As an essential part of the central dogma of
molecular biology, mRNA and other forms of RNA serve crucial roles
in biological systems by passing on genetic information. Although
research on chemical modifications of RNAs began in 1965 (1), there is limited knowledge regarding the
underlying regulatory mechanisms of RNA modifications in biological
processes. According to the MODOMICS database (https://iimcb.genesilico.pl/modomics),
172 different RNA chemical modifications, such as 5-methylcytosine,
1-methylguanosine, N6-methyladenosine (m6A) and N1-methyladenosine,
have been observed in all organisms at present. Among these
modifications, m6A methylation is considered the most abundant and
conserved internal transcriptional modification (2). Research on m6A methylation has been
limited in the past due to a lack of accurate detection methods;
however, with the development of high-throughput m6A sequencing
methods (3), the understanding of the
biological functions of m6A has advanced.
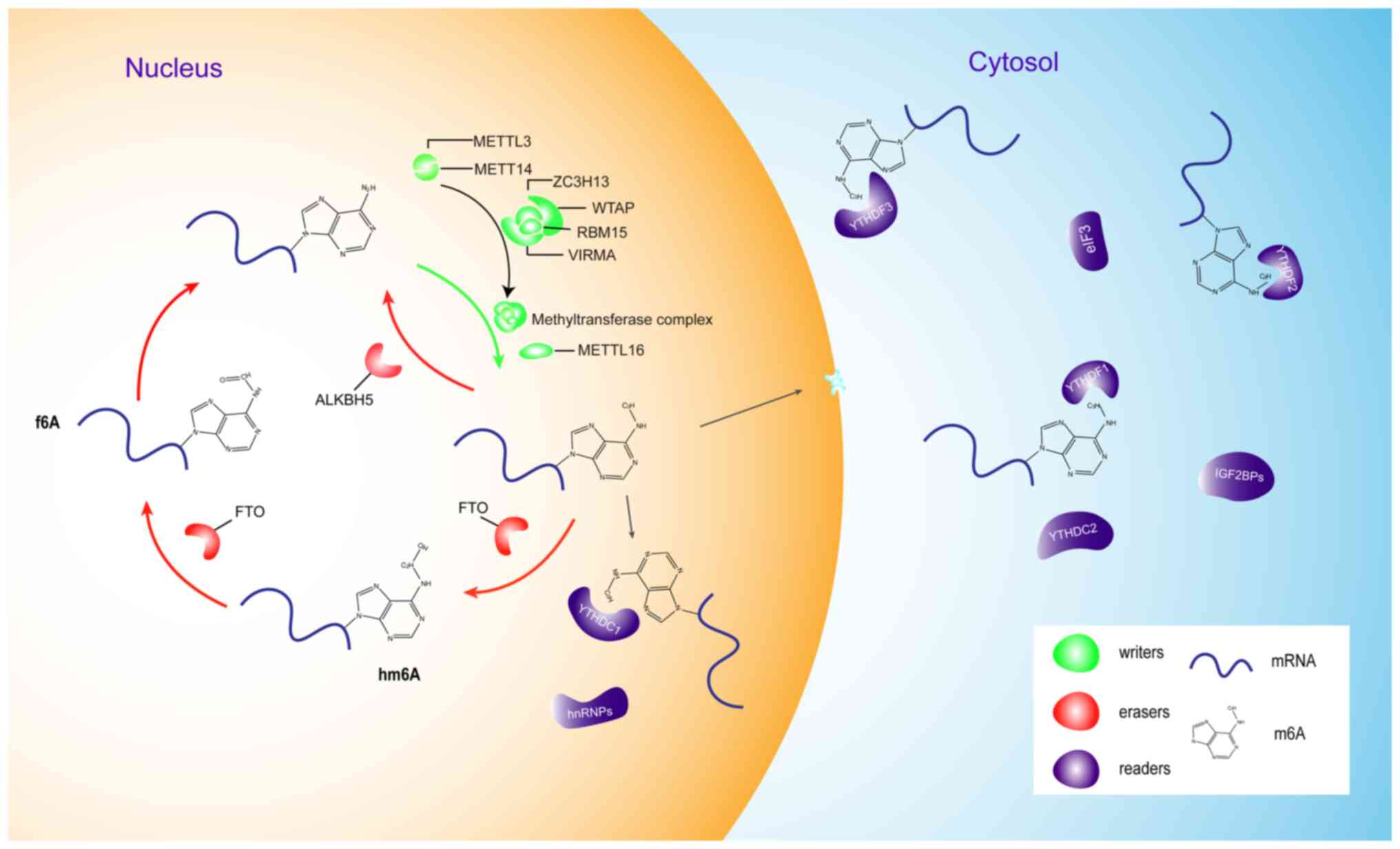
The process of m6A methylation is regulated by
several enzymes, including writers, erasers and readers (Fig. 1) (4,5). Writers
promote the formation of m6A (6–8), erasers
specifically remove the methylated group from mRNAs, and readers
recognize and bind m6A modifications to exert biological functions
(9,10). The observation of the demethylation
functions of fat mass and fat mass and obesity-associated protein
(FTO) (11), and alkB homologue
(ALKBH)5 (12) as an eraser,
demonstrated that m6A methylation is a dynamic and reversible
process. Malignant tumors are a group of abnormal cells with
distinctly different functions and gene expression compared with
normal cells. Research on the mechanisms of m6A in cancer has
recently advanced due to improvements in the understanding of the
roles of m6A in post-transcriptional modifications (4). The present review summarizes the
molecular functions and mechanisms of m6A and its three regulators
in human cancer, and discusses their roles in the regulation of
malignant tumor signaling pathways.
Since its discovery in the 1970s, m6A has been the
most prevalent modification in polyadenylated mRNAs (2). It has been estimated to be present in
three m6A residues per mRNA on average (13). Since it is ubiquitous in nature, m6A
can be found in yeast (14), fruit
flies (15), mammals (2,16) and
bacteria (17). Since m6A can undergo
reverse transcription to form thymine and cannot be detected by
chemical modifications, transcriptome-wide mapping of m6A remains
difficult (18). In 2012, a
high-throughput sequencing method based on antibodies was developed
by two independent groups to map m6A distribution in the entire RNA
sequence, which improved the detection efficiency of m6A (3,19).
It was originally hypothesized that the process of
m6A was static; however, in 2011, FTO (11) and ALKBH5 (12) were demonstrated to be able to function
as demethylases, indicating that the process of m6A is reversible.
Subsequently, various proteins, including Vir like m6A
methyltransferase associated (VIRMA) (8,16), insulin
like growth factor 2 (IGF2BP) (10)
and heterogeneous nuclear ribonucleoprotein (7), were demonstrated to function as writers
and readers.
It is well-known that writers and erasers regulate
m6A via methylation and demethylation, respectively (5,20,21). Furthermore, m6A groups exert
biological functions by being recognized by readers, which are a
type of specific binding protein (22,23).
In mammals, writers catalyze the methylation of m6A
in the form of a methyltransferase complex consisting of
methyltransferase-like 3 (METTL3), methyltransferase-like 14
(METTL14) (7) and Wilms' tumor
1-associating protein (WTAP) (7).
METTL14 has a greater effect on m6A than METTL3 although their
proportion in the complex is 1:1 (7).
Previous studies have identified more writers, including
methyltransferase-like protein 16 (METTL16) (24,25), zinc
finger CCCH domain-containing protein 13 (ZC3H13) (26), VIRMA (8,16) and
RNA-binding motif protein 15 (RBM15) (27). METTL16 is a methyltransferase which
binds to the conserved U6 small nuclear RNA, non-coding RNA and
precursor messenger RNA, and is involved in regulating
intracellular homeostasis and mRNA splicing in response to
intracellular S-adenosyl-L-methionine levels (24,25). VIRMA
(also referred to as KIAA1429) can promote m6A modification and
knockdown of VIRMA, resulting in a more conspicuous decrease of m6A
content than the effect of METTL3 and METTL14 knockdown in A549
cells (8). RBM15 catalyzes m6A
modification by binding to the U-rich region in long non-coding RNA
X inactive specific transcript (27).
In addition, ZC3H13 has been identified as a novel m6A writer in
mice and Drosophila (26). The
first eraser was identified in 2011 by Jia et al (11), who revealed that FTO could demethylate
m6A. ALKBH5 was identified as the second eraser (12) as it demethylates m6A in a different
way compared with FTO. The two intermediates,
N6-hydroxymethyladenosine and N6-formyladenosine are first oxidized
by FTO during the process of demethylation, while ALKBH5 catalyzes
the direct removal of m6A (12,28).
Readers can identify m6A modifications and bind to methylated RNA
to transfer biological signals to downstream signaling pathways
(21,22). Proteins containing the YT521-B
homology (YTH) domain, such as the YTH domain-containing family
(YTHDF) proteins, have been classed as readers (9). Notably, these recognition proteins of
m6A exhibit distinct mechanisms. For example, Wang et al
(29) reported the
translation-promoting role of YTHDF1 and the mRNA-destabilizing
role of YTHDF2. By interacting with initiation factors, including
IGF2BP1 and stress granule assembly factor 1, YTHDF1 enhances the
translation efficiency of target RNAs and ensures efficient protein
expression from these shared transcripts. By contrast, YTHDF2
accelerates the degradation of m6A-modified transcripts to control
the lifetime of the methylated transcripts (29,30).
YTHDF3 serves as a hub to regulate the RNA accessibility of YTHDF1
and YTHDF2 (31).
m6A is widely expressed in eukaryotes and serves a
crucial role in the regulation of various biological processes. In
mammals, m6A modifications affect development (12), metabolism (11,32–34) and
immunity (35–37). Furthermore, previous studies have
indicated that m6A has effects on stem cell differentiation
(38,39), human metabolic diseases (40), viral infections (41–44) and
inflammation (45).
Pluripotent mouse embryonic stem cells (mESCs)
undergo two different states during differentiation, naive and
primed (46). m6A modifications serve
key roles in the regulation of pluripotency during the transition
from the naive state to the primed state (38). METTL3 depletion has a different effect
on naïve and primed pluripotent stem cells. The depletion of METTL3
in naïve cells blocks differentiation and amplifies the highly
expressed naïve pluripotency genes, which boosts naïve circuitry
stability (38). When METTL3 and m6A
are inhibited in epiblast stem cells, which are in a primed state,
the expression levels of pluripotent genes are reduced, whereas the
expression levels of lineage commitment markers are increased
(38). By knocking out METTL3, Geula
et al (38) revealed m6A as a
timely maintainer of the balance between pluripotency and lineage
priming factors, thus ensuring the orderly differentiation of
mESCs. However, Batista et al (47) reported that the deletion of METTL3
maintains the self-renewal capacity of mESCs and mouse embryonic
fibroblasts. These contradictory results may be due to the cell
state. For example, different transcripts are expressed and
methylated in naïve and primed embryonic stem cells (ESCs)
(48). Therefore, METTL3 inactivation
regulates the expression levels of genes that affect cell fate and
identity, and this activity maintains pluripotency in naïve stem
cells but promotes differentiation in primed stem cells (38).
Other erasers and readers of m6A have also been
demonstrated to regulate the development and differentiation of
ESCs. Knocking out YTHDF2 enhances the proliferation of mouse and
human hematopoietic stem cells, highlighting its potential role in
transplantation-related applications (51). Notably, m6A modification has not only
been demonstrated to regulate differentiation in ESCs (38), but also in developmental cancer cells
(52). Lobo et al (52) revealed that abundance of m6A and
expression of its writer VIRMA/reader YTHDF3 are different among
testicular germ cell tumor (TGCT) subtypes, with higher levels in
seminomas. Higher VIRMA and YTHDF3 mRNA levels in seminomas
maintain a low differentiation level compared with teratoma, which
represents more differentiated TGCTs. However, Lobo et al
(52) observed a stronger m6A
immunostaining intensity in teratoma, suggesting that other writers
may be responsible for establishing m6A in teratoma and/or that m6A
modification may target other RNAs and even impart them a different
fate.
m6A is involved in metabolism and regulation of
metabolic genes. It has been demonstrated that the demethylase FTO
is involved in the metabolism of glucose and lipids in mammals
(33,40). As a classic target of fat metabolism,
FTO can induce mRNA expression of FOXO1, glucose-6-phosphatase
catalytic subunit and diacylglycerol O-acyltransferase 2, and is
closely associated with glucose metabolism in type 2 diabetes
(40). FTO has also been demonstrated
to regulate the expression levels of activating transcription
factor 4 to control glucose production in the liver (53). Wu et al (54) demonstrated that FTO modulates the
deposition of triglycerides and the accumulation of lipids by
regulating the m6A-YTHDF2 signaling pathway. At present, the
specific sites and complete mechanisms in glucose or fat production
are unknown, and thus, future studies are required to address
this.
Researchers have highlighted the roles of m6A in
anti-inflammatory immunity, antitumor immunity and adaptive
immunity (36,37). Yu et al (45) have demonstrated that YTHDF2 is
involved in the inflammatory response of macrophages. Knockdown of
YTHDF2 markedly increased the expression levels of IL-6, TNF-α and
IL-12, which were induced by lipopolysaccharide, and the
phosphorylation levels of p65, p38 and ERK1/2 in macrophages were
also upregulated. Furthermore, silencing of YTHDF2 could induce
upregulation of mitogen-activated protein kinase 4 and
mitogen-activated protein kinase 4 by stabilizing mRNA, activating
MAPK and NF-κB signaling pathways, and this aggravates the
inflammatory response in macrophages. Liu et al (55) reported that YTHDF2 recognized and
degraded, long non-coding RNA Dpf3 in dendritic cells specifically,
which markedly inhibited C-C motif chemokine receptor 7-mediated
dendritic cell migration and contributed to inflammatory responses.
Studies of the m6A-induced effect on antitumor immunity are
emerging and still in their infancy. Han et al (56) demonstrated that the antigen-specific
CD8+ T cell antitumor response was improved in
YTHDF1-deficient mice compared with mice in the wild-type group.
Blocking programmed death-ligand 1 could promote tumor regression
in YTHDF1-deficient mice (57). In
addition, the mechanisms by which m6A regulates adaptive immunity
is an emerging field of investigation (58). Li et al (58) first elucidated the function of m6A in
CD4+ T helper cells. The result suggested that deletion
of METTL3 in mouse T cells disrupted T cell homeostasis and
differentiation. The mRNAs of the suppressor of cytokine signaling
(SOCS) family, which are involved in STAT signaling, exhibit slower
mRNA decay and increased expression levels in Mettl3-deficient
naïve T cells (58). This increased
SOCS family activity consequently inhibits IL-7 mediated STAT5
activation and T cell homeostatic proliferation and differentiation
(58).
m6A modifications are involved in viral infections.
Human immunodeficiency virus type 1 (HIV-1) RNA is methylated by
m6A in infected cells, and readers, including YTHDF1-3, bind to
methylated HIV-1 RNA to inhibit viral reverse transcription and
translation (41,42). Partial knockout of m6A writers
decreases HIV-1 Gag synthesis and viral release, whereas knockout
of FTO has the opposite effect (42).
This indicates that m6A can enhance HIV-1 protein synthesis and
viral release, thereby contributing to the infection. Additionally,
the proteins regulated by m6A are known to modulate the life cycle
of hepatitis C virus (HCV) (43).
Depletion of METTL3 and METTL14 can increase the levels of HCV
infection by promoting infectious viral particle production without
affecting viral RNA replication (43,59). By
contrast, inhibition of the m6A demethylase FTO, but not ALKBH5,
has the opposite effect (26).
Furthermore, m6A has been demonstrated to serve important roles in
other Flaviviridae, such as Zika virus (44). Lichinchi et al (44) revealed that the depletion or
overexpression of the RNA methyltransferase could impact viral
replication, demonstrating that the host RNA methyltransferase
machinery acts as a key post-transcriptional regulator of Zika
virus. Furthermore, YTHDF proteins binding to Zika RNA indicates
another regulatory aspect of m6A readers, which serves a role in
viral RNA metabolism (44). Both RNA
modification layers may act as pro- or anti-viral factors in the
host (44).
Consistent with the regulation of m6A modifications
in normal biological processes, m6A is associated with a variety of
human cancer types. However, the catalysis of m6A in cancer is not
unitary. Numerous studies have demonstrated that m6A serves an
important role in various cancers, often via the actions of
regulators that influence m6A modifications and expression of
oncogenes or tumor suppressor genes. The special roles of m6A
regulators in human cancer types are summarized in Table I; however, the mechanisms by which m6A
regulators contribute to carcinogenesis remain to be elucidated.
The present review summarizes how the three types of m6A regulatory
proteins function in human cancer and discusses the role of m6A in
several classic signaling pathways.
Writers positively regulate m6A modifications. The
aberrant expression of writer proteins in tumors affects oncogenes
and tumor suppressors, thus influencing tumorigenesis (66), invasion (66) and metastasis (67). Interestingly, the mechanisms of
writers in different types of cancer are not uniform. METTL3 is
highly expressed in acute myeloid leukemia (AML) (68), and contributes to the translation of
oncogenes. In gastrointestinal cancer, METTL3 has been demonstrated
to be closely associated with the processes involved in the
progression of cancer, including tumor cell proliferation,
apoptosis, metastasis, angiogenesis and cancer stem cell
maintenance (69). A number of
studies have demonstrated that METTL3 generally acts as an oncogene
in gastrointestinal cancer types, such as gastric cancer (GC)
(70,71), colorectal cancer (CRC) (72), hepatocellular carcinoma (HCC)
(73) and pancreatic cancer (74,75).
Furthermore, the modified mRNA targets of METTL3 are diverse. For
example, METTL3-mediated m6A modification can increase the
expression levels of mRNA targets, including zinc finger MYM-type
containing 1 (ZMYM1) (70), SEC62
homolog, preprotein translocation factor (76) and MYC (71), in a way of enhancing mRNA stability in
GC, and promotes tumor cell proliferation, migration and invasion.
Similarly, other writers, including METTL16 (24,25),
ZC3H13 (26), VIRMA (8,16) and
RBM15 (27), have been reported to
have a complicated effect in other malignancies, such as
hepatocellular carcinoma (77),
colorectal cancer (78), prostate
cancer (79) and breast cancer
(80). It was hypothesized that
induction mechanisms other than m6A regulation cause this
phenomenon.
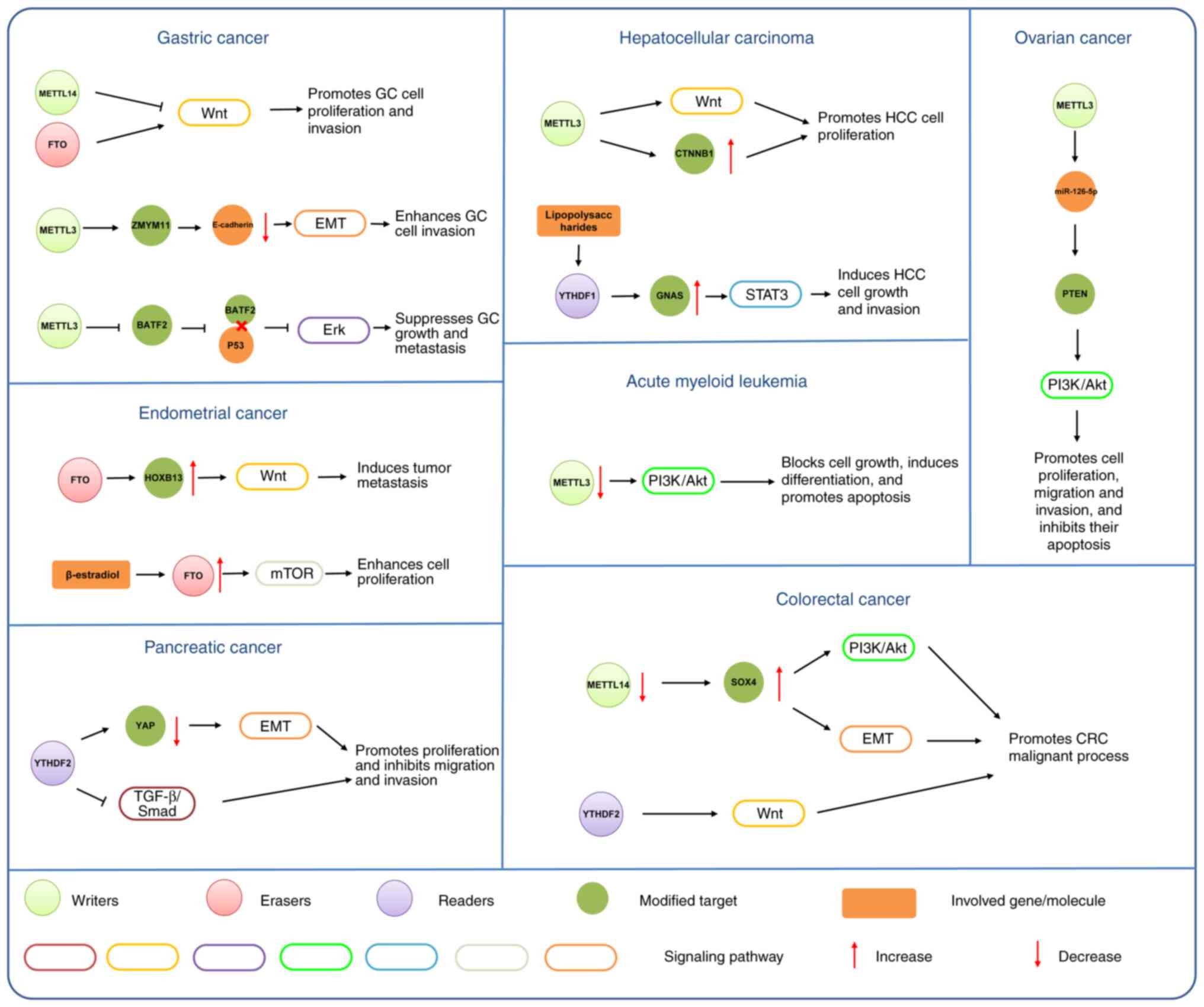
Increasing numbers of studies of erasers in cancer
are being performed. These studies have identified that the m6A
demethylase, FTO, serves a critical oncogenic role in AML (57,81,82).
Specifically, its high expression in AMLs with mixed lineage
leukemia rearrangements and fms related receptor tyrosine kinase
3-internal tandem duplication and/or nucleophosmin 1 mutations is
associated with increased tumorigenesis and invasion of AML cells
(81). Enhancing the expression
levels of FTO can reduce the levels of m6A and mRNA transcription
of ankyrin repeat and SOCS box containing 2 (ASB2) and retinoic
acid receptor α (RARA) (81). ASB2
and RARA are known to regulate the differentiation of leukemia
cells by inhibiting all-trans retinoic acid (81). In addition, FTO serves a crucial role
in cholangiocarcinoma (83) and
glioblastoma stem cells (84). In
contrast to FTO, an AML study based on The Cancer Genome Atlas
(TCGA) has suggested that ALKBH5, another m6A demethylase, exhibits
frequent copy number loss that results in non-carcinogenic effects
in AML (85). Furthermore, Zhang
et al (86) demonstrated that
ALKBH5 methylated FoxM1 to maintain proliferation and development
in glioblastoma stem-like cells.
The characterization of m6A readers has provided
valuable insight into to the mechanisms of m6A-mediated
post-transcriptional gene regulation in cancer. It has been
demonstrated that YTHDF1 is expressed at higher levels in CRC
tissues, and that it contributes to malignant phenotypes and poor
patient prognosis (87). A further
study has indicated that YTHDF1 is induced by the oncogene c-MYC,
and high YTHDF1 expression in malignant tumors can enhance the
resistance to anticancer drugs, including oxaliplatin and
fluorouracil (88). As another member
of the YTH domain-containing family, YTHDF2 recognizes m6A
modifications in the cytoplasm (31).
A previous study has identified that YTHDF2 could directly bind to
the 3′ end of the SOCS2 transcript, and that knockdown of YTHDF2
augmented SOCS2 expression in HCC cells (73). The SOCS family of proteins are
essential tumor suppressors in different cancer types, suggesting
an important role of YTHDF2 in human cancer (73). YTHDC2 is known to promote the mRNA
translation of hypoxia inducible factor α1 (HIF-1α) to induce the
metastasis of CRC (89). Knockdown of
YTHDC2 attenuates the protein expression of metastasis-related
genes, such as HIF-1α, and inhibits the metastasis in vitro
and in vivo (89). IGF2BP has
been demonstrated to be highly expressed in a variety of malignant
tumors, such as HCC, cervical cancer and AML (10,90). Huang
et al (90) reported that
IGF2BP has a positive effect on the stability and translation
levels of c-MYC, indicating the potential latent relationship
between IGF2BP and other readers.
As more research on m6A in cancer is being
conducted, several studies have examined whether m6A can regulate
cancer by affecting signaling pathways, and explored the specific
mechanisms of m6A. As a result, studies have demonstrated that m6A
can promote or inhibit malignant tumors by regulating different
signaling pathways (Figs. 2 and
3).
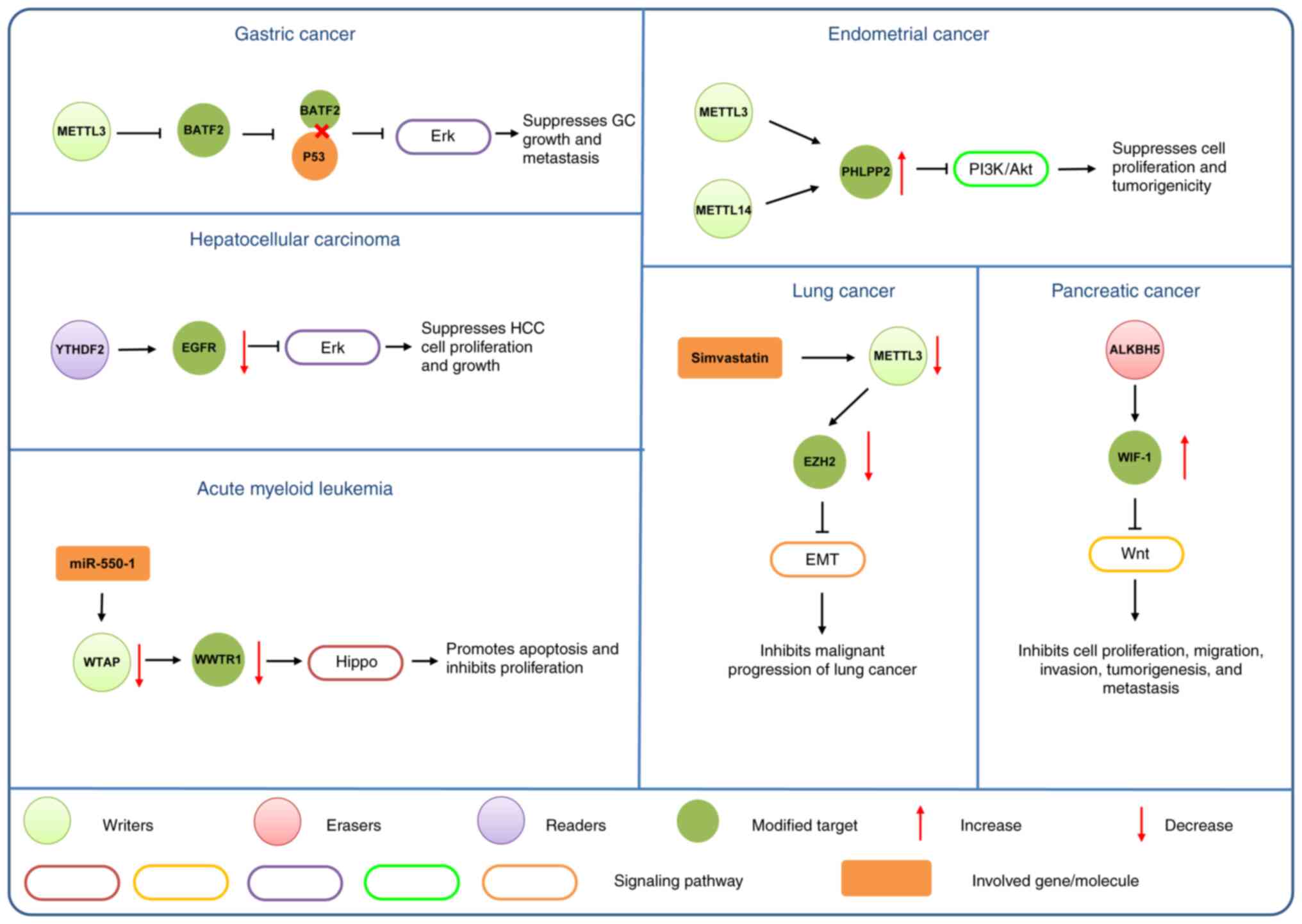
Wnt signaling is a pivotal regulatory signaling
pathway that has diverse roles in cancer progression. The m6A
modification targeting Wnt signaling has been a focus of cancer
research. According to a study conducted by Zhang et al
(91), the Wnt signaling pathway is
activated after the levels of m6A are reduced by inhibiting METTL14
in GC. By contrast, FTO knockout exhibits the opposite effect on
the Wnt signaling pathway (91). This
suggests that m6A can affect the activity of the Wnt signaling
pathway in GC. Similarly, E-cadherin is modulated by m6A; however,
more studies are required to improve the understanding of these
mechanisms (92). In endometrial
cancer, FTO promotes tumor metastasis and invasion (93). FTO catalyzes demethylation
modification in the 3′-untranslated region (3′-UTR) of HOXB13 mRNA,
thereby inhibiting m6A modification recognition by the YTHDF2
protein (93). This leads to
decreased HOXB13 mRNA decay and increased HOXB13 protein expression
and activation of the Wnt signaling pathway (93). Enhanced m6A modification is also
considered to be an oncogenic mechanism in hepatocellular
carcinoma; METTL3 expression is upregulated and Wnt/β-catenin
signaling pathway activity is induced via promotion of catenin β1
expression, which ultimately accelerates hepatocellular carcinoma
development (94). The Wnt signaling
pathway activates several cancer-related markers, including key
regulators of the cell cycle, proliferation, invasion, angiogenesis
and drug resistance (95). Therefore,
examining the effect of m6A on the Wnt signaling pathway will
provide guidance to explore the detailed mechanisms in cancer.
The EMT signaling pathway is a hot spot for cancer
research due to its role in the initial process of tissue
carcinogenesis. Furthermore, the markers of EMT are closely
associated with tumor progression processes, such as migration,
invasion, proliferation, anti-apoptosis, stemness and tumor
radio/chemosensitivity of cancer cells (96,97). Yue
et al (70) revealed the
METTL3-mediated m6A modification process in GC cells and identified
ZMYM1 as a target of METTL3. The elevated expression levels of
ZMYM1 repress the activation of E-cadherin promoter by recruiting
C-Terminal Binding Protein/Human lysine specific demethylase
l/CoREST complex, thus facilitating the EMT process. YTHDF2 is
highly expressed in various cancer types and is involved in dual
regulation (60,98). In pancreatic cancer, YTHDF2 knockdown
increases the expression levels of YAP, which is a key protein of
the TGF-β/Smad signaling pathway (98). A previous study has demonstrated that
there are two m6A sites in YY1 associated protein 1 (YAP), which
suggests that YTHDF2 directly binds to YAP mRNA to decrease the
stability of mRNA and regulate EMT via YAP signaling inhibition
(98). Progress has also been
achieved in the development of novel drug targets based on m6A
modifications. Chen et al (99) reported that simvastatin induced METTL3
downregulation in lung cancer tissues, which further influenced EMT
via m6A modification on EZH2 mRNA and inhibited the malignant
progression of lung cancer.
The PI3K/Akt signaling pathway is important for
cancer progression. Although aberrant activity of the PI3K/Akt
signaling pathway could be associated with tumorigenesis, it also
has a great impact on the proliferation, adhesion, invasion and
angiogenesis of malignant tumors (100). Increasing evidence suggests that m6A
modification is involved in carcinogenesis by targeting the
PI3K/Akt signaling pathway (101–105).
In renal cell carcinoma, METTL3 inhibits the PI3K/Akt/mTOR
signaling pathway and serves a role as a tumor suppressor gene
(101). Zhao et al (102) conducted an analysis for sequencing
data of gastrointestinal cancer from TCGA and Gene Expression
Omnibus, and demonstrated that m6A modification directly modulates
PI3K/Akt and mTOR signaling pathway activity by regulating critical
kinases in human gastrointestinal cancer. This conclusion was
supported and validated by a study by Chen et al (103). According to Chen et al
(103), knockdown of METTL14
markedly abolished SOX4 mRNA m6A modification and elevated SOX4
mRNA expression, whereas METTL14-mediated SOX4 mRNA degradation
stimulated PI3K/Akt signaling and inhibited CRC malignant process.
A study revealed that m6A modification can affect the activity of
the PI3K/Akt signaling pathway by regulating miRNA (104). Bi et al (104) demonstrated that METTL3 promoted
miR-126-5p maturation by modifying pri-miR-126-5p in ovarian
cancer. METTL3 knockdown inhibits the effect of miR-126-5p to
upregulate PTEN, which prevents PI3K/Akt/mTOR signaling pathway
activation. Furthermore, Liu et al (105) demonstrated that reductions of m6A
methylation mediated by METTL14 mutation or reduced expression
levels of METTL3 lead to the activation of the Akt signaling
pathway by decreasing PHLPP2 expression and increasing mTORC2
expression, which promotes cell proliferation in endometrial
cancer.
The ERK signaling pathway has been demonstrated to
be important for cancer progression. The substrates of ERK
signaling are broad, which make ERKs key regulators of
proliferation, migration, apoptosis and chemo-immune-resistance, as
well as appealing therapeutic targets in cancer (106). Zhong et al (107) revealed that YTHDF2 directly bound to
the m6A modification site of the EGFR 3′-UTR to promote the
degradation of EGFR mRNA in HCC cells, and this mechanism
suppressed MEK and ERK activation, cell proliferation and tumor
growth. However, previous studies, have revealed the interaction
between m6A modification and the ERK signaling pathway (108,109).
Xie et al (109) demonstrated
that basic leucine zipper ATF-like transcription factor 2 (BATF2)
could bind to p53 and enhanced its protein stability, thereby
inhibiting the phosphorylation of ERK in GC, and m6A modification
mediated by METTL3 could repress BATF2 mRNA expression, which
provides potential prognostic and therapeutic targets for GC
treatment. Conversely, the ERK signaling pathway has been
demonstrated to have a positive effect on m6A deposition (108). Sun et al (108) demonstrated that ERK could
phosphorylate METTL3 at S43/S50/S525 and WTAP at S306/S341, thus
stabilizing the m6A methyltransferase complex in ESC and malignant
tumor cells.
In addition to the aforementioned representative
signaling pathways, researchers have reported that m6A modification
also serves a crucial role in other classic signaling pathways.
Ghazi et al (110)
investigated the effects of fusaric acid on p53 expression and its
epigenetic regulation via promoter methylation and m6A modification
in HCC cells. The results revealed that fusaric acid epigenetically
decreased p53 expression by altering its m6A modification (110). Similarly, Ding et al
(111) reported that
lipopolysaccharides stimulation promotes GNAS complex locus (GNAS)
expression by increasing the m6A methylation levels of GNAS mRNA,
thus inducing HCC cell proliferation and invasion by interacting
with the STAT3 signaling pathway in HCC. In addition, Zhang et
al (112) demonstrated that
β-estradiol can accelerate FTO nuclear localization and increase
the proliferation of endometrial cancer cells by modulating the
mTOR signaling pathway; however, the mechanism by which estrogen
receptor-α mediates FTO nuclear accumulation is unclear (113).
At present, m6A modification is mechanistically
linked to the progression and prognosis of several types of cancer.
Given the complicated process of m6A catalysis in cancer, m6a and
its regulatory proteins may be novel therapeutic targets for cancer
diagnosis and prognosis. For example, METTL3 has been considered as
an oncogenic factor in numerous human types of cancer. According to
recent studies, METTL3 may be an independent prognostic factor for
patients with GC (114), CRC
(115) and HCC (73). Similarly, evidence also supports the
proliferative roles of FTO in cancer (81). FB23-2, a promising FTO inhibitor, has
been demonstrated to negatively regulate proliferation and
progression of human AML cell lines by inhibiting FTO (57). Furthermore, R-2HG, another
small-molecule inhibitor of FTO, exhibits anticancer activity in
AML (18). Additionally, clinical
data have demonstrated that the expression levels of ALKBH1 are
negatively associated with tumor size and TNM stage, and that the
expression levels of FTO are associated with improved overall
survival in patients with GC (116).
Despite extensive efforts being devoted to study m6A in cancer, a
number of issues associated with the function and mechanism of m6A
remain unknown. Considering that novel m6A readers and writers are
constantly emerging, m6A-mediated biological functions require
further exploration. Additionally, multifarious modification
targets and sites suggest that the specific mechanisms of m6A is
not unitary even in the same type of malignant tumor. This should
be clarified. The rapid development of detection methods and
several novel inhibitors of m6A-related factors will provide
practical assistance for researchers.
Increasing studies suggest that m6A is deeply
involved in the regulation of gene expression. m6A can determine
the fate during development and differentiation, and aberrant m6A
modifications can affect classic signaling pathways, including the
PI3K/AKT (91,101,105,112),
Wnt (91,92) and mTOR (101,113)
signaling pathways, in cancer. The dual role of m6A regulation in
cancer is unclear, and may be due to differences in cell types and
states. At present, m6A is gaining attention in cancer research,
and may provide promising targets for cancer therapies. Future
research may focus more on the specific mechanism of m6A
methyltransferases and demethylases, or the specificity and
sensitivity of readers. The regulatory role of m6A modification in
cancer is described as a ‘double-edged sword’ implying that
clinical applications require further investigation (117). Furthermore, the development of
methods for the detection and analysis of m6A is required to
improve the understanding of the underlying mechanisms.
Not applicable.
This work was supported by a grant from the
National Natural Science Foundation of China (grant no.
81660468).
All data generated or analyzed during this study
are included in this published article.
FL searched the literature and drafted the
manuscript for the study. FL designed the figures. XS and FL
revised the manuscript and assessed all the raw data. XS and FL are
responsible for confirming the authenticity of the data. All
authors read and approved the final manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Holley RW, Everett GA, Madison JT and
Zamir A: Nucleotide sequences in the yeast alanine transfer
ribonucleic acid. J Biol Chem. 240:2122–2128. 1965. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Desrosiers R, Friderici K and Rottman F:
Identification of methylated nucleosides in messenger RNA from
Novikoff hepatoma cells. Proc Natl Acad Sci USA. 71:3971–3975.
1974. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Dominissini D, Moshitch-Moshkovitz S,
Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K,
Jacob-Hirsch J, Amariglio N, Kupiec M, et al: Topology of the human
and mouse m6A RNA methylomes revealed by m6A-seq. Nature.
485:201–206. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yang Y, Hsu PJ, Chen YS and Yang YG:
Dynamic transcriptomic m(6)A decoration: Writers, erasers, readers
and functions in RNA metabolism. Cell Res. 28:616–624. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Meyer KD and Jaffrey SR: Rethinking
m6A readers, writers, and erasers. Annu Rev Cell Dev
Biol. 33:319–342. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Schumann U, Shafik A and Preiss T: METTL3
gains R/W access to the epitranscriptome. Mol Cell. 62:323–324.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ping XL, Sun BF, Wang L, Xiao W, Yang X,
Wang WJ, Adhikari S, Shi Y, Lv Y, Chen YS, et al: Mammalian WTAP is
a regulatory subunit of the RNA N6-methyladenosine
methyltransferase. Cell Res. 24:177–189. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Schwartz S, Mumbach MR, Jovanovic M, Wang
T, Maciag K, Bushkin GG, Mertins P, Ter-Ovanesyan D, Habib N,
Cacchiarelli D, et al: Perturbation of m6A writers reveals two
distinct classes of mRNA methylation at internal and 5′ sites. Cell
Rep. 8:284–296. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Haussmann IU, Bodi Z, Sanchez-Moran E,
Mongan NP, Archer N, Fray RG and Soller M: m6A
potentiates Sxl alternative pre-mRNA splicing for robust
Drosophila sex determination. Nature. 540:301–304. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Muller S, Glaß M, Singh AK, Haase J, Bley
N, Fuchs T, Lederer M, Dahl A, Huang H, Chen J, et al: IGF2BP1
promotes SRF-dependent transcription in cancer in a m6A- and
miRNA-dependent manner. Nucleic Acids Res. 47:375–390. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang
Y, Yi C, Lindahl T, Pan T, Yang YG and He C: N6-methyladenosine in
nuclear RNA is a major substrate of the obesity-associated FTO. Nat
Chem Biol. 7:885–887. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zheng G, Dahl JA, Niu Y, Fedorcsak P,
Huang CM, Li CJ, Vågbø CB, Shi Y, Wang WL, Song SH, et al: ALKBH5
is a mammalian RNA demethylase that impacts RNA metabolism and
mouse fertility. Mol Cell. 49:18–29. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Narayan P and Rottman FM: An in vitro
system for accurate methylation of internal adenosine residues in
messenger RNA. Science. 242:1159–1162. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bodi Z, Button JD, Grierson D and Fray RG:
Yeast targets for mRNA methylation. Nucleic Acids Res.
38:5327–5335. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hongay CF and Orr-Weaver TL:
Drosophila inducer of MEiosis 4 (IME4) is required for Notch
signaling during oogenesis. Proc Natl Acad Sci USA.
108:14855–14860. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yoon KJ, Ringeling FR, Vissers C, Jacob F,
Pokrass M, Jimenez-Cyrus D, Su Y, Kim NS, Zhu Y, Zheng L, et al:
Temporal control of Mammalian Cortical Neurogenesis by
m6A Methylation. Cell. 171:877–889 e817. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
McIntyre ABR, Alexander N, Grigorev K,
Bezdan D, Sichtig H, Chiu CY and Mason CE: Single-molecule
sequencing detection of N6-methyladenine in microbial reference
materials. Nat Commun. 10:5792019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hong T, Yuan Y, Chen Z, Xi K, Wang T, Xie
Y, He Z, Su H, Zhou Y, Tan ZJ, et al: Precise Antibody-Independent
m6A Identification via 4SedTTP-Involved and FTO-Assisted Strategy
at Single-Nucleotide Resolution. J Am Chem Soc. 140:5886–5889.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Meyer KD, Saletore Y, Zumbo P, Elemento O,
Mason CE and Jaffrey SR: Comprehensive analysis of mRNA methylation
reveals enrichment in 3′ UTRs and near stop codons. Cell.
149:1635–1646. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Shi H, Wei J and He C: Where, when, and
how: Context-Dependent functions of RNA methylation writers,
readers, and erasers. Mol Cell. 74:640–650. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zaccara S, Ries RJ and Jaffrey SR:
Reading, writing and erasing mRNA methylation. Nat Rev Mol Cell
Biol. 20:608–624. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhao BS, Roundtree IA and He C:
Post-transcriptional gene regulation by mRNA modifications. Nat Rev
Mol Cell Biol. 18:31–42. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Fu Y, Dominissini D, Rechavi G and He C:
Gene expression regulation mediated through reversible m6A RNA
methylation. Nat Rev Genet. 15:293–306. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Warda AS, Kretschmer J, Hackert P, Lenz C,
Urlaub H, Höbartner C, Sloan KE and Bohnsack MT: Human METTL16 is a
N6-methyladenosine (m6A) methyltransferase
that targets pre-mRNAs and various non-coding RNAs. EMBO Rep.
18:2004–2014. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Doxtader KA, Wang P, Scarborough AM, Seo
D, Conrad NK and Nam Y: Structural basis for regulation of METTL16,
an S-adenosylmethionine homeostasis factor. Mol Cell.
71:1001–1011.e4. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Guo J, Tang HW, Li J, Perrimon N and Yan
D: Xio is a component of the Drosophila sex determination
pathway and RNA N6-methyladenosine methyltransferase
complex. Proc Natl Acad Sci USA. 115:3674–3679. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Patil DP, Chen CK, Pickering BF, Chow A,
Jackson C, Guttman M and Jaffrey SR: m6A RNA methylation
promotes XIST-mediated transcriptional repression. Nature.
537:369–373. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Fu Y, Jia G, Pang X, Wang RN, Wang X, Li
CJ, Smemo S, Dai Q, Bailey KA, Nobrega MA, et al: FTO-mediated
formation of N6-hydroxymethyladenosine and N6-formyladenosine in
mammalian RNA. Nat Commun. 4:17982013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang X, Zhao BS, Roundtree IA, Lu Z, Han
D, Ma H, Weng X, Chen K, Shi H and He C: N(6)-methyladenosine
modulates messenger RNA translation efficiency. Cell.
161:1388–1399. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han
D, Fu Y, Parisien M, Dai Q, Jia G, et al:
N6-methyladenosine-dependent regulation of messenger RNA stability.
Nature. 505:117–120. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Shi H, Wang X, Lu Z, Zhao BS, Ma H, Hsu
PJ, Liu C and He C: YTHDF3 facilitates translation and decay of
N6-methyladenosine-modified RNA. Cell Res. 27:315–328.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhao X, Yang Y, Sun BF, Shi Y, Yang X,
Xiao W, Hao YJ, Ping XL, Chen YS, Wang WJ, et al: FTO-dependent
demethylation of N6-methyladenosine regulates mRNA splicing and is
required for adipogenesis. Cell Res. 24:1403–1419. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Peng S, Xiao W, Ju D, Sun B, Hou N, Liu Q,
Wang Y, Zhao H, Gao C, Zhang S, et al: Identification of entacapone
as a chemical inhibitor of FTO mediating metabolic regulation
through FOXO1. Sci Transl Med. 11:eaau71162019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ben-Haim MS, Moshitch-Moshkovitz S and
Rechavi G: FTO: Linking m6A demethylation to adipogenesis. Cell
Res. 25:3–4. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhang C, Fu J and Zhou Y: A review in
research progress concerning m6A methylation and immunoregulation.
Front Immunol. 10:9222019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ma Z, Gao X, Shuai Y, Xing X and Ji J: The
m6A epitranscriptome opens a new charter in immune system logic.
Epigenetics. 1–19. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Shulman Z and Stern-Ginossar N: The RNA
modification N(6)-methyladenosine as a novel regulator of the
immune system. Nat Immunol. 21:501–512. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Geula S, Moshitch-Moshkovitz S,
Dominissini D, Mansour AA, Kol N, Salmon-Divon M, Hershkovitz V,
Peer E, Mor N, Manor YS, et al: Stem cells. m6A mRNA methylation
facilitates resolution of naive pluripotency toward
differentiation. Science. 347:1002–1006. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Roundtree IA, Evans ME, Pan T and He C:
Dynamic RNA modifications in gene expression regulation. Cell.
169:1187–1200. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Xiao S, Zeng X, Fan Y, Su Y, Ma Q, Zhu J
and Yao H: Gene polymorphism association with type 2 diabetes and
related gene-gene and gene-environment interactions in a uyghur
population. Med Sci Monit. 22:474–487. 2016.PubMed/NCBI
|
|
41
|
Kennedy EM, Bogerd HP, Kornepati AV, Kang
D, Ghoshal D, Marshall JB, Poling BC, Tsai K, Gokhale NS, Horner SM
and Cullen BR: Posttranscriptional m(6)A editing of HIV-1 mRNAs
enhances viral gene expression. Cell host microbe. 19:675–685.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Tirumuru N, Zhao BS, Lu W, Lu Z, He C and
Wu L: N(6)-methyladenosine of HIV-1 RNA regulates viral infection
and HIV-1 Gag protein expression. Elife. 5:e155282016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Gokhale NS, McIntyre ABR, McFadden MJ,
Roder AE, Kennedy EM, Gandara JA, Hopcraft SE, Quicke KM, Vazquez
C, Willer J, et al: N6-Methyladenosine in flaviviridae viral RNA
genomes regulates infection. Cell host microbe. 20:654–665. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Lichinchi G, Zhao BS, Wu Y, Lu Z, Qin Y,
He C and Rana TM: Dynamics of human and viral RNA methylation
during Zika virus infection. Cell Host Microbe. 20:666–673. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Yu R, Li Q, Feng Z, Cai L and Xu Q: M6A
Reader YTHDF2 Regulates LPS-Induced inflammatory response. Int J
Mol Sci. 20:13232019. View Article : Google Scholar
|
|
46
|
Nichols J and Smith A: Naive and primed
pluripotent states. Cell Stem Cell. 4:487–492. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Batista PJ, Molinie B, Wang J, Qu K, Zhang
J, Li L, Bouley DM, Lujan E, Haddad B, Daneshvar K, et al:
m6A RNA modification controls cell fate transition in
mammalian embryonic stem cells. Cell stem cell. 15:707–719. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Buecker C, Srinivasan R, Wu Z, Calo E,
Acampora D, Faial T, Simeone A, Tan M, Swigut T and Wysocka J:
Reorganization of enhancer patterns in transition from naive to
primed pluripotency. Cell stem cell. 14:838–853. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Xu C, Wang X, Liu K, Roundtree IA, Tempel
W, Li Y, Lu Z, He C and Min J: Structural basis for selective
binding of m6A RNA by the YTHDC1 YTH domain. Nat Chem Biol.
10:927–929. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Kasowitz SD, Ma J, Anderson SJ, Leu NA, Xu
Y, Gregory BD, Schultz RM and Wang PJ: Nuclear m6A reader YTHDC1
regulates alternative polyadenylation and splicing during mouse
oocyte development. PLoS Genet. 14:e10074122018. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Li Z, Qian P, Shao W, Shi H, He XC, Gogol
M, Yu Z, Wang Y, Qi M, Zhu Y, et al: Suppression of m(6)A reader
Ythdf2 promotes hematopoietic stem cell expansion. Cell Res.
28:904–917. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Lobo J, Costa AL, Cantante M, Guimarães R,
Lopes P, Antunes L, Braga I, Oliveira J, Pelizzola M, Henrique R
and Jerónimo C: m6A RNA modification and its
writer/reader VIRMA/YTHDF3 in testicular germ cell tumors: A role
in seminoma phenotype maintenance. J Transl Med. 17:792019.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Zhou J, Wan J, Shu XE, Mao Y, Liu XM, Yuan
X, Zhang X, Hess ME, Brüning JC and Qian SB:
N6-Methyladenosine guides mRNA alternative translation
during integrated stress response. Mol Cell. 69:636–647.e7. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Wu R, Liu Y, Yao Y, Zhao Y, Bi Z, Jiang Q,
Liu Q, Cai M, Wang F, Wang Y and Wang X: FTO regulates adipogenesis
by controlling cell cycle progression via m6A-YTHDF2
dependent mechanism. Biochim Biophys Acta Mol Cell Biol Lipids.
1863:1323–1330. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Liu J, Zhang X, Chen K, Cheng Y, Liu S,
Xia M, Chen Y, Zhu H, Li Z and Cao X: CCR7 chemokine
receptor-inducible lnc-Dpf3 restrains dendritic cell migration by
inhibiting HIF-1α-mediated glycolysis. Immunity. 50:600–615.e15.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Han D, Liu J, Chen C, Dong L, Liu Y, Chang
R, Huang X, Liu Y, Wang J, Dougherty U, et al: Anti-tumour immunity
controlled through mRNA m(6)A methylation and YTHDF1 in dendritic
cells. Nature. 566:270–274. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Huang Y, Su R, Sheng Y, Dong L, Dong Z, Xu
H, Ni T, Zhang ZS, Zhang T, Li C, et al: Small-molecule targeting
of oncogenic FTO demethylase in acute myeloid leukemia. Cancer
Cell. 35:677–691.e10. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Li HB, Tong J, Zhu S, Batista PJ, Duffy
EE, Zhao J, Bailis W, Cao G, Kroehling L, Chen Y, et al:
m6A mRNA methylation controls T cell homeostasis by
targeting the IL-7/STAT5/SOCS pathways. Nature. 548:338–342. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Kim GW, Imam H, Khan M and Siddiqui A:
N6-Methyladenosine modification of hepatitis B and C
viral RNAs attenuates host innate immunity via RIG-I signaling. J
Biol Chem. 295:13123–13133. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Mapperley C, van de Lagemaat LN, Lawson H,
Tavosanis A, Paris J, Campos J, Wotherspoon D, Durko J, Sarapuu A,
Choe J, et al: The mRNA m6A reader YTHDF2 suppresses
proinflammatory pathways and sustains hematopoietic stem cell
function. J Exp Med. 218:e202008292021. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Hou J, Zhang H, Liu J, Zhao Z, Wang J, Lu
Z, Hu B, Zhou J, Zhao Z, Feng M, et al: YTHDF2 reduction fuels
inflammation and vascular abnormalization in hepatocellular
carcinoma. Mol Cancer. 18:1632019. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Lin Z, Hsu PJ, Xing X, Fang J, Lu Z, Zou
Q, Zhang KJ, Zhang X, Zhou Y, Zhang T, et al:
Mettl3-/Mettl14-mediated mRNA N6-methyladenosine
modulates murine spermatogenesis. Cell Res. 27:1216–1230. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Zhao BS and He C: ‘Gamete On’ for
m6A: YTHDF2 exerts essential functions in female
fertility. Mol Cell. 67:903–905. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Livneh I, Moshitch-Moshkovitz S, Amariglio
N, Rechavi G and Dominissini D: The m6A
epitranscriptome: Transcriptome plasticity in brain development and
function. Nat Rev Neurosci. 21:36–51. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Lence T, Akhtar J, Bayer M, Schmid K,
Spindler L, Ho CH, Kreim N, Andrade-Navarro MA, Poeck B, Helm M and
Roignant JY: m6A modulates neuronal functions and sex
determination in Drosophila. Nature. 540:242–247. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Lin S, Choe J, Du P, Triboulet R and
Gregory RI: The m(6)A methyltransferase METTL3 promotes translation
in human cancer cells. Mol Cell. 62:335–345. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Ma JZ, Yang F, Zhou CC, Liu F, Yuan JH,
Wang F, Wang TT, Xu QG, Zhou WP and Sun SH: METTL14 suppresses the
metastatic potential of hepatocellular carcinoma by modulating
N6 -methyladenosine-dependent primary MicroRNA
processing. Hepatology. 65:529–543. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Vu LP, Pickering BF, Cheng Y, Zaccara S,
Nguyen D, Minuesa G, Chou T, Chow A, Saletore Y, MacKay M, et al:
The N6-methyladenosine (m6A)-forming enzyme
METTL3 controls myeloid differentiation of normal hematopoietic and
leukemia cells. Nat Med. 23:1369–1376. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Wang Q, Geng W, Guo H, Wang Z, Xu K, Chen
C and Wang S: Emerging role of RNA methyltransferase METTL3 in
gastrointestinal cancer. J Hematol Oncol. 13:572020. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Yue B, Song C, Yang L, Cui R, Cheng X,
Zhang Z and Zhao G: METTL3-mediated N6-methyladenosine modification
is critical for epithelial-mesenchymal transition and metastasis of
gastric cancer. Mol Cancer. 18:1422019. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Yang DD, Chen ZH, Yu K, Lu JH, Wu QN, Wang
Y, Ju HQ, Xu RH, Liu ZX and Zeng ZL: METTL3 promotes the
progression of gastric cancer via targeting the MYC pathway. Front
Oncol. 10:1152020. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Shen C, Xuan B, Yan T, Ma Y, Xu P, Tian X,
Zhang X, Cao Y, Ma D, Zhu X, et al: m6A-dependent
glycolysis enhances colorectal cancer progression. Mol Cancer.
19:722020. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Chen M, Wei L, Law CT, Tsang FH, Shen J,
Cheng CL, Tsang LH, Ho DW, Chiu DK, Lee JM, et al: RNA
N6-methyladenosine methyltransferase-like 3 promotes liver cancer
progression through YTHDF2-dependent posttranscriptional silencing
of SOCS2. Hepatology. 67:2254–2270. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Zhang J, Bai R, Li M, Ye H, Wu C, Wang C,
Li S, Tan L, Mai D, Li G, et al: Excessive miR-25-3p maturation via
N6-methyladenosine stimulated by cigarette smoke
promotes pancreatic cancer progression. Nat Commun. 10:18582019.
View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Taketo K, Konno M, Asai A, Koseki J,
Toratani M, Satoh T, Doki Y, Mori M, Ishii H and Ogawa K: The
epitranscriptome m6A writer METTL3 promotes chemo- and
radioresistance in pancreatic cancer cells. Int J Oncol.
52:621–629. 2018.PubMed/NCBI
|
|
76
|
Liu T, Yang S, Sui J, Xu SY, Cheng YP,
Shen B, Zhang Y, Zhang XM, Yin LH, Pu YP and Liang GY: Dysregulated
N6-methyladenosine methylation writer METTL3 contributes to the
proliferation and migration of gastric cancer. J Cell Physiol.
235:548–562. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Wang P, Wang X, Zheng L and Zhuang C: Gene
signatures and prognostic values of m6A regulators in
hepatocellular carcinoma. Front Genet. 11:5401862020. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Zhu D, Zhou J, Zhao J, Jiang G, Zhang X,
Zhang Y and Dong M: ZC3H13 suppresses colorectal cancer
proliferation and invasion via inactivating Ras-ERK signaling. J
Cell Physiol. 234:8899–8907. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Barros-Silva D, Lobo J, Guimaraes-Teixeira
C, Carneiro I, Oliveira J, Martens-Uzunova ES, Henrique R and
Jerónimo C: VIRMA-Dependent N6-Methyladenosine modifications
regulate the expression of long non-coding RNAs CCAT1 and CCAT2 in
prostate cancer. Cancers (Basel). 12:7712020. View Article : Google Scholar
|
|
80
|
Qian JY, Gao J, Sun X, Cao MD, Shi L, Xia
TS, Zhou WB, Wang S, Ding Q and Wei JF: KIAA1429 acts as an
oncogenic factor in breast cancer by regulating CDK1 in an
N6-methyladenosine-independent manner. Oncogene. 38:6123–6141.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Li Z, Weng H, Su R, Weng X, Zuo Z, Li C,
Huang H, Nachtergaele S, Dong L, Hu C, et al: FTO plays an
oncogenic role in acute myeloid leukemia as a
N6-Methyladenosine RNA demethylase. Cancer Cell.
31:127–141. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Van Der Werf I and Jamieson C: The yin and
yang of RNA methylation: An imbalance of erasers enhances
sensitivity to FTO demethylase small-molecule targeting in leukemia
stem cells. Cancer Cell. 35:540–541. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Zhu T, Yong XLH, Xia D, Widagdo J and
Anggono V: Ubiquitination regulates the proteasomal degradation and
nuclear translocation of the fat mass and obesity-associated (FTO)
protein. J Mol Biol. 430:363–371. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Cui Q, Shi H, Ye P, Li L, Qu Q, Sun G, Sun
G, Lu Z, Huang Y, Yang CG, et al: m6A RNA methylation
regulates the self-renewal and tumorigenesis of glioblastoma stem
cells. Cell Rep. 18:2622–2634. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Kwok CT, Marshall AD, Rasko JE and Wong
JJ: Genetic alterations of m(6)A regulators predict poorer survival
in acute myeloid leukemia. J Hematol Oncol. 10:392017. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Zhang S, Zhao BS, Zhou A, Lin K, Zheng S,
Lu Z, Chen Y, Sulman EP, Xie K, Bögler O, et al: m6A
demethylase ALKBH5 maintains tumorigenicity of glioblastoma
stem-like cells by sustaining FOXM1 expression and cell
proliferation program. Cancer Cell. 31:591–606 e6. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Bai Y, Yang C, Wu R, Huang L, Song S, Li
W, Yan P, Lin C, Li D and Zhang Y: YTHDF1 regulates tumorigenicity
and cancer stem cell-like activity in human colorectal carcinoma.
Front Oncol. 9:3322019. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Nishizawa Y, Konno M, Asai A, Koseki J,
Kawamoto K, Miyoshi N, Takahashi H, Nishida N, Haraguchi N, Sakai
D, et al: Oncogene c-Myc promotes epitranscriptome m6A
reader YTHDF1 expression in colorectal cancer. Oncotarget.
9:7476–7486. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Tanabe A, Tanikawa K, Tsunetomi M, Takai
K, Ikeda H, Konno J, Torigoe T, Maeda H, Kutomi G, Okita K, et al:
RNA helicase YTHDC2 promotes cancer metastasis via the enhancement
of the efficiency by which HIF-1α mRNA is translated. Cancer Lett.
376:34–42. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Huang H, Weng H, Sun W, Qin X, Shi H, Wu
H, Zhao BS, Mesquita A, Liu C, Yuan CL, et al: Recognition of RNA
N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and
translation. Nat Cell Biol. 20:285–295. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Zhang C, Zhang M, Ge S, Huang W, Lin X,
Gao J, Gong J and Shen L: Reduced m6A modification predicts
malignant phenotypes and augmented Wnt/PI3K-Akt signaling in
gastric cancer. Cancer Med. 8:4766–4781. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Korshunov A, Sahm F, Zheludkova O, Golanov
A, Stichel D, Schrimpf D, Ryzhova M, Potapov A, Habel A, Meyer J,
et al: DNA methylation profiling is a method of choice for
molecular verification of pediatric WNT-activated medulloblastomas.
Neuro Oncol. 21:214–221. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Zhang L, Wan Y, Zhang Z, Jiang Y, Lang J,
Cheng W and Zhu L: FTO demethylates m6A modifications in HOXB13
mRNA and promotes endometrial cancer metastasis by activating the
WNT signalling pathway. RNA Biol. Nov 5–2020.(Epub ahead of print).
doi: 10.1080/15476286.2020.1841458. View Article : Google Scholar
|
|
94
|
Liu L, Wang J, Sun G, Wu Q, Ma J, Zhang X,
Huang N, Bian Z, Gu S, Xu M, et al: m6A mRNA methylation
regulates CTNNB1 to promote the proliferation of hepatoblastoma.
Mol Cancer. 18:1882019. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Le PN, McDermott JD and Jimeno A:
Targeting the Wnt pathway in human cancers: Therapeutic targeting
with a focus on OMP-54F28. Pharmacol Ther. 146:1–11. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Li Z, Chen Y, An T, Liu P, Zhu J, Yang H,
Zhang W, Dong T, Jiang J, Zhang Y, et al: Nuciferine inhibits the
progression of glioblastoma by suppressing the SOX2-AKT/STAT3-Slug
signaling pathway. J Exp Clin Cancer Res. 38:1392019. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Li M, Bu X, Cai B, Liang P, Li K, Qu X and
Shen L: Biological role of metabolic reprogramming of cancer cells
during epithelialmesenchymal transition (Review). Oncol Rep.
41:727–741. 2019.PubMed/NCBI
|
|
98
|
Chen J, Sun Y, Xu X, Wang D, He J, Zhou H,
Lu Y, Zeng J, Du F, Gong A and Xu M: YTH domain family 2
orchestrates epithelial-mesenchymal transition/proliferation
dichotomy in pancreatic cancer cells. Cell Cycle. 16:2259–2271.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Chen WW, Qi JW, Hang Y, Wu JX, Zhou XX,
Chen JZ, Wang J and Wang HH: Simvastatin is beneficial to lung
cancer progression by inducing METTL3-induced m6A modification on
EZH2 mRNA. Eur Rev Med Pharmacol Sci. 24:4263–4270. 2020.PubMed/NCBI
|
|
100
|
Aoki M and Fujishita T: Oncogenic roles of
the PI3K/AKT/mTOR axis. Curr Top Microbiol Immunol. 407:153–189.
2017.PubMed/NCBI
|
|
101
|
Li X, Tang J, Huang W, Wang F, Li P, Qin
C, Qin Z, Zou Q, Wei J, Hua L, et al: The M6A methyltransferase
METTL3: Acting as a tumor suppressor in renal cell carcinoma.
Oncotarget. 8:96103–96116. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Zhao Q, Zhao Y, Hu W, Zhang Y, Wu X, Lu J,
Li M, Li W, Wu W, Wang J, et al: m6A RNA modification
modulates PI3K/Akt/mTOR signal pathway in gastrointestinal cancer.
Theranostics. 10:9528–9543. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Chen X, Xu M, Xu X, Zeng K, Liu X, Pan B,
Li C, Sun L, Qin J, Xu T, et al: METTL14-mediated
N6-methyladenosine modification of SOX4 mRNA inhibits tumor
metastasis in colorectal cancer. Mol Cancer. 19:1062020. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Bi X, Lv X, Liu D, Guo H, Yao G, Wang L,
Liang X and Yang Y: METTL3-mediated maturation of miR-126-5p
promotes ovarian cancer progression via PTEN-mediated PI3K/Akt/mTOR
pathway. Cancer Gene Ther. Sep 16–2020.(Epub ahead of print). doi:
10.1038/s41417-020-00222-3. View Article : Google Scholar
|
|
105
|
Liu J, Eckert MA, Harada BT, Liu SM, Lu Z,
Yu K, Tienda SM, Chryplewicz A, Zhu AC, Yang Y, et al:
m6A mRNA methylation regulates AKT activity to promote
the proliferation and tumorigenicity of endometrial cancer. Nat
Cell Biol. 20:1074–1083. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Salaroglio IC, Mungo E, Gazzano E, Kopecka
J and Riganti C: ERK is a pivotal player of chemo-immune-resistance
in cancer. Int J Mol Sci. 20:25052019. View Article : Google Scholar
|
|
107
|
Zhong L, Liao D, Zhang M, Zeng C, Li X,
Zhang R, Ma H and Kang T: YTHDF2 suppresses cell proliferation and
growth via destabilizing the EGFR mRNA in hepatocellular carcinoma.
Cancer Lett. 442:252–261. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Sun HL, Zhu AC, Gao Y, Terajima H, Fei Q,
Liu S, Zhang L, Zhang Z, Harada BT, He YY, et al: Stabilization of
ERK-Phosphorylated METTL3 by USP5 Increases m6A
methylation. Mol Cell. 80:633–647.e7. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Xie JW, Huang XB, Chen QY, Ma YB, Zhao YJ,
Liu LC, Wang JB, Lin JX, Lu J, Cao LL, et al: m6A
modification-mediated BATF2 acts as a tumor suppressor in gastric
cancer through inhibition of ERK signaling. Mol Cancer. 19:1142020.
View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Ghazi T, Nagiah S and Chuturgoon AA:
Fusaric acid decreases p53 expression by altering promoter
methylation and m6A RNA methylation in human hepatocellular
carcinoma (HepG2) cells. Epigenetics. 1–13. 2020.(Epub ahead of
print).
|
|
111
|
Ding H, Zhang X, Su Y, Jia C and Dai C:
GNAS promotes inflammation-related hepatocellular carcinoma
progression by promoting STAT3 activation. Cell Mol Biol Lett.
25:82020. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Zhang Z, Zhou D, Lai Y, Liu Y, Tao X, Wang
Q, Zhao G, Gu H, Liao H, Zhu Y, et al: Estrogen induces endometrial
cancer cell proliferation and invasion by regulating the fat mass
and obesity-associated gene via PI3K/AKT and MAPK signaling
pathways. Cancer Lett. 319:89–97. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Zhu Y, Shen J, Gao L and Feng Y: Estrogen
promotes fat mass and obesity-associated protein nuclear
localization and enhances endometrial cancer cell proliferation via
the mTOR signaling pathway. Oncol Rep. 35:2391–2397. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Wang Q, Chen C, Ding Q, Zhao Y, Wang Z,
Chen J, Jiang Z, Zhang Y, Xu G, Zhang J, et al: METTL3-mediated
m6A modification of HDGF mRNA promotes gastric cancer
progression and has prognostic significance. Gut. 69:1193–1205.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Li T, Hu PS, Zuo Z, Lin JF, Li X, Wu QN,
Chen ZH, Zeng ZL, Wang F, Zheng J, et al: METTL3 facilitates tumor
progression via an m6A-IGF2BP2-dependent mechanism in
colorectal carcinoma. Mol Cancer. 18:1122019. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Li Y, Zheng D, Wang F, Xu Y, Yu H and
Zhang H: Expression of demethylase genes, FTO and ALKBH1, is
associated with prognosis of gastric cancer. Dig Dis Sci.
64:1503–1513. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Wang S, Chai P and Jia R and Jia R: Novel
insights on m6A RNA methylation in tumorigenesis: A
double-edged sword. Mol Cancer. 17:1012018. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Cheng M, Sheng L, Gao Q, Xiong Q, Zhang H,
Wu M, Liang Y, Zhu F, Zhang Y, Zhang X, et al: The m(6)A
methyltransferase METTL3 promotes bladder cancer progression via
AFF4/NF-kappaB/MYC signaling network. Oncogene. 38:3667–3680. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Li F, Yi Y, Miao Y, Long W, Long T, Chen
S, Cheng W, Zou C, Zheng Y, Wu X, et al:
N6-Methyladenosine modulates nonsense-mediated mRNA
decay in human glioblastoma. Cancer Res. 79:5785–5798. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Wang K, Jiang L, Zhang Y and Chen C:
Progression of thyroid carcinoma is promoted by the m6A
methyltransferase METTL3 through regulating m6A
methylation on TCF1. Onco Targets Ther. 13:1605–1612. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Weng H, Huang H, Wu H, Qin X, Zhao BS,
Dong L, Shi H, Skibbe J, Shen C, Hu C, et al: METTL14 inhibits
hematopoietic stem/progenitor differentiation and promotes
leukemogenesis via mRNA m6A modification. Cell Stem
Cell. 22:191–205.e9. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Bansal H, Yihua Q, Iyer SP, Ganapathy S,
Proia DA, Penalva LO, Uren PJ, Suresh U, Carew JS, Karnad AB, et
al: WTAP is a novel oncogenic protein in acute myeloid leukemia.
Leukemia. 28:1171–1174. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Chen Y, Peng C, Chen J, Chen D, Yang B, He
B, Hu W, Zhang Y, Liu H, Dai L, et al: WTAP facilitates progression
of hepatocellular carcinoma via m6A-HuR-dependent epigenetic
silencing of ETS1. Mol Cancer. 18:1272019. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Cheng X, Li M, Rao X, Zhang W, Li X, Wang
L and Huang G: KIAA1429 regulates the migration and invasion of
hepatocellular carcinoma by altering m6A modification of ID2 mRNA.
Onco Targets Ther. 12:3421–3428. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Xu D, Shao W, Jiang Y, Wang X, Liu Y and
Liu X: FTO expression is associated with the occurrence of gastric
cancer and prognosis. Oncol Rep. 38:2285–2292. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Li J, Han Y, Zhang H, Qian Z, Jia W, Gao
Y, Zheng H and Li B: The m6A demethylase FTO promotes the growth of
lung cancer cells by regulating the m6A level of USP7 mRNA. Biochem
Biophys Res Commun. 512:479–485. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Li J, Zhu L, Shi Y, Liu J, Lin L and Chen
X: m6A demethylase FTO promotes hepatocellular carcinoma
tumorigenesis via mediating PKM2 demethylation. Am J Transl Res.
11:6084–6092. 2019.PubMed/NCBI
|
|
128
|
Chao Y, Shang J and Ji W:
ALKBH5-m6A-FOXM1 signaling axis promotes proliferation
and invasion of lung adenocarcinoma cells under intermittent
hypoxia. Biochem Biophys Res Commun. 521:499–506. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Lin X, Chai G, Wu Y, Li J, Chen F, Liu J,
Luo G, Tauler J, Du J, Lin S, et al: RNA m(6)A methylation
regulates the epithelial mesenchymal transition of cancer cells and
translation of Snail. Nat Commun. 10:20652019. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Li J, Xie H, Ying Y, Chen H, Yan H, He L,
Xu M, Xu X, Liang Z, Liu B, et al: YTHDF2 mediates the mRNA
degradation of the tumor suppressors to induce AKT phosphorylation
in N6-methyladenosine-dependent way in prostate cancer. Mol Cancer.
19:1522020. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Dixit D, Prager BC, Gimple RC, Poh HX,
Wang Y, Wu Q, Qiu Z, Kidwell RL, Kim LJ, Xie Q, et al: The RNA m6A
reader YTHDF2 maintains oncogene expression and is a targetable
dependency in glioblastoma stem cells. Cancer Discov. 11:480–499.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Chang G, Shi L, Ye Y, Shi H, Zeng L,
Tiwary S, Huse JT, Huo L, Ma L, Ma Y, et al: YTHDF3 induces the
translation of m6A-enriched gene transcripts to promote
breast cancer brain metastasis. Cancer Cell. 38:857–871 e7. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Ma L, Chen T, Zhang X, Miao Y, Tian X, Yu
K, Xu X, Niu Y, Guo S, Zhang C, et al: The m6A reader
YTHDC2 inhibits lung adenocarcinoma tumorigenesis by suppressing
SLC7A11-dependent antioxidant function. Redox Biol. 38:1018012021.
View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Wu Y, Yang X, Chen Z, Tian L, Jiang G,
Chen F, Li J, An P, Lu L, Luo N, et al: m6A-induced
lncRNA RP11 triggers the dissemination of colorectal cancer cells
via upregulation of Zeb1. Mol Cancer. 18:872019. View Article : Google Scholar : PubMed/NCBI
|