–113. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Somorácz A, Korompay A, Törzsök P, Patonai
A, Erdélyi-Belle B, Lotz G, Schaff Z and Kiss A: Tricellulin
expression and its prognostic significance in primary liver
carcinomas. Pathol Oncol Res. 20:755–764. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Naugler WE and Karin M: NF-kappaB and
cancer-identifying targets and mechanisms. Curr Opin Genet Dev.
18:19–26. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Gilmore TD: Introduction to NF-kappaB:
Players, pathways, perspectives. Oncogene. 25:6680–6684. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Escarcega RO, Fuentes-Alexandro S,
Garcia-Carrasco M, Gatica A and Zamora A: The transcription factor
nuclear factor-kappa B and cancer. Clin Oncol (R Coll Radiol).
19:154–161. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Kojima T, Fuchimoto J, Yamaguchi H, Ito T,
Takasawa A, Ninomiya T, Kikuchi S, Ogasawara N, Ohkuni T, Masaki T,
et al: c-Jun N-terminal kinase is largely involved in the
regulation of tricellular tight junctions via tricellulin in human
pancreatic duct epithelial cells. J Cell Physiol. 225:720–733.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Xia Y, Shen S and Verma IM: NF-κB, an
active player in human cancers. Cancer Immunol Res. 2:823–830.
2014. View Article : Google Scholar : PubMed/NCBI
|