Ferroptosis, a novel form of regulated cell death
(RCD), first proposed by Dixon et al (1) in 2012 and is characterized by the
overwhelming iron-dependent accumulation of lethal lipid reactive
oxygen species (ROS). The morphological hallmarks of ferroptotic
death are a reduction or loss of mitochondrial cristae (1), condensation of the mitochondrial
membrane (2) and rupture of the
outer mitochondrial membrane (3).
An initial characterization of ferroptotic biochemical demonstrated
that cysteine depletion or inactivation of glutathione peroxidase 4
(GPX4) activity, which causes exhaustion of the intracellular pool
of glutathione (GSH), iron accumulation and lipid peroxidation,
specifically triggers this form of cell death (4). The genetic features of ferroptosis
shows that it primarily dysregulates ferroptotic molecular on
antioxidant metabolism, iron and lipid metabolism, such as SLC7A11,
GPX4, TfR1, ACSL4, which are involved in the initiation of
ferroptosis (5–7). As shown in Table I, there are no forms of
morphological, biochemical, or genetic crosstalk between
ferroptosis and other types of RCD, including apoptosis, autosis,
pyroptosis, autophagy, necroptosis and various other forms of
RCD.
As a cellular process, ferroptosis can be triggered
by various pathological conditions in humans and animals (4,8–10).
Notably, emerging evidence has indicated that ferroptosis likely
prevents tumorigenesis, such as gastric cancer (11), non-small-cell lung carcinoma
(12), glioblastoma (13) and colorectal cancer (14). Ferroptosis is now accepted as an
adaptive process in biological systems that acts as a tumor
suppressive mechanism to eradicate the malignant cells, but the
activation of oxidative stress pathways when metabolism is
dysregulated leads to tumorigenesis (15). Interestingly, recent evidence has
suggested that non-coding RNAs (ncRNAs), particularly micro RNAs
(miRNAs/miRs), long non-coding RNAs (lncRNAs) and circular RNAs
(circRNAs), serve vital roles in regulating ferroptosis (16). These ncRNAs are involved in iron
metabolism, ROS metabolism and ferroptosis-related amino-acid
metabolism, which regulates the process of ferroptosis initiation
(17). Of particular interest, the
accumulation of abundant lipid ROS in cells is the most critical
factor for triggering ferroptosis (18). Conversely, ncRNAs can directly or
indirectly regulating lipid ROS-related molecules to maintain redox
dynamics during periods of high levels of ROS generation, and work
to reduce ROS levels below toxic thresholds, which allows tumor
cells to exhibit tolerances to relatively high levels of cellular
ROS and avoids initiating ferroptosis (19). A moderate increase in cellular ROS
levels promotes cell proliferation, survival and malignant
transformation (19). These
findings highlight the potential targets for anticancer treatments
via genetic or pharmacological interference in ncRNA-regulated
ferroptotic cell death. In the present review, the primary
mechanism of ferroptosis initiation and the involvement of ncRNAs
in ferroptosis in various types of cancer cells is summarized, with
the aim of highlighting potentially novel strategies for
personalized cancer treatment.
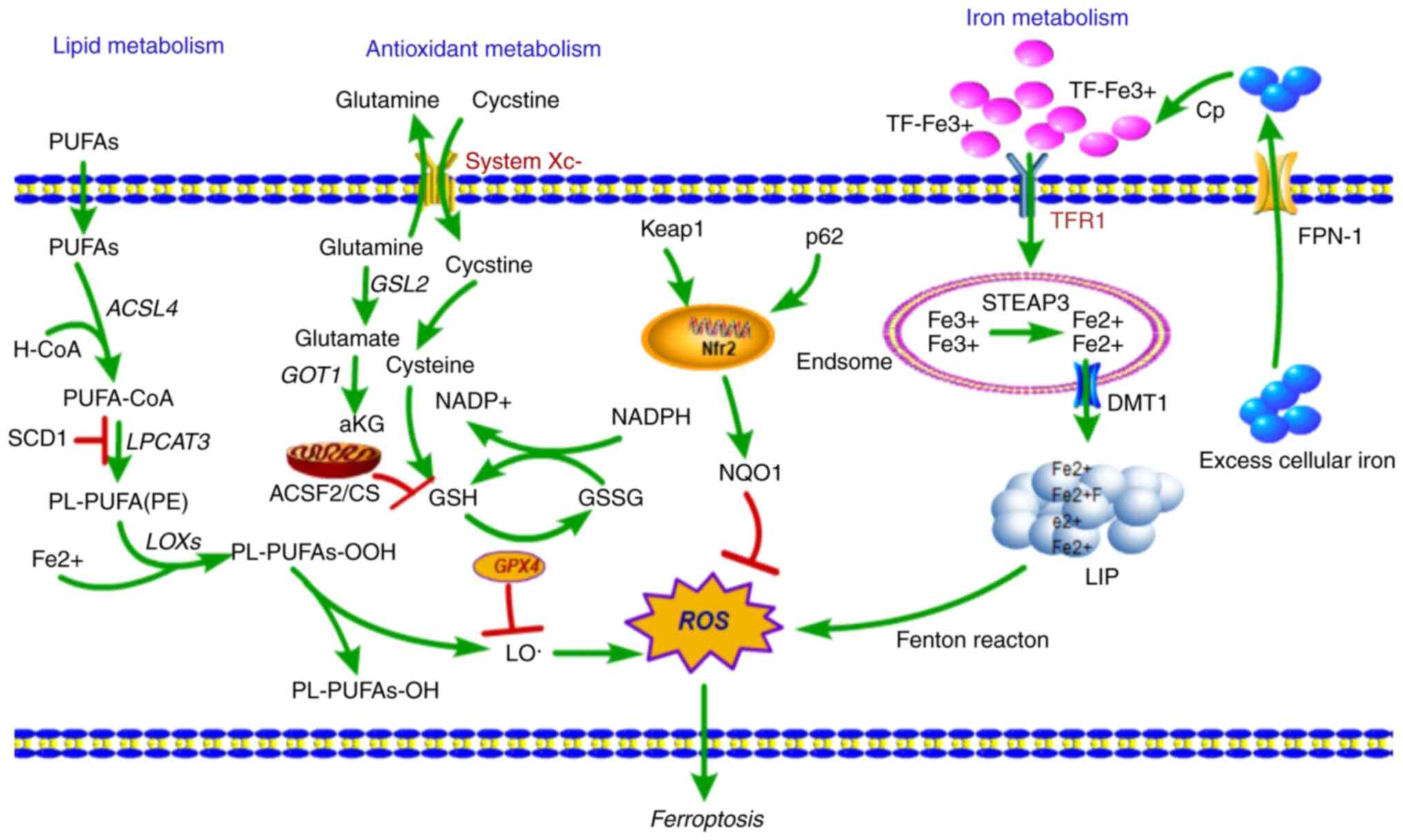
Iron is an essential nutrient, as it is necessary
for the maintenance of cellular metabolism and all several
important physiological activities, such as oxygen transport, DNA
synthesis and ATP production (20).
As iron is ubiquitously present, cellular iron homeostasis is a
complex and tightly regulated process though the acquisition,
utilization, storage and recycling of iron (5). The cellular iron balance is maintained
through the redox cycle and iron intake (Fig. 1). The cellular iron redox cycle is
primarily dependent on the Fenton reaction (21). In the cellular Fenton reaction,
ferrous iron (Fe2+) is oxidized to ferric iron
(Fe3+) during the conversion of
H2O2 into reactive hydroxyl radicals;
conversely, Fe3+ is then reduced back to Fe2+
through superoxide radicals (22).
In of iron intake, transferrin receptor 1 (TfR1) is expressed on
the surface of the majority of cells, where it primarily takes up
transferrin (TF)-bound iron into cells. The
TfR1/TF-(Fe3+)2 complex is endocytosed
(23), and Fe3+ is
released from TF (24), reduced to
Fe2+ by ferric reductase six-transmembrane epithelial
antigen of the prostate 3 (STEAP3), and then transported across the
endosomal membrane by divalent metal transporter 1 (DMT1) (25).
The imported cellular iron enters the transient
cytosolic labile iron pool, a pool of chelatable and redox-active
iron (26), which is utilized by
cells for various metabolic processes or stored in ferritin
(27). Excess cellular iron is
exported out of the cell and transported into circulation by
ferroportin 1 (FPN-1), after which it is oxidized by the
ferroxidase-ceruloplasmin and binds to serum TF (28). Furthermore, cellular iron balance is
also regulated by a network of iron-dependent proteins: The
iron-responsive elements (IREs) and iron-regulatory proteins
(IRPs). IRPs are cytosolic proteins that regulate the expression of
genes involved in iron import (TfR1, DMT1), storage [ferritin
(FTH), FTH1 and FTL] and export (FPN-1) by binding IREs (29).
Iron metabolism is an indispensable component of
ferroptosis that distinguishes it from other types of RCD. Iron can
gain and lose electrons, rendering it capable of contributing to
free radical formation. When cellular iron is overloaded, the free
radicals accumulate aberrantly, causing increased production of
ROS. This effect leads to oxidative stress, which results in
ferroptotic cell death (30).
However, dysregulation of iron metabolism also serves an active
role in carcinogenesis and promotes tumor growth (5,31).
TfR1 is a major regulator of intracellular iron
uptake, and researchers found that abnormal accumulation of TfR1 on
the cell surface is a specific marker of ferroptosis (32). In hepatocellular carcinoma, TfR1 and
FTH1 are upregulated in erastin and sorafenib induced ferroptotic
cell death (33), and TfR1 is also
upregulated in erastin-induced cell death in myeloid leukemia cell
lines (34). Furthermore, in Calu-1
lung cancer cells and HT-1080 fibrosarcoma cells, IRE-binding
protein 2 (IREB2) is an essential gene for erastin-induced
ferroptosis by regulating TFRC, FTH1 and FTL (1). Furthermore, several studies have
suggested that inhibition of DMT1 may prevent iron translocation,
leading to lysosomal iron overload, ROS production and ferroptotic
cell death in cancer stem cells (35), and sulfasalazine induced ferroptosis
is reduced by the inhibitory effect of estrogen receptor on TFRC
and DMT1 in breast cancer cells (36). Artemisinin compounds sensitize
cancer cells to ferroptosis by regulating IRP/IRE-controlled iron
homeostasis (37). Therefore,
targeting iron metabolic pathways may offer novel therapeutic
options for cancer therapy.
Fatty acid (FA) metabolism provides specific lipid
precursors for energy storage, membrane biosynthesis, generation of
signaling molecules and lipid oxidation that result in an
accumulation of an abundance of lipid ROS (38). Although ferroptosis is induced by
multiple stimuli, the accumulation of abundant lipid ROS in cells
is the most critical factor causing ferroptotic cell death. In
addition to iron-generated ROS production via the Fenton reaction,
ROS from lipid oxidation appears to serve a role in ferroptosis
(Fig. 1). Therefore, lipid
peroxidation is crucial for induction of ferroptosis.
In the process of lipid metabolism, arachidonic acid
(AA), a fatty acid substrate, is activated by acyl-CoA synthetase
long-chain family member 4 (ACSL4) to produce AA-CoA, and then
AA-CoA is esterified by lysophosphatidylcholine acyltransferase 3
(LPCAT3) to phosphatidyl-(PE)-AA (39). PE-AA is oxidized to cytotoxic
PE-AA-OOH by lipoxygenases (LOXs) that are activated during
catalysis of Fe2+ (40).
Under physiological conditions, glutathione peroxidase 4 (GPX4)
reduces cytotoxic PE-AA-OOH to non-cytotoxic PE-AA-OH, which
protects cells from oxidative damage. When GPX4 is inactivated or
depleted, PE-AA-OOH accumulates in the cell, and this induces
ferroptosis (40). Thus, lipid
peroxidation accounts for a large proportion of ferroptosis
initiation.
ACSL4 is a key enzyme involved in the synthesis of
long chain unsaturated fatty acids. ACSL4 was found to sensitize
RSL3-induced ferroptosis through altering the cellular lipid
composition (8). In hepatocellular
carcinoma patients who had complete or partial responses to
sorafenib-induced ferroptosis, and had higher ACSL4 expression in
the pretreated tumor tissues than those who did not respond, ACSL4
was a predictive biomarker for sensitivity of sorafenib in
hepatocellular carcinoma (41).
Consistently, ACSL4 suppresses the proliferation of tumor cells
through activation of ferroptosis in glioma cells (42). Furthermore, a CRISPR-based genetic
screen identified ACSL4 and LPCAT3 as promoting of RSL3- and
DPI7-induced ferroptosis, but they did not affect erastin-induced
ferroptosis (39). Several studies
have supported the conclusion that PUFAs can be oxidized, producing
the lipid peroxides that promote the induction of ferroptosis
(43). Therefore, targeting the
lipid metabolism pathway may also be a novel means of tumor
therapy.
GSH, a thiol-containing tripeptide, is a potent
antioxidant whose synthesis is limited by the constant import of
cysteine and the availability of cystine/cysteine. The system
Xc− antiporter is a cystine/glutamate transporter that
takes up extracellular cystine in exchange for intracellular
glutamate (44). SLC7A11, expressed
at the cell surface, is a regulatory light chain component of the
system Xc− transporter and is essential for cystine
cellular uptake and serves a role in intracellular GSH synthesis
(19). Once imported into cells,
intracellular cystine is reduced to cysteine, a precursor of GSH
used in GSH biosynthesis. GPX4, a central mediator of ferroptosis,
which has phospholipid peroxidase activity, catalyzes the reduction
of lipid peroxides to lipid alcohols using GSH as an essential
co-factor, thus preventing cells from undergoing too much lipid
peroxidation (45). Blockade of a
member of the system Xc− antiporter, SLC7A11, and
inhibition of GPX4 were shown to induce ferroptosis (1). Both interventions impaired cellular
antioxidant defenses, thereby facilitating toxic ROS accumulation,
suggesting antioxidant pathways as potential regulators of
ferroptosis.
Erastin, a RAS-selective lethal compound, triggers
ferroptosis by directly inhibiting system Xc− activity
to reduce GSH levels in cancer cells (1,2).
Similarly, sulfasalazine, a drug used to treat chronic
inflammation, also triggers ferroptosis through directly inhibiting
SLC7A11 activity (46). Similar to
the above two compounds, p53, a well-characterized tumor
suppressor, was also shown to sensitize cells to ferroptosis
through the repression of SLC7A11 (47,48).
Furthermore, the tumor suppressor BRCA1-associated protein 1
suppresses SLC7A11 transcription by decreasing H2Aub, leading to
elevated lipid peroxidation and thus, increased ferroptosis
(49). kelch-like ECH-associated
protein 1 (Keap1) can also suppress the expression of SLC7A11
through degrading the transcription factor nuclear factor erythroid
2-related factor 2 (Nrf2), which is a master transcription factor
of the antioxidant response (50).
Another molecular mechanism of ferroptosis is the direct
suppression of GPX4 by promoting its degradation or the loss of its
activity. GPX4 was identified as a target protein of the classical
ferroptosis inducer RSL3 (51),
which directly binds to GPX4 to inactivate the peroxidase activity
of GPX4 and induce ferroptosis (52). Several ferroptosis inducers directly
inhibit GPX4 function including DPI7, DPI10, DPI12, DPI13, DPI17,
DPI18, DPI19 and ML162 (52,53),
and several ferroptosis inducers have an indirect effect on GPX4
function, including SRS13–45 (46),
SRS13-60 (46), buthionine
(54), sulfoximine (52), DPI2 (52), lanperisone (55), sorafenib (56) and erastin derivatives (52). Taken together, these studies show
that the SLC7A11-GSH-GPX4 axis primarily mediates the initiation of
ferroptosis, and that GPX4 serves a central role in regulating
ferroptosis.
Well-established regulatory mechanisms that regulate
changes in iron and ROS metabolism in cancer have recently been
identified. ncRNAs are being increasingly recognized as vital
regulatory mediators of ferroptosis.
A set of miRNAs that post-transcriptionally regulate
gene expression by RNA silencing have been demonstrated to be
involved in the regulation of iron and ROS metabolism. The levels
of these miRNAs are directly or indirectly correlated with
ferroptosis.
lncRNAs are a class of non-coding RNAs >200
nucleotides in length that function to regulate gene expression by
epigenetic, transcriptional and translational modulation. lncRNAs
have been implicated in various biological processes. Recent
studies have shown dysregulation of several lncRNAs is also
involved in the ferroptotic process (Table II).
lncRNA P53RRA is downregulated in lung cancer and
acts as a tumor suppressor. In the cytoplasm, P53RRA interacts with
G3BP1 to activate the p53 signaling pathway, which in-turn promotes
erastin-induced ferroptosis by increasing lipid ROS and altering
the iron concentration (67).
lncRNA LINC00336 is upregulated in lung cancer and functions as an
oncogene. LINC00336 competes with miR-6852 for CBS, inhibiting
ferroptosis by decreasing iron concentrations, ROS and
mitochondrial superoxide levels, as well as the mitochondrial
membrane potential (58). lncRNA
GABPB1-AS1 is an antisense lncRNA of GABPB1 that downregulates
GABPB1 levels by blocking GABPB1 translation, leading to
peroxiredoxin-5 peroxidase suppression and increased lipid ROS
concentrations, ultimately promoting erastin-induced ferroptosis
(68).
CircRNAs are class of non-coding RNA characterized
by a covalently closed loop structure leaving no free ends and have
been demonstrated to be involved in tumorigenesis. CircTTBK2 is
upregulated in glioma and functions as a master regulator of CPEB4
by sponging miR-217. Knockdown of circTTBK2 promoted
erastin-induced ferroptosis accompanied with an increase in the
intracellular concentrations of ROS, iron and ferrous iron by
competing with miR-217 for CBS in glioma cells (66).
Previous studies have demonstrated that cellular
iron overload causes ferroptosis. TfR1 is a critical transporter
involved in iron uptake and a specific ferroptosis marker, which
imports Tf-iron from the extracellular environment into cells,
contributing to the cellular iron pool required for ferroptosis
(32). miR-320 (69), miR-107 (70), miR-148a (71), miR-7-5p/miR-141-3p (72), miR-152 (73) and miR-210 (74) are all involved in suppression of
TfR1 by directly targeting TfR1. Therefore, it has been reasonably
shown that these miRNAs can suppress ferroptosis by targeting
TfR1.
FTH1, a major intracellular iron storage protein, is
an iron regulators involved in iron storage. Expression levels of
FTH1 are regulated by oncogenic RAS signaling, which controls the
cellular iron pool and ferroptosis sensitivity in tumor cells
(51). FTH1 is regulated by NRF2 in
ferroptosis, knockdown of FTH1 enhances erastin or
sorafenib-induced ferroptosis sensitivity in hepatocellular
carcinoma, suggesting that reduced iron storage may contribute to
cellular iron overload causing ferroptosis and that FTH1 may serve
as a specific marker of ferroptosis marker as well (54). miR-200b is involved in the
repression of FTH1 by directly targeting FTH1, which transforms
H2O2 and O2 into the reactive •OH
radical, thus inducing tumor cell death (75). Oncogenic miR-638 and miR-362 have
been identified as targets of FTH1 transcript or multiple FTH1
pseudogenes by an unbiased screen in prostate cancer (76). lncRNA H19 is the pre-miRNA template
of miR-675, and knockdown of FTH1 upregulates H19 expression and
thus its cognate miR-675, and H19/miR-675 activation primarily
contributes to altered iron metabolism induced by FTH1 silencing
(77). Therefore, it has been
reasonably confirmed that these miRNAs may suppress ferroptosis by
targeting TfR1. Together, these studies have shown that these
ncRNAs may be involved in regulating the process of ferroptosis
through iron storage.
IREB2 is an intra-cellular iron metabolism
RNA-binding protein which regulates the translation and the
stability of iron homeostasis related genes. Knock down of IREB2
suppresses erastin-induced ferroptosis by amino acid/cystine
deprivation (1). miR-29 regulates
IREB2 directly, thus affecting both energy production and redox
status of the cell (78).
Furthermore, miR-29a-related genetic variants alter the expression
of IREB2 and may modify the risk of lung cancer together with
dietary iron intake (79).
Oncogenic miR-935 is elevated in renal cell carcinoma, and miR-935
directly suppresses the transcription of IREB2 by binding to the
3′-UTRs of IREB2 (80). Therefore,
these miRNAs may suppress ferroptosis by targeting IREB2.
DMT1 is a widely expressed key iron transporter
located within the plasma membrane and membranes of lysosomes and
endosomes, which enables the uptake of Fe2+ to the
cytosol following iron endocytosis. DMT1 inhibitors were selected
as a target in cancer stem cells by blocking lysosomal iron
translocation, which leads to lysosomal iron accumulation, and thus
production of ROS and induction of ferroptotic cell death (35). DMT1 is also involved in
sulfasalazine-induced ferroptosis via activation of iron metabolism
in breast cancer cells (36).
miR-Let-7d binds to the 3′-UTR of DMT1-IRE decreasing its
expression at both the mRNA and protein levels in K562 and HEL
cells (81). miR-16 family members
miR-16, miR-195, miR-497 and miR-15b have been shown to suppress
intestinal DMT1 expression by targeting DMT1 3′-UTR in HCT116 cells
(82). These miRNAs may be involved
in ferroptosis by targeting DMT1.
ACSL is expressed on the mitochondrial outer
membrane and endoplasmic reticulum, where they catalyze fatty acids
to form acyl-CoAs, which are lipid metabolic intermediates that
facilitate fatty acid metabolism and membrane modifications
(83). According to genome-wide
recessive genetic screening, ACSL4 has been identified as an
essential pro-ferroptotic gene and as a critical determinant of
ferroptosis sensitivity by shaping cellular lipid composition
(8). Another study also showed that
ACSL4 is a biomarker and contributor of ferroptosis via
ACSL4-mediated production of 5-hydroxyeicosatetraenoic acid
(5-HETE) (84).
miR-34a-5p/miR-204-5p (85),
miR-141 (86), miR-3595 (87), miR-34a/c (88,89),
miR-548p (90), miR-205 (91), miR-224-5p (92) and
miR-19b-3p/miR-17-5p/miR-130a-3p/miR-150-5p/miR-7a-5p/miR-144-3p/miR-16-5p
(93) can suppress the
transcription of ACSL4. These miRNAs may inhibit ferroptosis by
targeting ACSL4. In addition, a recent study reported that lncRNA
NEAT1 promotes the transcription of ACSL4 by competing with
miR-34a-5p and miR-204-5p, which may suppress ferroptosis (85).
LOXs are a family of iron-containing enzymes,
including six LOX genes in humans; LOX5, LOX12, LOX12B, LOX15,
LOX15B and LOXE3 (94). These genes
can catalyze dioxygenation of PUFAs to produce fatty acid
hydroperoxides in a stereospecific manner (94). Oxidation of PUFAs by LOXs had been
implicated in erastin-induced ferroptosis (94). LOX15-driven enzymatic generation of
lipid peroxidation is a hallmark of ferroptotic signals (95). In the miR-17 family, miR-18a and
miR-203 bind to four sites of the 3′-UTR in 15-LOX1, and miR-17,
miR-20a, miR-20b, miR-106a, miR-106b, miR-93 and miR-590-3p bind to
four sites of the 3′-UTR of 15-LOX2 (96). Oncogenic miR-219-2 (97) directly targets the 3′-UTR of 15-LOX,
whereas miR-674-5p (98),
miR-216a-3p (99) and
miR-19a-3p/miR-125b-5p (100)
regulate 5-LOX through directly targeting the 3′-UTR of 5-LOX.
GPX4, unlike other members of the GPX family, serve
a unique role in physiology; they catalyze the reduction of lipid
peroxides in a complex cellular membrane environment.
Overexpression or knockdown of GPX4 modulates the lethality of
ferroptosis inducers, indicating that GPX4 is an essential
regulator of ferroptotic cell death (52). miR-181a-5p decreases the expression
of GPX4 by targeting SBP2 or SECISBP2 and reduces the ability to
counter oxidation, which may promote ferroptosis (101,102).
Stearoyl-CoA desaturase 1 (SCD1) is a rate-limiting
step catalytic enzyme in mono-unsaturated fatty acid (MUFA)
synthesis that serves a central role in FA metabolism by converting
the saturated fatty acids palmitate and stearate to the MUFAs
palmitoleate (PMA) and oleate. SCD1, as an inhibitor of
ferroptosis, serves an important role in the negative regulation of
ferroptosis through the products of MUFAs (103). miR-27a (104), miR-212-5p (105), miR-103 (106), miR-192* (107), miR-378 (108), miR-4668 (109), miR-600 (110) and let-7c (111) significantly suppress the relative
expression of SCD1 by directly binding to its 3′-UTR. Moreover,
lncRNA uc.372 promotes the transcription of SCD1 by competing with
miR-4668 (109).
Citrate synthases (CSs) are implicated in the
regulation of mitochondrial fatty acid metabolism, which supply a
specific lipid precursor necessary for ferroptotic cell death
(1). Silencing CS suppresses
erastin-induced ferroptosis (1).
miR-122 suppresses the expression of mRNAs and proteins related to
CS (112), whereas miR-19 only
regulates the expression of proteins related to CS (113). Therefore, these ncRNAs have been
implicated in promoting ferroptosis by targeting lipid
metabolism-related genes.
Nrf2 is a pivotal inhibitor of ferroptosis due to
its ability to inhibit cellular iron uptake, limit ROS production,
and upregulate SLC7A11 expression by regulating the Nrf2-targeted
genes FTH1, HO-1 and NQO1. Certain miRNAs can directly or
indirectly suppress the transcription of Nrf2 or Nrf2 signaling to
promote ferroptosis. For example, miR-675 (114), miR-181 (115), miR-302b-3p (116), miR-141 (117,118), miR-1225 (119), miR-25 (120), miR-128-3p (121), miR-19b (122), miR-125b (123) and miR-494 (124) restrain Nrf2 signaling by targeting
Nrf2-related genes. In contrast, miR-365 (125), miR-495 (126), miR-136 (127), miR-34a (128), miR-340-5p (129), miR-125b (130), miR-101-3p (131,132), miR-155 (133), miR-380-3p (134), miR-144 (135–137), miR-153 (138), miR-28/miR-708 (139), miR-129-3p (140), miR-27b (141), miR-140-5p (142), miR-93 (143) and miR-365-1/miR-193b/miR-29-b1
(144) have been shown to decrease
Nrf2 levels through directly binding to the 3′-UTR of Nrf2.
Additionally, certain miRNAs activate Nrf2 signaling via a variety
of mechanisms, ultimately resulting in inhibition of ferroptosis.
For example, miR-152-3p (145),
miR-101 (146), miR-455 (147), miR-601 (148), miR-7 (149), miR-200a (150), miR-873-5p (151), miR-24-3p (152), miR-34b (153), miR-223 (154), miR-146b-5p (155) and miR-98-5p (156) activate Nrf2 signaling by targeting
Nrf2-related genes. It is thus hypothesized that these miRNAs can
regulate ferroptosis by targeting Nrf2, but this has not yet been
demonstrated.
SLC7A11, the subunit of cystine-glutamate
antiporter, is a crucial mediator in the process of ferroptosis.
Studies have shown that miR-27a (183), miR-375 (184) and miR-26b (185) directly suppress the transcription
of SLC7A11 by binding to its 3′-UTR. Therefore, these miRNAs have
been implicated in promoting ferroptosis by directly targeting
SLC7A11. Furthermore, lncRNAs SLC7A11-AS1 (186) and AS-SLC7A11 (187), the antisense lncRNAs of SLC7A11,
suppress the transcription of SLC7A11. Therefore, these two
SLC7A11-antisense lncRNAs have been hypothesized to suppress
ferroptosis by downregulating SLC7A11 levels.
Keap1 is a member of the BTB-kelch protein family,
which are primarily located in the perinuclear region of the
cytoplasm (188). Keap1 represses
Nrf2 transcriptional activity, a transcriptional target of Keap1.
Overexpression of Keap1 enhanced erastin- and RSL3-induced
ferroptosis, while knockdown conferred resistance to ferroptosis
(189). Studies have shown that
overexpression of miR-7 (149),
miR-873-5p (151), miR-24-3p
(152), miR-34b (153), miR-223 (154), miR-26b (190), miR-941 (191), miR-200a (192,193), miRNA-421 (194), miR-626 (195), miR-1225 (119), miR-141 (118) and miR-432 (196) suppressed Keap1 3′-UTR expression
and downregulated its mRNA and protein expression. Notably, lncRNA
MALAT1 could epigenetically downregulate Keap1 expression (161). lncRNA KRAL functions as a ceRNA by
effectively binding to miR-141 and then restoring Keap1 expression
(117). These studies suggest that
Keap1 related-ncRNAs are involved in the process of
ferroptosis.
GOT1 is essential for cell sustaining proliferation
and maintenance of redox homeostasis. Reduced GOT1 suppresses
erastin-induced ferroptosis by amino acid/cystine deprivation
(197). According to previous
studies, both in pancreatic cancer and melanoma, miR-9-5p inhibited
the expression of GOT1 by directly binding to its 3′-UTR,
ultimately resulting in decreased proliferation, glutamine
metabolism and redox homeostasis, which suppresses the process of
ferroptosis (57,198).
Collectively, the modulators of ferroptotic markers
are their related ncRNAs, which serve critical roles in the
regulation of ferroptosis. As discussed above, ncRNAs possess tumor
suppressor or oncogenic roles in the process of ferroptosis during
the course of tumorigenesis and progression. Thus, targeting ncRNAs
may be a viable strategy in the development of novel cancer
treatments.
Ferroptosis likely inhibits tumor development
and/or progression, thus inducing ferroptosis is a promising
strategy for anticancer therapy. ncRNA expression patterns show
specificity for specific tumor and tissue types, highlighting
ncRNAs as potential therapeutic targets in cancer. With advances in
biotechnologies, such as genome editing, high-throughput sequencing
and nanotechnology, ncRNAs can be theoretically used as molecular
targets for cancer therapy. Therefore, ncRNAs are considered as an
emerging and viable candidates for precision medicine depending on
its property of tissue-specific expression.
Thus far, among the annotated ncRNAs, miRNAs,
lncRNAs and circRNAs are the most extensively investigated. They
function as either oncogenes or tumor suppressors, which induce or
inhibit ferroptosis by targeting their mRNAs, respectively.
Previously, several preclinical studies have investigated
RNA-guided precision medicine for cancer treatment (161,199–201). For example, miR-34a mimic-mediated
tumor suppression was the first miRNA-based therapy to be used in
the clinic (202). lncRNA MALAT1
with antisense oligonucleotide-conjugated nanostructure inhibited
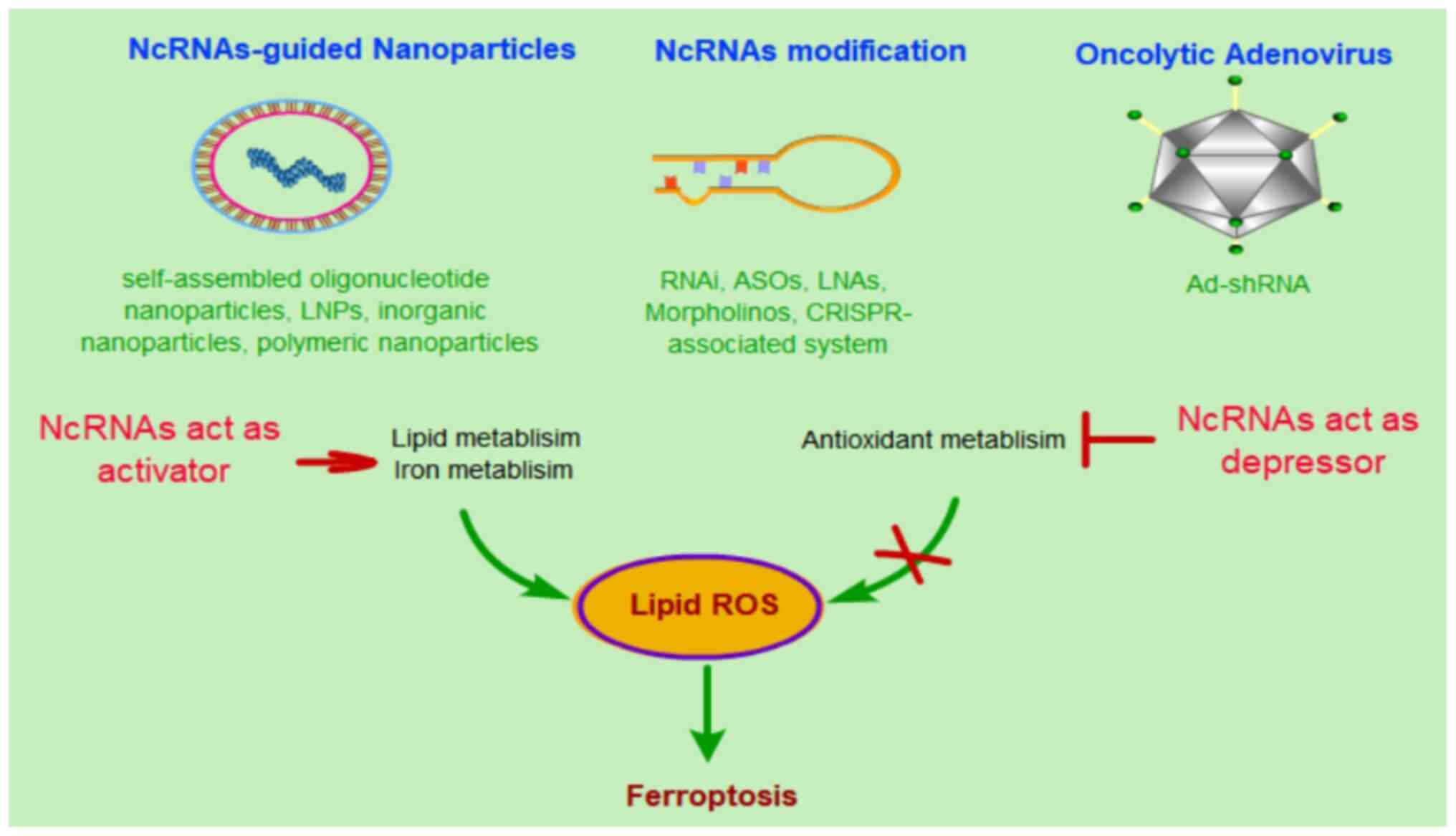
metastasis of lung cancer cells (203). In total, three strategies have
been proposed for ncRNA-based therapy: i) ncRNA-guided
nanoparticles, ii) ncRNA modification and iii) an oncolytic
adenovirus strategy (204).
The methods described above are currently the most
promising ncRNA-based treatment strategies for cancer. These
therapeutic approaches can also be used in ncRNAs targeting
ferroptosis for cancer treatment. Most of the ncRNAs regulate lipid
ROS-related molecules and antioxidant metabolism-related molecules,
which leads to increased tumor cell tolerance for relatively higher
ROS levels and thus reduced possibility of initiating ferroptosis.
At same time, high levels of cellular ROS promote tumor cell
growth. To initiate ferroptotic cell death, stimulating ncRNAs need
to activate lipid and iron metabolism or otherwise activate
antioxidant metabolism, which in turn leads to an accumulation of
cellular ROS and eventually cell death (Fig. 2). Thus, ncRNAs have been considered
not only as therapeutic targets for cancer therapy, but also as
potentially promising therapeutic tools for precision medicine.
However, the majority of studies regarding the use of ncRNAs
therapeutically are still in their early stages. Several problems
need to be overcome before they can be used clinically, such as the
off-target effects, short half-life, severe toxicity and low
transfection efficiency in ncRNA guided strategies (204). A large number of further studies
are still required.
Ferroptosis is a novel type of cell death with
distinct functions intricately involved in numerous physiological
processes and various diseases. Substantial progress in exploring
the mechanisms of ferroptosis and understanding on how oncogenic
states drive sensitivity to ferroptosis has been made.
Collectively, these studies have demonstrated ferroptosis as a
tumor suppressive mechanism that inhibits tumor growth and
contributes to chemotherapy sensitivity, and that induction of
ferroptosis is a viable anticancer therapeutic strategy,
particularly for drug-resistant tumors.
However, cellular sensitivity to ferroptosis likely
depends on the cell type and physiological conditions. What types
of physiological processes are associated with ferroptosis? Under
what context do cells benefit from ferroptotic cell death? Studies
exploring the association between cancer and ferroptosis are still
limited. Although several candidate primary markers of ferroptosis
have been identified, and the pathways they target are known,
several candidates fail to acquire their special cellular
conditions and exhibit poor pharmacokinetics. A large number of
recent studies have demonstrated that miRNAs, lncRNAs and circRNAs
serve an important role in the process of ferroptosis, and that
these ncRNAs may affect the regulation of ferroptosis in a cell
type-dependent or tissue type-dependent manner. Due to the
heterogeneity of gene expression on a per individual basis,
ncRNA-based treatment strategies can be used for personalized
cancer treatment and may eventually exhibit more specificity than
ferroptosis-inducing drugs such as erastin, sulfasalazine and RSL3.
Thus, targeting ncRNAs may at present be considered a prototypic
intervention which has the potential to be superior in terms of
precision compared with established anti-tumor drugs. Moreover,
with the development of gene related technologies, ncRNAs
constitute promising potential targets for gene therapy. However, a
deeper understanding of the mechanisms by which ncRNAs regulate
ferroptosis is still required, and tissue specific expression of
ncRNAs and the variety of off-target effects are major
challenges.
In summary, ncRNAs may serve as anticancer targets
by regulating ferroptosis, which is a novel and promising means of
treating drug-resistant cancer. Targeting key ncRNA-related
ferroptotic molecules may create novel opportunities for gene
therapy for the treatment of cancer.
Not applicable.
This study was supported by National Natural
Science Foundation of China (NSFC) through grants no. 81270561 and
Program of High-level Talents Introduction in the First Affiliated
Hospital of Chengdu Medical College through grants no.
CYFY-GQ17.
Not applicable.
YL and QH wrote the manuscript. YL, QH, BH, YL and
SH created the figures and tables. YL and JX conceived the topic of
this review. All authors read and approved the final
manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Dixon SJ, Lemberg KM, Lamprecht MR, Skouta
R, Zaitsev EM, Gleason CE, Patel DN, Bauer AJ, Cantley AM, Yang WS,
et al: Ferroptosis: An iron-dependent form of nonapoptotic cell
death. Cell. 149:1060–1072. 2012. View Article : Google Scholar
|
|
2
|
Yagoda N, von Rechenberg M, Zaganjor E,
Bauer AJ, Yang WS, Fridman DJ, Wolpaw AJ, Smukste I, Peltier JM,
Boniface JJ, et al: RAS-RAF-MEK-dependent oxidative cell death
involving voltage-dependent anion channels. Nature. 447:864–868.
2007. View Article : Google Scholar
|
|
3
|
Friedmann Angeli JP, Schneider M, Proneth
B, Tyurina YY, Tyurin VA, Hammond VJ, Herbach N, Aichler M, Walch
A, Eggenhofer E, et al: Inactivation of the ferroptosis regulator
Gpx4 triggers acute renal failure in mice. Nat Cell Biol.
16:1180–1191. 2014. View Article : Google Scholar
|
|
4
|
Dixon SJ: Ferroptosis: Bug or feature?
Immunol Rev. 277:150–157. 2017. View Article : Google Scholar
|
|
5
|
Manz DH, Blanchette NL, Paul BT, Torti FM
and Torti SV: Iron and cancer: Recent insights. Ann NY Acad Sci.
1368:149–161. 2016. View Article : Google Scholar
|
|
6
|
Carbone M and Melino G: Lipid metabolism
offers anticancer treatment by regulating ferroptosis. Cell Death
Differ. 26:2516–2519. 2019. View Article : Google Scholar
|
|
7
|
Desideri E, Ciccarone F and Ciriolo MR:
Targeting glutathione metabolism: Partner in crime in anticancer
therapy. Nutrients. 11:19262019. View Article : Google Scholar
|
|
8
|
Doll S, Proneth B, Tyurina YY, Panzilius
E, Kobayashi S, Ingold I, Irmler M, Beckers J, Aichler M, Walch A,
et al: ACSL4 dictates ferroptosis sensitivity by shaping cellular
lipid composition. Nat Chem Biol. 13:91–98. 2017. View Article : Google Scholar
|
|
9
|
Chen X, Xu S, Zhao C and Liu B: Role of
TLR4/NADPH oxidase 4 pathway in promoting cell death through
autophagy and ferroptosis during heart failure. Biochem Biophys Res
Commun. 516:37–43. 2019. View Article : Google Scholar
|
|
10
|
Wang N, Zeng GZ, Yin JL and Bian ZX:
Artesunate activates the ATF4-CHOP-CHAC1 pathway and affects
ferroptosis in Burkitt's Lymphoma. Biochem Biophys Res Commun.
519:533–539. 2019. View Article : Google Scholar
|
|
11
|
Wang C, Shi M, Ji J, Cai Q, Zhao Q, Jiang
J, Liu J, Zhang H, Zhu Z and Zhang J: Stearoyl-CoA desaturase 1
(SCD1) facilitates the growth and anti-ferroptosis of gastric
cancer cells and predicts poor prognosis of gastric cancer. Aging
(Albany NY). 12:15374–15391. 2020. View Article : Google Scholar
|
|
12
|
Liu P, Wu D, Duan J, Xiao H, Zhou Y, Zhao
L and Feng Y: NRF2 regulates the sensitivity of human NSCLC cells
to cystine deprivation-induced ferroptosis via FOCAD-FAK signaling
pathway. Redox Biol. 37:1017022020. View Article : Google Scholar
|
|
13
|
Zhang Y, Fu X, Jia J, Wikerholmen T, Xi K,
Kong Y, Wang J, Chen H, Ma Y, Li Z, et al: Glioblastoma therapy
using codelivery of cisplatin and glutathione peroxidase targeting
siRNA from iron oxide nanoparticles. ACS Appl Mater Interfaces.
12:43408–43421. 2020. View Article : Google Scholar
|
|
14
|
Sharma P, Shimura T, Banwait JK and Goel
A: Andrographis-mediated chemosensitization through activation of
ferroptosis and suppression of β-catenin/Wnt-signaling pathways in
colorectal cancer. Carcinogenesis. 41:1385–1394. 2020. View Article : Google Scholar
|
|
15
|
Fanzani A and Poli M: Iron, oxidative
damage and ferroptosis in rhabdomyosarcoma. Int J Mol Sci.
18:17182017. View Article : Google Scholar
|
|
16
|
Mou Y, Wang J, Wu J, He D, Zhang C, Duan C
and Li B: Ferroptosis, a new form of cell death: Opportunities and
challenges in cancer. J Hematol Oncol. 12:342019. View Article : Google Scholar
|
|
17
|
Fearnhead HO, Vandenabeele P and Vanden
Berghe T: How do we fit ferroptosis in the family of regulated cell
death? Cell Death Differ. 24:1991–1998. 2017. View Article : Google Scholar
|
|
18
|
Badgley MA, Kremer DM, Maurer HC,
DelGiorno KE, Lee HJ, Purohit V, Sagalovskiy IR, Ma A, Kapilian J,
Firl CEM, et al: Cysteine depletion induces pancreatic tumor
ferroptosis in mice. Science. 368:85–89. 2020. View Article : Google Scholar
|
|
19
|
Trachootham D, Alexandre J and Huang P:
Targeting cancer cells by ROS-mediated mechanisms: A radical
therapeutic approach? Nat Rev Drug Discov. 8:579–591. 2009.
View Article : Google Scholar
|
|
20
|
Altamura S, Marques O, Colucci S, Mertens
C, Alikhanyan K and Muckenthaler MU: Regulation of iron
homeostasis: Lessons from mouse models. Mol Aspects Med.
75:1008722020. View Article : Google Scholar
|
|
21
|
Koppenol WH and Hider RH: Iron and redox
cycling. Do's and don'ts. Free Radic Biol Med. 133:3–10. 2019.
View Article : Google Scholar
|
|
22
|
Kajarabille N and Latunde-Dada GO:
Programmed cell-death by ferroptosis: Antioxidants as mitigators.
Int J Mol Sci. 20:49682019. View Article : Google Scholar
|
|
23
|
Frazer DM and Anderson GJ: The regulation
of iron transport. Biofactors. 40:206–214. 2014. View Article : Google Scholar
|
|
24
|
El Hage Chahine JM, Hemadi M and Ha-Duong
NT: Uptake and release of metal ions by transferrin and interaction
with receptor 1. Biochim Biophys Acta. 1820:334–347. 2012.
View Article : Google Scholar
|
|
25
|
Ohgami RS, Campagna DR, Greer EL,
Antiochos B, McDonald A, Chen J, Sharp JJ, Fujiwara Y, Barker JE
and Fleming MD: Identification of a ferrireductase required for
efficient transferrin-dependent iron uptake in erythroid cells. Nat
Genet. 37:1264–1269. 2005. View
Article : Google Scholar
|
|
26
|
Kakhlon O and Cabantchik ZI: The labile
iron pool: Characterization, measurement, and participation in
cellular processes. Free Radic Biol Med. 33:1037–1046. 2002.
View Article : Google Scholar
|
|
27
|
Philpott CC, Ryu MS, Frey A and Patel S:
Cytosolic iron chaperones: Proteins delivering iron cofactors in
the cytosol of mammalian cells. J Biol Chem. 292:12764–12771. 2017.
View Article : Google Scholar
|
|
28
|
Harris ZL, Durley AP, Man TK and Gitlin
JD: Targeted gene disruption reveals an essential role for
ceruloplasmin in cellular iron efflux. Proc Natl Acad Sci USA.
96:10812–10817. 1999. View Article : Google Scholar
|
|
29
|
Zhang DL, Ghosh MC and Rouault TA: The
physiological functions of iron regulatory proteins in iron
homeostasis-an update. Front Pharmacol. 5:1242014. View Article : Google Scholar
|
|
30
|
Dixon SJ and Stockwell BR: The role of
iron and reactive oxygen species in cell death. Nat Chem Biol.
10:9–17. 2014. View Article : Google Scholar
|
|
31
|
Recalcati S, Correnti M, Gammella E, Raggi
C, Invernizzi P and Cairo G: Iron metabolism in liver cancer stem
cells. Front Oncol. 9:1492019. View Article : Google Scholar
|
|
32
|
Feng H, Schorpp K, Jin J, Yozwiak CE,
Hoffstrom BG, Decker AM, Rajbhandari P, Stokes ME, Bender HG, Csuka
JM, et al: Transferrin receptor is a specific ferroptosis marker.
Cell Rep. 30:3411–3423.e7. 2020. View Article : Google Scholar
|
|
33
|
Bai T, Lei P, Zhou H, Liang R, Zhu R, Wang
W, Zhou L and Sun Y: Sigma-1 receptor protects against ferroptosis
in hepatocellular carcinoma cells. J Cell Mol Med. 23:7349–7359.
2019. View Article : Google Scholar
|
|
34
|
Ye F, Chai W, Xie M, Yang M, Yu Y, Cao L
and Yang L: HMGB1 regulates erastin-induced ferroptosis via
RAS-JNK/p38 signaling in HL-60/NRASQ61L cells. Am J
Cancer Res. 9:730–739. 2019.
|
|
35
|
Turcu AL, Versini A, Khene N, Gaillet C,
Cañeque T, Müller S and Rodriguez R: DMT1 inhibitors kill cancer
stem cells by blocking lysosomal iron translocation. Chemistry.
26:7369–7373. 2020. View Article : Google Scholar
|
|
36
|
Yu H, Yang C, Jian L, Guo S, Chen R, Li K,
Qu F, Tao K, Fu Y, Luo F and Liu S: Sulfasalazine-induced
ferroptosis in breast cancer cells is reduced by the inhibitory
effect of estrogen receptor on the transferrin receptor. Oncol Rep.
42:826–838. 2019.
|
|
37
|
Chen GQ, Benthani FA, Wu J, Liang D, Bian
ZX and Jiang X: Artemisinin compounds sensitize cancer cells to
ferroptosis by regulating iron homeostasis. Cell Death Differ.
27:242–254. 2020. View Article : Google Scholar
|
|
38
|
de Carvalho CCCR and Caramujo MJ: The
various roles of fatty acids. Molecules. 23:25832018. View Article : Google Scholar
|
|
39
|
Dixon SJ, Winter GE, Musavi LS, Lee ED,
Snijder B, Rebsamen M, Superti-Furga G and Stockwell BR: Human
haploid cell genetics reveals roles for lipid metabolism genes in
nonapoptotic cell death. ACS Chem Biol. 10:1604–1609. 2015.
View Article : Google Scholar
|
|
40
|
Kagan VE, Mao G, Qu F, Angeli JP, Doll S,
Croix CS, Dar HH, Liu B, Tyurin VA, Ritov VB, et al: Oxidized
arachidonic and adrenic PEs navigate cells to ferroptosis. Nat Chem
Biol. 13:81–90. 2017. View Article : Google Scholar
|
|
41
|
Feng J, Lu PZ, Zhu GZ, Hooi SC, Wu Y,
Huang XW, Dai HQ, Chen PH, Li ZJ, Su WJ, et al: ACSL4 is a
predictive biomarker of sorafenib sensitivity in hepatocellular
carcinoma. Acta Pharmacol Sin. Jun 15–2020.(Epub ahead of print).
doi: 10.1038/s41401-020-0439-x. View Article : Google Scholar
|
|
42
|
Cheng J, Fan YQ, Liu BH, Zhou H, Wang JM
and Chen QX: ACSL4 suppresses glioma cells proliferation via
activating ferroptosis. Oncol Rep. 43:147–158. 2020.
|
|
43
|
Richard D, Kefi K, Barbe U, Bausero P and
Visioli F: Polyunsaturated fatty acids as antioxidants. Pharmacol
Res. 57:451–455. 2008. View Article : Google Scholar
|
|
44
|
Lewerenz J, Hewett SJ, Huang Y, Lambros M,
Gout PW, Kalivas PW, Massie A, Smolders I, Methner A, Pergande M,
et al: The cystine/glutamate antiporter system x(c)(−) in health
and disease: From molecular mechanisms to novel therapeutic
opportunities. Antioxid Redox Signal. 18:522–555. 2013. View Article : Google Scholar
|
|
45
|
Stockwell BR, Friedmann Angeli JP, Bayir
H, Bush AI, Conrad M, Dixon SJ, Fulda S, Gascón S, Hatzios SK,
Kagan VE, et al: Ferroptosis: A regulated cell death nexus linking
metabolism, redox biology, and disease. Cell. 171:273–285. 2017.
View Article : Google Scholar
|
|
46
|
Dixon SJ, Patel DN, Welsch M, Skouta R,
Lee ED, Hayano M, Thomas AG, Gleason CE, Tatonetti NP, Slusher BS
and Stockwell BR: Pharmacological inhibition of cystine-glutamate
exchange induces endoplasmic reticulum stress and ferroptosis.
Elife. 3:e025232014. View Article : Google Scholar
|
|
47
|
Vousden KH and Prives C: Blinded by the
light: The growing complexity of p53. Cell. 137:413–431. 2009.
View Article : Google Scholar
|
|
48
|
Jiang L, Kon N, Li T, Wang SJ, Su T,
Hibshoosh H, Baer R and Gu W: Ferroptosis as a p53-mediated
activity during tumour suppression. Nature. 520:57–62. 2015.
View Article : Google Scholar
|
|
49
|
Zhang Y, Zhuang L and Gan B: BAP1
suppresses tumor development by inducing ferroptosis upon SLC7A11
repression. Mol Cell Oncol. 6:15368452018. View Article : Google Scholar
|
|
50
|
Rojo de la Vega M, Chapman E and Zhang DD:
NRF2 and the hallmarks of cancer. Cancer Cell. 34:21–43. 2018.
View Article : Google Scholar
|
|
51
|
Yang WS and Stockwell BR: Synthetic lethal
screening identifies compounds activating iron-dependent,
nonapoptotic cell death in oncogenic-RAS-harboring cancer cells.
Chem Biol. 15:234–245. 2008. View Article : Google Scholar
|
|
52
|
Yang WS, SriRamaratnam R, Welsch ME,
Shimada K, Skouta R, Viswanathan VS, Cheah JH, Clemons PA, Shamji
AF, Clish CB, et al: Regulation of ferroptotic cancer cell death by
GPX4. Cell. 156:317–331. 2014. View Article : Google Scholar
|
|
53
|
Weiwer M, Bittker JA, Lewis TA, Shimada K,
Yang WS, MacPherson L, Dandapani S, Palmer M, Stockwell BR,
Schreiber SL and Munoz B: Development of small-molecule probes that
selectively kill cells induced to express mutant RAS. Bioorg Med
Chem Lett. 22:1822–1826. 2012. View Article : Google Scholar
|
|
54
|
Sun X, Ou Z, Chen R, Niu X, Chen D, Kang R
and Tang D: Activation of the p62-Keap1-NRF2 pathway protects
against ferroptosis in hepatocellular carcinoma cells. Hepatology.
63:173–184. 2016. View Article : Google Scholar
|
|
55
|
Shaw AT, Winslow MM, Magendantz M, Ouyang
C, Dowdle J, Subramanian A, Lewis TA, Maglathin RL, Tolliday N and
Jacks T: Selective killing of K-ras mutant cancer cells by small
molecule inducers of oxidative stress. Proc Natl Acad Sci USA.
108:8773–8778. 2011. View Article : Google Scholar
|
|
56
|
Louandre C, Marcq I, Bouhlal H, Lachaier
E, Godin C, Saidak Z, François C, Chatelain D, Debuysscher V,
Barbare JC, et al: The retinoblastoma (Rb) protein regulates
ferroptosis induced by sorafenib in human hepatocellular carcinoma
cells. Cancer Lett. 356:971–977. 2015. View Article : Google Scholar
|
|
57
|
Zhang K, Wu L, Zhang P, Luo M, Du J, Gao
T, O'Connell D, Wang G, Wang H and Yang Y: miR-9 regulates
ferroptosis by targeting glutamic-oxaloacetic transaminase GOT1 in
melanoma. Mol Carcinog. 57:1566–1576. 2018. View Article : Google Scholar
|
|
58
|
Wang M, Mao C, Ouyang L, Liu Y, Lai W, Liu
N, Shi Y, Chen L, Xiao D, Yu F, et al: Long noncoding RNA LINC00336
inhibits ferroptosis in lung cancer by functioning as a competing
endogenous RNA. Cell Death Differ. 26:2329–2343. 2019. View Article : Google Scholar
|
|
59
|
Luo M, Wu L, Zhang K, Wang H, Zhang T,
Gutierrez L, O'Connell D, Zhang P, Li Y, Gao T, et al: miR-137
regulates ferroptosis by targeting glutamine transporter SLC1A5 in
melanoma. Cell Death Differ. 25:1457–1472. 2018. View Article : Google Scholar
|
|
60
|
Gomaa A, Peng D, Chen Z, Soutto M,
Abouelezz K, Corvalan A and El-Rifai W: Epigenetic regulation of
AURKA by miR-4715-3p in upper gastrointestinal cancers. Sci Rep.
9:169702019. View Article : Google Scholar
|
|
61
|
Niu Y, Zhang J, Tong Y, Li J and Liu B:
Physcion 8-O-β-glucopyranoside induced ferroptosis via regulating
miR-103a-3p/GLS2 axis in gastric cancer. Life Sci. 237:1168932019.
View Article : Google Scholar
|
|
62
|
Tomita K, Fukumoto M, Itoh K, Kuwahara Y,
Igarashi K, Nagasawa T, Suzuki M, Kurimasa A and Sato T: miR-7-5p
is a key factor that controls radioresistance via intracellular
Fe2+ content in clinically relevant radioresistant
cells. Biochem Biophys Res Commun. 518:712–718. 2019. View Article : Google Scholar
|
|
63
|
Qin Z, Freitas E, Sullivan R, Mohan S,
Bacelieri R, Branch D, Romano M, Kearney P, Oates J, Plaisance K,
et al: Upregulation of xCT by KSHV-encoded microRNAs facilitates
KSHV dissemination and persistence in an environment of oxidative
stress. PLoS Pathog. 6:e10007422010. View Article : Google Scholar
|
|
64
|
Xiao FJ, Zhang D, Wu Y, Jia QH, Zhang L,
Li YX, Yang YF, Wang H, Wu CT and Wang LS: miRNA-17-92 protects
endothelial cells from erastin-induced ferroptosis through
targeting the A20-ACSL4 axis. Biochem Biophys Res Commun.
515:448–454. 2019. View Article : Google Scholar
|
|
65
|
Bai T, Liang R, Zhu R, Wang W, Zhou L and
Sun Y: MicroRNA-214-3p enhances erastin-induced ferroptosis by
targeting ATF4 in hepatoma cells. J Cell Physiol. 235:5637–5648.
2020. View Article : Google Scholar
|
|
66
|
Zhang HY, Zhang BW, Zhang ZB and Deng QJ:
Circular RNA TTBK2 regulates cell proliferation, invasion and
ferroptosis via miR-761/ITGB8 axis in glioma. Eur Rev Med Pharmacol
Sci. 24:2585–2600. 2020.
|
|
67
|
Mao C, Wang X, Liu Y, Wang M, Yan B, Jiang
Y, Shi Y, Shen Y, Liu X, Lai W, et al: A G3BP1-interacting lncRNA
promotes ferroptosis and apoptosis in cancer via nuclear
sequestration of p53. Cancer Res. 78:3484–3496. 2018.
|
|
68
|
Qi W, Li Z, Xia L, Dai J, Zhang Q, Wu C
and Xu S: lncRNA GABPB1-AS1 and GABPB1 regulate oxidative stress
during erastin-induced ferroptosis in HepG2 hepatocellular
carcinoma cells. Sci Rep. 9:161852019. View Article : Google Scholar
|
|
69
|
Schaar DG, Medina DJ, Moore DF, Strair RK
and Ting Y: miR-320 targets transferrin receptor 1 (CD71) and
inhibits cell proliferation. Exp Hematol. 37:245–255. 2009.
View Article : Google Scholar
|
|
70
|
Fu Y, Lin L and Xia L: miR-107 function as
a tumor suppressor gene in colorectal cancer by targeting
transferrin receptor 1. Cell Mol Biol Lett. 24:312019. View Article : Google Scholar
|
|
71
|
Babu KR and Muckenthaler MU: miR-148a
regulates expression of the transferrin receptor 1 in
hepatocellular carcinoma. Sci Rep. 9:15182019. View Article : Google Scholar
|
|
72
|
Miyazawa M, Bogdan AR, Hashimoto K and
Tsuji Y: Regulation of transferrin receptor-1 mRNA by the interplay
between IRE-binding proteins and miR-7/miR-141 in the 3′-IRE
stem-loops. RNA. 24:468–479. 2018. View Article : Google Scholar
|
|
73
|
Kindrat I, Tryndyak V, de Conti A,
Shpyleva S, Mudalige TK, Kobets T, Erstenyuk AM, Beland FA and
Pogribny IP: MicroRNA-152-mediated dysregulation of hepatic
transferrin receptor 1 in liver carcinogenesis. Oncotarget.
7:1276–1287. 2016. View Article : Google Scholar
|
|
74
|
Yoshioka Y, Kosaka N, Ochiya T and Kato T:
Micromanaging iron homeostasis: Hypoxia-inducible micro-RNA-210
suppresses iron homeostasis-related proteins. J Biol Chem.
287:34110–34119. 2012. View Article : Google Scholar
|
|
75
|
Xu D, Liu D, Wang B, Chen C, Chen Z, Li D,
Yang Y, Chen H and Kong MG: In Situ OH Generation from
O2- and H2O2 plays a critical role
in plasma-induced cell death. PLoS One. 10:e01282052015. View Article : Google Scholar
|
|
76
|
Chan JJ, Kwok ZH, Chew XH, Zhang B, Liu C,
Soong TW, Yang H and Tay Y: A FTH1 gene:pseudogene: microRNA
network regulates tumorigenesis in prostate cancer. Nucleic Acids
Res. 46:1998–2011. 2018. View Article : Google Scholar
|
|
77
|
Di bSanzo M, Chirillo R, Aversa I,
Biamonte F, Santamaria G, Giovannone ED, Faniello MC, Cuda G and
Costanzo F: shRNA targeting of ferritin heavy chain activates
H19/miR-675 axis in K562 cells. Gene. 657:92–99. 2018. View Article : Google Scholar
|
|
78
|
Ripa R, Dolfi L, Terrigno M, Pandolfini L,
Savino A, Arcucci V, Groth M, Terzibasi Tozzini E, Baumgart M and
Cellerino A: MicroRNA miR-29 controls a compensatory response to
limit neuronal iron accumulation during adult life and aging. BMC
Biol. 15:92017. View Article : Google Scholar
|
|
79
|
Zhang L, Ye Y, Tu H, Hildebrandt MA, Zhao
L, Heymach JV, Roth JA and Wu X: MicroRNA-related genetic variants
in iron regulatory genes, dietary iron intake, microRNAs and lung
cancer risk. Ann Oncol. 28:1124–1129. 2017. View Article : Google Scholar
|
|
80
|
Liu F, Chen Y, Chen B, Liu C and Xing J:
miR-935 promotes clear cell renal cell carcinoma migration and
invasion by targeting IREB2. Cancer Manag Res. 11:10891–10900.
2019. View Article : Google Scholar
|
|
81
|
Andolfo I, De Falco L, Asci R, Russo R,
Colucci S, Gorrese M, Zollo M and Iolascon A: Regulation of
divalent metal transporter 1 (DMT1) non-IRE isoform by the microRNA
Let-7d in erythroid cells. Haematologica. 95:1244–1252. 2010.
View Article : Google Scholar
|
|
82
|
Jiang S, Guo S, Li H, Ni Y, Ma W and Zhao
R: Identification and functional verification of MicroRNA-16 family
targeting intestinal divalent metal transporter 1 (DMT1) in vitro
and in vivo. Front Physiol. 10:8192019. View Article : Google Scholar
|
|
83
|
Soupene E and Kuypers FA: Mammalian
long-chain acyl-CoA synthetases. Exp Biol Med (Maywood).
233:507–521. 2008. View Article : Google Scholar
|
|
84
|
Yuan H, Li X, Zhang X, Kang R and Tang D:
Identification of ACSL4 as a biomarker and contributor of
ferroptosis. Biochem Biophys Res Commun. 478:1338–1343. 2016.
View Article : Google Scholar
|
|
85
|
Jiang X, Guo S, Zhang Y, Zhao Y, Li X, Jia
Y, Xu Y and Ma B: lncRNA NEAT1 promotes docetaxel resistance in
prostate cancer by regulating ACSL4 via sponging miR-34a-5p and
miR-204-5p. Cell Signal. 65:1094222020. View Article : Google Scholar
|
|
86
|
Park S, Oh J, Kim YI, Choe SK, Chun CH and
Jin EJ: Suppression of ABCD2 dysregulates lipid metabolism via
dysregulation of miR-141:ACSL4 in human osteoarthritis. Cell
Biochem Funct. 36:366–376. 2018. View Article : Google Scholar
|
|
87
|
Wu X, Zhi F, Lun W, Deng Q and Zhang W:
Baicalin inhibits PDGF-BB-induced hepatic stellate cell
proliferation, apoptosis, invasion, migration and activation via
the miR-3595/ACSL4 axis. Int J Mol Med. 41:1992–2002. 2018.
|
|
88
|
Bai C, Gao Y, Zhang X, Yang W and Guan W:
MicroRNA-34c acts as a bidirectional switch in the maturation of
insulin-producing cells derived from mesenchymal stem cells.
Oncotarget. 8:106844–106857. 2017. View Article : Google Scholar
|
|
89
|
Ooi J, Bernardo BC, Singla S, Patterson
NL, Lin RCY and McMullen JR: Identification of miR-34 regulatory
networks in settings of disease and antimiR-therapy: Implications
for treating cardiac pathology and other diseases. RNA Biol.
14:500–513. 2017. View Article : Google Scholar
|
|
90
|
Zhou L and Hussain MM: Human MicroRNA-548p
decreases hepatic apolipoprotein B secretion and lipid synthesis.
Arterioscler Thromb Vasc Biol. 37:786–793. 2017. View Article : Google Scholar
|
|
91
|
Cui M, Xiao Z, Sun B, Wang Y, Zheng M, Ye
L and Zhang X: Involvement of cholesterol in hepatitis B virus X
protein-induced abnormal lipid metabolism of hepatoma cells via
up-regulating miR-205-targeted ACSL4. Biochem Biophys Res Commun.
445:651–655. 2014. View Article : Google Scholar
|
|
92
|
Peng Y, Xiang H, Chen C, Zheng R, Chai J,
Peng J and Jiang S: miR-224 impairs adipocyte early differentiation
and regulates fatty acid metabolism. Int J Biochem Cell Biol.
45:1585–1593. 2013. View Article : Google Scholar
|
|
93
|
Park S, Oh J, Kim M and Jin EJ: Bromelain
effectively suppresses Kras-mutant colorectal cancer by stimulating
ferroptosis. Anim Cells Syst (Seoul). 22:334–340. 2018. View Article : Google Scholar
|
|
94
|
Yang WS, Kim KJ, Gaschler MM, Patel M,
Shchepinov MS and Stockwell BR: Peroxidation of polyunsaturated
fatty acids by lipoxygenases drives ferroptosis. Proc Natl Acad Sci
USA. 113:E4966–E4975. 2016. View Article : Google Scholar
|
|
95
|
Stoyanovsky DA, Tyurina YY, Shrivastava I,
Bahar I, Tyurin VA, Protchenko O, Jadhav S, Bolevich SB, Kozlov AV,
Vladimirov YA, et al: Iron catalysis of lipid peroxidation in
ferroptosis: Regulated enzymatic or random free radical reaction?
Free Radic Biol Med. 133:153–161. 2019. View Article : Google Scholar
|
|
96
|
Li MY, Liu LZ, Li W, Ng CSH, Liu Y, Kong
AWY, Zhao Z, Wang S, Qi H, Jia H, et al: Ambient fine particulate
matter inhibits 15-lipoxygenases to promote lung carcinogenesis. J
Exp Clin Cancer Res. 38:3592019. View Article : Google Scholar
|
|
97
|
Fredman G, Li Y, Dalli J, Chiang N and
Serhan CN: Self-limited versus delayed resolution of acute
inflammation: Temporal regulation of pro-resolving mediators and
microRNA. Sci Rep. 2:6392012. View Article : Google Scholar
|
|
98
|
Su K, Wang Q, Qi L, Hua D, Tao J, Mangan
CJ, Lou Y and Li L: MicroRNA-674-5p/5-LO axis involved in
autoimmune reaction of Concanavalin A-induced acute mouse liver
injury. Toxicol Lett. 258:101–107. 2016. View Article : Google Scholar
|
|
99
|
Wang D, Li Y, Zhang C, Li X and Yu J:
miR-216a-3p inhibits colorectal cancer cell proliferation through
direct targeting COX-2 and ALOX5. J Cell Biochem. 119:1755–1766.
2018. View Article : Google Scholar
|
|
100
|
Busch S, Auth E, Scholl F, Huenecke S,
Koehl U, Suess B and Steinhilber D: 5-lipoxygenase is a direct
target of miR-19a-3p and miR-125b-5p. J Immunol. 194:1646–1653.
2015. View Article : Google Scholar
|
|
101
|
Xue J, Min Z, Xia Z, Cheng B, Lan B, Zhang
F, Han Y, Wang K and Sun J: The hsa-miR-181a-5p reduces oxidation
resistance by controlling SECISBP2 in osteoarthritis. BMC
Musculoskelet Disord. 19:3552018. View Article : Google Scholar
|
|
102
|
Min Z, Guo Y, Sun M, Hussain S, Zhao Y,
Guo D, Huang H, Heng L, Zhang F, Ning Q, et al: Selenium-sensitive
miRNA-181a-5p targeting SBP2 regulates selenoproteins expression in
cartilage. J Cell Mol Med. 22:5888–5898. 2018. View Article : Google Scholar
|
|
103
|
Konstorum A, Tesfay L, Paul BT, Torti FM,
Laubenbacher RC and Torti SV: Systems biology of ferroptosis: A
modeling approach. J Theor Biol. 493:1102222020. View Article : Google Scholar
|
|
104
|
Zhang M, Sun W, Zhou M and Tang Y:
MicroRNA-27a regulates hepatic lipid metabolism and alleviates
NAFLD via repressing FAS and SCD1. Sci Rep. 7:144932017. View Article : Google Scholar
|
|
105
|
Guo Y, Yu J, Wang C, Li K, Liu B, Du Y,
Xiao F, Chen S and Guo F: miR-212-5p suppresses lipid accumulation
by targeting FAS and SCD1. J Mol Endocrinol. 59:205–217. 2017.
View Article : Google Scholar
|
|
106
|
Zhang M, Tang Y, Tang E and Lu W:
MicroRNA-103 represses hepatic de novo lipogenesis and alleviates
NAFLD via targeting FASN and SCD1. Biochem Biophys Res Commun.
524:716–722. 2020. View Article : Google Scholar
|
|
107
|
Mysore R, Zhou Y, Sädevirta S,
Savolainen-Peltonen H, Nidhina Haridas PA, Soronen J, Leivonen M,
Sarin AP, Fischer-Posovszky P, Wabitsch M, et al: MicroRNA-192*
impairs adipocyte triglyceride storage. Biochim Biophys Acta.
1861:342–351. 2016. View Article : Google Scholar
|
|
108
|
Zhang Y, Li C, Li H, Song Y, Zhao Y, Zhai
L, Wang H, Zhong R, Tang H and Zhu D: miR-378 activates the
pyruvate-PEP futile cycle and enhances lipolysis to ameliorate
obesity in mice. EbioMedicine. 5:93–104. 2016. View Article : Google Scholar
|
|
109
|
Guo J, Fang W, Sun L, Lu Y, Dou L, Huang
X, Tang W, Yu L and Li J: Ultraconserved element uc.372 drives
hepatic lipid accumulation by suppressing miR-195/miR4668
maturation. Nat Commun. 9:6122018. View Article : Google Scholar
|
|
110
|
El Helou R, Pinna G, Cabaud O, Wicinski J,
Bhajun R, Guyon L, Rioualen C, Finetti P, Gros A, Mari B, et al:
miR-600 acts as a bimodal switch that regulates breast cancer stem
cell fate through WNT signaling. Cell Rep. 18:2256–2268. 2017.
View Article : Google Scholar
|
|
111
|
Zhou Z, Lu Y, Wang Y, Du L, Zhang Y and
Tao J: Let-7c regulates proliferation and osteodifferentiation of
human adipose-derived mesenchymal stem cells under oxidative stress
by targeting SCD-1. Am J Physiol Cell Physiol. 316:C57–C69. 2019.
View Article : Google Scholar
|
|
112
|
Zeng Y, Lv Y, Tao L, Ma J, Zhang H, Xu H,
Xiao B, Shi Q, Ma K and Chen L: G6PC3, ALDOA and CS induction
accompanies miR-122 down-regulation in the mechanical asphyxia and
can serve as hypoxia biomarkers. Oncotarget. 7:74526–74536. 2016.
View Article : Google Scholar
|
|
113
|
Pinto SK, Lamon S, Stephenson EJ, Kalanon
M, Mikovic J, Koch LG, Britton SL, Hawley JA and Camera DM:
Expression of microRNAs and target proteins in skeletal muscle of
rats selectively bred for high and low running capacity. Am J
Physiol Endocrinol Metab. 313:E335–E343. 2017. View Article : Google Scholar
|
|
114
|
Luo H, Wang J, Liu D, Zang S, Ma N, Zhao
L, Zhang L, Zhang X and Qiao C: The lncRNA H19/miR-675 axis
regulates myocardial ischemic and reperfusion injury by targeting
PPARα. Mol Immunol. 105:46–54. 2019. View Article : Google Scholar
|
|
115
|
Zhao MW, Yang P and Zhao LL: Chlorpyrifos
activates cell pyroptosis and increases susceptibility on oxidative
stress-induced toxicity by miR-181/SIRT1/PGC-1α/Nrf2 signaling
pathway in human neuroblastoma SH-SY5Y cells: Implication for
association between chlorpyrifos and Parkinson's disease. Environ
Toxicol. 34:699–707. 2019. View Article : Google Scholar
|
|
116
|
Zhang Z, Wang N, Zhang Y, Zhao J and Lv J:
Downregulation of microRNA-302b-3p relieves oxygen-glucose
deprivation/re-oxygenation induced injury in murine hippocampal
neurons through up-regulating Nrf2 signaling by targeting
fibroblast growth factor 15/19. Chem Biol Interact. 309:1087052019.
View Article : Google Scholar
|
|
117
|
Wu L, Pan C, Wei X, Shi Y, Zheng J, Lin X
and Shi L: lncRNA KRAL reverses 5-fluorouracil resistance in
hepatocellular carcinoma cells by acting as a ceRNA against
miR-141. Cell Commun Signal. 16:472018. View Article : Google Scholar
|
|
118
|
Zhou B, Liu HY, Zhu BL and Yue AX:
MicroRNA-141 protects PC12 cells against
hypoxia/reoxygenation-induced injury via regulating Keap1-Nrf2
signaling pathway. J Bioenerg Biomembr. 51:291–300. 2019.
View Article : Google Scholar
|
|
119
|
Reziwan K, Sun D, Zhang B and Zhao Z:
MicroRNA-1225 activates Keap1-Nrf2-HO-1 signalling to inhibit
TNFα-induced osteoclastogenesis by mediating ROS generation. Cell
Biochem Funct. 37:256–265. 2019. View Article : Google Scholar
|
|
120
|
Duan Q and Si E: MicroRNA-25 aggravates
Aβ1-42-induced hippocampal neuron injury in Alzheimer's disease by
downregulating KLF2 via the Nrf2 signaling pathway in a mouse
model. J Cell Biochem. 120:15891–15905. 2019. View Article : Google Scholar
|
|
121
|
Zhao X, Jin Y, Li L, Xu L, Tang Z, Qi Y,
Yin L and Peng J: MicroRNA-128-3p aggravates doxorubicin-induced
liver injury by promoting oxidative stress via targeting Sirtuin-1.
Pharmacol Res. 146:1042762019. View Article : Google Scholar
|
|
122
|
Liu X, Zhao H, Luo C, Du D, Huang J, Ming
Q, Jin F, Wang D and Huang W: Acetaminophen responsive miR-19b
modulates SIRT1/Nrf2 signaling pathway in drug-induced
hepatotoxicity. Toxicol Sci. 170:476–488. 2019. View Article : Google Scholar
|
|
123
|
Chen YF, Wei YY, Yang CC, Liu CJ, Yeh LY,
Chou CH, Chang KW and Lin SC: miR-125b suppresses oral oncogenicity
by targeting the anti-oxidative gene PRXL2A. Redox Biol.
22:1011402019. View Article : Google Scholar
|
|
124
|
Ling Y, Li ZZ, Zhang JF, Zheng XW, Lei ZQ,
Chen RY and Feng JH: MicroRNA-494 inhibition alleviates acute lung
injury through Nrf2 signaling pathway via NQO1 in sepsis-associated
acute respiratory distress syndrome. Life Sci. 210:1–8. 2018.
View Article : Google Scholar
|
|
125
|
Gao M, Li C, Xu M, Liu Y, Cong M and Liu
S: lncRNA MT1DP aggravates cadmium-induced oxidative stress by
repressing the function of Nrf2 and is dependent on interaction
with miR-365. Adv Sci (Weinh). 5:18000872018. View Article : Google Scholar
|
|
126
|
Geng JF, Liu X, Zhao HB, Fan WF, Geng JJ
and Liu XZ: lncRNA UCA1 inhibits epilepsy and seizure-induced brain
injury by regulating miR-495/Nrf2-ARE signal pathway. Int J Biochem
Cell Biol. 99:133–139. 2018. View Article : Google Scholar
|
|
127
|
Wang X and Wang J: High-content hydrogen
water-induced downregulation of miR-136 alleviates non-alcoholic
fatty liver disease by regulating Nrf2 via targeting MEG3. Biol
Chem. 399:397–406. 2018. View Article : Google Scholar
|
|
128
|
Huang X, Gao Y, Qin J and Lu S: The
mechanism of long non-coding RNA MEG3 for hepatic
ischemia-reperfusion: Mediated by miR-34a/Nrf2 signaling pathway. J
Cell Biochem. 119:1163–1172. 2018. View Article : Google Scholar
|
|
129
|
Wu LL, Cai WP, Lei X, Shi KQ, Lin XY and
Shi L: NRAL mediates cisplatin resistance in hepatocellular
carcinoma via miR-340-5p/Nrf2 axis. J Cell Commun Signal.
13:99–112. 2019. View Article : Google Scholar
|
|
130
|
Zhang X, Chu X, Gong X, Zhou H and Cai C:
The expression of miR-125b in Nrf2-silenced A549 cells exposed to
hyperoxia and its relationship with apoptosis. J Cell Mol Med.
24:965–972. 2020. View Article : Google Scholar
|
|
131
|
Qin Z, Zhu K, Xue J, Cao P, Xu L, Xu Z,
Liang K, Zhu J and Jia R: Zinc-induced protective effect for
testicular ischemia-reperfusion injury by promoting antioxidation
via microRNA-101-3p/Nrf2 pathway. Aging (Albany NY). 11:9295–9309.
2019. View Article : Google Scholar
|
|
132
|
Dong XQ, Zhang YH, Shang XQ and Zeng YJ:
Effects of miR-101 on the proliferation and apoptosis of gastric
mucosal epithelial cells via Nrf2/ARE signaling pathway. Eur Rev
Med Pharmacol Sci. 23:5187–5194. 2019.
|
|
133
|
Chen J, Li C, Liu W, Yan B, Hu X and Yang
F: miRNA-155 silencing reduces sciatic nerve injury in diabetic
peripheral neuropathy. J Mol Endocrinol. 63:227–238. 2019.
View Article : Google Scholar
|
|
134
|
Cai Z, Zheng F, Ding Y, Zhan Y, Gong R, Li
J, Aschner M, Zhang Q, Wu S and Li H: Nrf2-regulated miR-380-3p
blocks the translation of Sp3 protein and its mediation of
paraquat-induced toxicity in mouse neuroblastoma N2a cells. Toxicol
Sci. 171:515–529. 2019. View Article : Google Scholar
|
|
135
|
Srinoun K, Sathirapongsasuti N,
Paiboonsukwong K, Sretrirutchai S, Wongchanchailert M and Fucharoen
S: miR-144 regulates oxidative stress tolerance of thalassemic
erythroid cell via targeting NRF2. Ann Hematol. 98:2045–2052. 2019.
View Article : Google Scholar
|
|
136
|
Yin Y, Liu H, Xu J, Shi D, Zhai L, Liu B,
Wang L, Liu G and Qin J: miR-144-3p regulates the resistance of
lung cancer to cisplatin by targeting Nrf2. Oncol Rep.
40:3479–3488. 2018.
|
|
137
|
Li B, Zhu X, Ward CM, Starlard-Davenport
A, Takezaki M, Berry A, Ward A, Wilder C, Neunert C, Kutlar A and
Pace BS: MIR-144-mediated NRF2 gene silencing inhibits fetal
hemoglobin expression in sickle cell disease. Exp Hematol.
70:85–96, e5. 2019. View Article : Google Scholar
|
|
138
|
Zhu X, Zhao Y, Hou W and Guo L: miR-153
regulates cardiomyocyte apoptosis by targeting Nrf2/HO-1 signaling.
Chromosome Res. 27:167–178. 2019. View Article : Google Scholar
|
|
139
|
Khadrawy O, Gebremedhn S, Salilew-Wondim
D, Taqi MO, Neuhoff C, Tholen E, Hoelker M, Schellander K and
Tesfaye D: Endogenous and exogenous modulation of Nrf2 mediated
oxidative stress response in bovine granulosa cells: Potential
implication for ovarian function. Int J Mol Sci. 20:16352019.
View Article : Google Scholar
|
|
140
|
Sun W, Yi Y, Xia G, Zhao Y, Yu Y, Li L,
Hua C, He B, Yang B, Yu C, et al: Nrf2-miR-129-3p-mTOR axis
controls an miRNA regulatory network involved in HDACi-induced
autophagy. Mol Ther. 27:1039–1050. 2019. View Article : Google Scholar
|
|
141
|
Huang Y, Huang L, Zhu G, Pei Z and Zhang
W: Downregulated microRNA-27b attenuates lipopolysaccharide-induced
acute lung injury via activation of NF-E2-related factor 2 and
inhibition of nuclear factor κB signaling pathway. J Cell Physiol.
234:6023–6032. 2019. View Article : Google Scholar
|
|
142
|
Liu QQ, Ren K, Liu SH, Li WM, Huang CJ and
Yang XH: MicroRNA-140-5p aggravates hypertension and oxidative
stress of atherosclerosis via targeting Nrf2 and Sirt2. Int J Mol
Med. 43:839–849. 2019.
|
|
143
|
Singh B, Ronghe AM, Chatterjee A, Bhat NK
and Bhat HK: MicroRNA-93 regulates NRF2 expression and is
associated with breast carcinogenesis. Carcinogenesis.
34:1165–1172. 2013. View Article : Google Scholar
|
|
144
|
Chorley BN, Campbell MR, Wang X, Karaca M,
Sambandan D, Bangura F, Xue P, Pi J, Kleeberger SR and Bell DA:
Identification of novel NRF2-regulated genes by ChIP-Seq: Influence
on retinoid X receptor alpha. Nucleic Acids Res. 40:7416–7429.
2012. View Article : Google Scholar
|
|
145
|
Zhang A, Qian Y and Qian J:
MicroRNA-152-3p protects neurons from
oxygen-glucose-deprivation/reoxygenation-induced injury through
upregulation of Nrf2/ARE antioxidant signaling by targeting PSD-93.
Biochem Biophys Res Commun. 517:69–76. 2019. View Article : Google Scholar
|
|
146
|
Kim JH, Lee KS, Lee DK, Kim J, Kwak SN, Ha
KS, Choe J, Won MH, Cho BR, Jeoung D, et al: Hypoxia-responsive
microRNA-101 promotes angiogenesis via heme oxygenase-1/vascular
endothelial growth factor axis by targeting cullin 3. Antioxid
Redox Signal. 21:2469–2482. 2014. View Article : Google Scholar
|
|
147
|
Xu D, Zhu H, Wang C, Zhu X, Liu G, Chen C
and Cui Z: microRNA-455 targets cullin 3 to activate Nrf2 signaling
and protect human osteoblasts from hydrogen peroxide. Oncotarget.
8:59225–59234. 2017. View Article : Google Scholar
|
|
148
|
Chen ZJ, Rong L, Huang D and Jiang Q:
Targeting cullin 3 by miR-601 activates Nrf2 signaling to protect
retinal pigment epithelium cells from hydrogen peroxide. Biochem
Biophys Res Commun. 515:679–687. 2019. View Article : Google Scholar
|
|
149
|
Kabaria S, Choi DC, Chaudhuri AD, Jain MR,
Li H and Junn E: MicroRNA-7 activates Nrf2 pathway by targeting
Keap1 expression. Free Radic Biol Med. 89:548–556. 2015. View Article : Google Scholar
|
|
150
|
Eades G, Yang M, Yao Y, Zhang Y and Zhou
Q: miR-200a regulates Nrf2 activation by targeting Keap1 mRNA in
breast cancer cells. J Biol Chem. 286:40725–40733. 2011. View Article : Google Scholar
|
|
151
|
Wang J, Ishfaq M, Xu L, Xia C, Chen C and
Li J: METTL3/m6A/miRNA-873-5p attenuated oxidative
stress and apoptosis in colistin-induced kidney injury by
modulating Keap1/Nrf2 pathway. Front Pharmacol. 10:5172019.
View Article : Google Scholar
|
|
152
|
Xiao X, Lu Z, Lin V, May A, Shaw DH, Wang
Z, Che B, Tran K, Du H and Shaw PX: MicroRNA miR-24-3p reduces
apoptosis and regulates Keap1-Nrf2 pathway in mouse cardiomyocytes
responding to ischemia/reperfusion injury. Oxid Med Cell Longev.
2018:70421052018. View Article : Google Scholar
|
|
153
|
Huang R, Ma J, Niu B, Li J, Chang J, Zhang
Y, Liu P and Luan X: miR-34b protects against focal cerebral
ischemia-reperfusion (I/R) injury in rat by targeting Keap1. J
Stroke Cerebrovasc Dis. 28:1–9. 2019. View Article : Google Scholar
|
|
154
|
Ding X, Jian T, Wu Y, Zuo Y, Li J, Lv H,
Ma L, Ren B, Zhao L, Li W and Chen J: Ellagic acid ameliorates
oxidative stress and insulin resistance in high glucose-treated
HepG2 cells via miR-223/keap1-Nrf2 pathway. Biomed Pharmacother.
110:85–94. 2019. View Article : Google Scholar
|
|
155
|
Li X, Zhang W, Xiao M, Wang F, Zhou P,
Yang J and Chen X: MicroRNA-146b-5p protects oligodendrocyte
precursor cells from oxygen/glucose deprivation-induced injury
through regulating Keap1/Nrf2 signaling via targeting
bromodomain-containing protein 4. Biochem Biophys Res Commun.
513:875–882. 2019. View Article : Google Scholar
|
|
156
|
Sun X, Li X, Ma S, Guo Y and Li Y:
MicroRNA-98-5p ameliorates oxygen-glucose deprivation/reoxygenation
(OGD/R)-induced neuronal injury by inhibiting Bach1 and promoting
Nrf2/ARE signaling. Biochem Biophys Res Commun. 507:114–121. 2018.
View Article : Google Scholar
|
|
157
|
Feng X, Zhao J, Ding J, Shen X, Zhou J and
Xu Z: lncRNA Blnc1 expression and its effect on renal fibrosis in
diabetic nephropathy. Am J Transl Res. 11:5664–5672. 2019.
|
|
158
|
Li H, Zhu X, Hu L, Li Q, Ma J and Yan J:
Loss of exosomal MALAT1 from ox-LDL-treated vascular endothelial
cells induces maturation of dendritic cells in atherosclerosis
development. Cell Cycle. 18:2255–2267. 2019. View Article : Google Scholar
|
|
159
|
Fan JB, Zhang Y, Liu W, Zhu XH, Xu DW,
Zhao JN and Cui ZM: Long non-coding RNA MALAT1 protects human
osteoblasts from dexamethasone-induced injury via activation of
PPM1E-AMPK signaling. Cell Physiol Biochem. 51:31–45. 2018.
View Article : Google Scholar
|
|
160
|
Chen J, Ke S, Zhong L, Wu J, Tseng A,
Morpurgo B, Golovko A, Wang G, Cai JJ, Ma X, et al: Long noncoding
RNA MALAT1 regulates generation of reactive oxygen species and the
insulin responses in male mice. Biochem Pharmacol. 152:94–103.
2018. View Article : Google Scholar
|
|
161
|
Amodio N, Stamato MA, Juli G, Morelli E,
Fulciniti M, Manzoni M, Taiana E, Agnelli L, Cantafio MEG, Romeo E,
et al: Drugging the lncRNA MALAT1 via LNA gapmeR ASO inhibits gene
expression of proteasome subunits and triggers anti-multiple
myeloma activity. Leukemia. 32:1948–1957. 2018. View Article : Google Scholar
|
|
162
|
Zeng R, Zhang R, Song X, Ni L, Lai Z, Liu
C and Ye W: The long non-coding RNA MALAT1 activates Nrf2 signaling
to protect human umbilical vein endothelial cells from hydrogen
peroxide. Biochem Biophys Res Commun. 495:2532–2538. 2018.
View Article : Google Scholar
|
|
163
|
Joo MS, Shin SB, Kim EJ, Koo JH, Yim H and
Kim SG: Nrf2-lncRNA controls cell fate by modulating p53-dependent
Nrf2 activation as an miRNA sponge for Plk2 and p21cip1.
FASEB J. 33:7953–7969. 2019. View Article : Google Scholar
|
|
164
|
Liu M, Song Y and Han Z: Study on the
effect of lncRNA AK094457 on OX-LDL induced vascular smooth muscle
cells. Am J Transl Res. 11:5623–5633. 2019.
|
|
165
|
Porsch M, Özdemir E, Wisniewski M, Graf S,
Bull F, Hoffmann K, Ignatov A, Haybaeck J, Grosse I, Kalinski T and
Nass N: Time resolved gene expression analysis during tamoxifen
adaption of MCF-7 cells identifies long non-coding RNAs with
prognostic impact. RNA Biol. 16:661–674. 2019. View Article : Google Scholar
|
|
166
|
Xiao X, Yuan Q, Chen Y, Huang Z, Fang X,
Zhang H, Peng L and Xiao P: lncRNA ENST00000453774.1 contributes to
oxidative stress defense dependent on autophagy mediation to reduce
extracellular matrix and alleviate renal fibrosis. J Cell Physiol.
234:9130–9343. 2019. View Article : Google Scholar
|
|
167
|
Gao M, Zhao B, Chen M, Liu Y, Xu M, Wang
Z, Liu S and Zhang C: Nrf-2-driven long noncoding RNA ODRUL
contributes to modulating silver nanoparticle-induced effects on
erythroid cells. Biomaterials. 130:14–27. 2017. View Article : Google Scholar
|
|
168
|
Dong H, Wang W, Mo S, Liu Q, Chen X, Chen
R, Zhang Y, Zou K, Ye M, He X, et al: Long non-coding RNA SNHG14
induces trastuzumab resistance of breast cancer via regulating
PABPC1 expression through H3K27 acetylation. J Cell Mol Med.
22:4935–4947. 2018. View Article : Google Scholar
|
|
169
|
Luzon-Toro B, Fernández RM,
Martos-Martínez JM, Rubio-Manzanares-Dorado M, Antiñolo G and
Borrego S: lncRNA LUCAT1 as a novel prognostic biomarker for
patients with papillary thyroid cancer. Sci Rep. 9:143742019.
View Article : Google Scholar
|
|
170
|
Sun Z, Huang G and Cheng H: Transcription
factor Nrf2 induces the up-regulation of lncRNA TUG1 to promote
progression and adriamycin resistance in urothelial carcinoma of
the bladder. Cancer Manag Res. 11:6079–6090. 2019. View Article : Google Scholar
|
|
171
|
Zhang Z, Xiong R, Li C, Xu M and Guo M:
lncRNA TUG1 promotes cisplatin resistance in esophageal squamous
cell carcinoma cells by regulating Nrf2. Acta Biochim Biophys Sin
(Shanghai). 51:826–833. 2019. View Article : Google Scholar
|
|
172
|
Gong W, Li J, Zhu G, Wang Y, Zheng G and
Kan Q: Chlorogenic acid relieved oxidative stress injury in retinal
ganglion cells through IncRNA-TUG1/Nrf2. Cell Cycle. 18:1549–1559.
2019. View Article : Google Scholar
|
|
173
|
Wu XC, Wang SH, Ou HH, Zhu B, Zhu Y, Zhang
Q, Yang Y and Li H: The NmrA-like family domain containing 1
pseudogene Loc344887 is amplified in gallbladder cancer and
promotes epithelial-mesenchymal transition. Chem Biol Drug Des.
90:456–463. 2017. View Article : Google Scholar
|
|
174
|
Zheng ZG, Xu H, Suo SS, Xu XL, Ni MW, Gu
LH, Chen W, Wang LY, Zhao Y, Tian B and Hua YJ: The essential role
of H19 contributing to cisplatin resistance by regulating
glutathione metabolism in high-grade serous ovarian cancer. Sci
Rep. 6:260932016. View Article : Google Scholar
|
|
175
|
Li HQ, Wu YB, Yin CS, Chen L, Zhang Q and
Hu LQ: Obestatin attenuated doxorubicin-induced cardiomyopathy via
enhancing long noncoding Mhrt RNA expression. Biomed Pharmacother.
81:474–481. 2016. View Article : Google Scholar
|
|
176
|
Zhou L, Xu DY, Sha WG, Shen L, Lu GY and
Yin X: Long non-coding MIAT mediates high glucose-induced renal
tubular epithelial injury. Biochem Biophys Res Commun. 468:726–732.
2015. View Article : Google Scholar
|
|
177
|
Yuan X, Wang J, Tang X, Li Y, Xia P and
Gao X: Berberine ameliorates nonalcoholic fatty liver disease by a
global modulation of hepatic mRNA and lncRNA expression profiles. J
Transl Med. 13:242015. View Article : Google Scholar
|
|
178
|
Zhao F, Lin T, He W, Han J, Zhu D, Hu K,
Li W, Zheng Z, Huang J and Xie W: Knockdown of a novel lincRNA
AATBC suppresses proliferation and induces apoptosis in bladder
cancer. Oncotarget. 6:1064–1078. 2015. View Article : Google Scholar
|
|
179
|
Zhang L, Liu Z, Li X, Zhang P, Wang J, Zhu
D, Chen X and Ye L: Low long non-coding RNA HOTAIR expression is
associated with down-regulation of Nrf2 in the spermatozoa of
patients with asthenozoospermia or oligoasthenozoospermia. Int J
Clin Exp Pathol. 8:14198–14205. 2015.
|
|
180
|
Li CP, Wang SH, Wang WQ, Song SG and Liu
XM: Long noncoding RNA-Sox2OT knockdown alleviates diabetes
mellitus-induced retinal ganglion cell (RGC) injury. Cell Mol
Neurobiol. 37:361–369. 2017. View Article : Google Scholar
|
|
181
|
Wang Y, Wang J, Wei LJ, Zhu DM and Zhang
JS: Biological function and mechanism of lncRNA-MEG3 in Tenon's
capsule fibroblasts proliferation: By MEG3-Nrf2 protein
interaction. Biomed Pharmacother. 87:548–554. 2017. View Article : Google Scholar
|
|
182
|
Li Y, Gao X, Wang Z, Liu W, Xu F, Hu Y, Li
Y and Shi L: Circular RNA 4099 aggravates hydrogen peroxide-induced
injury by down-regulating microRNA-706 in L02 cells. Life Sci.
241:1168262020. View Article : Google Scholar
|
|
183
|
Drayton RM, Dudziec E, Peter S, Bertz S,
Hartmann A, Bryant HE and Catto JW: Reduced expression of miRNA-27a
modulates cisplatin resistance in bladder cancer by targeting the
cystine/glutamate exchanger SLC7A11. Clin Cancer Res. 20:1990–2000.
2014. View Article : Google Scholar
|
|
184
|
Wu Y, Sun X, Song B, Qiu X and Zhao J:
miR-375/SLC7A11 axis regulates oral squamous cell carcinoma
proliferation and invasion. Cancer Med. 6:1686–1697. 2017.
View Article : Google Scholar
|
|
185
|
Liu XX, Li XJ, Zhang B, Liang YJ, Zhou CX,
Cao DX, He M, Chen GQ, He JR and Zhao Q: MicroRNA-26b is
underexpressed in human breast cancer and induces cell apoptosis by
targeting SLC7A11. FEBS Lett. 585:1363–1367. 2011. View Article : Google Scholar
|
|
186
|
Luo Y, Wang C, Yong P, Ye P, Liu Z, Fu Z,
Lu F, Xiang W, Tan W and Xiao J: Decreased expression of the long
non-coding RNA SLC7A11-AS1 predicts poor prognosis and promotes
tumor growth in gastric cancer. Oncotarget. 8:112530–112549. 2017.
View Article : Google Scholar
|
|
187
|
Yuan J, Liu Z and Song R: Antisense lncRNA
As-SLC7A11 suppresses epithelial ovarian cancer progression mainly
by targeting SLC7A11. Pharmazie. 72:402–407. 2017.
|
|
188
|
Watai Y, Kobayashi A, Nagase H, Mizukami
M, McEvoy J, Singer JD, Itoh K and Yamamoto M: Subcellular
localization and cytoplasmic complex status of endogenous Keap1.
Genes Cells. 12:1163–1178. 2007. View Article : Google Scholar
|
|
189
|
Fan Z, Wirth AK, Chen D, Wruck CJ, Rauh M,
Buchfelder M and Savaskan N: Nrf2-Keap1 pathway promotes cell
proliferation and diminishes ferroptosis. Oncogenesis. 6:e3712017.
View Article : Google Scholar
|
|
190
|
Xian S, Li J and Zhang Z: miR-26b inhibits
isoproterenol-induced cardiac fibrosis via the Keap1/Nrf2 signaling
pathway. Exp Ther Med. 19:2067–2074. 2020.
|
|
191
|
Li SP, Cheng WN, Li Y, Xu HB, Han H, Li P
and Zhang DX: Keap1-targeting microRNA-941 protects endometrial
cells from oxygen and glucose deprivation-re-oxygenation via
activation of Nrf2 signaling. Cell Commun Signal. 18:322020.
View Article : Google Scholar
|
|
192
|
Jiang Z, Wu J, Ma F, Jiang J, Xu L, Du L,
Huang W, Wang Z, Jia Y, Lu L and Wu H: MicroRNA-200a improves
diabetic endothelial dysfunction by targeting KEAP1/NRF2. J
Endocrinol. 245:129–140. 2020. View Article : Google Scholar
|
|
193
|
Wang X, Ye L, Zhang K, Gao L, Xiao J and
Zhang Y: Upregulation of microRNA-200a in bone marrow mesenchymal
stem cells enhances the repair of spinal cord injury in rats by
reducing oxidative stress and regulating Keap1/Nrf2 pathway. Artif
Organs. 44:744–752. 2020. View Article : Google Scholar
|
|
194
|
Duan FG, Wang MF, Cao YB, Dan Li, Li RZ,
Fan XX, Khan I, Lai HL, Zhang YZ, Hsiao WW, et al: MicroRNA-421
confers paclitaxel resistance by binding to the KEAP1 3′UTR and
predicts poor survival in non-small cell lung cancer. Cell Death
Dis. 10:8212019. View Article : Google Scholar
|
|
195
|
Xu XZ, Tang Y, Cheng LB, Yao J, Jiang Q,
Li KR and Zhen YF: Targeting Keap1 by miR-626 protects retinal
pigment epithelium cells from oxidative injury by activating Nrf2
signaling. Free Radic Biol Med. 143:387–396. 2019. View Article : Google Scholar
|
|
196
|
Akdemir B, Nakajima Y, Inazawa J and Inoue
J: miR-432 induces NRF2 stabilization by directly targeting KEAP1.
Mol Cancer Res. 15:1570–1578. 2017. View Article : Google Scholar
|
|
197
|
Gao M, Monian P, Quadri N, Ramasamy R and
Jiang X: Glutaminolysis and transferrin regulate ferroptosis. Mol
Cell. 59:298–308. 2015. View Article : Google Scholar
|
|
198
|
Wang J, Wang B, Ren H and Chen W: miR-9-5p
inhibits pancreatic cancer cell proliferation, invasion and
glutamine metabolism by targeting GOT1. Biochem Biophys Res Commun.
509:241–248. 2019. View Article : Google Scholar
|
|
199
|
Moskalev EA, Schubert M and Hoheisel JD:
RNA-directed epigenomic reprogramming: An emerging principle of a
more targeted cancer therapy? Genes Chromosomes Cancer. 51:105–110.
2012. View Article : Google Scholar
|
|
200
|
Li Y, Duo Y, Zhai P, He L, Zhong K, Zhang
Y, Huang K, Luo J, Zhang H and Yu X: Dual targeting delivery of
miR-328 by functionalized mesoporous silica nanoparticles for
colorectal cancer therapy. Nanomedicine (Lond). 13:1753–1772. 2018.
View Article : Google Scholar
|
|
201
|
Li F, Wang F, Zhu C, Wei Q, Zhang T and
Zhou YL: miR-221 suppression through nanoparticle-based miRNA
delivery system for hepatocellular carcinoma therapy and its
diagnosis as a potential biomarker. Int J Nanomedicine.
13:2295–2307. 2018. View Article : Google Scholar
|
|
202
|
Bader AG: miR-34-a microRNA replacement
therapy is headed to the clinic. Front Genet. 3:1202012. View Article : Google Scholar
|
|
203
|
Gong N, Teng X, Li J and Liang XJ:
Antisense oligonucleotide-conjugated nanostructure-targeting lncRNA
MALAT1 inhibits cancer metastasis. ACS Appl Mater Interfaces.
11:37–42. 2019. View Article : Google Scholar
|
|
204
|
Wang WT, Han C, Sun YM, Chen TQ and Chen
YQ: Noncoding RNAs in cancer therapy resistance and targeted drug
development. J Hematol Oncol. 12:552019. View Article : Google Scholar
|
|
205
|
De Duve C and Wattiaux R: Functions of
lysosomes. Annu Rev Physiol. 28:435–492. 1966. View Article : Google Scholar
|
|
206
|
Kerr JF, Wyllie AH and Currie AR:
Apoptosis: A basic biological phenomenon with wide-ranging
implications in tissue kinetics. Br J Cancer. 26:239–257. 1972.
View Article : Google Scholar
|
|
207
|
Cookson BT and Brennan MA:
Pro-inflammatory programmed cell death. Trends Microbiol.
9:113–114. 2001. View Article : Google Scholar
|
|
208
|
Degterev A, Huang Z, Boyce M, Li Y, Jagtap
P, Mizushima N, Cuny GD, Mitchison TJ, Moskowitz MA and Yuan J:
Chemical inhibitor of nonapoptotic cell death with therapeutic
potential for ischemic brain injury. Nat Chem Biol. 1:112–119.
2005. View Article : Google Scholar
|
|
209
|
Overholtzer M, Mailleux AA, Mouneimne G,
Normand G, Schnitt SJ, King RW, Cibas ES and Brugge JS: A
nonapoptotic cell death process, entosis, that occurs by
cell-in-cell invasion. Cell. 131:966–979. 2007. View Article : Google Scholar
|