Introduction
Smoking is the leading cause of chronic obstructive
pulmonary disease, a progressive and eventually debilitating lung
disease (1). Furthermore, smokers
of tobacco are 20–40 times more at risk of developing lung cancer
in comparison to non-smokers (2).
Tobacco can be smoked using different ways, two
common ways include the use of cigarettes or waterpipes. Several
studies have investigated the chemical composition of cigarette
smoke and more than 5,000 bioactive chemical compounds have been
isolated, including over 60 carcinogens (3). Waterpipe smoke analysis has revealed
that it contains significant concentrations of toxicants including
27 known or suspected carcinogens thought to cause dependence,
heart disease, lung disease and cancer (4). Despite the adverse health effects
among users, waterpipe usage is growing in popularity and public
health interventions remain below what is present for cigarette
smoking (5).
Several studies have addressed the in vivo
effects of WPS on waterpipe smokers health. Smokers are found to
have high urinary concentrations of several toxins including
carcinogens (6), resulting in
profound effects on lung function (7). Waterpipe smokers were also observed to
have 6-fold greater risk of developing lung cancer (8). At the molecular level, DNA repair gene
expression was reported to be decreased in the blood of waterpipe
smokers, while DNA damage-related gene expression was increased
(9). It has also been reported that
WPS induces endothelial cell dysfunction, inflammation, and
impaired repair mechanisms with implication in vascular disease
(10). In this respect, nicotine,
present in WPS, induces bronchial epithelial cell apoptosis and
senescence via ROS-mediated autophagy-impairment (11). WPSC also induces cell cycle arrest
and cellular senescence mediated by the p53-p21 pathway in alveolar
type 2 cell disease (10), whereas
it induces apoptosis in human aortic endothelial cells (10,12).
All these data highlight the damaging effects of WPS. More
importantly, WPS may contribute towards EMT, tumor heterogeneity
and immune escape. These processes are known to play critical roles
in tumor plasticity and are important factors impacting both the
diagnosis and treatment of cancer patients (13).
The aim of the present study examined the changes in
tumor lung cell gene expression related to DNA damage,
inflammation, EMT and stemness. In addition, the consequence of
WPSC treatment on immune recognition and killing by NK cells was
investigated. Our results emphasized the potential impact of WPSC
on tumor lung cell behavior and provide insights into their
associated transcriptomic response including DNA damage,
inflammation, and cell plasticity.
Materials and methods
Waterpipe smoke sampling and
analysis
Waterpipe smoke collection was performed as
previously described (14).
Briefly, 17.5 g double apple flavor tobacco (mouassal) was placed
in the head piece of the waterpipe which was then tightly wrapped
using a perforated aluminum foil. Two pieces of quick lighting
charcoal briquettes were used to heat the tobacco. The generated
smoke was collected using a robotic machine (IREADY LLC) that
simulates the human smoking process. The puff duration was set at 5
sec per puff with 15 sec inter-puff duration, for a total of 80
puffs per session. Collection of the smoke condensate was carried
out on pre-conditioned glass wool fibers packed inside a T-shaped
tube. It is important to note that under our experimental
conditions, the cells were exposed to the waterpipe smoke
condensate samples. To identify the chemical composition of the
condensate and to eliminate any masking effect of the large
glycerin peak during gas chromatography-mass spectrometry (GC-MS),
successive extraction steps were performed. The extraction
procedure was carried out by mixing 72.6 mg of the extract in 4 ml
of toluene. The mixture was stirred for 24 h and allowed to
separate. In this step, glycerin is not expected to move into the
toluene layer. Then, 0.15 ml of the remaining components of the
extract were dissolved in 15 ml of ethanol followed by a dilution
of 1:40 in ethanol prior to gas chromatography mass spectrometry
(GCMS) analysis to eliminate detector saturation. Specifically, 2
ml of toluene was added to the smoke condensate (20 ml), agitated
for 2 h on an orbital shaker (Z206A), and spun at 100 × g for 5 min
at room temperature. The toluene was then separated prior to
dissolving in 2 ml ethanol which was used for the GCMS analysis. It
is worth noting that reproducibility was assured by using internal
standards. Tridecane and 1-octadecene were used as internal
standards for toluene extract while dibutyl phthalate was used for
ethanol solution prior to GCMS analysis. The toluene and ethanol
extract were labelled as ‘toluene’ and ‘toluene-ethanol’,
respectively. The toluene and ethanol solutions were then analyzed
using GCMS-QP2010 Ultra instrument (Shimadzu Corporation). Rtx-5MS
capillary column (30 m in length, 0.25 µm in thickness, and a
diameter of 0.25 mm) was used. The column oven temperature was held
at 40°C for 3 min then ramped at a rate of 5°C per min to 300°C
where it was held constant for 15 min. Helium was used as the
carrier gas at a column flow of 1.0 ml/min. The safety, hazard and
toxicity evaluation of the toluene and ethanol extracts was
performed using ToxNet and PubChem resources.
Cell culture and proliferation
assay
A549 (gift from Professor Fathia Mami Chouaib,
Gustave Roussy, Villejuif Cedex, France; RRID:CVCL_0023) and H460
cells (AddexBio C0016003 RRID:CVCL_0459) were grown in complete
RPMI-1640 medium, (cat. no. 61870010; Gibco; Thermo Fisher
Scientific, Inc.) supplemented with 10% heat-inactivated fetal
bovine serum (cat. no. 10270-106; Gibco; Thermo Fisher Scientific,
Inc.), 1% penicillin-streptomycin (cat. no. 15140-122; Gibco;
Thermo Fisher Scientific, Inc.) and 1% sodium pyruvate (cat. no.
11360-039; Gibco; Thermo Fisher Scientific, Inc.). BEAS-2B cells
(ECACC 95102433 RRID: CVCL_0168) were grown in BEGM media (cat. no.
CC-3170; Lonza) on collagen (cat. no. A1048301; Gibco; Thermo
Fisher Scientific, Inc.)-coated tissue culture dishes according to
the manufacturers protocol. NK92 cells (gift from Professor Fathia
Mami Chouaib) were cultured in complete RPMI-1640 medium
supplemented with 200 IU of IL2 recombinant human protein (cat. no.
PHC0021; Gibco; Thermo Fisher Scientific, Inc.). H460 and BEAS-2B
were purchased from a commercial source that guarantees cell line
authenticity. We tested and confirmed that all the cell lines were
mycoplasma-free.
Trypan blue exclusion assay was used to determine
the cell number and viability of A549, H460 and BEAS-2B cells
exposed to various concentrations and durations of WPSC as
described earlier in the text. Subsequently, 50,000 cells (A549 and
H460) or 100,000 cells (BEAS-2B) were plated on 3.5-cm dishes for
24 h after which the cells were treated with 1 ml of complete media
containing WPSC at the indicated concentrations. Plates were
counted, every day for 7 days and the number of live and dead cells
was determined.
RNA extraction/cDNA and qPCR
RNA was purified using Easy Blue (cat. no. 17061;
Intron Biotechnology, Inc.) according to the manufacturers
protocol. RNA quality and quantity were assessed by nanodrop and 1%
agarose gel electrophoresis. cDNA synthesis was performed using
high capacity cDNA reverse transcription kit (cat. no. 4374966;
Thermo Fisher Scientific Inc.). qPCR was performed using SYBR-Green
(cat. no. 4309155; Thermo Fisher Scientific Inc.) using AB 7500
FAST Real-Time PCR system. Analysis was performed using the ∆∆Cq
method (15). Forward (F) and
reverse (R) primers used in this study were: GAPDH forward,
(5′-GCCACATCGCTCAGACAC-3′) and GAPDH reverse,
(5′-CCAGAGTTAAAAGCAGCC-3′); CCL2 forward,
(5′-CAGCCAGATGCAATCAATGCC-3′) and CCL2 reverse,
(5′-TGGAATCCTGAACCCACTTCT-3′), CASP1 forward,
(5′-TTTCCGCAAGGTTCGATTTTCA-3′) and CASP1 reverse,
(5′-GGCATCTGCGCTCTACCATC-3′), IL1B forward,
(5′-TTCGACACATGGGATAACGAGG-3′) and IL1B reverse,
(5′-TTTTTGCTGTGAGTCCCGGAG-3′); CD44 forward,
(5′-TGCCGCTTTGCAGGTGTATT-3′) and CD44 reverse,
(5′-CCGATGCTCAGAGCTTTCTCC-3′); SERPINE2 forward,
(5′-TGGTGATGAGATACGGCGTAA-3′) and SERPINE2 reverse,
(5′-GTTAGCCACTGTCACAATGTCTT-3′); SNAI2 forward,
(5′-TAGGAAGAGATCTGCCAGAC-3′) and SNAI2 reverse,
(5′-CCCCAAGGCACATACTGTTA-3′); ACTA2 forward,
(5′-CTATGAGGGCTATGCCTTGCC-3′) and ACTA2 reverse,
(5′-GCTCAGCAGTAGTAACGAAGGA-3′); IL6 forward,
(5′-ACTCACCTCTTCAGAACGAATTG-3′) and IL6 reverse,
(5′-CCATCTTTGGAAGGTTCAGGTTG-3′).
Alkaline comet assay
Cells were harvested and prepared as described in
Olive and Banáth (16). Briefly,
the treated cells were centrifuged at 100 × g at room temperature
for 5 min. Cell pellet was re-suspended in 1X PBS and approximately
5,000 cells per slide were considered for the assay. Cells were
mixed with 0.75% low-melting agarose (LMA) at 37°C, and the mixture
was loaded onto glass slides pre-coated with 1.5% normal melting
agarose. Coverslips were then placed gently on the slides to allow
even spreading of the gel and then placed at 4°C for gelling. A
third layer of the 0.75% LMA was added onto the slide to fill any
residual holes in the second layer, and was allowed to solidify.
After gelling, the slides were immersed in chilled lysing solution
containing 2.5 M NaCl, 100 mM EDTA, 10 mM Tris base (pH 10) with 1%
Triton X-100 and 10% DMSO and were kept overnight at 4°C. After
lysis, the slides were placed in a horizontal electrophoresis tank
and soaked in cold alkaline electrophoresis buffer (1 mM EDTA, 300
mM NaOH). Slides were left for 30 min for DNA unwinding and
electrophoresis was performed at 0.7 V/cm and 300 mA for 35 min.
The slides were then removed from the electrophoresis buffer and
were neutralized using 0.4 M Tris (pH 7.4). To capture images, the
slides were stained with 100 ml of ethidium bromide (0.2 mg/100 ml)
and were visualized at ×20 magnification on Zeiss LSM 800 with
Airyscan. In total, 50 images were taken per slide and the data
were analyzed using OpenComet (17)
pluggin on ImageJ software. The tail length was considered as a
suitable parameter for measuring the extent of DNA damage.
Immunofluorescence
Cells were fixed in 4% paraformaldehyde (cat. no.
28906; Thermo Fisher Scientific Inc.) in 1X PBS for 10 min at room
temperature. Cells were then washed with 1X PBS and permeabilized
with 0.1% TX-100 in PBS for 15 min at room temperature. Prior to
staining, the cells were blocked in 2% BSA in 1X PBS for 1 h at
room temperature. The cells were then stained with a primary and
secondary antibodies as described below with 3×5 min washes after
each antibody staining. Cells were then mounted on glass slides
using Prolong gold antifade reagent (cat. no. P36930; Thermo Fisher
Scientific, Inc.) and visualized on Zeiss LSM 800 with Airyscan.
The comet assays were visualized using a 20× plan apochromat
objective, with Zeiss Axio cam 305 mono (Zeiss LSM 800).
Immunological synapse formation
assay
For visualizing immunological synapse 100,000 lung
cells were incubated with 300,000 NK92 cells in the presence of
IL-2 for 30 min in a 37°C incubator. Cells were subsequently fixed
and stained for the markers described in the text.
Antibodies used in this study
Mouse monoclonal anti-human Phospho-histone H2AX
(cat. no. 05-636; 1:1,000 dilution; Merck Millipore), rabbit
anti-human histone H2AX (cat. no. P16104; 1:2,000 dilution; Ray
Biotech), rabbit anti-human p21 (cat. no. 2147; 1:1,000 dilution;
Cell Signaling Technology), rabbit anti-human 53BP1 (cat. no. 4937;
1:1,000 dilution; Cell Signaling Technology), mouse anti-human p53
(cat. no. 48818; 1:1,000 dilution; Cell Signaling Technology),
mouse monoclonal anti-gizzard β-actin (cat. no. sc-47778; 1:5,000
dilution; Santa Cruz Biotechnology), phalloidin (cat. no. A12379;
1:1,000 dilution; Thermo Fisher Scientific Inc.), DAPI (cat. no.
D1306; 1:36,000 dilution; Thermo Fisher Scientific Inc.). Secondary
antibodies used are goat anti-mouse Alexa Fluor 568 (A11004;
1:1,000 dilution; Thermo Fisher Scientific, Inc.), goat anti-rabbit
Alexa Fluor 488 (cat. no. A11034; 1:1,000 dilution; Thermo Fisher
Scientific Inc.), goat anti-mouse Alexa Fluor 488 (cat. no. A11001;
1:1,000 dilution; Thermo Fisher Scientific Inc.), goat anti-rabbit
Alexa Fluor 568 (cat. no. A11011; 1:1.000 dilution; Thermo Fisher
Scientific Inc.).
Immunoblotting
Cells grown in 3.5-cm plates were washed once with
1X ice-cold PBS and lysed in 100 ml of RIPA (150 mM NaCl, 0.1%
TX-100, 0.5% NaDOC, 0.1% SDS, 50 mM Tris-CL pH 8.0) with protease
inhibitor cocktail (cat. no. P2714; Sigma-Aldrich; Merck KGaA).
Proteins were quantified following brief sonication by Pierce BCA
protein assay kit (cat. no. 23225; Thermo Fisher Scientific Inc.).
Then, 8–12 mg of proteins were loaded on 10 or 12% SDS-PAGE, and
transferred onto a nitrocellulose membrane (cat. no. GE10600004;
Sigma-Aldrich; Merck KGaA) at 80 V for 3 h for low molecular weight
proteins or 80 V for 8 h for high molecular weight proteins. After
incubation with 5% BSA in TBST (10 mM Tris, pH 8.0, 150 mM NaCl,
0.5% Tween-20) for 60 min at room temperature, the membrane was
washed once with TBST and incubated with the listed antibodies
overnight at 4°C according to their data sheets.
β-galactosidase senescence assay
Cells were stained for β-galactosidase using
Senescence β-galactosidase Staining Kit (cat. no. 9860S; Cell
Signaling Technology) according to manufacturers protocol.
NK92-mediated cytotoxicity assay
Cytotoxicity assay was performed as previously
described (18) with the following
modifications: IL-2-activated NK92 effector cells were incubated
with target A549 or H460 cells with the E/T ratios, 1:10, and 1:20
in a 5% CO2 incubator at 37°C in a 96-well plate in
triplicates. After 6 h of co-culture, NK activity was measured
using the LDH Cytotoxicity Assay Kit (cat. no. 88954; Pierce™)
according to the manufacturers protocol.
Microarray analysis
Total RNA concentrations were determined using the
Qubit® RNA HS Assay Kit (cat. no. Q32852; Invitrogen;
Thermo Fisher Scientific, Inc.) using Qubit® 3.0 (cat.
no. Q33216; Invitrogen; Thermo Fisher Scientific, Inc.) and RNA
quality was assessed by running a 2% agarose gel and checking the
18S and 28S rRNA bands. Subsequently, 100 ng of good quality RNA
was prepared for transcriptome profiling using the Human Clariom™ D
Assay (cat. no. 902922; Applied Biosystems). All steps were
performed following the manufacturers protocol, where in brief, the
GeneChip™ WT PLUS Reagent Kit (cat. no. 902281; Applied Biosystems)
was first used to generate amplified and biotinylated sense-strand
DNA that was then hybridized on the Human Clariom™ D Arrays
(Applied Biosystems™). These arrays include over 6,765,500 probes
that can detect genes, exons, and alternative splicing events
giving rise to coding and long non-coding RNA isoforms. The arrays
were washed and stained on the GeneChip Fluidics Station 450 (cat.
no. 00-0079; Applied Biosystems) and scanned with the GeneChip™
Scanner 3,000 7G (cat. no. 00-0213; Applied Biosystems). The
scanned images and DAT files were inspected for the absence of
bubbles and proper scanner alignment, respectively, while the
generated raw files (CELL) were imported to the Transcriptome
Analysis Console (TAC) 4.0 software (Applied Biosystems) for data
analysis. Expression (Gene + Exon) analysis was carried out on TAC
by applying the Gene + Exon-Signal Space Transformation-Robust
Multi-Array Average (SST-RMA) algorithm. Default settings were used
for determining relative gene expression levels of each transcript,
which included applying the ebayes (Empirical Bayes Statistics for
Differential Expression) ANOVA method for statistical testing, as
well as setting the threshold for gene expression fold change (FC)
between the treated and untreated samples at ≤ −2 or ≥2. Only
coding and multiple complex loci with P-value <0.05 were
considered for further analysis. A heatmap was generated by
hierarchical clustering of common deregulated genes using complete
linkage with Euclidean distance on Heatmapper (19). Venn diagram was plotted using Venny
(20). Pathway enrichment analysis
was performed using Gene Set Enrichment Analysis (GSEA) software
(21), wherein overlaps between
common deregulated genes in the microarray data and Hallmark gene
sets H were computed (22).
Pathways with P-value <0.05 and FDR q-value <0.05 were
considered significantly enriched.
Statistical analysis
Statistical analyses were carried out using GraphPad
Software version 6 (GraphPad Software, Inc.). All data are
expressed as means ± SEM. Significant differences were found using
two-way analysis of variance (ANOVA) followed by Bonferroni post
hoc test.
Results
Toxic compounds identified in
WPSC
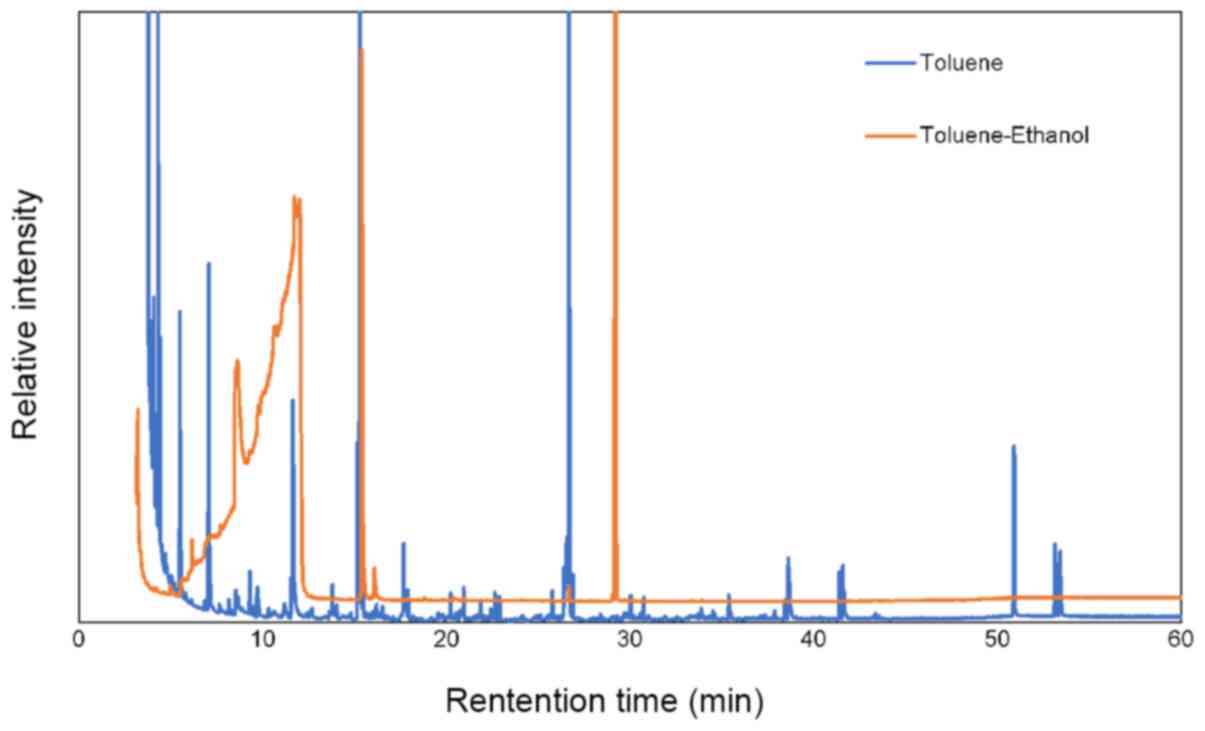
We first attempted to identify chemical compounds in
WPSC by analyzing the toluene extract and the ethanol solution
using GCMS. Many peaks were identified in the chromatograms
including some that are present at trace levels. Major peaks are
shown in Fig. 1. An internal
standard was used to evaluate the reproducibility of the analysis
and to allow the calculation of relative concentrations. Table I summarizes the major chemical
compounds identified in both the toluene and ethanol solutions.
Most of these compounds are known to cause serious health problems.
The safety, hazard and toxicity evaluation of the toluene and
ethanol extracts were performed using ToxNet and PubChem resources.
Several compounds were identified as carcinogens or may cause
various toxicities and health hazards. Several harmful compounds
were identified and may be associated with the burning of coal as
reported in a previous study (14).
For example, ethyl benzene, a component previously identified in
coal smoke was also found in WPSC. It is classified by the
International Agency for Research on Cancer (IARC) as a possible
human carcinogen and may cause kidney failure as a result of
prolonged exposure at low concentrations (14). Indene, another compound found in
WPSC, is a polycyclic aromatic hydrocarbon which was identified in
coal smoke (14). Furthermore,
several identified compounds are known to cause irritation to the
skin, eyes and respiratory tract, including
3-ethoxy-4-hydroxy-benzaldehyde, 1,3-dimethyl-benzene, benzyl
alcohol, docosane, ethylbenzene, 3-ethoxypropionaldehyde, and
2,3-dihydro-3,5-dihydroxy-6-methyl-4H-pyran-4-one. Many of the
identified compounds are known to interfere with and alter the
functions of the central nervous system including 1,3-dimethyl
benzene, benzyl alcohol, ethyl cyclohexane, ethyl benzene and
nicotine. In addition, several compounds are known to induce DNA
damage including benzaldehyde, 1,3-dimethyl Benzene, and
5-hydroxymethyl furfural (23).
 | Table I.Major chemical compounds identified
in the toluene and ethanol extracts. |
Table I.
Major chemical compounds identified
in the toluene and ethanol extracts.
| Toluene | Ethanol |
|---|
| Benzaldehyde | Hexadecane,
2,6,10,14-tetramethyl- (CAS) |
| Benzaldehyde,
3-ethoxy-4-hydroxy- (CAS) | Hexadecanoic acid,
1-(hydroxymethyl)-1,2-ethanediyl ester |
| Benzene,
1,3-dimethyl- | 4H-Pyran-4-one,
2,3-dihydro-3,5-dihydroxy-6-methyl- |
| Benzyl alcohol |
5-Hydroxymethylfurfural |
| Benzenepropanoic
acid, 3,5-bis(1,1-dimethylethyl)-4-hydroxy-, octadecyl ester | 1H-Indene,
1-hexadecyl-2,3-dihydro- |
| Cyclopentane,
1-ethyl-2-methyl-, cis- | Nonyl tetradecyl
ether |
| Cyclohexane,
ethyl- | Nicotine |
| Decane,
3,3,8-trimethyl- | octadecanoic acid,
3-oxo-, ethyl ester |
| DECANE,
3,3,7-TRIMETHYL- | Octadecanoic acid,
2,3-dihydroxypropyl ester |
| Dihydro methyl
jasmonate | Phenol,
2,4-bis(1,1-dimethylethyl)-, phosphite (3:1) |
| 2,3-Dihydroxypropyl
icosanoate, 2TMS derivative | Pyrrolidine-D4 |
| Docosane | Ethanol,
2-[(triethylsilyl)oxy]- |
| Dodecane,
4,6-dimethyl- | 1,2,3-Propanetriol,
1-acetate |
| Ethylbenzene | Pyridine,
3-(1-methyl-2-pyrrolidinyl)-, (S)- (CAS) |
| Hexadecane,
2,6,10,14-tetramethyl- (CAS) | Propanal, 3-ethoxy-
(CAS) |
| 1-Hexadecanol | Trans-Anethole |
|
|
Tris(2,4-di-tert-butylphenyl)
phosphate |
| Hexane,
3,3-dimethyl- (CAS) |
Trans-4-hydroxymethyl-2-methyl-1,3-dioxolane |
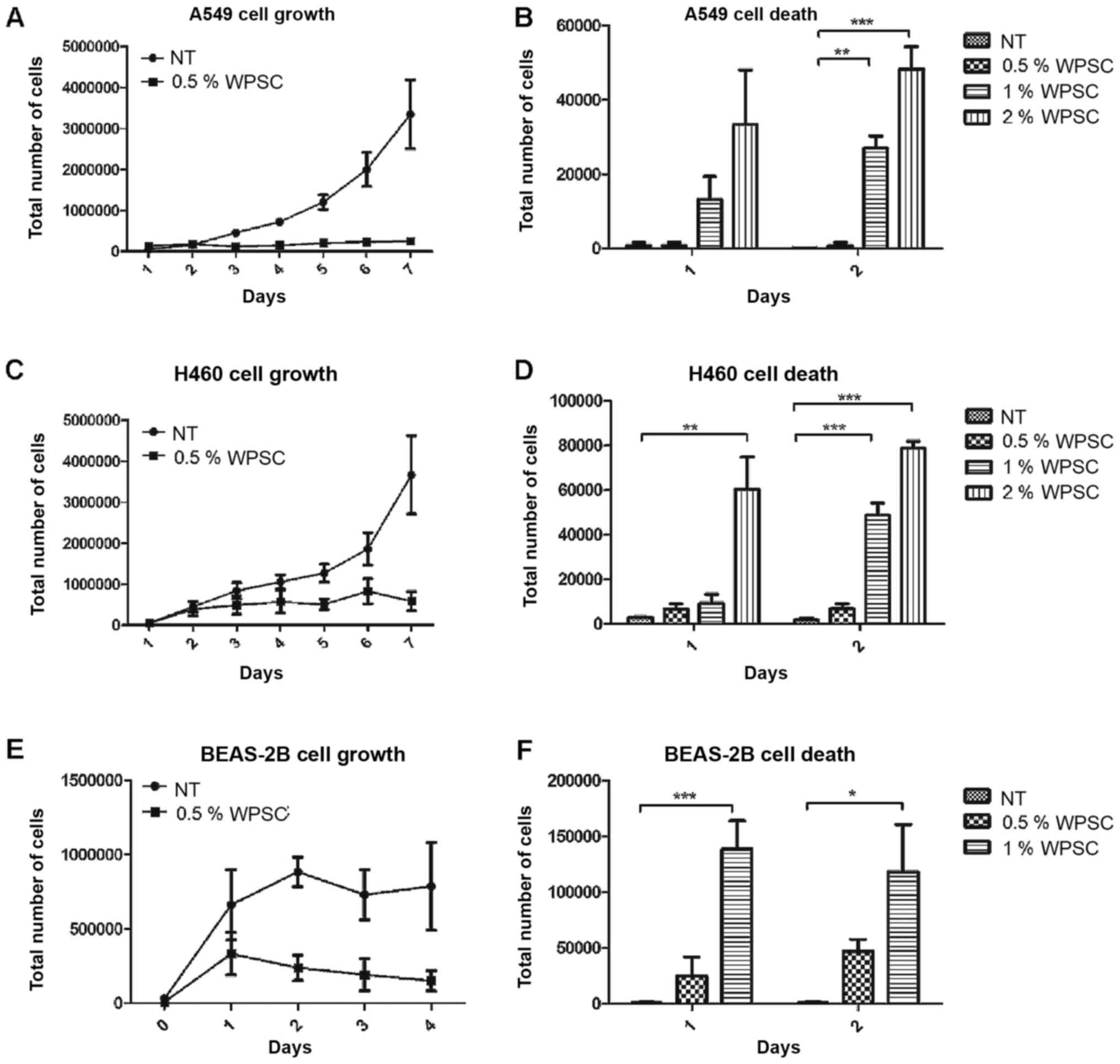
Effect of WPSC on lung cancer cell
proliferation
We first tested the cytotoxic effect of WPSC on two
non-small cell lung cancer cells, A549, and H460 as well as on a
normal bronchial epithelial cell line, BEAS-2B. Both cell number
and viability of tested cells were measured in the presence of
various concentrations of WPSC condensate (0.5, 1 and 2%) for 8
consecutive days with re-administration of fresh WPSC at day 4.
Additionally, WPSC treatment resulted in a dose- and time-dependent
decrease in cancer cell proliferation (Fig. 2A-D). Hemocytometer images are shown
in Fig. S1. At higher doses of 1
and 2% an increase in cell death was observed. By contrast, the
lowest concentration of 0.5% resulted in a significant increase in
the cell death of BEAS-2B normal lung cells following 2 days of
treatment (Fig. 2E and F);
consequently, these cells were not analyzed further. Subsequent
experiments on lung cancer cells were performed using 0.5% WPSC,
considering the absence of cell death.
WPSC increases DNA damage and cellular
senescence in lung cancer cells
DNA damage often arises as a result of normal
cellular processes, as a by-product of the cells own metabolic
activity. It can also be induced as a result of environmental
exposure to chemical agents. We therefore investigated whether WPSC
induced DNA damage and used the alkaline comet assay to detect and
quantify the degree of gross DNA damage. As shown in Fig. S2A and B, WPSC induced an increase
in tail length in the cancer cell lines A549 and H460 when compared
to untreated cells, indicating an increase in the frequency of DNA
breaks. Comet assay reflects the physical status of genomic DNA
while 53BP1/γH2AX staining represents processes related to the
biological DNA damage response (DDR) (24). We investigated the effect of WPSC on
the DDR using two markers: Phosphorylated histone H2AX at Ser139
(γH2AX) and 53BP1. Foci formation indicates the aggregation of
these proteins induced by DNA damage. Consistent with the comet
tail formation, in response to WPSC treatment, an increase in the
nuclear foci for both γH2AX and 53BP1 in A549 and H460 cells was
observed (Fig. S2C-E).
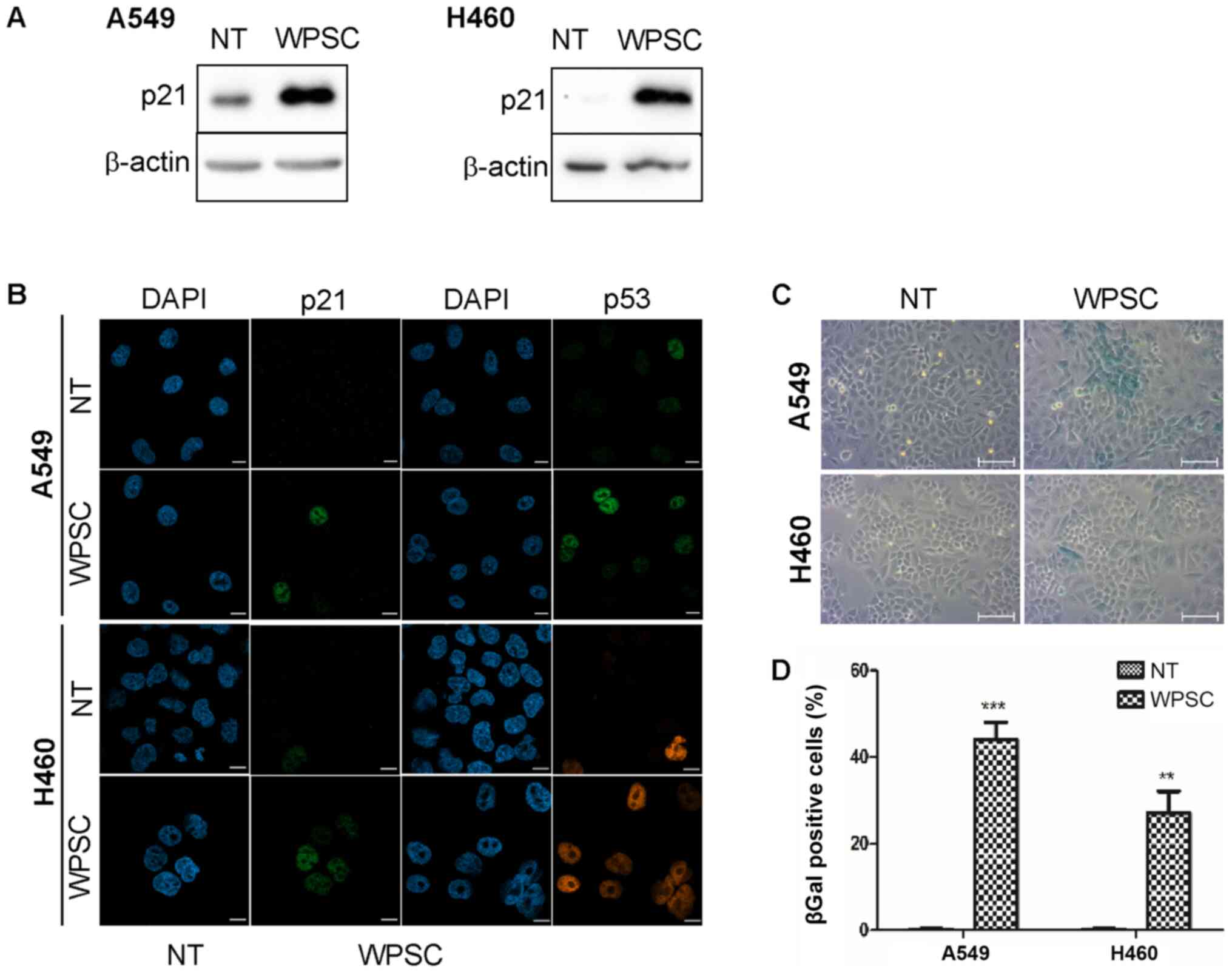
Since waterpipe smoke condensate resulted in
decreased cell growth, we tested whether the lung cancer cells were
undergoing apoptosis or cell cycle arrest and senescence.
Consistent with cell cycle arrest and induction of senescence, we
found that WPSC induced an increase in p21 protein expression as
revealed by western blot analysis (Fig.
3A). Using confocal microscopy analysis, we demonstrated an
accumulation of both p21 and p53 proteins in the nuclei of both
cancer cells (Fig. 3B). This was
also supported by an increase in β-galactosidase activity in these
cells indicating the induction of cellular senescence (Fig. 3C and D).
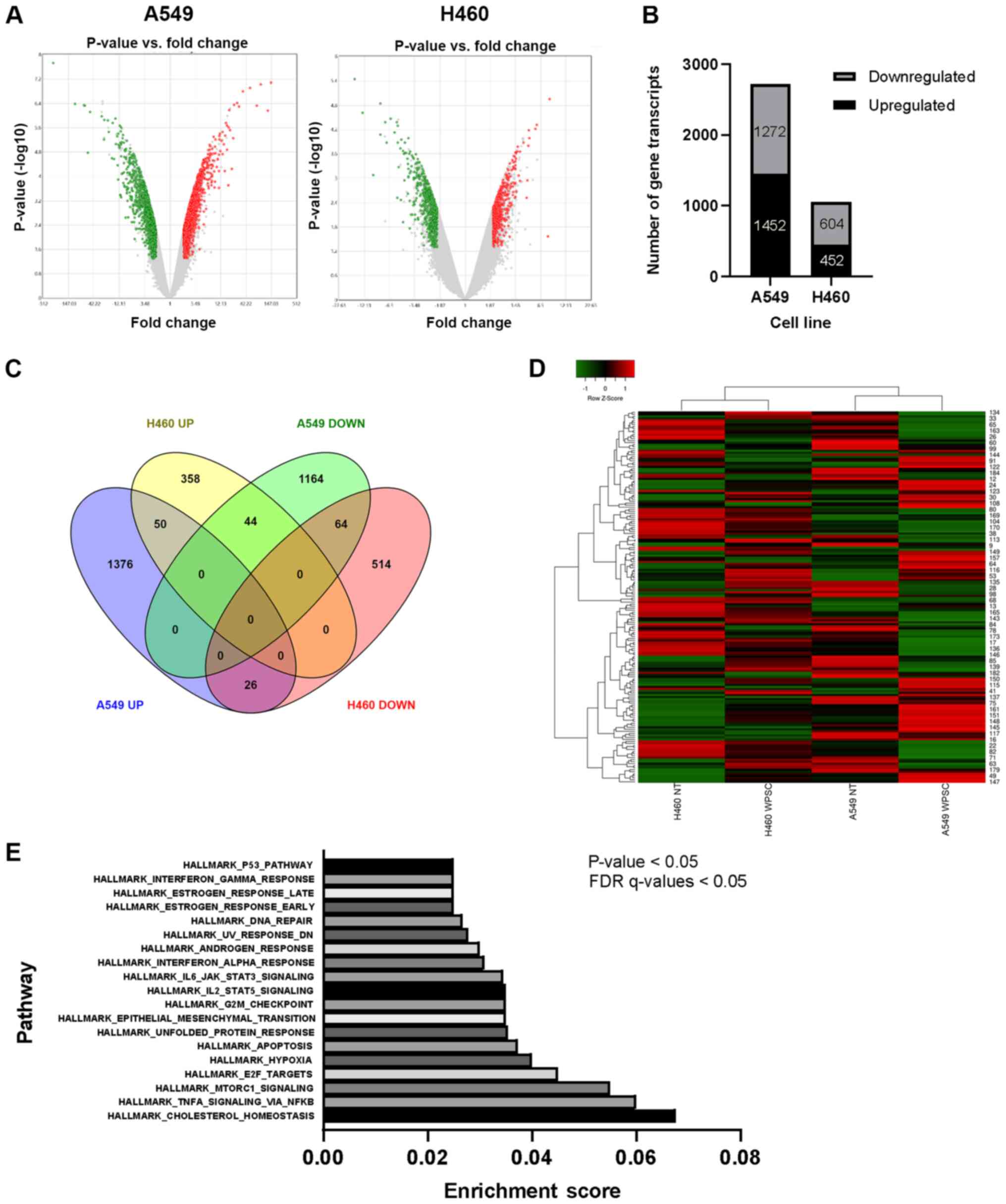
Transcriptomic changes associated with
WPSC treatment
To determine whether WPSC treatment impacts the
molecular pathways associated with EMT, stemness and immune evasion
and to identify novel gene expression patterns in response to WPSC
treatment, a microarray analysis was performed. RNA derived from
biological duplicates of A549 and H460 cell lines 8 days
post-treatment with WPSC, as well as their untreated counterparts
was analyzed. Considering only coding and multiple complex loci
with fold change of ≤ - 2 and ≥2 and P-value of <0.05, volcano
plots were generated. A greater number of differentially expressed
gene transcripts in A549 (1452 upregulated and 1272 downregulated)
in comparison with H460 (452 upregulated and 604 downregulated)
(Fig. 4A and B) was observed. Among
these differentially expressed transcripts, 184 were common to both
A549 and H460 (Fig. 4C and D). Of
these 70 transcripts showed opposite patterns of deregulation:
upregulated in one cell line and downregulated in the other or vice
versa. Additionally, 50 transcripts were upregulated, and 64
transcripts were downregulated in the two cell lines, respectively
(Fig 4D). Pathway enrichment
analysis revealed that the commonly deregulated genes clustered in
pathways involved EMT, the cell cycle, apoptosis, DNA repair and
the inflammatory response, among others (Fig. 4E).
WPSC activates the inflammatory
response in lung cancer cells
Waterpipe smoking has been shown to trigger the
inflammatory response in humans as measured in the blood of
waterpipe smokers (25). Using
microarray analysis, gene expression in the inflammatory pathway
(Fig. 5A and B) was detected. IL-1β
is a key mediator of inflammation and promotes invasiveness
(26), whereas the chemokine CCL2
regulates inflammatory responses and also plays a role in tumor
progression and metastasis (27).
In addition, IL-6 is a pro-inflammatory cytokine with key roles in
EMT (28). Fig. 5A and B shows a significant increase
in expression levels of these genes in response to WPSC. We also
investigated the expression levels of caspase-1, the cysteine
protease that converts the inactive proform of IL-1β to the active
inflammatory cytokine (29). The
results showed that the expression level was slightly increased in
the two cells (Fig. 5A and B),
consistent with the increase in IL-1β.
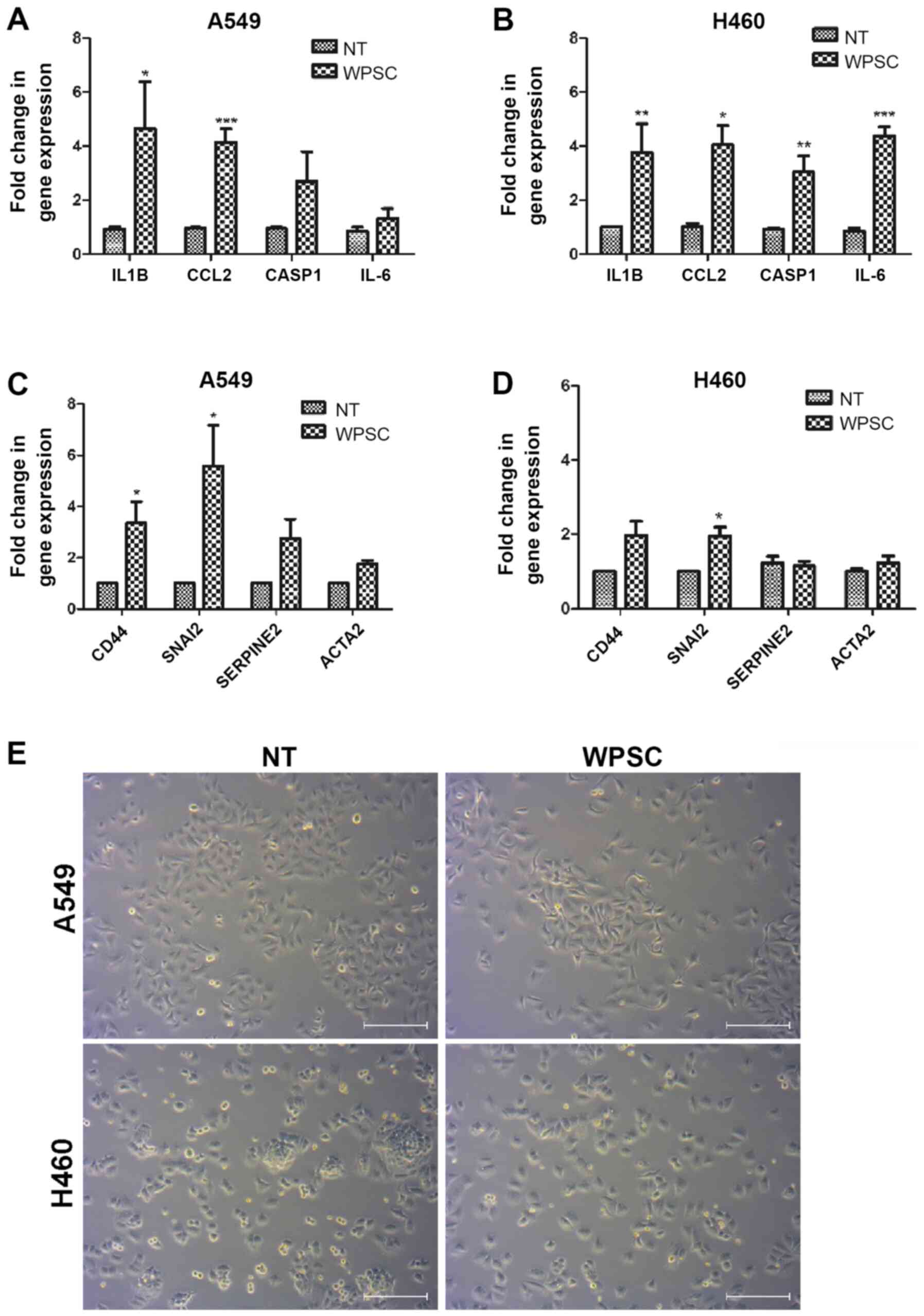 | Figure 5.WPSC effects on the inflammatory
response and on the expression of EMT- and stemness-related genes.
(A and C) A549 and (B and D) H460 gene expression was analyzed by
qPCR for caspase-1, IL-1β, CCL2, IL-6, CD44, SNAI2, SERPINE2 and
ACTA2. Results represent means of three independent experiments,
and were run in duplicates. *P≤0.05, **P≤0.01, ***P≤0.001. (E)
Bright-field images for the A549 and H460 cells cultured with WPSC.
Scale bar, 100 µm. |
WPSC induces EMT and stemness
properties
EMT and stem cell-like traits are intertwined
processes that dictate tumor aggressiveness. It is well established
that EMT generates cells with stem cell properties (30). Using microarray analysis, several
genes involved in EMT were found to be enriched including IL-6,
IL-15, GLIPR1, CD44 and COL5A. Using qPCR analysis, we
demonstrated that both EMT and stemness genes are regulated
(Fig. 5C and D), an increase in the
cancer stem cell marker CD44 was observed. CD44 is also known to
promote EMT (31). Furthermore, an
increase in SNAI2, an EMT key transcription factor (32), SERPINE2, a protease that promotes
ECM deposition and invasion (33)
and ACTA2 coding for alpha smooth muscle actin that promotes
motility and invasion (34) were
also increased in A549 cells. Cell morphological changes in
response to WPSC were observed. WPSC treatment converted the cells
from a ‘cuboidal’ epithelial structure into an elongated
mesenchymal shape (Fig. 5E).
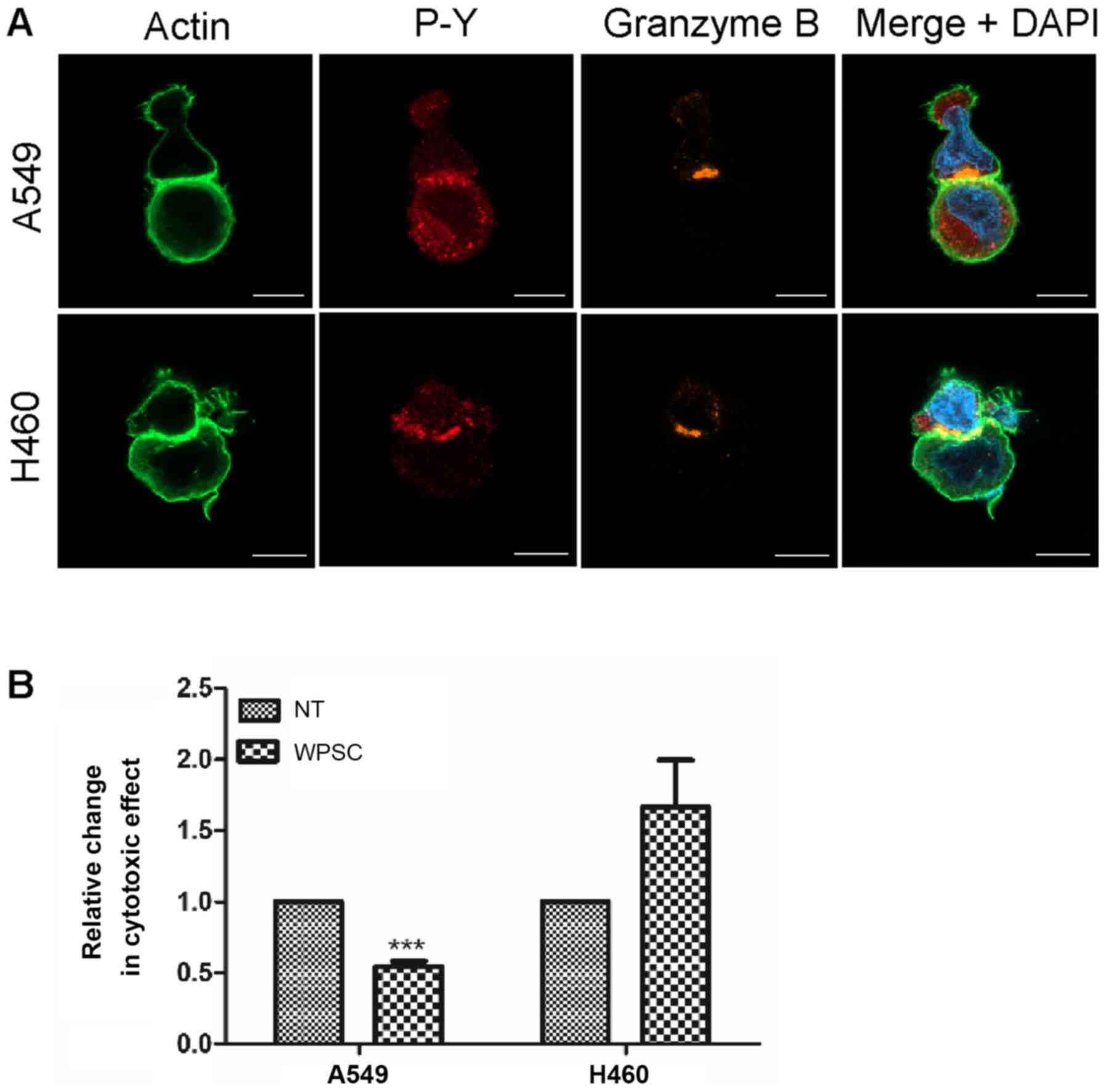
WPSC effect on synapse formation and
NK cell-mediated killing
We examined whether WPSC treatment of cancer cells
interferes with tumor cell recognition and killing. As is evident
in Fig. 6A, the immunological
synapse between NK cells and WPSC-treated A549 and H460 cancer cell
was not affected. Nevertheless, as shown in Fig. 6B, WPSC induced a significant
decrease in the killing of A549 cells but had no significant effect
on the H460 cells.
Discussion
In the present study, we investigated the effects of
WPSC, using a new method based on a smoking topography stimulating
machine on non-small-cell lung cancer cells. We demonstrated that
WPSC treatment resulted in a dose-dependent decrease in cell
proliferation with cell death at higher concentrations. Both
cellular senescence and apoptosis were induced in cancer cells at
the lower WPSC concentration of 0.5%. However, such treatment
resulted in cell death of normal lung cells. As such, we are in the
process of addressing the effects of short-term duration treatment
with various concentrations of WPSC on normal lung cells.
Furthermore, we did not explore doses lower than 0.5% as this did
not inhibit growth in cancer cells. However, the molecular changes
described in this study may be induced at lower doses. Adaptive
cellular responses are activated upon microenvironmental stress,
such as WPS exposure, and are critical in the prevention of tissue
damage and transformation. The net effect of these responses
depends on the duration, nature, and intensity of the stress.
Indeed, at shorter duration treatment of 72 h we did not observe
any cell death (data not shown), which is consistent with previous
findings showing that only senescence was generated at this time
point in A549 cells (10).
Prolonged exposure to environmental stresses may
become deleterious in promoting cell death, excessive inflammation,
and tissue remodeling. Indeed, we observed that WPSC treatment
resulted in increased DNA damage and a subsequent increase in γH2AX
and 53BP1, which is consistent with the recent finding of Yoshida
et al, indicating that tobacco smoking resulted in the
increase in mutational burden of lung cells (35), and with a known increase in the
expression of both γH2Ax and 53BP1 in several cancers including
breast, bladder, lung, head and neck (36). By contrast, cigarette smoke has been
shown to induce a decrease in nucleotide excision repair
mechanisms, and does induce DNA damage as measured by COMET assay
(37,38), which raises the interesting
possibility that other factors present in WPS may induce a toxic
response independent of DNA damage in these cells.
We then analyzed the effects of 8-day treatment of
WPSC on the transcriptomic profile of A549 and H460 cells. The
results showed an increase in the expression of genes involved in
inflammation, EMT and in the generation of cancer stem cells. It is
well known that inflammation plays an important role in the
tumor-specific microenvironment, which is thought to facilitate all
phases of tumorigenesis, from initiation to metastasis (39). Inflammatory mediators also exhibit
the potential to induce the acquisition of CSCs properties
(40). In the course of this study,
we demonstrated an increase in IL-1β, IL-6, CCL2 and caspase-1
levels, known to be associated with the induction of EMT (41–43).
IL-6 and IL-1β are also found to be senescence-associated secreted
factors (44). WPS in humans has a
significant association with systemic inflammation (25,45,46).
Our results are consistent with this role for WPSC and further
highlight the importance of inhibiting inflammatory processes in WP
smokers as a means for more effective cancer treatment.
Of the EMT-related genes, we observed an increase in
the expression of SNAI2, SERPINE2, ACTA2 as well as
CD44, also is a regulator of cancer stemness. WPS was shown
to induce EMT in breast cancer cell lines through FAK and Erk1/Erk2
expression (47) and resulted in an
increase in invasion and migration properties of breast cancer
cells (48). Our results therefore
suggest that WPSC exposure to lung cancer cells may change their
physiology to promote EMT processes and acquire CSCs
properties.
NK cell-mediated immune surveillance strengthens
host defense against certain microbial agents and cells undergoing
malignant transformation. We investigated the effects of this WPS
treatment on the ability of NK cells to recognize and kill the lung
cancer cell targets. To date, no data are available as to the
effect of WPS on NK cell function. Nevertheless, in mice, chronic
exposure to WPS has been reported to promote immune suppression
(49). In addition, in cigarette
smokers lower populations of NK cells are found (50), and patients with lung cancer had a
marked decrease in NK activity compared to non-smokers (51). However, other reports indicate that
cigarette smoking was associated with an increase in NK cell
tumoricidal activity without any alteration in the absolute number
of NK cells in blood (52).
Furthermore, NK cells from cigarette-exposed mice produced more
IFN-γ following stimulation with IL-12, IL-18, or both (53). Under our experimental conditions,
the results showed that A549 cell killing was reduced as compared
to non-treated cells, whereas H460 cell survival was not affected.
However, the capacity of the NK cells to form synapses with the
WPSC-treated cancer cells was not affected in the two cell lines.
This is most likely due to genetic variations in the two cell
lines, engaging communication networks that could also include the
release of inflammatory mediators that signal to the immune system.
This is interesting as one possibility is that WPSC could induce
several pathways that affect susceptibility to lysis. Nevertheless,
we did not observe a correlation between the EMT/CSC induction in
A549 cells and a decrease in their NK-mediated cell lysis.
As a limitation of the study, we did not explore the
molecular mechanisms of cell apoptosis, as we found WPSC affecting
other hallmark properties of cancer including EMT-, stemness- and
genomic instability (DNA damage)-related changes. These
characteristics are not unique to waterpipe smoke as cigarette
smoke has also been shown to induce EMT cell properties (54) in addition to apoptosis (55).
WPSC can induce apoptosis (12). At the same time, WPSC also drives
hallmark cancer properties (senescence and EMT, stemness and
genomic instability) that promote survival of these cells. In
addition, these cells also acquire properties to escape immune
surveillance, which complicates the situation further. These
characteristics are not unique to WPS as cigarette smoke has also
been shown to impair NK- mediated immune surveillance (54) and to induce EMT cell properties
(54) as well as apoptosis
(55). We are currently culturing
the cells in WPSC media for longer durations (up to 4–5 months) and
we see that these tumor cells keep dividing (unpublished data). As
DNA damage contributes to increased genomic instability and
mutational burden, WPSC can also contribute to genetic
heterogeneity of tumors.
Our results indicate that WPSC is a contributing
factor in the pathogenesis of lung cancer, through impairing cell
growth, inducing inflammation and DNA damage. If we are to assume
that a cancer patient continues to smoke, WPSC may serve as fuel to
the cancer cells and may contribute to metastases. In fact,
continued smoking is considered a strong adverse predictor of
survival and increases the risk of a second lung cancer compared to
those patients who stopped smoking (56). Thus, therapy modalities can be more
effective by eliminating smoke exposure to cancer patients and by
targeting the inflammatory mechanisms in order to control the
emergence of aggressive cancer clones with EMT/CSCs features.
Supplementary Material
Supporting Data
Acknowledgements
The authors would like to convey appreciation to
Angele Fauvel, to Abderemane Abdou (INSERM U1186) and to Ziad Sara
(AUS), for their technical assistance.
Funding
The present study was supported by Al Jalila
Foundation (AJF 2018009) to Zaarour. The smoke sampling and
analysis research were funded by the office of Research and
Graduate Studies at the American University of Sharjah
(FRG19-L-S11).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available with the corresponding author on reasonable
request.
Authors contributions
SC conceived, significantly guided in the design,
analysis, and interpretation of the findings of this study. RFZ,
PP, NZ, AR and ST conceived and designed experiments, acquired and
interpreted data. RAK, FA performed the microarray-related
experiments and analysis with contributions from GHV. YES performed
the WPSC purification and analysis. HN conducted the confocal
imaging. All authors discussed the results and contributed to the
final manuscript.
Ethical approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
All authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
WPS
|
waterpipe smoke
|
|
WPSC
|
waterpipe smoke condensate
|
|
EMT
|
epithelial to mesenchymal
transition
|
|
NK
|
natural killer
|
References
|
1
|
Polverino F, Sam A and Guerra S: COPD: To
Be or Not to Be, That is the Question. Am J Med. 132:1271–1278.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Montazeri Z, Nyiraneza C, El-Katerji H and
Little J: Waterpipe smoking and cancer: Systematic review and
meta-analysis. Tob Control. 26:92–97. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Talhout R, Schulz T, Florek E, van Benthem
J, Wester P and Opperhuizen A: Hazardous compounds in tobacco
smoke. Int J Environ Res Public Health. 8:613–628. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Shihadeh A, Schubert J, Klaiany J, El
Sabban M, Luch A and Saliba NA: Toxicant content, physical
properties and biological activity of waterpipe tobacco smoke and
its tobacco-free alternatives. Tob Control. 24 (Suppl 1):i22–i30.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Maziak W, Jawad M, Jawad S, Ward KD,
Eissenberg T and Asfar T: Interventions for waterpipe smoking
cessation. Cochrane Database Syst Rev (7). CD0055492015.
|
|
6
|
Etemadi A, Poustchi H, Chang CM, Blount
BC, Calafat AM, Wang L, De Jesus VR, Pourshams A, Shakeri R, Shiels
MS, et al: Urinary biomarkers of carcinogenic exposure among
cigarette, waterpipe, and smokeless tobacco users and never users
of tobacco in the Golestan Cohort Study. Cancer Epidemiol
Biomarkers Prev. 28:337–347. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Boskabady MH, Farhang L, Mahmodinia M,
Boskabady M and Heydari GR: Comparison of pulmonary function and
respiratory symptoms in water pipe and cigarette smokers.
Respirology. 17:950–956. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Awan KH, Siddiqi K, Patil Sh and Hussain
QA: Assessing the affect of waterpipe smoking on cancer outcome - a
systematic review of current evidence. Asian Pac J Cancer Prev.
18:495–502. 2017.PubMed/NCBI
|
|
9
|
Alsaad AM, Al-Arifi MN, Maayah ZH, Attafi
IM, Alanazi FE, Belali OM, Alhoshani A, Asiri YA and Korashy HM:
Genotoxic impact of long-term cigarette and waterpipe smoking on
DNA damage and oxidative stress in healthy subjects. Toxicol Mech
Methods. 29:119–127. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Rammah M, Dandachi F, Salman R, Shihadeh A
and El-Sabban M: In vitro cytotoxicity and mutagenicity of
mainstream waterpipe smoke and its functional consequences on
alveolar type II derived cells. Toxicol Lett. 211:220–231. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bodas M, Van Westphal C,
Carpenter-Thompson R, K Mohanty D and Vij N: Nicotine exposure
induces bronchial epithelial cell apoptosis and senescence via ROS
mediated autophagy-impairment. Free Radic Biol Med. 97:441–453.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Rammah M, Dandachi F, Salman R, Shihadeh A
and El-Sabban M: In vitro effects of waterpipe smoke condensate on
endothelial cell function: A potential risk factor for vascular
disease. Toxicol Lett. 219:133–142. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gerlinger M, Rowan AJ, Horswell S, Math M,
Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N,
Stewart A, et al: Intratumor heterogeneity and branched evolution
revealed by multiregion sequencing. N Engl J Med. 366:883–892.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Elsayed Y, Dalibalta S and Abu-Farha N:
Chemical analysis and potential health risks of hookah charcoal.
Sci Total Environ. 569-570:262–268. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Olive PL and Banáth JP: The comet assay: A
method to measure DNA damage in individual cells. Nat Protoc.
1:23–29. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gyori BM, Venkatachalam G, Thiagarajan PS,
Hsu D and Clement MV: OpenComet: An automated tool for comet assay
image analysis. Redox Biol. 2:457–465. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Terry S, Abdou A, Engelsen AST, Buart S,
Dessen P, Corgnac S, Collares D, Meurice G, Gausdal G, Baud V, et
al: AXL Targeting overcomes human lung cancer cell resistance to
NK- and CTL-mediated cytotoxicity. Cancer Immunol Res. 7:1789–1802.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Babicki S, Arndt D, Marcu A, Liang Y,
Grant JR, Maciejewski A and Wishart DS: Heatmapper: Web-enabled
heat mapping for all. Nucleic Acids Res. 44((W1)): W147–53. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Oliveros JC: An interactive tool for
comparing lists with Venns diagrams. Publicly available at.
http://bioinfogp.cnb.csic.es/tools/venny/index.html
|
|
21
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES, et al: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liberzon A, Birger C, Thorvaldsdóttir H,
Ghandi M, Mesirov JP and Tamayo P: The Molecular Signatures
Database (MSigDB) hallmark gene set collection. Cell Syst.
1:417–425. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Durling LJ, Busk L and Hellman BE:
Evaluation of the DNA damaging effect of the heat-induced food
toxicant 5-hydroxymethylfurfural (HMF) in various cell lines with
different activities of sulfotransferases. Food Chem Toxicol.
47:880–884. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kurashige T, Shimamura M and Nagayama Y:
Differences in quantification of DNA double-strand breaks assessed
by 53BP1/γH2AX focus formation assays and the comet assay in
mammalian cells treated with irradiation and N-acetyl-L-cysteine. J
Radiat Res (Tokyo). 57:312–317. 2016. View Article : Google Scholar
|
|
25
|
Kumari B, Aslam SK, Zaheer S, Adil SO and
Shafique K: Systemic inflammatory markers among waterpipe smokers,
cigarette smokers, and nonsmokers. J Addict Med. 13:55–60. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Apte RN, Dotan S, Elkabets M, White MR,
Reich E, Carmi Y, Song X, Dvozkin T, Krelin Y and Voronov E: The
involvement of IL-1 in tumorigenesis, tumor invasiveness,
metastasis and tumor-host interactions. Cancer Metastasis Rev.
25:387–408. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li M, Knight DA, A Snyder L, Smyth MJ and
Stewart TJ: A role for CCL2 in both tumor progression and
immunosurveillance. OncoImmunology. 2:e254742013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Browning L, Patel MR, Horvath EB, Tawara K
and Jorcyk CL: IL-6 and ovarian cancer: Inflammatory cytokines in
promotion of metastasis. Cancer Manag Res. 10:6685–6693. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Siegmund B, Lehr HA, Fantuzzi G and
Dinarello CA: IL-1 beta -converting enzyme (caspase-1) in
intestinal inflammation. Proc Natl Acad Sci USA. 98:13249–13254.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan
A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, et al: The
epithelial-mesenchymal transition generates cells with properties
of stem cells. Cell. 133:704–715. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Bhattacharya R, Mitra T, Ray Chaudhuri S
and Roy SS: Mesenchymal splice isoform of CD44 (CD44s) promotes
EMT/invasion and imparts stem-like properties to ovarian cancer
cells. J Cell Biochem. 119:3373–3383. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Jayachandran A, Königshoff M, Yu H,
Rupniewska E, Hecker M, Klepetko W, Seeger W and Eickelberg O: SNAI
transcription factors mediate epithelial-mesenchymal transition in
lung fibrosis. Thorax. 64:1053–1061. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hindriksen S and Bijlsma MF: Cancer stem
cells, EMT, and developmental pathway activation in pancreatic
tumors. Cancers (Basel). 4:989–1035. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Tomaskovic-Crook E, Thompson EW and Thiery
JP: Epithelial to mesenchymal transition and breast cancer. Breast
Cancer Res. 11:2132009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yoshida K, Gowers KHC, Lee-Six H,
Chandrasekharan DP, Coorens T, Maughan EF, Beal K, Menzies A,
Millar FR, Anderson E, et al: Tobacco smoking and somatic mutations
in human bronchial epithelium. Nature. 578:266–272. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Willers H, Gheorghiu L, Liu Q, Efstathiou
JA, Wirth LJ, Krause M and von Neubeck C: DNA damage response
assessments in human tumor samples provide functional biomarkers of
radiosensitivity. Semin Radiat Oncol. 25:237–250. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Holcomb N, Goswami M, Han SG, Clark S,
Orren DK, Gairola CG and Mellon I: Exposure of human lung cells to
tobacco smoke condensate inhibits the nucleotide excision repair
pathway. PLoS One. 11:e01588582016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Hang B, Sarker AH, Havel C, Saha S, Hazra
TK, Schick S, Jacob P III, Rehan VK, Chenna A, Sharan D, et al:
Thirdhand smoke causes DNA damage in human cells. Mutagenesis.
28:381–391. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Lowe DB and Storkus WJ: Chronic
inflammation and immunologic-based constraints in malignant
disease. Immunotherapy. 3:1265–1274. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Jeong YJ, Oh HK, Park SH and Bong JG:
Association between inflammation and cancer stem cell phenotype in
breast cancer. Oncol Lett. 15:2380–2386. 2018.PubMed/NCBI
|
|
41
|
Masola V, Carraro A, Granata S, Signorini
L, Bellin G, Violi P, Lupo A, Tedeschi U, Onisto M, Gambaro G, et
al: In vitro effects of interleukin (IL)-1 beta inhibition on the
epithelial-to-mesenchymal transition (EMT) of renal tubular and
hepatic stellate cells. J Transl Med. 17:122019. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Ling Z, Yang X, Chen X, Xia J, Cheng B and
Tao X: CCL2 promotes cell migration by inducing
epithelial-mesenchymal transition in oral squamous cell carcinoma.
J Oral Pathol Med. 48:477–482. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Li J, Zhou Y, Liu Y, Dai B, Zhang YH,
Zhang PF and Shi XL: Sorafenib inhibits caspase-1 expression
through suppressing TLR4/stat3/SUMO1 pathway in hepatocellular
carcinoma. Cancer Biol Ther. 19:1057–1064. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Coppé JP, Patil CK, Rodier F, Sun Y, Muñoz
DP, Goldstein J, Nelson PS, Desprez PY and Campisi J:
Senescence-associated secretory phenotypes reveal
cell-nonautonomous functions of oncogenic RAS and the p53 tumor
suppressor. PLoS Biol. 6:2853–2868. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Khan NA, Lawyer G, McDonough S, Wang Q,
Kassem NO, Kas-Petrus F, Ye D, Singh KP, Kassem NOF and Rahman I:
Systemic biomarkers of inflammation, oxidative stress and tissue
injury and repair among waterpipe, cigarette and dual tobacco
smokers. Tob Control. 2019.PubMed/NCBI
|
|
46
|
Qasim H, Alarabi AB, Alzoubi KH, Karim ZA,
Alshbool FZ and Khasawneh FT: The effects of hookah/waterpipe
smoking on general health and the cardiovascular system. Environ
Health Prev Med. 24:582019. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Sadek KW, Haik MY, Ashour AA, Baloch T,
Aboulkassim T, Yasmeen A, Vranic S, Zeidan A and Al Moustafa AE:
Water-pipe smoking promotes epithelial-mesenchymal transition and
invasion of human breast cancer cells via ERK1/ERK2 pathways.
Cancer Cell Int. 18:1802018. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Patil S, Subbannayya T, Mohan SV, Babu N,
Advani J, Sathe G, Rajagopalan P, Patel K, Bhandi S, Solanki H, et
al: Proteomic changes in oral keratinocytes chronically exposed to
Shisha (Water Pipe). OMICS. 23:86–97. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Reyes-Caballero H, Park B, Loube J,
Sanchez I, Vinayachandran V, Choi Y, Woo J, Edwards J, Brinkman MC,
Sussan T, et al: Immune modulation by chronic exposure to waterpipe
smoke and immediate-early gene regulation in murine lungs. Tob
Control. 2019.PubMed/NCBI
|
|
50
|
Tollerud DJ, Clark JW, Brown LM, Neuland
CY, Mann DL, Pankiw-Trost LK, Blattner WA and Hoover RN:
Association of cigarette smoking with decreased numbers of
circulating natural killer cells. Am Rev Respir Dis. 139:194–198.
1989. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Phillips B, Marshall ME, Brown S and
Thompson JS: Effect of smoking on human natural killer cell
activity. Cancer. 56:2789–2792. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Newman LS, Kreiss K and Campbell PA:
Natural killer cell tumoricidal activity in cigarette smokers and
in silicotics. Clin Immunol Immunopathol. 60:399–411. 1991.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Motz GT, Eppert BL, Wortham BW,
Amos-Kroohs RM, Flury JL, Wesselkamper SC and Borchers MT: Chronic
cigarette smoke exposure primes NK cell activation in a mouse model
of chronic obstructive pulmonary disease. J Immunol. 184:4460–4469.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Vu T, Jin L and Datta PK: Effect of
cigarette smoking on epithelial to mesenchymal transition (EMT) in
lung cancer. J Clin Med. 5:E442016. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Hoshino Y, Mio T, Nagai S, Miki H, Ito I
and Izumi T: Cytotoxic effects of cigarette smoke extract on an
alveolar type II cell-derived cell line. Am J Physiol Lung Cell Mol
Physiol. 281:L509–L516. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Jassem J: Tobacco smoking after diagnosis
of cancer: Clinical aspects. Transl Lung Cancer Res. 8 (Suppl
1):S50–S58. 2019. View Article : Google Scholar : PubMed/NCBI
|