Introduction
Primary liver cancer ranks as the sixth most
frequently diagnosed cancer and the second most common cause of
tumor-related deaths in the world (1). Due to the poor prognosis of this
disease, its mortality has increased, reaching 30,160 deaths in
2020 (2). As the most common
subtype of primary liver cancer, hepatocellular carcinoma comprises
more than 90% of all primary liver cancer cases (3). Liver cirrhosis is the predominant
predisposing factor of liver cancer and usually occurs during
chronic liver injury, resulting in subsequent death of liver cells
(4). Common reasons of cirrhosis
include metabolic liver disorders, autoimmune diseases, chronic
viral infections and environmental factors (4). The incidence of liver cancer is
unevenly distributed in the world due to the wide variability of
the diverse pathogenic causes of cirrhosis. More than 80% of liver
cancer cases are reported in Eastern Asia and Africa, where chronic
hepatitis B viral infection is endemic (5). In contrast to hepatitis B, hepatitis C
viral infection is considered the most common cause of liver cancer
in Europe and in the United States (6). In general, a variety of treatment
methods that mainly include surgical resection and chemotherapy can
effectively reduce tumor progression when the patients are
diagnosed at the early stages, which increases considerably the
five-year survival rate of the early-stage liver cancer patients
(7). Unfortunately, a considerable
number of liver cancer patients are diagnosed at the advanced stage
of the disease, where the five-year survival rate remains dismal
(8,9). Understanding the etiology of liver
cancer may be helpful in developing novel effective therapeutic
targets for liver cancer patients.
As one of the most important members of non-coding
RNAs, circular RNAs (circRNAs) have a unique circular structure
that is completely different from other linear RNAs (10). A previous study has revealed that
circRNAs are widely present in eukaryotic cells and play a key role
during cell proliferation, apoptosis and differentiation (11). circRNAs exist mainly in the
cytoplasm of eukaryotic cells and they act as sponges to specific
microRNAs (miRNAs) in order to modulate the expression of target
genes (12). Aberrant expression of
circRNAs can be usually observed in pathological conditions, such
as human cancers and neurodegenerative and autoimmune disorders
(13–15). These aberrantly expressed circRNAs
may be used as promising markers in disease screening. Manipulation
of their expression is considered a popular research strategy in
disease studies. In recent years, an increasing number of circRNAs
have been identified that are dysregulated in liver cancer and play
an important role during tumor growth as determined by in
vitro and in vivo models, revealing the complex essence
of liver carcinogenesis (16,17).
The present study aimed to investigate the
biological functions and potential mechanisms of hsa_circ_0008537
(circ_0008537) in liver tumorigenesis. The data aimed to provide a
novel therapeutic axis for liver cancer treatment.
Materials and methods
Liver cancer specimens and cell
lines
A total of 70 liver cancer tissues and matched
non-tumor samples were obtained from patients (aged 42–78 years; 39
male and 31 female patients) who were diagnosed with liver cancer
at the Affiliated Neijiang Second People's Hospital of Southwest
Medical University (Neijiang, China) from March 2009 to December
2019. The present study was approved (approval no.
NSH-2019-005-020) by the Ethics Committee of the Affiliated
Neijiang Second People's Hospital of the Southwest Medical
University. The subjects provided written informed consent for
their participation in the study protocol. The tissues were
immersed into liquid nitrogen immediately following resection from
the liver cancer patients and were subsequently transferred to
−80°C until further use. Four liver cancer cell lines (Huh-7,
SNU449, SK-Hep-1 and HepG2), one normal immortalized liver cell
line (THLE-3) and 293 cells were obtained from the American Type
Culture Collection (ATCC) and maintained in 10% fetal bovine serum
(FBS; product no. SH30406.05; HyClone; Cytiva) with Dulbecco's
modified Eagle's medium (DMEM; product no. D5796; Sigma-Aldrich;
Merck KGaA) at 37°C, 95% O2 and 5% CO2.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from liver cancer tissues
and cells following lysis using the TRIzol reagent (Invitrogen;
Thermo Fisher Scientific, Inc.). Its quality was verified using
spectrophotometric methods. Subsequently, 2 µg RNA samples were
subjected for reverse transcription with the Bestar™ qPCR RT kit
(DBI Biosciences). To examine the resistance of circ_0008537 to
RNase R, RT-qPCR was conducted on RNA with or without RNase R
treatment. RT-qPCR was performed using specific divergent primers
and the Power SYBR green master mix (Thermo Fisher Scientific,
Inc.) on a 7500 Fast Real-Time PCR system (Applied Biosystems;
Thermo Fisher Scientific, Inc.). The conditions of PRC
amplification were enzyme activation at 95°C for 15 min, followed
by 40 cycles of denaturation at 95°C for 15 sec, annealing at 58°C
for 30 sec, and extension at 72°C for 30 sec. The gene expression
was quantitated using the 2−ΔΔCq method from 3
independent repetitions (18). The
primers used in the present study are listed in Table I.
 | Table I.The sequences of primers in the
RT-qPCR assay. |
Table I.
The sequences of primers in the
RT-qPCR assay.
| ID | Sequence
(5′-3′) |
|---|
| GAPDH | F:
TGTTCGTCATGGGTGTGAAC |
|
| R:
ATGGCATGGACTGTGGTCAT |
| circ_0008537 | F:
TTACATGGAGCCCCACTACC |
|
| R:
TCGTCACTCGATGTTGAAGG |
| miR-153-3p | F:
GGCTCAAGTGTGATTCATACT |
|
| R:
GAGTCAATACTCTTAAGGCATC |
| U6 | F:
CTTCGGCAGCACATATAC |
|
| R:
GAACGCTTCACGAATTTGC |
Sanger sequencing
As described in a previous study (19), the extracted total RNAs were used
for Sanger sequencing, which was conducted by BGI Genomics Co.,
Ltd.
Oligonucleotide transfection
Two circ_0008537 siRNAs (si-circ-1 and si-circ-2),
si-control (si-ctrl), circ_000853-overexpressed plasmids, empty
vector (pcDNA3.1, EV), miR-153-3p mimic and its scramble control
were all purchased from Integrated Biotech Solutions. Lipofectamine
3000 (Invitrogen; Thermo Fisher Scientific, Inc.), which were
applied to complete the transfection of oligonucleotides. The
sequences of the oligonucleotides included miR-153-3p mimic,
5′-UUGCAUAGUCACAAAAGUGAUC-3′, scrambled miRNA scramble control,
5′-CAGUACUUUUGUGUAGUACAA-3′; si-circ-1 target sequences,
5′-AGCCCCACUACCAGGAUGAAA-3′; si-circ-2 target sequences,
5′-GAGCCCCACUACCAGGAUGAA-3′; si-ctrl, 5′-UCUGAGAGGAUUCUAGGU-3′.
HepG2 or SK-Hep-1 cells were transfected with si-circ-1 (50 nM),
si-circ-2 (50 nM), si-ctrl (50 nM), circ_0008537-overexpressed
plasmids (50 ng), EV (pcDNA3.1, 50 ng), miR-153-3p mimics (100 nM)
or scramble control (100 nM) at 37°C for 48 h using Lipofectamine
3000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.) based on
the experimental instructions. After transfection for 48 h, the
cells were applied fort the subsequent experiments.
Colony formation assay
The treated liver cancer cells (2,000 cells/well)
were seeded in 6-well plates containing 2 ml DMEM and subsequently
maintained in a cell incubator with 95% O2 and 5%
CO2 for two weeks. Subsequently, the colonies were fixed
in 75% ethanol at room temperature for 15 min and stained with
Giemsa solution at room temperature for 20 min. The visible
colonies (>10 cells) were counted manually and visualized by a
light microscope using a magnification of ×20.
EdU incorporation assay
The treated liver cancer cells (1×104
cells/well) were seeded into sterile 24-well plates and the EdU kit
(Invitrogen; Thermo Fisher Scientific, Inc.) was used to examine
cell proliferation following the instructions of the manufacturer.
Cell imaging was conducted using a laser confocal microscope
(Olympus Corporation) with a magnification of ×20. DAPI (1 mg/ml)
was utilized to stain the cell nucleus at room temperature for 10
min.
Transwell assay
The Transwell chambers (8 µm; Corning Inc.) were
coated with or without Matrigel and were used to examine the
abilities of invasion or migration, individually. The treated HepG2
and SK-Hep-1 cells (1×104 cells/well) were seeded into
the upper chamber containing serum-free DMEM, and DMEM with FBS was
placed into the lower chamber. Following 24 h of cell incubation,
the cells invading or migrating the membrane were stained with 1%
crystal violet at room temperature for 30 min after fixation in 4%
paraformaldehyde at room temperature for 30 min. Six random fields
were selected for cell number counting using a light microscope at
a magnification of ×20.
Wound-healing assay
A total of 2 ml DMEM containing treated liver cancer
cells (1×105 cells) were added into 35-mm petri dishes
and cultured at 37°C until 100% confluence. Subsequently, a
straight scratch was made by a sterile pipette tip (100 µl) on the
surface of the cell layer. The cells were allowed to grow at 37°C
for an additional 24 h. The images were obtained using an Olympus
digital camera at the 0- and 24-h time-points following cell
scratching.
Bioinformatics
GLI3 was identified as the host gene of circ_0008537
through University of California Santa Cruz (UCSC) (http://genome.ucsc.edu/). The potential target miRNAs
of circ_0008537 were predicted via starBase v2.0 (http://starbase.sysu.edu.cn/) and circular RNA
Interactome (https://circinteractome.nia.nih.gov/).
Plasmid construction and dual
luciferase activity assay
The wild-type (WT) or mutant (Mut) miR-153-3p
binding sequence of circ_0008537 was inserted into the psi-CHECK-2
plasmid to establish the recombinant luciferase reporter plasmids
(Luc-circ WT and Luc-circ Mut). 293 cells (ATCC) were
co-transfected with Luc-circ WT or Luc-circ Mut and miR-153-3p or
its scramble control with Lipofectamine 3000. Following 48 h of
incubation after co-transfection, the firefly and Renilla
luciferase activities of 293 cells were examined using the
Dual-Luciferase Assay System (Promega Corporation) following the
supplier's protocol. The relative luciferase activity was counted
by normalizing the firefly luciferase activity against the
Renilla luciferase activity.
RNA immunoprecipitation (RIP)
In line with the manufacturer's instructions, RIP
was conducted using the EZMagna RIP kit (EMD Millipore). Briefly,
liver cancer cells were lysed in RIPA lysis buffer (cat no. 89900;
Thermo Fisher Scientific, Inc.) containing RNase repressor and the
cell extracts were incubated with magnetic beads conjugated with
Ago antibody (product code ab32381; Abcam) or IgG (product code
ab207995; Abcam) overnight at 4°C, after being stirred for 1 h.
Following washing with RIPA buffer three times, the RNA of the
RNA/antibody complex was extracted using Trizol and its expression
was analyzed using RT-qPCR.
Western blot analysis
Treated liver cancer cells were lysed in RIPA buffer
(cat no. 89900; Thermo Fisher Scientific, Inc.) for total protein
extraction. The protein concentration was determined using a BCA
kit (Beyotime Institute of Biotechnology) and the protein samples
(40 µg) in each lane were separated with 10% SDS-PAGE. The target
proteins were transferred to a PVDF membrane (EMD Millipore), which
was incubated in 5% non-fat milk at room temperature for 90 min to
block non-specific binding sites. Subsequently, the membranes were
incubated at 4°C overnight with primary antibodies against
pro-survival protein myeloid cell leukemia 1 (MCL1; rat product
code ab243136; 1:1,000), Snail1 (rabbit; 1:1,000; product code
ab216347) and actin (rabbit; product code ab179467; 1:5,000; all
from Abcam). The membranes were probed with HRP-conjugated donkey
anti-rabbit secondary antibodies (1:2,000; product code ab6802;
Abcam) at room temperature for 1 h. The signals were visualized
using an ECL system (Thermo Fisher Scientific, Inc.).
Kyoto encyclopedia of genes and
genomes (KEGG) analysis and ceRNA network
The biological pathways of miR-153-3p were analyzed
using KEGG (http://www.genome.jp/kegg/) (20). The circ_0008537-miR-153-3p-mRNA
regulatory network was established using the Cytoscape software
(Version 3.4.0, http://www.cytoscape.org/).
Statistical analysis
The data are presented as the mean ± SEM. The data
were statistically analyzed using SPSS 21.0 (IBM Corp.) and the
statistical graphs were drawn using GraphPad Prism (version 8;
GraphPad Prism Software, Inc.). Paired t-tests were employed to
analyze the statistical significance between tumor and non-tumor
samples, and unpaired t-tests were used for the comparison of the
other two groups. For non-parametric, data in which the medians
were compared, Wilcoxon rank-sum test was used for paired samples
and Mann-Whitney U test for unpaired samples. One-way ANOVA with
post hoc Tukey's test was applied for the multiple comparisons. The
correlations were analyzed using Pearson correlation analysis.
Kaplan-Meier curves and the log-rank test were applied for the
differences in survival. P<0.05 was considered to indicate a
statistically significant difference.
Results
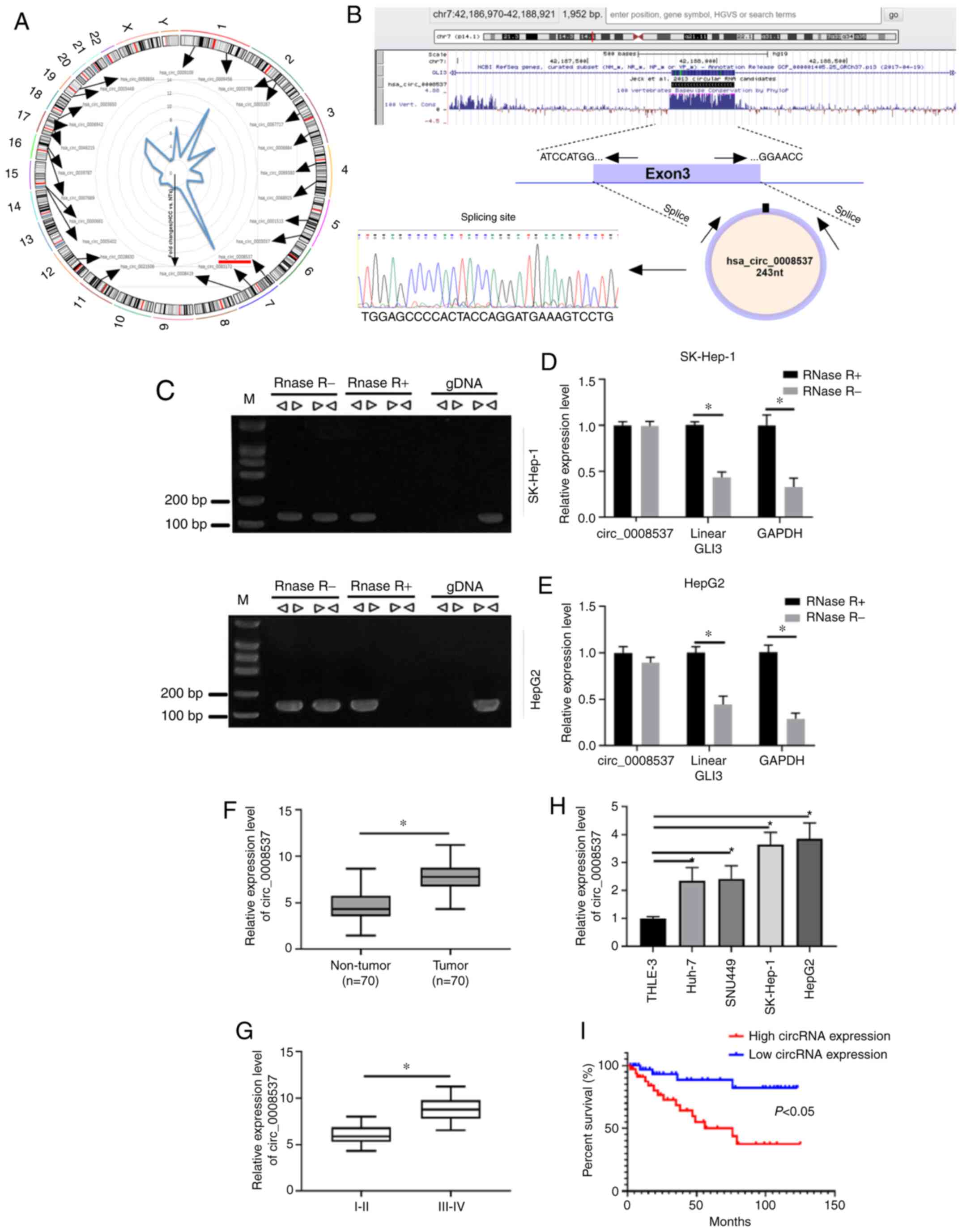
circ_00085377 is highly expressed in
liver cancer
A previous study has demonstrated using microarray
analysis that clusters of circRNAs are aberrantly expressed in
liver cancer compared with the corresponding expression in adjacent
non-tumorous tissues (21). Among
these dysregulated circRNAs, the expression levels of 24 circRNAs
were upregulated in liver cancer samples compared to the
corresponding levels noted in normal samples. These circRNAs were
widely distributed in chromosomes 1–7 and 11–19 among which
circ_0008537 exhibited the highest-fold changes with regard to
differential expression (Fig. 1A).
By searching UCSC, GLI3 was identified as the host gene of
circ_0008537, whereas the splicing site of circ_0008537 was
determined by Sanger sequencing (Fig.
1B). Convergent and divergent primers were adopted to amplify
circ_0008537 based on cDNA or gDNA in the presence of RNase R. The
results indicated that circ_0008537 could be amplified by divergent
primers based on cDNA but not on gDNA (Fig. 1C). Moreover, RNase R exposure
degraded linear GLI3, while no effects were observed on the
expression of circ_0008537 in SK-Hep-1 and HepG2 cells (Fig. 1D and E). RT-qPCR analysis indicated
higher expression levels of circ_0008537 in liver cancer tissue
samples than those observed in matched non-tumor samples
(P<0.001; Fig. 1F). Moreover,
the data indicated that circ_0008537 expression was increased in
stage III–IV compared with that observed in stage I–II tumors
(P<0.001; Fig. 1G). In addition,
four liver cancer cell lines (Huh-7, SNU449, SK-Hep-1 and HepG2)
and one normal immortalized liver cell line (THLE-3) were used for
circ_0008537 level detection. The results demonstrated that
circ_0008537 expression was significantly upregulated in Huh-7,
SNU449, SK-Hep-1 and HepG2 cell lines compared with the expression
observed in the THLE-3 cell line (P<0.01 and P<0.001;
Fig. 1H). The analysis of the
overall survival demonstrated that liver cancer patients with high
expression of circ_0008537 exhibited a worse survival rate than
those with low expression of circ_0008537 (P<0.05; Fig. 1I). These findings indicated that
circ_0008537 expression was increased in liver cancer and that it
was associated with a worse prognosis.
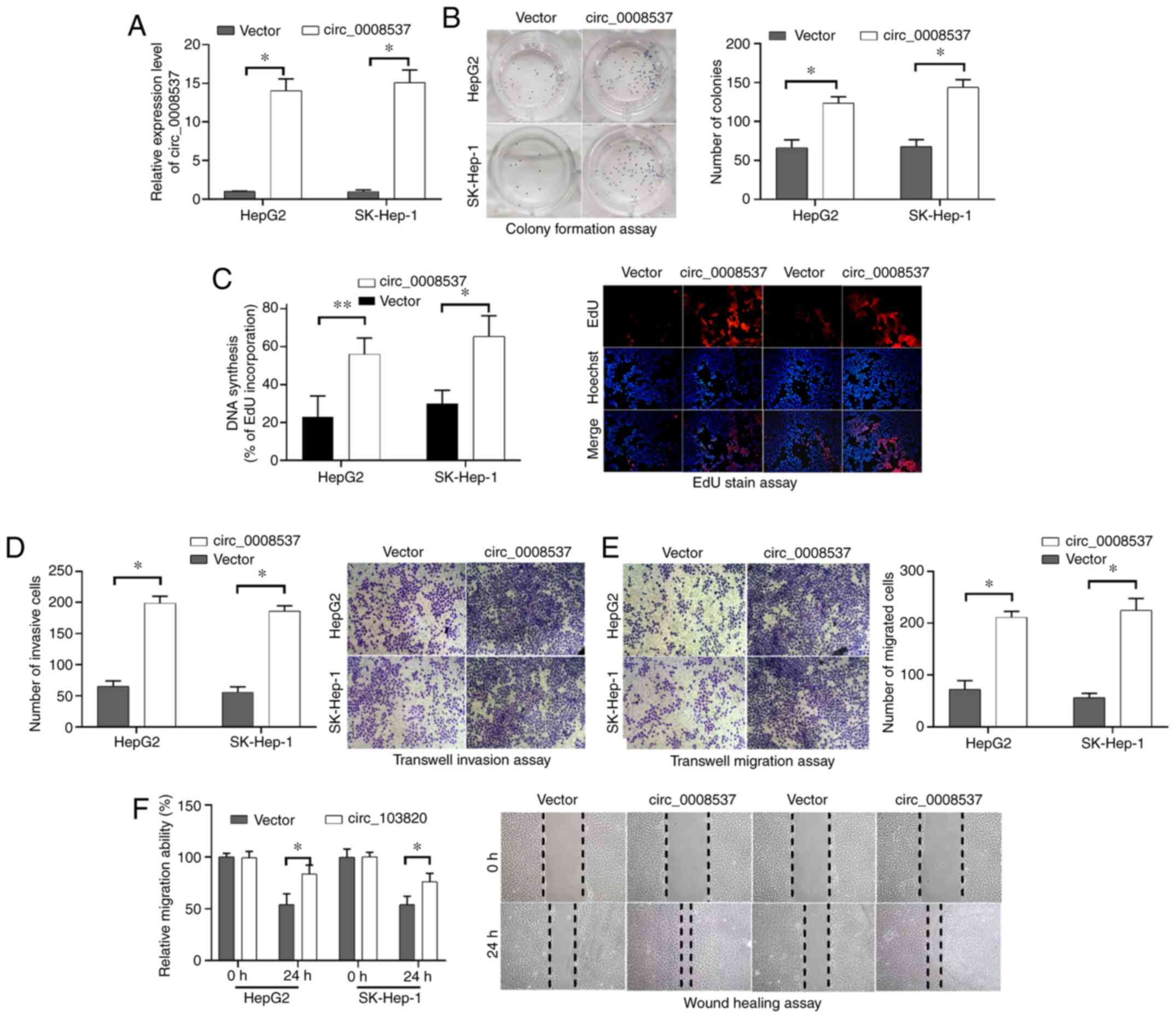
circ_0008537 overexpression
facilitates liver cancer cell proliferation invasion and
migration
To investigate the function of circ_0008537 in liver
cancer, the effects of circ_0008537 overexpression on liver cancer
cell proliferation, invasion and migration were investigated in
vitro. Following transfection of circ_0008537, its expression
levels were significantly increased in HepG2 and SK-Hep-1 cells
compared to those observed in cells transfected with empty vector
(P<0.001; Fig. 2A). The colony
formation assay demonstrated that circ_0008537 overexpression
significantly increased the number of HepG2 and SK-Hep-1 cell
colonies (P<0.01; Fig. 2B),
indicating that liver cancer cell proliferation was induced by
circ_0008537 overexpression. The results derived from EdU staining
further confirmed this conclusion (P<0.01; Fig. 2C). By using Transwell chambers
coated with or without Matrigel, the experiments demonstrated that
circ_0008537 overexpression facilitated the invasive and migratory
abilities of HepG2 and SK-Hep-1 cells (P<0.001; Fig. 2D and E). The effects of circ_0008537
overexpression on liver cancer cell migratory ability were further
supported by the wound-healing assay in HepG2 and SK-Hep-1 cells
(P<0.05; Fig. 2F). The results
indicated that circ_0008537 overexpression facilitated liver cancer
cell proliferation, invasion and migration in vitro.
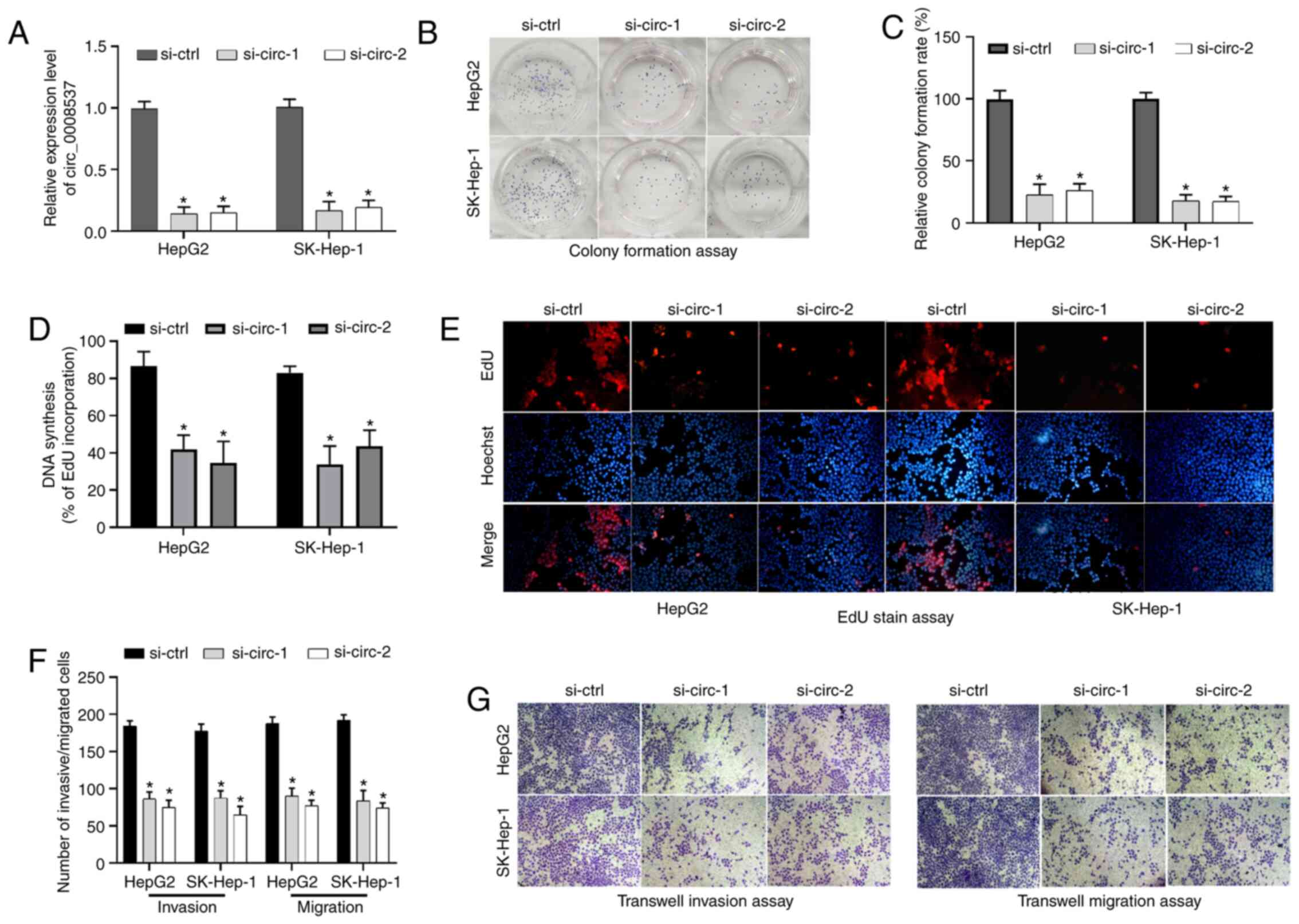
circ_0008537 silencing suppresses
liver cancer cell proliferation, invasion and migration
circ_0008537 silencing was carried out in HepG2 and
SK-Hep-1 cells to further study its functions in liver cancer cell
proliferation, invasion and migration. circ_0008537 expression was
significantly decreased in HepG2 and SK-Hep-1 cells following
transfection of siRNAs against circ_0008537 (si-circ-1 and
si-circ-2) compared with that observed in the si-control group
(P<0.05; Fig. 3A). By using the
colony formation assay and EdU staining, the analysis demonstrated
that circ_0008537 silencing resulted in a significant inhibition of
cell proliferative activity in HepG2 and SK-Hep-1 cells (P<0.01
and P<0.01; Fig. 3B-E). In
addition, circ_0008537 silencing significantly attenuated the
invasive and migratory activities of HepG2 and SK-Hep-1 cells
(P<0.001; Fig. 3F and G).
Therefore, circ_0008537 silencing repressed liver cancer cell
proliferation, invasion and migration in vitro.
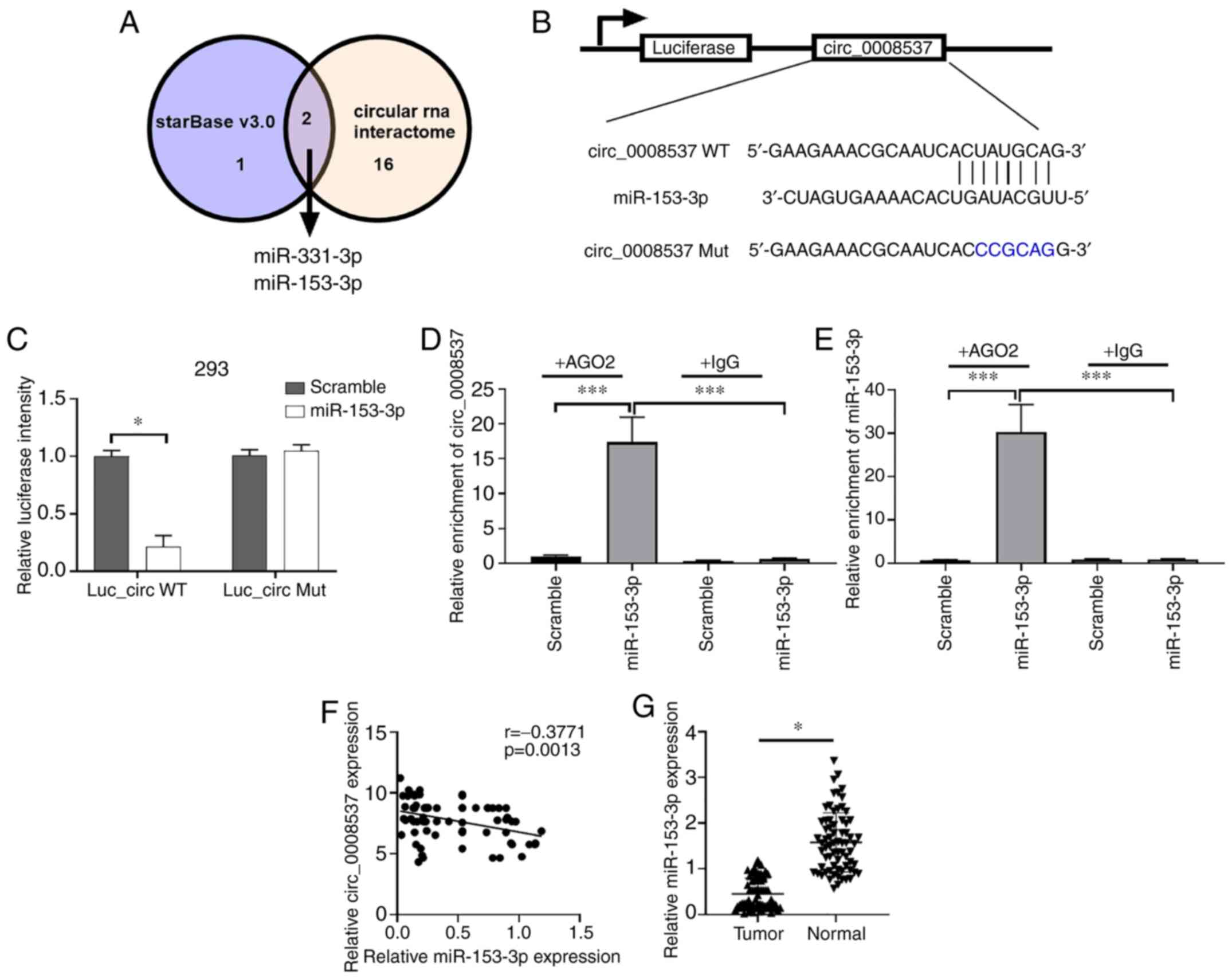
circ_0008537 serves as a sponge of
miR-153-3p
To explore the mechanism of circ_0008537 in liver
carcinogenesis, starBase and Circular RNA Interactome were used to
screen the potential target miRNAs of circ_0008537. Only miR-153-3p
and miR-331-3p were predicted by both the starBase and the Circular
RNA Interactome (Fig. 4A).
Dual-luciferase assay was adopted to validate the interplay between
miR-151-3p and circ_0008537. Transfection of the cells with
miR-153-3p caused a significant reduction of luciferase intensity
of 293 cells driven by wild-type circ_0008537 (Luc_Circ WT), while
these effects were not observed in 293 cells when the binding
sequence was mutated (P<0.001; Fig.
4B and C). Since Ago2 is an essential protein in the formation
of RNA-induced silencing complex (RISC) (22), an RNA-binding protein
immunoprecipitation assay was performed using Ago2 to further
assess whether miR-153-3p and circ_0008537 could form a RISC.
circ_0008537 was enriched by miR-153-3p in the presence of Ago2
(P<0.001; Fig. 4D and E). A
negative correlation was observed in the expression levels of
miR-153-3p and circ_0008537 in liver cancer (P<0.001; Fig. 4F). In addition, the relative
expression of miR-153-3p in liver cancer tissue samples was
revealed to be decreased compared with matched normal samples
(P<0.001; Fig. 4G). Overall, the
data indicated that circ_0008537 served as a sponge of miR-153-3p
and revealed a negative correlation with the expression of the
latter in liver cancer.
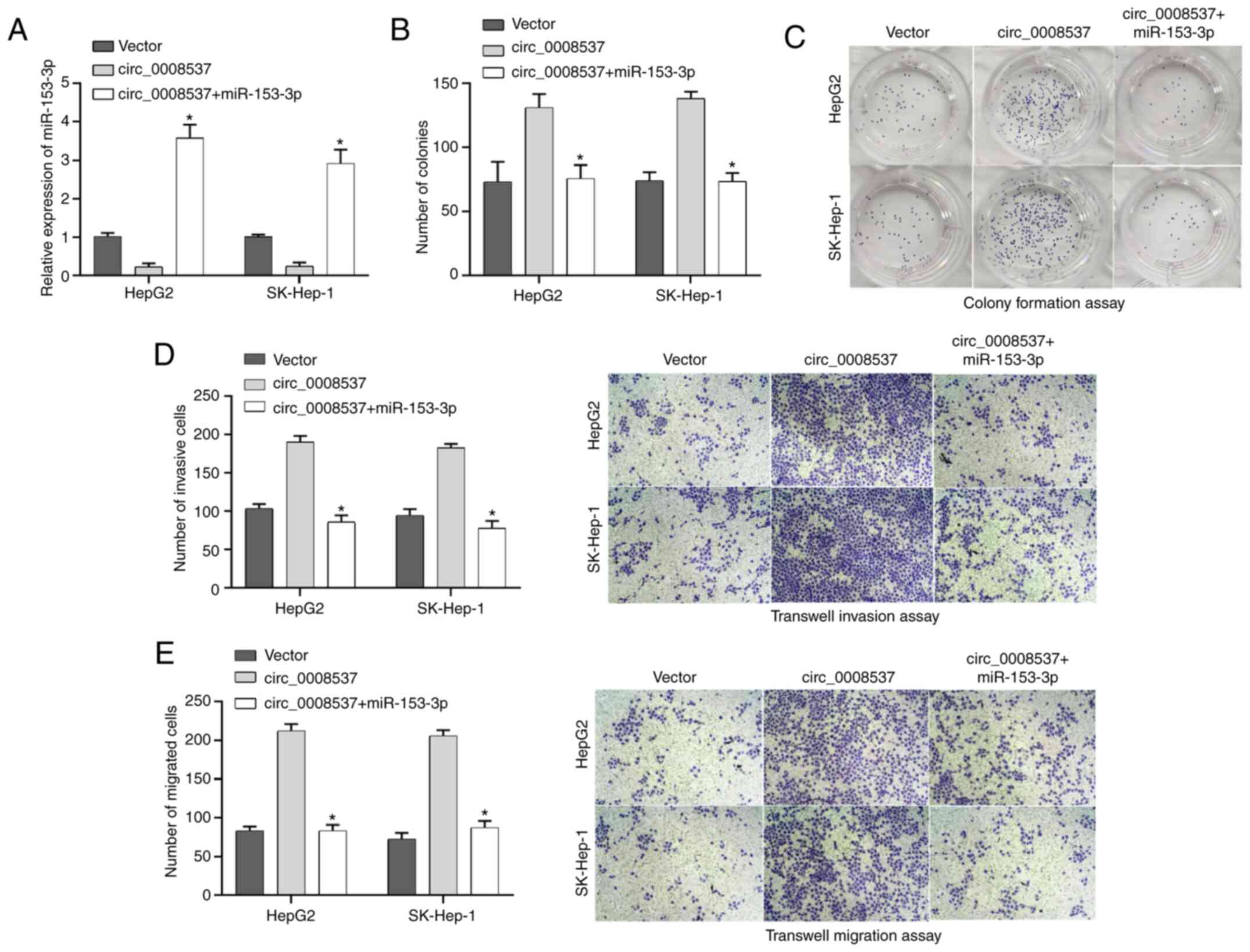
miR-153-3p reverses the facilitated
effects of circ_000857 on liver cancer cell proliferation, invasion
and migration
Subsequently, a rescue assay was performed to
investigate whether miR-153-3p was involved in liver cancer
progression mediated by circ_0008537. RT-qPCR analysis indicated
that miR-153-3p transfection reversed the downregulation of
miR-153-3p expression induced by circ_0008537 overexpression in
both HepG2 and SK-Hep-1 cells (P<0.05; Fig. 5A). The colony formation assay
demonstrated that the circ_0008537 overexpression-induced effects
on cell proliferation were abolished by miR-153-3p transfection
(P<0.01 and P<0.001; Fig. 5B and
C). The enhanced migratory and invasive abilities in the
circ_0008537-overexpressed HepG2 and SK-Hep-1 cells were abrogated
by miR-153-3p (P<0.05; Fig. 5D and
E). Overall, miR-153-3p reversed the facilitated effects of
circ_000857 on liver cancer cell proliferation, invasion and
migration.
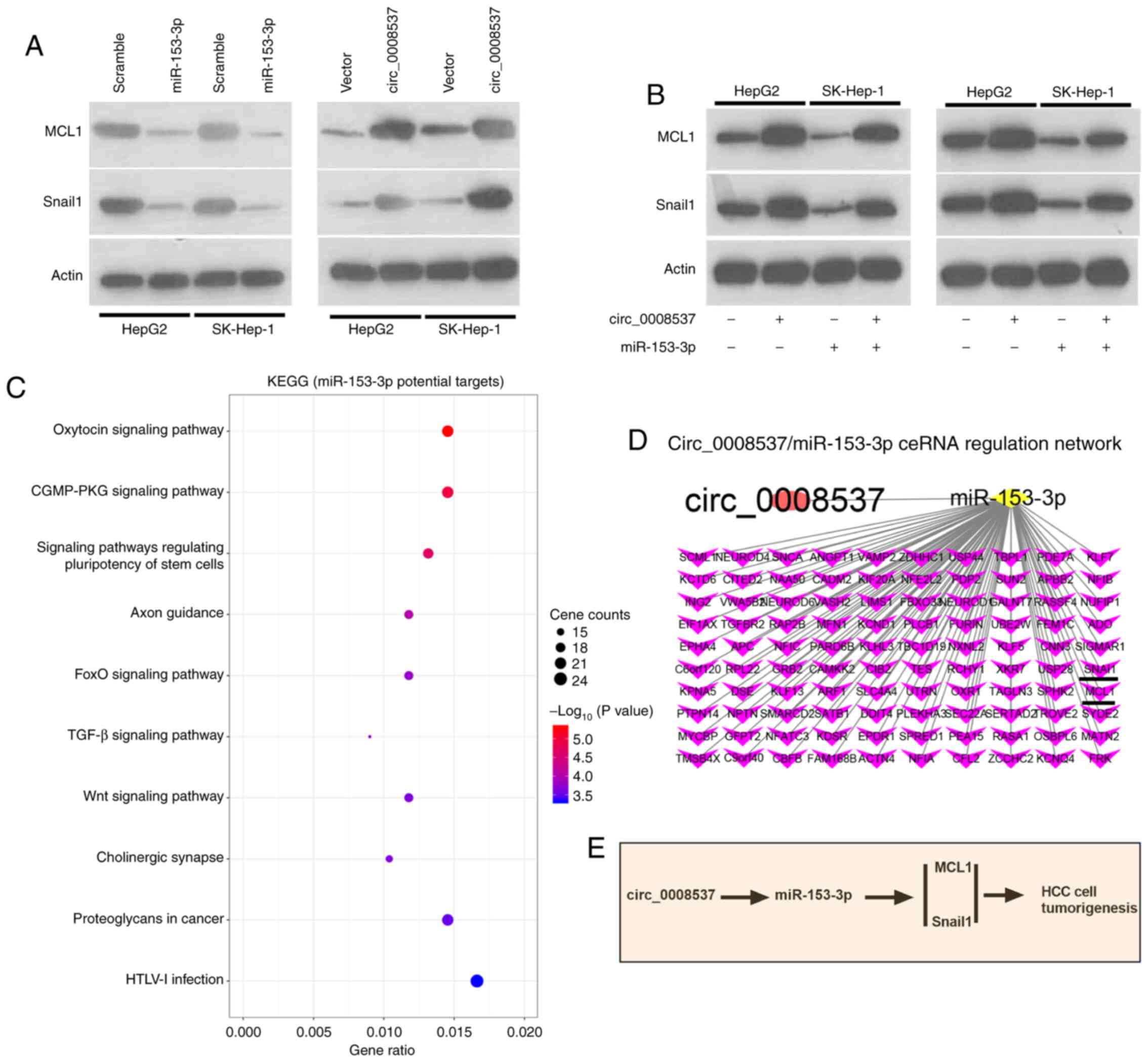
circ_0008537 indirectly regulates MCL1
and Snail1 through miR-153-3p
The bioinformatic analysis predicted multiple target
genes of miR-153-3p, including MCL1 and Snail1. In order to
investigate whether MCL1 and Snail1 were regulated by miR-153-3p,
their expression levels were investigated by western blotting
following overexpression of miR-153-3p. The results indicated that
miR-153-3p overexpression significantly decreased the expression
levels of MCL1 and Snail1 in HepG2 and SK-Hep-1 cells, while
circ_0008537 overexpression exhibited the opposite effects
(Fig. 6A). In view of the interplay
between circ_0008537 and miR-153-3p in liver cancer, further
investigations examined whether the regulatory effects of
circ_0008537 on MCL1 and Snail1 could be abolished by miR-153-3p.
Co-transfection of circ_0008537 and miR-153-3p reversed the
downregulation effect on MCL1 and Snail1 induced by miR-153-3p
overexpression (Fig. 6B). KEGG
pathway analysis was subsequently performed on the identified
target genes of miR-153-3p. The top ten pathways of miR-153-3p are
presented in Fig. 6C. A ceRNA
network indicated the interplay of circ_0008537 with miR-153-3p and
its target genes (Fig. 6D).
Overall, the data demonstrated that circ_0008537 indirectly
regulated the expression levels of MCL1 and Snail1 through
miR-153-3p (Fig. 6E).
Discussion
In view of the high possibility of tumor metastasis,
liver cancer has long been considered an aggressive malignant
tumor, which reduces the life expectancy of the affected population
(23). The understanding of the
potential molecular mechanisms of liver cancer pathogenesis has
long been considered the most effective strategy of developing
drugs for liver cancer treatment. Nevertheless, only a limited
number of effective drugs have been developed due to the complexity
of liver cancer etiology (24).
circRNAs have quickly become one of the main research focuses in
the field of liver cancer since they were initially reported to be
involved in liver tumorigenesis. In the present study,
differentially expressed circRNAs were screened between liver
cancer and matched normal tissue samples. The investigations
focused on the role and mechanisms of circ_0008537 increased
expression levels in liver cancer tumor growth.
The Hedgehog (Hh) signaling pathway, an extremely
conserved pathway in evolution, has been reported to be involved in
multiple aspects of embryonic development and stem cell maintenance
(25). Recently, the Hh pathway was
revealed to be abnormally activated in various types of
malignancies, including non-small cell lung, esophageal and liver
cancers (26–28). As a key mediator of the Hh cascade,
Gli3 has also been revealed to act as a tumor promoter in human
tumors (29). Gli3 inhibition by
miR-378 was reported to attenuate activation of hepatic stellate
cells and liver fibrosis (30). In
the present study, circ_0008537 interacted with the GLI3 gene and
was highly expressed in liver cancer tissues and cells.
The role of circRNAs in liver tumorigenesis has been
well documented by accumulating evidence, indicating both oncogenic
and inhibitory tumor progression effects (16). circHIAT1 has been identified as an
oncogene in liver cancer by modulating the PTEN pathway via
sponging miR-3171 (31). In
contrast to circHIAT1, circRNA-ABCB10 was confirmed to be a liver
cancer repressor by sponging miR-340-5p and miR-452-5p to release
NRP1 and ABL2 (32). The adverse
effects of different circRNAs in the pathogenesis of liver cancer
indicate the complex interactions of circRNAs with pathways that
lead to the development of liver cancer. In the present study,
overexpression and silencing experiments of circ_0008537 were
performed in liver cancer cells and the data demonstrated that
circ_0008537 acted as an oncogene in liver cancer. To the best of
our knowledge, the present study is the first to report the
biological functions of circ_0008537 in liver tumorigenesis.
miRNAs are a class of noncoding single stranded RNA
molecules of approximately 22 nucleotides in length, which are
involved in a range of crucial biological processes, including
development, hematopoiesis, organogenesis, apoptosis, proliferation
and even cancer development (33,34).
The miRNA-mRNA axis has been demonstrated by numerous studies to be
one of the most common pathways through which circRNAs play their
roles in regulating tumor growth at the transcriptional and post
transcriptional levels (14,35).
Therefore, the potential target miRNAs of circ_0008537 were
predicted using the following two bioinformatic software: starBase
and circular RNA interactome. Two miRNAs (miR-153-3p and
miR-331-3p) were identified and miR-153-3p was selected for further
analysis. miR-153-3p has been reported to participate in tumor
progression of multiple human tumors, such as gastric cancer,
esophageal squamous cell carcinoma and oral squamous cell carcinoma
(36–38). Nevertheless, the role of miR-153-3p
in liver cancer has not been previously investigated. In the
present study, a negative correlation between the expression levels
of miR-153-3p and circ_0008537 was revealed. Overexpression of
miR-153-3p could reverse the tumor promoting effects of
circ_0008537 on liver cancer progression. Moreover, miR-153-3p was
revealed to abolish the downregulation of MCL1 and Snail1
expression induced by miR-153-3p.
In conclusion, the present study provided in
vitro evidence demonstrating that circ_0008537 facilitated
liver carcinogenesis by indirectly regulating MCL1 and Snail1
through miR-153-3p. However, in vivo studies are required to
further confirm this conclusion. In addition, we will explore the
more detailed mechanisms of the circ_0008537/miR-153-3p axis in HCC
progression in a future study.
Acknowledgements
Not applicable.
Funding
The present work was supported by The Sichuan
Provincial Department of Science and Technology of China (grant no.
15089), the Sichuan Province Department of Medical Association
Scientific Research Subject of China (grant no. S16043), the Youth
Innovation of Scientific Research Subject of Sichuan Province
Medical Association of China (grant no. 2016, Q16044), the
Scientific Research Subject of Science and Technology Bureau of
Sichuan Province Neijiang City of China (grant no. Neijiang
2017021), and the Planning Project of Sichuan Provincial Science
and Technology Department (grant no. 2018JY0419).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
GY and OJ designed the experiments. GY, OJ, XL, JL,
SH, YW, JZ and DL performed the experiments. GY, XL, JL, SH
collected the data. OJ and JZ analyzed the data. OJ provided the
resource supports. GY, XL and OJ drafted the manuscript. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved (approval no.
NSH-2019-005-020) by the Ethics Committee of the Affiliated
Neijiang Second People's Hospital of the Southwest Medical
University. The subjects provided written informed consent for
their participation in the study protocol.
Patient consent for publication
Not applicable.
Competing interests
The authors have no commercial or other associations
that may pose a conflict of interest.
References
|
1
|
Kudo M, Kitano M, Sakurai T and Nishida N:
General rules for the clinical and pathological study of primary
liver cancer, nationwide follow-up survey and clinical practice
guidelines: The outstanding achievements of the liver cancer study
group of Japan. Dig Dis. 33:765–770. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2020. CA Cancer J Clin. 70:7–30. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lafaro KJ, Demirjian AN and Pawlik TM:
Epidemiology of hepatocellular carcinoma. Surg Oncol Clin N Am.
24:1–17. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Moradpour D and Blum HE: Pathogenesis of
hepatocellular carcinoma. Eur J Gastroenterol Hepatol. 17:477–483.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
El-Serag HB: Hepatocellular carcinoma. N
Engl J Med. 365:1118–1127. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chonprasertsuk S and Vilaichone RK:
Epidemiology and treatment of hepatocellular carcinoma in Thailand.
Jpn J Clin Oncol. 47:294–297. 2017.PubMed/NCBI
|
|
7
|
Levrero M: Viral hepatitis and liver
cancer: The case of hepatitis C. Oncogene. 25:3834–3847. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Mazzoccoli G, Miele L, Oben J, Grieco A
and Vinciguerra M: Biology, epidemiology, clinical aspects of
hepatocellular carcinoma and the role of sorafenib. Curr Drug
Targets. 17:783–799. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Worns MA and Galle PR: Hepatocellular
carcinoma in 2017: Two large steps forward, one small step back.
Nat Rev Gastroenterol Hepatol. 15:74–76. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Patop IL and Kadener S: circRNAs in
cancer. Curr Opin Genet Dev. 48:121–127. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yu T, Wang Y, Fan Y, Fang N, Wang T, Xu T
and Shu Y: CircRNAs in cancer metabolism: A review. J Hematol
Oncol. 12:902019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Szabo L and Salzman J: Detecting circular
RNAs: Bioinformatic and experimental challenges. Nat Rev Genet.
17:679–692. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Haque S and Harries LW: Circular RNAs
(circRNAs) in health and disease. Genes (Basel). 8:3532017.
View Article : Google Scholar
|
|
14
|
Han B, Chao J and Yao H: Circular RNA and
its mechanisms in disease: From the bench to the clinic. Pharmacol
Ther. 187:31–44. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Akhter R: Circular RNA and Alzheimer's
disease. Adv Exp Med Biol. 1087:239–243. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Fu L, Jiang Z, Li T, Hu Y and Guo J:
Circular RNAs in hepatocellular carcinoma: Functions and
implications. Cancer Med. 7:3101–3109. 2018. View Article : Google Scholar
|
|
17
|
Hu J, Li P, Song Y, Ge YX, Meng XM, Huang
C, Li J and Xu T: Progress and prospects of circular RNAs in
hepatocellular carcinoma: Novel insights into their function. J
Cell Physiol. 233:4408–4422. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zheng J, Zhang H, Banerjee S, Li Y, Zhou
J, Yang Q, Tan X, Han P, Fu Q, Cui X, et al: A comprehensive
assessment of next-generation sequencing variants validation using
a secondary technology. Mol Genet Genomic Med. 7:e007482019.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang Z, Shang P, Li Q, Wang L, Chamba Y,
Zhang B, Zhang H and Wu C: iTRAQ-based proteomic analysis reveals
key proteins affecting muscle growth and lipid deposition in pigs.
Sci Rep. 7:467172017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cui S, Qian Z, Chen Y, Li L, Li P and Ding
H: Screening of up- and downregulation of circRNAs in HBV-related
hepatocellular carcinoma by microarray. Oncol Lett. 15:423–432.
2018.PubMed/NCBI
|
|
22
|
Chen Y, Yang F, Fang E, Xiao W, Mei H, Li
H, Li D, Song H, Wang J, Hong M, et al: Circular RNA circAGO2
drives cancer progression through facilitating HuR-repressed
functions of AGO2-miRNA complexes. Cell Death Differ. 26:1346–1364.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Blum HE: Molecular therapy and prevention
of hepatocellular carcinoma. Hepatobiliary Pancreat Dis Int.
2:11–22. 2003.PubMed/NCBI
|
|
24
|
Grandhi MS, Kim AK, Ronnekleiv-Kelly SM,
Kamel IR, Ghasebeh MA and Pawlik TM: Hepatocellular carcinoma: From
diagnosis to treatment. Surg Oncol. 25:74–85. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Skoda AM, Simovic D, Karin V, Kardum V,
Vranic S and Serman L: The role of the Hedgehog signaling pathway
in cancer: A comprehensive review. Bosn J Basic Med Sci. 18:8–20.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhuang H, Cao G, Kou C and Liu T:
CCL2/CCR2 axis induces hepatocellular carcinoma invasion and
epithelial-mesenchymal transition in vitro through
activation of the Hedgehog pathway. Oncol Rep. 39:21–30.
2018.PubMed/NCBI
|
|
27
|
Liu Y, Huber RM, Kiefl R, Tufman A and
Kauffmann-Guerrero D: Hedgehog pathway activation might mediate
pemetrexed resistance in NSCLC cells. Anticancer Res. 40:1451–1458.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yang C, Zheng X, Ye K, Sun Y, Lu Y, Fan Q
and Ge H: miR-135a inhibits the invasion and migration of
esophageal cancer stem cells through the Hedgehog signaling pathway
by targeting Smo. Mol Ther Nucleic Acids. 19:841–852. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ma Y, Li G, Hu J, Liu X and Shi B:
MicroRNA-494 regulates Gli3 expression and inhibits pancreatic
cancer cells growth and migration. J Cell Biochem. 119:5324–5331.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hyun J, Wang S, Kim J, Rao KM, Park SY,
Chung I, Ha CS, Kim SW, Yun YH and Jung Y: MicroRNA-378 limits
activation of hepatic stellate cells and liver fibrosis by
suppressing Gli3 expression. Nat Commun. 7:109932016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liang Z, Liu Z, Cheng C, Wang H, Deng X,
Liu J, Liu C, Li Y and Fang W: VPS33B interacts with NESG1 to
modulate EGFR/PI3K/AKT/c-Myc/P53/miR-133a-3p signaling and induce
5-fluorouracil sensitivity in nasopharyngeal carcinoma. Cell Death
Dis. 10:3052019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yang W, Ju HY and Tian XF: Circular
RNA-ABCB10 suppresses hepatocellular carcinoma progression through
upregulating NRP1/ABL2 via sponging miR-340-5p/miR-452-5p. Eur Rev
Med Pharmacol Sci. 24:2347–2357. 2020.PubMed/NCBI
|
|
33
|
Shang H, Sun L, Braun T, Si Q and Tong J:
Association between miR-124 rs531564 and miR-100 rs1834306
polymorphisms and cervical cancer: A meta-analysis. Eur J Gynaecol
Oncol. 40:925–931. 2019.
|
|
34
|
Liu B, Shyr Y, Cai J and Liu Q: Interplay
between miRNAs and host genes and their role in cancer. Brief Funct
Genomics. 18:255–266. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chen Y, Li C, Tan C and Liu X: Circular
RNAs: A new frontier in the study of human diseases. J Med Genet.
53:359–365. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhi XH, Jiang K, Ma YY and Zhou LQ:
OIP5-AS1 promotes the progression of gastric cancer cells via the
miR-153-3p/ZBTB2 axis. Eur Rev Med Pharmacol Sci. 24:2428–2441.
2020.PubMed/NCBI
|
|
37
|
Zuo J, Zhao M, Fan Z, Liu B, Wang Y, Li Y,
Lv P, Xing L, Zhang X and Shen H: MicroRNA-153-3p regulates cell
proliferation and cisplatin resistance via Nrf-2 in esophageal
squamous cell carcinoma. Thorac Cancer. 11:738–747. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Chang AC, Lien MY, Tsai MH, Hua CH and
Tang CH: WISP-1 promotes epithelial-mesenchymal transition in oral
squamous cell carcinoma cells via the miR-153-3p/Snail axis.
Cancers (Basel). 11:19032019. View Article : Google Scholar
|