Introduction
Colorectal cancer (CRC) is one of the most common
types of cancer worldwide, especially in China within the last
decade (1). CRC progression, from
adenoma to adenocarcinoma, is a multistep process (2). CRC results from a series of genetic
and epigenetic alterations in key growth regulatory genes, which
endow the colorectal cells with proliferative, survival, and
metastatic advantages (3). The
median overall survival (OS) of metastatic CRC (mCRC) is less than
3 years. Chemotherapy, radiotherapy, targeted therapy, and
immunotherapy are mainly palliative treatments (4). Therefore, there is a need for more
efficacious strategies to prolong the OS of mCRC.
Telomere maintenance 2 (TELO2, also known as tel2)
was first identified in Saccharomyces cerevisiae and was required
in telomere length regulation and telomere position (5). TELO2 binds with phosphatidylinositol
3-kinase-related kinase (PIKK) family members, including ATM
(ataxia telangiectasia mutated), ATR (ATM- and rad3-related), and
mammalian target of rapamycin (mTOR) (6), and it maintains the stability of
PIKKs, which leads to the translation, growth, and autophagic
regulation of cells (7,8). The PI3K/Akt/mTOR pathway is frequently
dysregulated in CRC patients, and the therapeutic potential of
targeting mTOR in the treatment of CRC is rational and has been
shown to improve disease progression (9). With regard to mTOR, TELO2 is not only
critical for protein stability, but it is also essential for the
integrity of mTOR complexes (10,11).
Degradation of TELO2 promotes the survival of multiple myeloma
under growth factor withdrawal (12). Although there are several reports
indicating that TELO2 demonstrates an oncogenic profile in solid
tumors, such as breast cancer (13)
and high-grade gliomas (14), the
role and molecular mechanism of TELO2 in CRC tumorigenesis have yet
to be defined. A study on TELO2 in CRC demonstrates that a germline
mutation of TELO2 regulated the senescence pathway and is related
to sessile serrated adenomas and adenocarcinoma progression
(15). Additionally,
rapamycin-insensitive companion of mTOR (RICTOR), a specific
adaptor of mTORC2 (16), plays an
important role in cancer cell proliferation, autophagy, migration,
and invasion via activation of protein kinase B (Akt) (17,18)
and is positively correlated with prognosis in CRC (19). Thus, we hypothesized that TELO2 is
important in the development of CRC via mTOR and RICTOR. In this
study, we performed bioinformatics combined with in vitro
experiments and analysis of clinical characteristics of CRC
patients in order to characterize the effect of TELO2 on CRC
progression.
Materials and methods
Cell lines and tissue specimens
The anti-TELO2 antibody (ab122722, 1:500 dilution)
was purchased from Abcam (Cambridge). The anti-RICTOR antibody
(cat. no. A300-459A-M, 1:1,000 dilution) was purchased from Bethyl
(Montgomery). Antibodies against mTOR (cat. no. 2972, 1:1,000
dilution), RAPTOR (cat. no. 4978, 1:1,000 dilution),
phosphorylated-Akt (Ser473, cat. no. 4051, 1:500 dilution), Akt
(cat. no. 9272, 1:500 dilution), S6K1pT389 (cat. no. 9204, 1:500
dilution), S6K1 (cat. no. 9202, 1:500 dilution), 4E-BP1pS65 (no.
9451, 1:500 dilution), 4E-BP1 (no. 9452, 1:500 dilution), tubulin
(cat. no. 2146, 1:1,000 dilution), ubiquitin (cat. no. 43124,
1:1,000 dilution), and Myc-tag (cat. no. 2276, 1:1,000 dilution)
were obtained from Cell Signaling. The anti-β-actin antibody (cat.
no. A1978, 1:2,000 dilution) was purchased from Sigma (Milwaukee).
LoVo cells, a human colorectal cancer cell line (CBP60032, Cobioer,
Shanghai), was selected from the GENT2 database, authenticated by
STR analysis (September, 2016) and maintained in RPMI-1640 basic
media containing 10% fetal calf serum (FBS, no. 10100, Gibco,
Shanghai) at 37°C with 5% CO2. Serum-deprived conditions
were defined as maintaining cells in RPMI-1640 media with 0.02%
FBS. Four pairs of CRC tissues (on the edge of cancerous specimens
without necrosis) and normal adjacent tissues (2 cm away from the
cancer margin) were collected from resected surgical specimens in
our hospital from April, 2019 to December, 2019. There were 2 males
and 2 females, with an age range of 53 to 81. Tissue array slides
(no. HColA180Su10) were purchased from Superchip (Shanghai, China).
All of the patients signed informed consent for the use of their
tissues, and the Institutional Review Committee of Gannan Medical
University (Ganzhou, Jiangxi) approved this study (no.
2018023).
Tissue array and immunohistochemistry
(IHC)
For IHC experiments, tissue array slides were used
with 100 CRC tissue spots and 80 adjacent normal tissue spots.
Endogenous peroxidase activity was blocked by incubation with
hydrogen peroxide, and antigen retrieval was then performed by
incubation in a pepsin solution at 37°C. The sections were then
incubated with an anti-TELO2 antibody (1:500) or anti-RICTOR
(1:4,000) antibody at 4°C overnight, followed by incubation with
the biotin-linked anti-rabbit IgG (Dako) and then with the ABC
complex (ab8647; Cambridge). The staining sections were then
reviewed and scored as follows by a pathologist with over 15 years
of experience: Cells with <10% staining were scored as negative
staining (−, 1); cells with 10-49% staining were scored as (+, 2);
cells with 50-74% staining were scored as (++, 3); and cells with
75-100% staining were scored as (+++, 4). The staining color was
scored as light-yellow particle (1), brown-yellow particle (2), and brown particle (3). The final score was defined as staining
number score multiplied by staining color score (20). The scores of negative expression
were between 0 and 5, and the scores that exceeded 5 were
identified as positive expression.
RNA isolation and quantitative
PCR
Cells were harvested and total RNA was extracted
from LoVo cells (transfected with scramble siRNA or RICTOR siRNA)
using TRIzol reagent (no. 15596026; Gibco) as previously described
(21). qPCR was performed using
Power SYBR-Green PCR Master Mix (no. 4309155; Applied Biosystems).
The qPCR conditions were 5 min at 95°C, followed by 50 cycles of
95°C for 15 sec, 56°C for 30 sec, and 72°C for 40 sec, 72°C for 5
min was included for a final extension. The primer sequences used
in RT-PCR were as follows: TELO2 forward,
5′-GTCCCTGAAGCGGTATCTCG-3′ and reverse, 5′-TGCTGGCAAGACATCTGAGG-3′
(107 bp); GAPDH forward, 5′-GTCAACGGATTTGGTCGTATTG-3′ and reverse,
5′-CTCCTGGAAGATGGTGATGGG-3′ (216 bp).
Transient transfection and generation
of stably transfected cells
Knockdown of RICTOR expression was performed by
transfecting cells with siRNA duplexes (sc-61478; Santa Cruz
Biotechnology) using Lipofectamine 3000 (Thermo Fischer
Scientific). Scrambled RNA (scr-siRNA) was used as a negative
control. The TELO2 shRNA plasmid (sc-93308-SH) was purchased from
Santa Cruz Biotechnology. The pLPC-Myc-TELO2 (no. 22802) plasmid
was purchased from Addgene. The indicated plasmids were transfected
into LoVo cells. Transfects were cultured in RPMI-1640 medium
supplemented with puromycin (Medchemexpress LLC) to generate stable
cell lines.
Western blot analysis and
co-immunoprecipitation (Co-IP)
Whole cell lysates and cytoplasmic protein were
prepared following the instructions of a protein extraction kit
from Sangon Biotech (C006255 and C510001). For western blot
analysis, 30 µg of whole protein lysates was used to detect the
indicated protein, 4-12% NuPAGE Bis-Tris gel (NP0322BOX,
Invitrogen) was used for electrophoresis. STRING database
(https://string-db.org) was used for predicting
the protein combination before experiments. For Co-IP assay, 1%
Triton (strong lysis buffer which can depolymerize the mTOR
complex) or CHAPS (mild buffer which can maintain the integrated
mTOR complex) lysis buffer was used. After pre-incubation with
protein G PLUS-Agarose beads (20423, Thermo Fisher Scientific), an
equal amount of protein (500 µg) was incubated with the indicated
antibodies (1 µg, RICTOR or TELO2). 1% Triton or CHAPS buffer was
used to wash beads at 4°C, 200 × g three times. Then 1% of the
input was loaded to detect the protein level. PVDF (LC2002; Thermo
Fisher) was used as transmembrane. ImageJ software was used to scan
the grey scores of images. Cycloheximide (CHX; Calbiochem) was used
for protein chasing experiment.
WST-1 cell proliferation assay
Each group of isolated tumor cells was seeded into 5
wells of a 96-well plate and incubated for 24, 48, or 72 h in
RPMI-1640 containing 10% FBS. After washing, fixing, and
permeabilizing the cells, 10 µl of WST-1 was added to each well and
incubated at 37°C for 4 h. The absorbance at 490 nm was measured
with a microplate reader (Thermo Fisher Scientific, Vantaa,
Finland). The survival rate was calculated using the proportion
between the absorbance of different cells.
Anchorage-independent cell growth
assay
LoVo cells (1.25×103) transfected with
different plasmids were seeded into medium with 0.35% agar and
plated in triplicate on plates containing a 0.7% agar base.
Colonies were stained with Coomassie Blue (no. sc-24972; Santa Cruz
Biotechnology). Colonies containing at least 50 cells were counted
using Photoshop software (Adobe Systems).
Cell cycle analysis
At 48 h post-transfection, LoVo cells with scramble
shRNA or TELO2 shRNA was digested with trypsin (no. 25300054;
Thermo Fischer Scientific), washed twice with PBS, and collected by
centrifugating at 1,000 × g for 5 min. For cell cycle analysis, the
cells were fixed with pre-cooled 70% ethanol at 4°C overnight and
digested with 200 mg/ml ribonuclease A (KeyGen BioTech, KGA511) at
37°C for 30 min. The cells were then stained by the addition of 100
µl of propidium iodide (KeyGen BioTech, KGA511) at 4°C, 30 min in
the dark via flow cytometry (FCM) analysis.
Cell migration and invasion
assays
Cell migration was assessed by a wound healing assay
(22). Briefly, the indicated cells
were seeded, cultured overnight until over 95% confluency, and then
wounded with a pipette tip. The media were changed to remove cell
debris, and images were captured at 24 and 48 h post-wounding. Cell
invasion was assessed using the Matrigel Invasion Chamber (BD
Biosciences) according to the manufacturer's instructions. The
cells (1×105) were re-suspended in serum-free media and
placed on each Transwell membrane filter insert, with the lower
chamber filled with complete medium. After 24-h incubation, the
cells were stained with 0.005% crystal violet at room temperature
for 10 min and counted under a microscope.
Statistical analysis
Data were presented as the mean ± standard deviation
of at least three independent experiments, using SPSS 19.0 version
(SPSS, Inc.). Groups with different treatments were compared using
a two-tailed Student's t-test. ANOVA was used to compare more than
two groups, and Bonferroni test was used in the post-hoc
comparison. The expression of TELO2 between cancer tissue and
adjacent normal tissue was compared using a two-independent
non-parametric test (Mann-Whitney U test) in the IHC assay, a
heatmap was used to present the expression correlations between two
proteins. Kaplan-Meier and log rank tests were used to analyze the
survival difference. Correlation analyses for the quantification of
TELO2 and RICTOR staining were performed using Spearman's
correlation. The corresponding relationship between TELO2
expression and CRC clinical features was analyzed by a Chi-square
test. P<0.05 indicated a statistically significant
difference.
Results
TELO2 was expressed at higher levels
in CRC
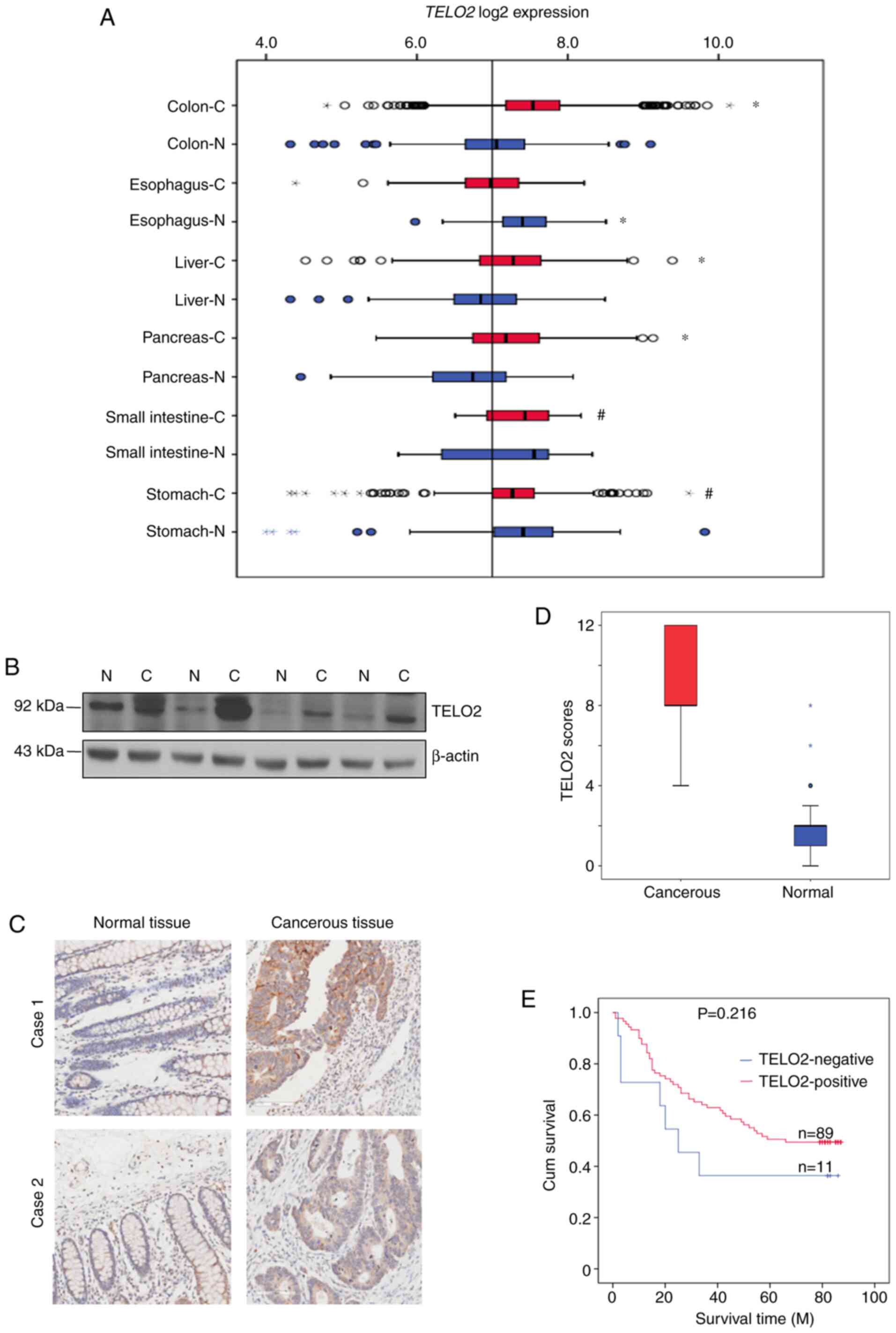
In the GPL570 platform (HG-U133_Plus_2) of the GENT2
database (23), we found that the
mRNA level of TELO2 was upregulated in adrenal gland, bladder,
bone, breast, colon, liver, lung, muscle, pancreas, and skin cancer
tissues in relation to corresponding normal tissues. The
fold-change expression of TELO2 in colon normal tissues was 0.509
lower than cancerous tissues (P<0.001). No similar results were
observed in esophageal and gastric cancers (Fig. 1A), indicating that TELO2 may be
specifically associated with colon cancer in the GI tract. We
collected four pairs of human CRC tissues and matched non-cancerous
mucosa and assessed the expression of TELO2 by western blotting. As
shown in Fig. 1B, all of the cancer
tissues presented a higher expression level of TELO2 as compared
with their corresponding non-cancerous controls. An IHC assay also
showed higher expression levels of TELO2 in a CRC tissue array, and
TELO2 was located in both the cytoplasm and nucleus (Fig. 1C). Fig.
1D shows the statistical difference in the expression scores
between colorectal cancer and normal tissues.
TELO2 correlated with age, lymph node
metastasis, and TNM stage in CRC patients
The correlation between TELO2 expression levels of
CRC samples and a set of clinicopathological characteristics,
including age, sex, tumor size, tumor location, histologic type,
histology, T stage, N stage, and TNM stage, were analyzed using a
Chi-square test (Table I). TELO2
was expressed at higher levels in CRC tissues. In our cohort of
patients, there was positive TELO2 expression in 89 cases, and
negative TELO2 expression in 11 cases. The expression of TELO2 was
higher in older patients, negative lymph node metastasis, or local
stages (I and II) of TNM (92.11, 96.15 and 96.08%, respectively) as
compared to younger patients, positive lymph node metastasis, or
advanced stages (III and IV) of TNM (79.17, 81.25 and 81.63%,
respectively) (P<0.05). TELO2-positive patients trended towards
a longer overall survival (OS) time than the TELO2-negative cohort,
although this distinction was not significantly different between
the two groups (Fig. 1E).
 | Table I.Difference of clinical
characteristics in TELO2-positive and -negative patients. |
Table I.
Difference of clinical
characteristics in TELO2-positive and -negative patients.
| Parameters | Total (N) | TELO2 (+) | TELO2 (−) | P-value |
|---|
| Age (y) | 100 | 68.31±10.810 | 59.55±7.118 | 0.010a |
| Sex | 100 |
|
| 0.757 |
|
Male | 58 | 51 | 7 |
|
|
Female | 42 | 38 | 4 |
|
| Tumor
sizes (cm) | 100 | 5.62±2.177 | 5.35±2.14 | 0.707 |
| Tumor location | 100 |
|
| 0.750 |
|
Left | 42 | 36 | 6 |
|
|
Right | 58 | 53 | 5 |
|
| Histologic
grade | 100 |
|
| 0.073 |
| Grade
1 | 17 | 15 | 2 |
|
| Grade
2 | 74 | 68 | 6 |
|
| Grade
3 | 9 | 6 | 3 |
|
| Histology | 100 |
|
| 0.392 |
|
Tubular | 83 | 75 | 8 |
|
|
Mucinous | 17 | 14 | 3 |
|
| T stage | 100 |
|
| 0.753 |
| T1 +
T2 | 0+4 | 0+4 | 0 |
|
| T3 +
T4 | 64+32 | 57+28 | 11 |
|
| N stage | 100 |
|
| 0.043a |
| N0 | 52 | 50 | 2 |
|
| N1 +
N2 | 36+12 | 30+9 | 9 |
|
| TNM stage | 100 |
|
| 0.045a |
| I + II
(local stage) | 4+47 | 4+45 | 2 |
|
| III +
IV (advanced stage) | 44+5 | 37+3 | 7 |
|
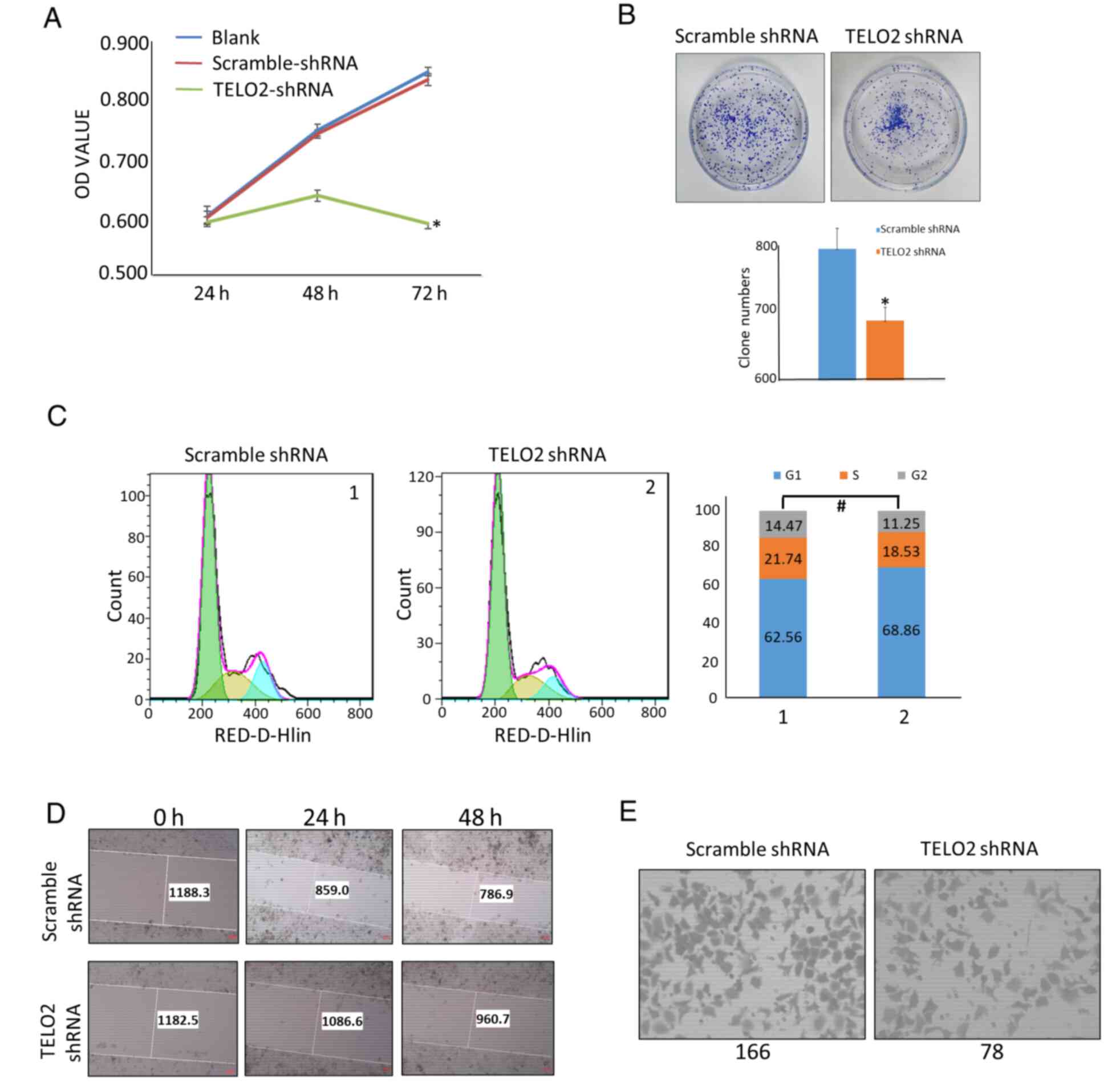
TELO2 induced malignant biological
behavior in CRC
We searched the TELO2 gene level in cancer cell
lines using the GENT2 database. LoVo cells expressed TELO2 at the
highest level out of six different CRC cell lines including CACO2,
COLO205, HCT116, HT29, LoVo, and SW480 (Table SI). To assess the effect of TELO2
on the malignant behavior of CRC cells in vitro, we
established a stable LoVo cell line transfected with TELO2 shRNA or
scrambled shRNA. A WST-1 and soft agar assay were performed to
detect the proliferation and anchorage-independent growth ability,
respectively. Our results showed that TELO2 downregulation
significantly inhibited the proliferation and colony-forming
capacity of LoVo CRC cells (Fig. 2A and
B). In order to assess the effect of TELO2 downregulation on
the cell cycle of CRC cells, flow cytometry was performed. Compared
with the cells transfected with scrambled shRNA, LoVo cells
transfected with TELO2-shRNA exhibited cells in the G1/S stage,
while no statistical difference was detected (Fig. 2C). To examine cell migration and
invasion in vitro, we used a wound healing assay and
Transwell matrix-coated cell culture inserts. After the
downregulation of TELO2, the mobility and invasiveness of LoVo
cells decreased as compared with the control cells (Fig. 2D and E). These data indicated that
TELO2 downregulation inhibited the malignant biological behavior of
CRC cells.
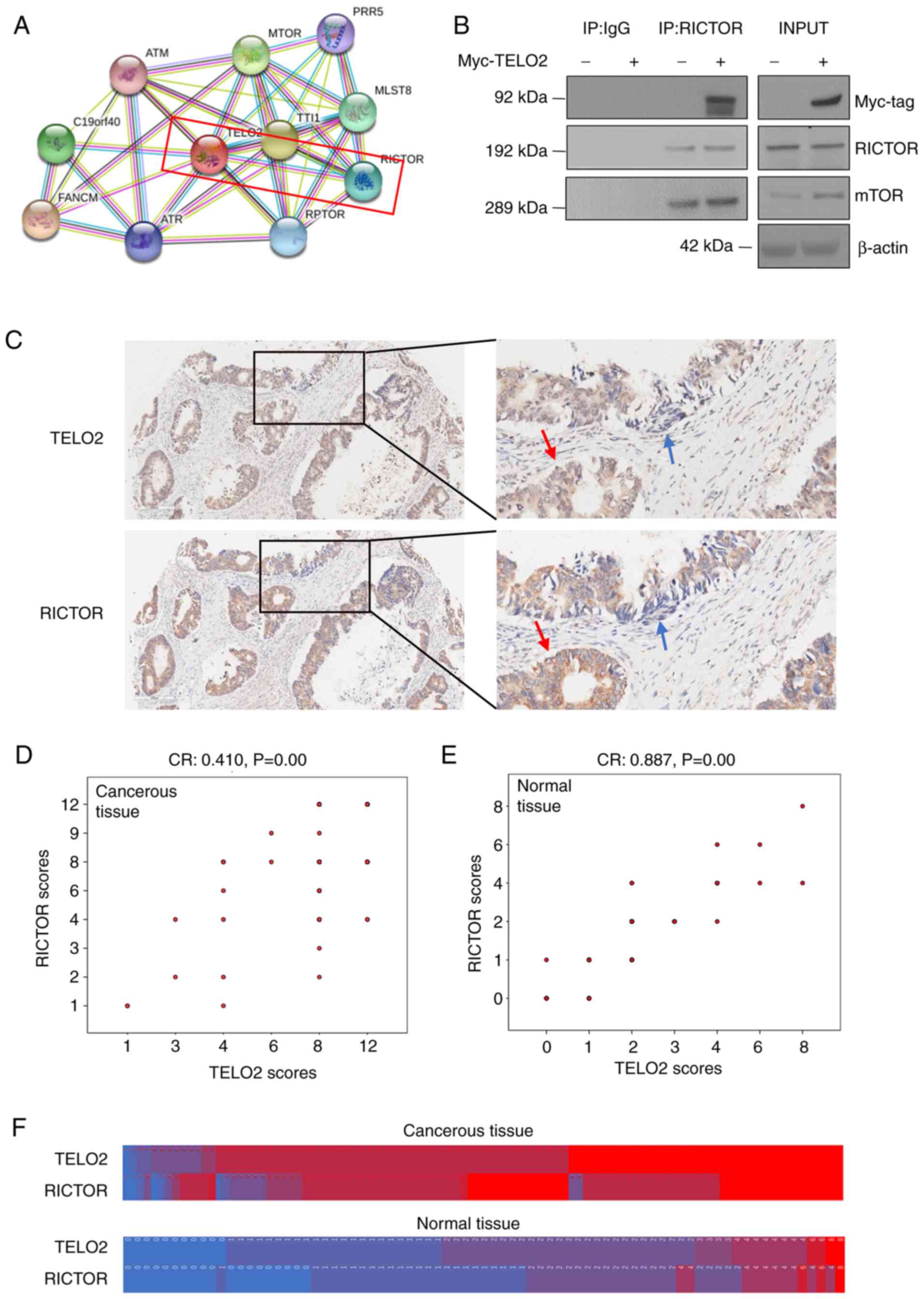
RICTOR bound to TELO2 and was
positively correlated with TELO2 in CRC
Using a STRING database, we found that >10
proteins probably bound with TELO2. The combined score between
RICTOR and TELO2 was 0.925 (Fig.
3A). Based on this analysis, a Co-IP assay was performed to
detect the binding between the two proteins. We demonstrated that
TELO2 bound with RICTOR in cells treated with CHAPS lysis buffer,
which maintained the integrity of the mTORC2 complex (Fig. 3B). The location of TELO2 was further
confirmed via an IHC assay and a tissue array. As shown in Fig. 3C, the expression of TELO2 was
located in both the nucleus and cytoplasm, while RICTOR expression
was mainly identified in the cytoplasm. Moreover, the expression
location of RICTOR and TELO2 in CRC tissue cells was similar, which
is indicated by the arrows. After scoring, we analyzed the data by
Spearman's correlation (Fig. 3D and
E), which showed that RICTOR was positively associated with
TELO2 in both CRC tissue and adjacent normal colonic mucosa. A
heatmap further confirmed the positive relationship between these
two proteins (Fig. 3F).
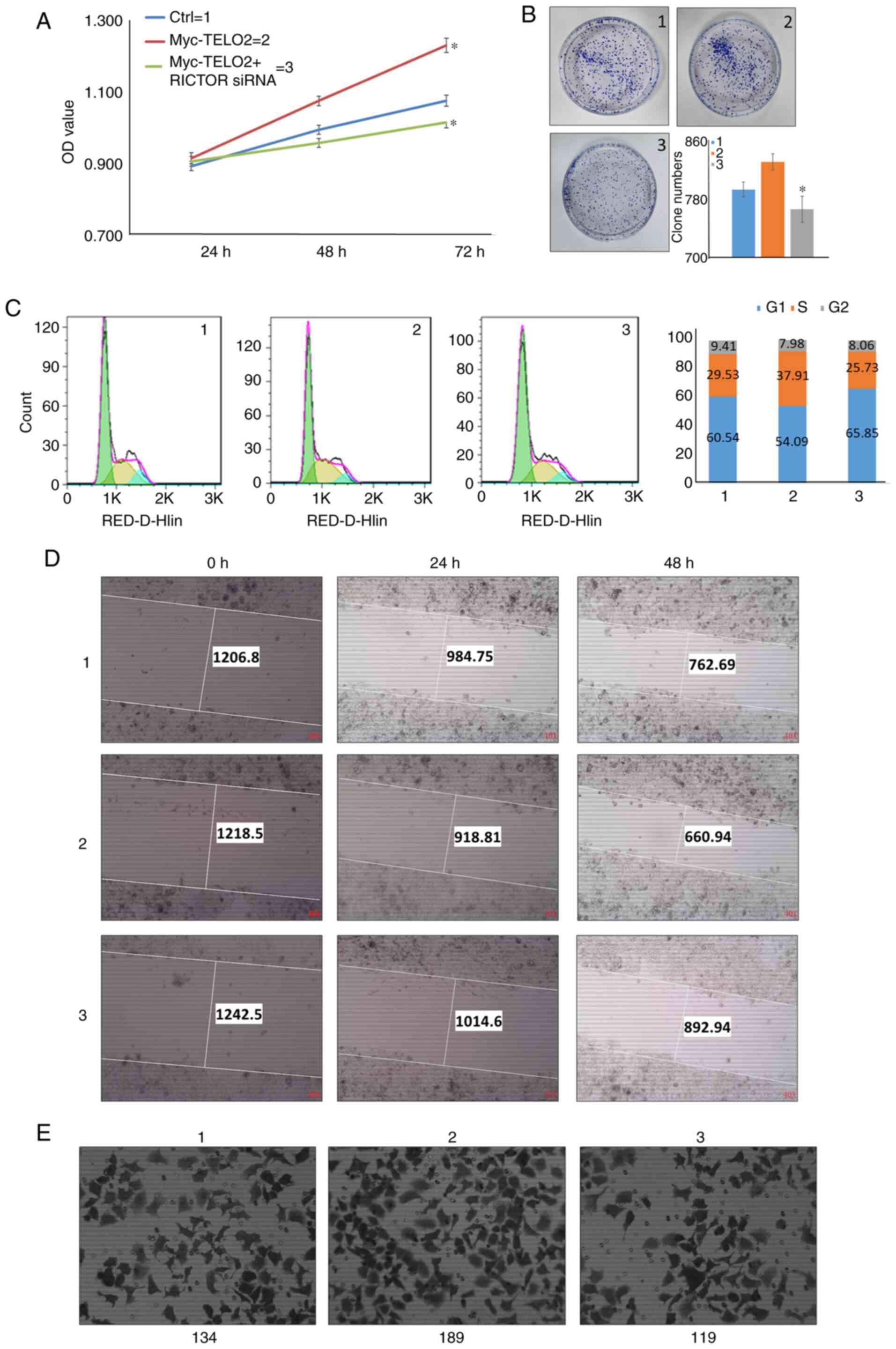
Inhibition of RICTOR reversed
TELO2-induced tumorigenesis in vitro
To verify whether RICTOR was required for
TELO2-induced malignant behavior in CRC, we employed a RICTOR
knockdown strategy in TELO2-overexpressed LoVo cells in
vitro. As shown in Fig. 4A and
B, TELO2 overexpression increased the proliferation and
anchorage-independent growth ability, while transient transfection
with RICTOR siRNA decreased the proliferation and
anchorage-independent growth ability. Knock-down of RICTOR also
blocked LoVo-Myc-TELO2 stable cells in the G1 phase of the cell
cycle, which led to cell cycle arrest or death (Fig. 4C). Similar results were observed
where the induction of metastatic phenotypes caused by the
overexpression of TELO2 was abolished by the inhibition of RICTOR,
as evidenced by the wound healing and Transwell chamber assays
(Fig. 4D and E).
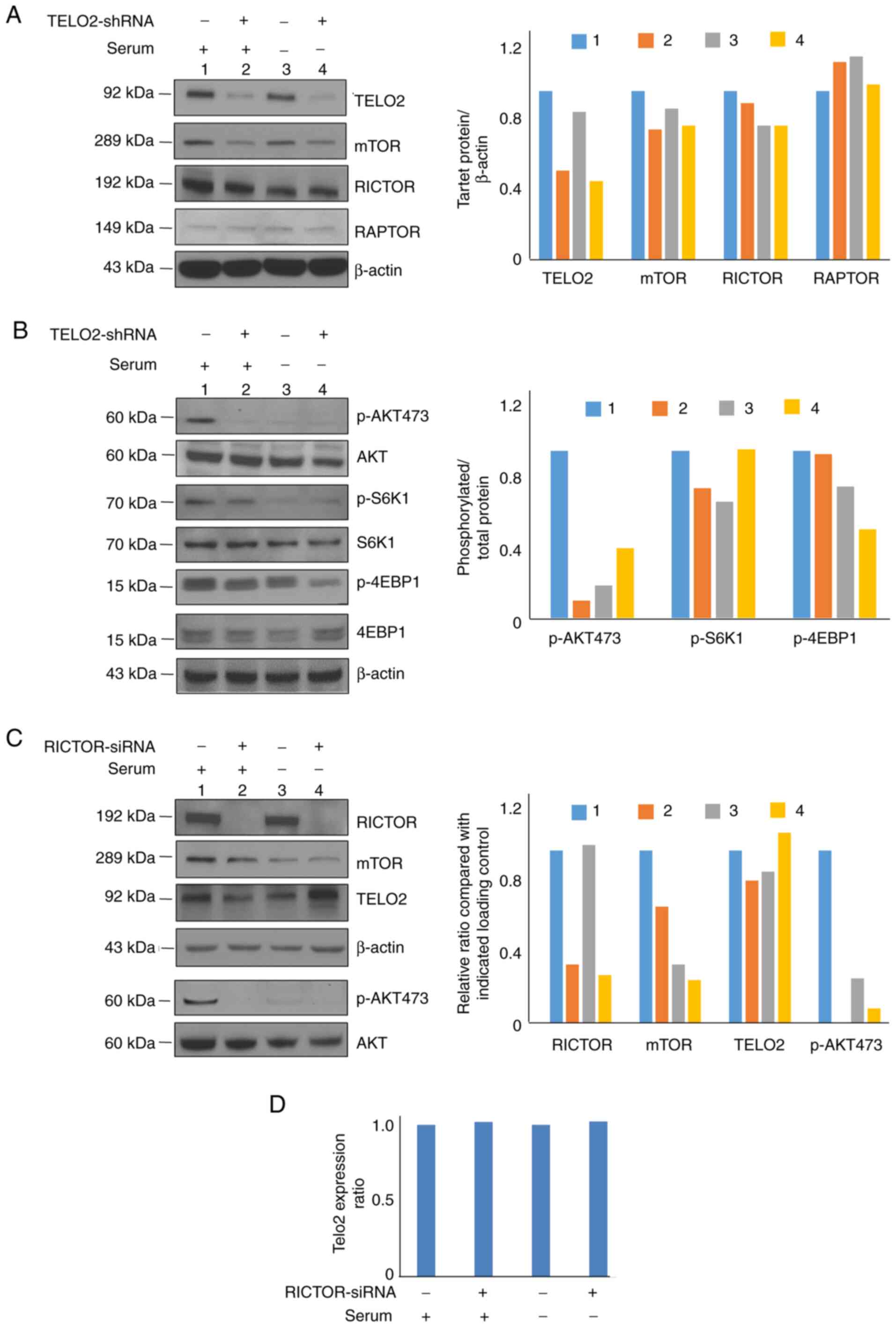
TELO2 induced tumorigenesis through
mTORC2 activity with serum supplement
LoVo cells were stably transfected with TELO2 shRNA
and cultured in conventional medium. Whole cell lysates were
harvested, and western blot analysis was performed. Inhibition of
TELO2 only decreased mTOR expression without changing RICTOR and
RAPTOR levels (Fig. 5A).
Additionally, levels of phosphorylated Akt Ser473, a protein
downstream of mTORC2, was decreased, while the target proteins of
mTORC1, p-S6K1 and p-4EBP1, were changed slightly under the
treatment. However, no significant change was demonstrated in those
cells without serum supplement, except for p-4EBP1 with a
distinctive decrease (Fig. 5B). We
then knocked down RICTOR using siRNA, and our results indicated
that RICTOR downregulation inhibited TELO2 slightly and pAktS473
when cultured under normal conditions, while the expression of
TELO2 was increased in RICTOR knockdown cells under serum
deprivation (Fig. 5C). RT-PCR
demonstrated that expression of RICTOR had no effect on the mRNA
level of TELO2 in both normal or serum-deprived cultures (Fig. 5D).
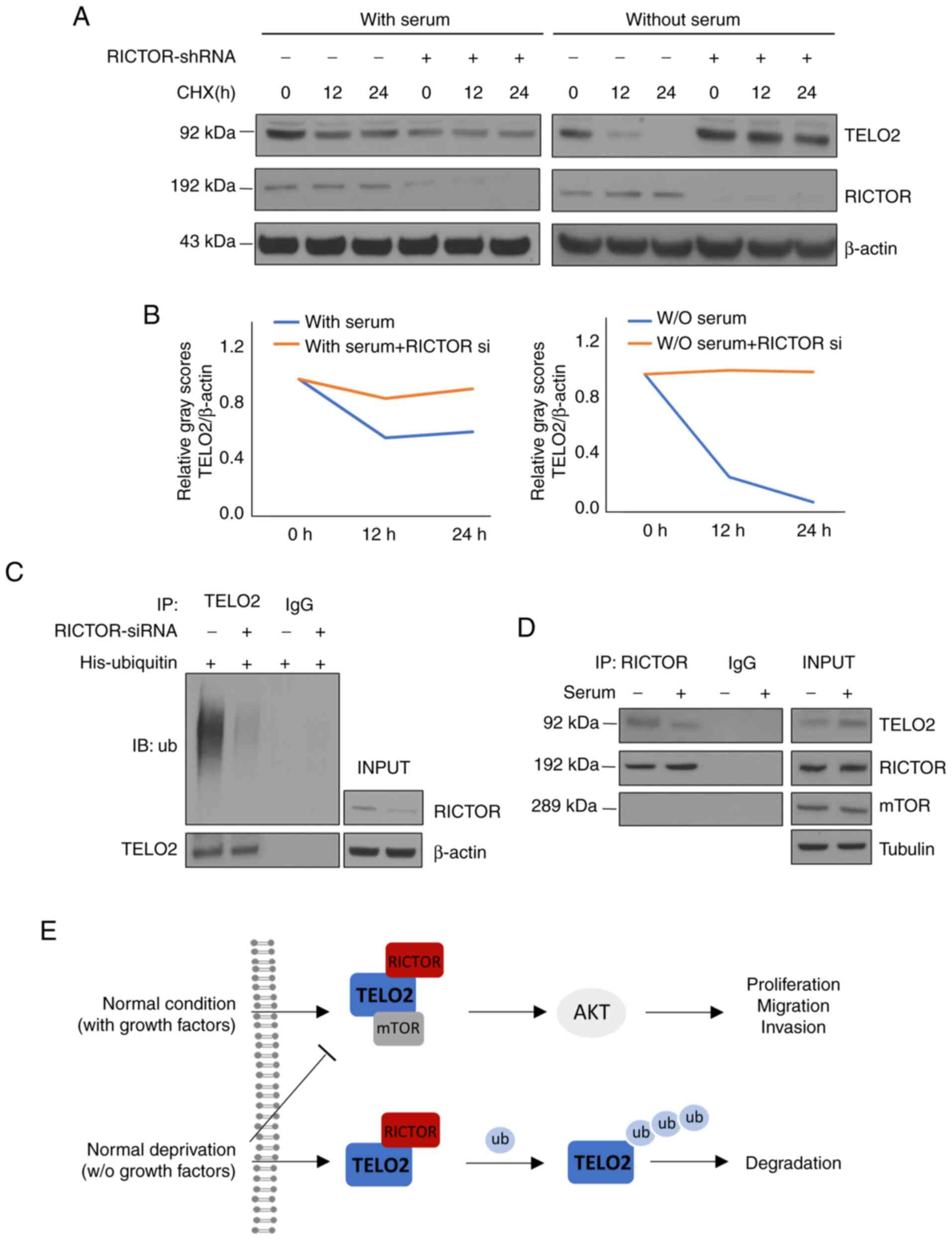
RICTOR degraded TELO2 by
ubiquitination under serum deprivation
To address the mechanism of how RICTOR affected the
protein level of TELO2, we investigated the stability of TELO2
protein using a CHX chase assay. Time-course experiments showed
that the stability of TELO2 was higher in cells with serum in the
culture as compared with cells under serum deprivation. The
half-life of TELO2 was further prolonged in LoVo cells transfected
with RICTOR siRNA as compared with cells transfected with scrambled
siRNA when cultured without serum. This function was reduced in
cells cultured with a serum supplement (Fig. 6A and B). We then performed in
vitro ubiquitination assays to further address whether RICTOR
promoted the ubiquitination of TELO2. His-ubiquitin and/or RICTOR
siRNA were transfected transiently, and knockdown of RICTOR reduced
the ubiquitination of both TELO2 with no serum culture (Fig. 6C). More importantly, the binding of
RICTOR and TELO2 was increased in serum-deprived cells as compared
with cells that were grown with serum in the culture, and mTOR was
not present in this compound (Fig.
6D).
TELO2 functioned as double-edged sword
in CRC
In Fig. 6E, we
concluded the two-sides functions of TELO2 under different
conditions in CRC. In normal condition with growth factors during
in the progression of CRC, TELO2 binds with RICTOR as mTORC2
complex in promoting proliferation, migration and invasion by AKT
pathway. However, in serum deprivation usually happened under a
heavy burden of tumor, TELO2 is ubiquitinated by RICTOR through an
mTORC2-independent manner.
Discussion
In the present study, we characterized the role and
partial mechanism of TELO2 in CRC progression. TELO2 was expressed
at high levels in CRC. Inhibition of TELO2 resulted in the
reduction of tumorigenesis in CRC cells, but no difference was
shown in the OS between TELO2-positive and -negative patients. The
TELO2 expression rate was positively correlated with age and
negatively correlated with lymph node metastasis and TNM stage at
the same time. To the best of our knowledge, these results
indicated, for the first time, that TELO2 expression is higher
during the local stages of CRC and leads to tumorigenesis and
metastasis, while the expression rate of TELO2 is decreased during
the advanced stages of CRC progression with lymph node and organ
metastasis. The differential expression of TELO2 during the various
stages of CRC offset its role in the prognosis suggesting that
TELO2 plays a distinctive role in the development of CRC. The role
of TELO2 in the prognosis of CRC, which differs from that in breast
cancer (14), is still unclear,
indicating that mechanism of TELO2 is different in various
cancers.
Next, we needed to confirm the mechanism of
TELO2-induced cancer progression in CRC. TELO2 and Tti1 play a
critical role in mTOR complex formation (10). mTORC1, mainly containing mTOR,
RAPTOR (regulatory protein associated with mTOR), and mLST8
(mammalian lethal with Sec13 protein 8), regulates cell growth and
metabolism through two key effectors, p70S6 Kinase 1 (S6K1) and
eIF4E binding protein (4EBP) (24,25).
mTORC2, including mTOR, RICTOR, and mLST8, controls proliferation,
survival, and migration by phosphorylating the AGC subfamily of
protein kinases (a subgroup of Ser/Thr protein kinases based on
sequence alignments of their catalytic kinase domain), including
Akt at Ser473 (16,26). It has been previously reported that
TELO2 stabilizes mTORC1-substrate interactions to activate T cell
and mitogenic signaling (7).
However, no studies have shown the role of TELO2 in mTORC2. In the
present study, a positive correlation between TELO2 and RICTOR
protein was identified. Inhibition of RICTOR attenuated
TELO2-induced proliferation, cell cycle progression, invasion, and
migration in CRC cells. These data indicated that TELO2 induces
tumorigenesis mainly via binding with RICTOR, a mTORC2-specific
member, and dissociation of this compound could repress the
oncogenic ability of TELO2.
Finally, we studied the binding pattern of TELO2 and
RICTOR in CRC under different conditions. Nutrients and growth
factors are usually supplied by the bloodstream in solid tumors,
and growth factors are the only well-defined stimulus for mTORC2
through the phosphorylation of Akt (27). Akt plays a strong carcinogenic role
by either inducing or inhibiting downstream transcription factors
(28,29). However, inhibition of mTOR by
nutrient- or serum-deprivation stimulates the ubiquitin proteasome
system to degrade ubiquitinated proteins (30). In addition, RICTOR was originally
identified as a specific binding partner of mTOR that regulates the
cytoskeleton and phosphorylates Akt. Multiple complexes containing
RICTOR have been identified as mTORC2-independent with oncogenic
actions (31,32). For example, RICTOR is proposed to be
a scaffold protein for integrin-linked kinase (ILK) and appears to
be mandatory for TGFb-1-mediated epithelial-mesenchymal transition
(33). Another RICTOR-containing
complex is composed of tetraspanin 8 and integrin a3, which is
required for the assembly and function of the mTORC2 complex in
glioma cells (34). Interestingly,
RICTOR forms a complex with Cullin-1 to enhance its E3 ligase
activity and promote serum/glucocorticoid-induced kinase (SGK)
ubiquitination (35). Our previous
study indicated that RICTOR and FBXW-7, an E3 ligase complex, exert
anti-oncogenic activities by promoting the degradation of c-Myc and
cyclin E in CRC cells without serum supplementation (36). Thus, this study investigated that
inhibition of TELO2 under normal culture conditions, which mainly
reduced the activity of Akt, with a slight change in 4EBP1 and
S6K1. This effect was reduced with serum deprivation, indicating
that the TELO2/RICTOR complex induced cell survival, proliferation,
and migration during CRC progression in a serum-dependent manner
via the mTORC2 pathway. In addition, RICTOR functioned as an E3
adaptor and degraded TELO2 under serum-deprived culture conditions,
further confirming that TELO2 was ubiquitinated in an
mTOR-independent manner. However, identification of the TELO2 and
RICTOR binding site would allow for further evaluation of the
mechanism of the TELO2/RICTOR complex during serum deprivation.
In conclusion (Fig.
6E), our study demonstrated that TELO2 functions as a trigger
of CRC progression through mTORC2, particularly RICTOR in normal
cultural conditions. However, this effect could be reversed with
serum deprivation, which is common in solid tumor progression and
partially supports the theory that TELO2 does not play a role in
CRC prognosis. Thus, the present study provides evidence that TELO2
acts as a vital role in CRC growth and progression, and as such,
may serve as a therapeutic target for CRC. This TELO2-driven
therapy should be further confirmed for the functions of TELO2 in
normal cells and its double-edged sword under different conditions
yet.
Supplementary Material
Supporting Data
Acknowledgements
We would like to thank LetPub (www.letpub.com) for its linguistic assistance during
the preparation of this manuscript.
Funding
This study was supported by National Nature Science
Foundation of China (no. 81860440) and Nature Science Foundation of
Jiangxi (no. 20202BAB206047).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author upon reasonable
request.
Authors' contributions
ZG, XZ, and HZ performed the experiments. XW, FT,
LH, JZ, and ZG designed the study. JH reviewed the IHC results. NZ,
XL, and YZ collected the patients' information and performed the
statistical analysis. ZG wrote the manuscript. All of the authors
approved the final version.
Ethics approval and consent to
participate
The proposal of this study was approved by the
Institutional Review Committee of Gannan Medical University. All of
the patients signed informed consent for the use of their
tissues.
Patient consent for publication
Not applicable.
Competing interests
There are no competing interests to be declared.
Glossary
Abbreviations
Abbreviations:
|
RICTOR
|
rapamycin-insensitive companion of
mTOR
|
|
TELO2
|
telomere maintenance 2
|
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2019. CA Cancer J Clin. 69:7–34. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lin SH, Raju GS, Huff C, Ye Y, Gu J, Chen
JS, Hildebrandt MAT, Liang H, Menter DG, Morris J, et al: The
somatic mutation landscape of premalignant colorectal adenoma. Gut.
67:1299–1305. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Marschner N, Zacharias S, Lordick F,
Hegewisch-Becker S, Martens U, Welt A, Hagen V, Gleiber W, Bohnet
S, Kruggel L, et al: Association of disease progression with
health-related quality of life among adults with breast, lung,
pancreatic, and colorectal cancer. JAMA Netw Open. 3:e2006432020.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chiorean EG, Nandakumar G, Fadelu T, Temin
S, Alarcon-Rozas AE, Bejarano S, Croitoru AE, Grover S, Lohar PV,
Odhiambo A, et al: Treatment of patients with late-stage colorectal
cancer: ASCO resource-stratified guideline. JCO Glob Oncol.
6:414–438. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Runge KW and Zakian VA: TEL2, an essential
gene required for telomere length regulation and telomere position
effect in Saccharomyces cerevisiae. Mol Cell Biol. 16:3094–3105.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Saxton RA and Sabatini DM: mTOR signaling
in growth, metabolism, and disease. Cell. 169:361–371. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Brown MC and Gromeier M: MNK controls
mTORC1: Substrate association through regulation of TELO2 binding
with mTORC1. Cell Rep. 18:1444–1457. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Takai H, Wang RC, Takai KK, Yang H and de
Lange T: Tel2 regulates the stability of PI3K-related protein
kinases. Cell. 131:1248–1259. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bahrami A, Khazaei M, Hasanzadeh M,
ShahidSales S, Joudi Mashhad M, Farazestanian M, Sadeghnia HR,
Rezayi M, Maftouh M, Hassanian SM and Avan A: Therapeutic potential
of targeting PI3K/AKT pathway in treatment of colorectal cancer:
Rational and progress. J Cell Biochem. 119:2460–2469. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kaizuka T, Hara T, Oshiro N, Kikkawa U,
Yonezawa K, Takehana K, Iemura S, Natsume T and Mizushima N: Tti1
and Tel2 are critical factors in mammalian target of rapamycin
complex assembly. J Biol Chem. 285:20109–20116. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Takai H, Xie Y, de Lange T and Pavletich
NP: Tel2 structure and function in the Hsp90-dependent maturation
of mTOR and ATR complexes. Genes Dev. 24:2019–2030. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fernandez-Saiz V, Targosz BS, Lemeer S,
Eichner R, Langer C, Bullinger L, Reiter C, Slotta-Huspenina J,
Schroeder S, Knorn AM, et al: SCFFbxo9 and CK2 direct the cellular
response to growth factor withdrawal via Tel2/Tti1 degradation and
promote survival in multiple myeloma. Nat Cell Biol. 15:72–81.
2013. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Morais-Rodrigues F, Silv Erio-Machado R,
Kato RB, Rodrigues DLN, Valdez-Baez J, Fonseca V, San EJ, Gomes
LGR, Dos Santos RG, Vinicius Canário Viana M, et al: Analysis of
the microarray gene expression for breast cancer progression after
the application modified logistic regression. Gene. 726:1441682020.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Feng SW, Chen Y, Tsai WC, Chiou HC, Wu ST,
Huang LC, Lin C, Hsieh CC, Yang YJ and Hueng DY: Overexpression of
TELO2 decreases survival in human high-grade gliomas. Oncotarget.
7:46056–46066. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gala MK, Mizukami Y, Le LP, Moriichi K,
Austin T, Yamamoto M, Lauwers GY, Bardeesy N and Chung DC: Germline
mutations in oncogene-induced senescence pathways are associated
with multiple sessile serrated adenomas. Gastroenterology.
146:520–529. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sarbassov DD, Guertin DA, Ali SM and
Sabatini DM: Phosphorylation and regulation of Akt/PKB by the
rictor-mTOR complex. Science. 307:1098–1101. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yang G, Murashige DS, Humphrey SJ and
James DE: A positive feedback loop between Akt and mTORC2 via SIN1
phosphorylation. Cell Rep. 12:937–943. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lampada A, O'Prey J, Szabadkai G, Ryan KM,
Hochhauser D and Salomoni P: mTORC1-independent autophagy regulates
receptor tyrosine kinase phosphorylation in colorectal cancer cells
via an mTORC2-mediated mechanism. Cell Death Differ. 24:1045–1062.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang L, Qi J, Yu J, Chen H, Zou Z, Lin X
and Guo L: Overexpression of rictor protein in colorectal cancer is
correlated with tumor progression and prognosis. Oncol Lett.
14:6198–6202. 2017.PubMed/NCBI
|
|
20
|
Creytens D: NKX2.2 immunohistochemistry in
the distinction of ewing sarcoma from cytomorphologic mimics:
Diagnostic utility and pitfalls-comment on russell-goldman et
al. Cancer Cytopathol. 127:2022019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhao Y, Zhang W, Guo Z, Ma F, Wu Y, Bai Y,
Gong W, Chen Y, Cheng T, Zhi F, et al: Inhibition of the
transcription factor Sp1 suppresses colon cancer stem cell growth
and induces apoptosis in vitro and in nude mouse xenografts.
Oncol Rep. 30:1782–1792. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chen Y, Liu J, Wang W, Xiang L, Wang J,
Liu S, Zhou H and Guo Z: High expression of hnRNPA1 promotes cell
invasion by inducing EMT in gastric cancer. Oncol Rep.
39:1693–1701. 2018.PubMed/NCBI
|
|
23
|
Park SJ, Yoon BH, Kim SK and Kim SY:
GENT2: An updated gene expression database for normal and tumor
tissues. BMC Med Genomics. 12 (Suppl 5):S1012019. View Article : Google Scholar
|
|
24
|
Carroll B: Spatial regulation of mTORC1
signalling: Beyond the Rag GTPases. Semin Cell Dev Biol.
107:103–111. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Rabanal-Ruiz Y and Korolchuk VI: mTORC1
and nutrient homeostasis: The central role of the lysosome. Int J
Mol Sci. 19:8182018. View Article : Google Scholar
|
|
26
|
Li X and Gao T: mTORC2 phosphorylates
protein kinase Cζ to regulate its stability and activity. EMBO Rep.
15:191–198. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Knudsen JR, Fritzen AM, James DE, Jensen
TE, Kleinert M and Richter EA: Growth factor-dependent and
-independent activation of mTORC2. Trends Endocrinol Metab.
31:13–24. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Manning BD and Toker A: AKT/PKB signaling:
Navigating the network. Cell. 169:381–405. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Jia R and Bonifacino JS: Lysosome
positioning influences mTORC2 and AKT signaling. Mol Cell.
75:26–38.e3. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhao J, Zhai B, Gygi SP and Goldberg AL:
mTOR inhibition activates overall protein degradation by the
ubiquitin proteasome system as well as by autophagy. Proc Natl Acad
Sci USA. 112:15790–15797. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Gkountakos A, Pilotto S, Mafficini A,
Vicentini C, Simbolo M, Milella M, Tortora G, Scarpa A, Bria E and
Corbo V: Unmasking the impact of Rictor in cancer: Novel insights
of mTORC2 complex. Carcinogenesis. 39:971–980. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhang S, Qian G, Zhang QQ, Yao Y, Wang D,
Chen ZG, Wang LJ, Chen M and Sun SY: mTORC2 suppresses
GSK3-dependent snail degradation to positively regulate cancer cell
invasion and metastasis. Cancer Res. 79:3725–3736. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Serrano I, McDonald PC, Lock FE and Dedhar
S: Role of the integrin-linked kinase (ILK)/Rictor complex in
TGFβ-1-induced epithelial-mesenchymal transition (EMT). Oncogene.
32:50–60. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Pan SJ, Zhan SK, Pan YX, Liu W, Bian LG,
Sun B and Sun QF: Tetraspanin 8-rictor-integrin α3 complex is
required for glioma cell migration. Int J Mol Sci. 16:5363–5374.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Gao D, Wan L, Inuzuka H, Berg AH, Tseng A,
Zhai B, Shaik S, Bennett E, Tron AE, Gasser JA, et al: Rictor forms
a complex with Cullin-1 to promote SGK1 ubiquitination and
destruction. Mol Cell. 39:797–808. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Guo Z, Zhou Y, Evers BM and Wang Q: Rictor
regulates FBXW7-dependent c-Myc and cyclin E degradation in
colorectal cancer cells. Biochem Biophys Res Commun. 418:426–432.
2012. View Article : Google Scholar : PubMed/NCBI
|