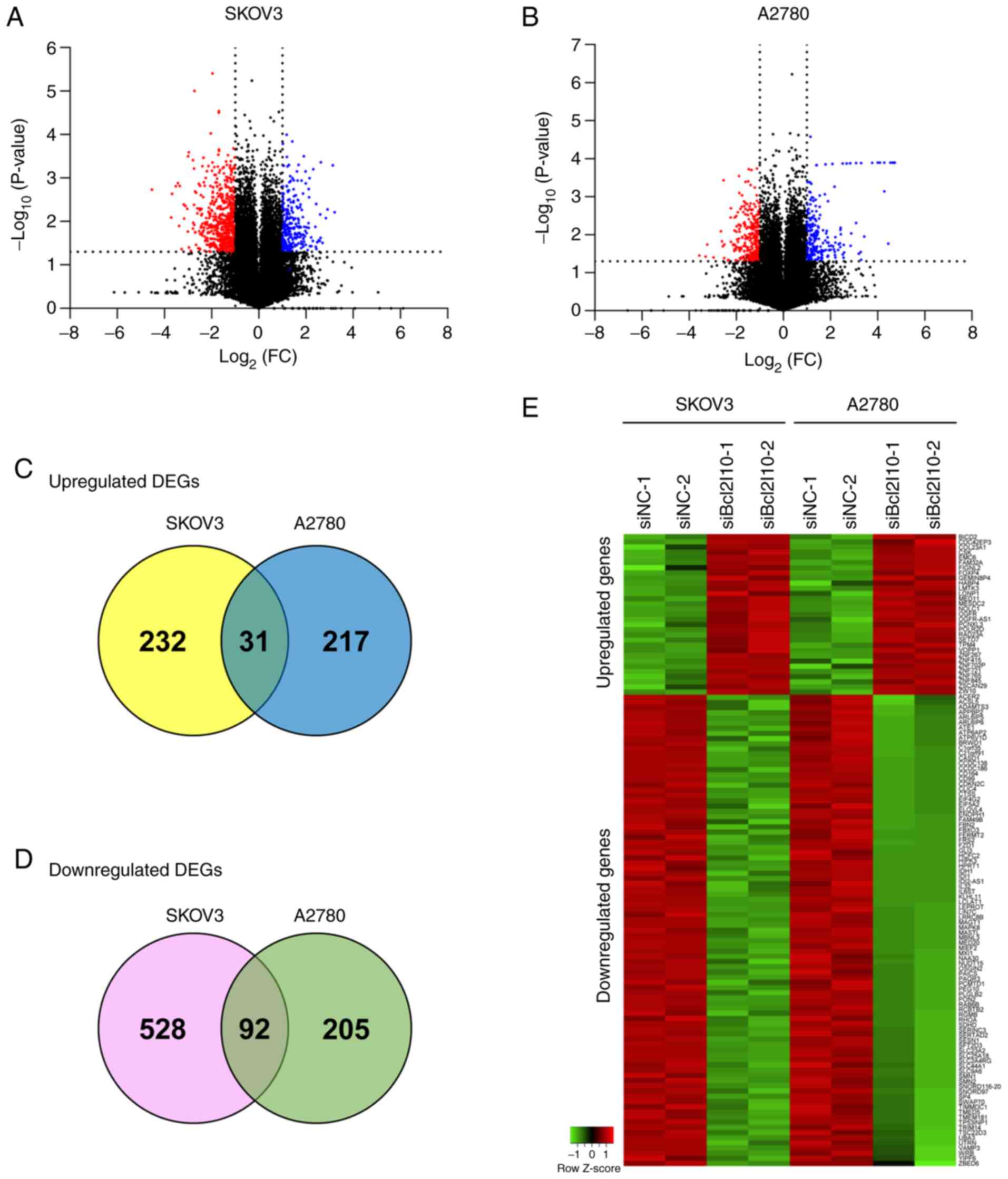
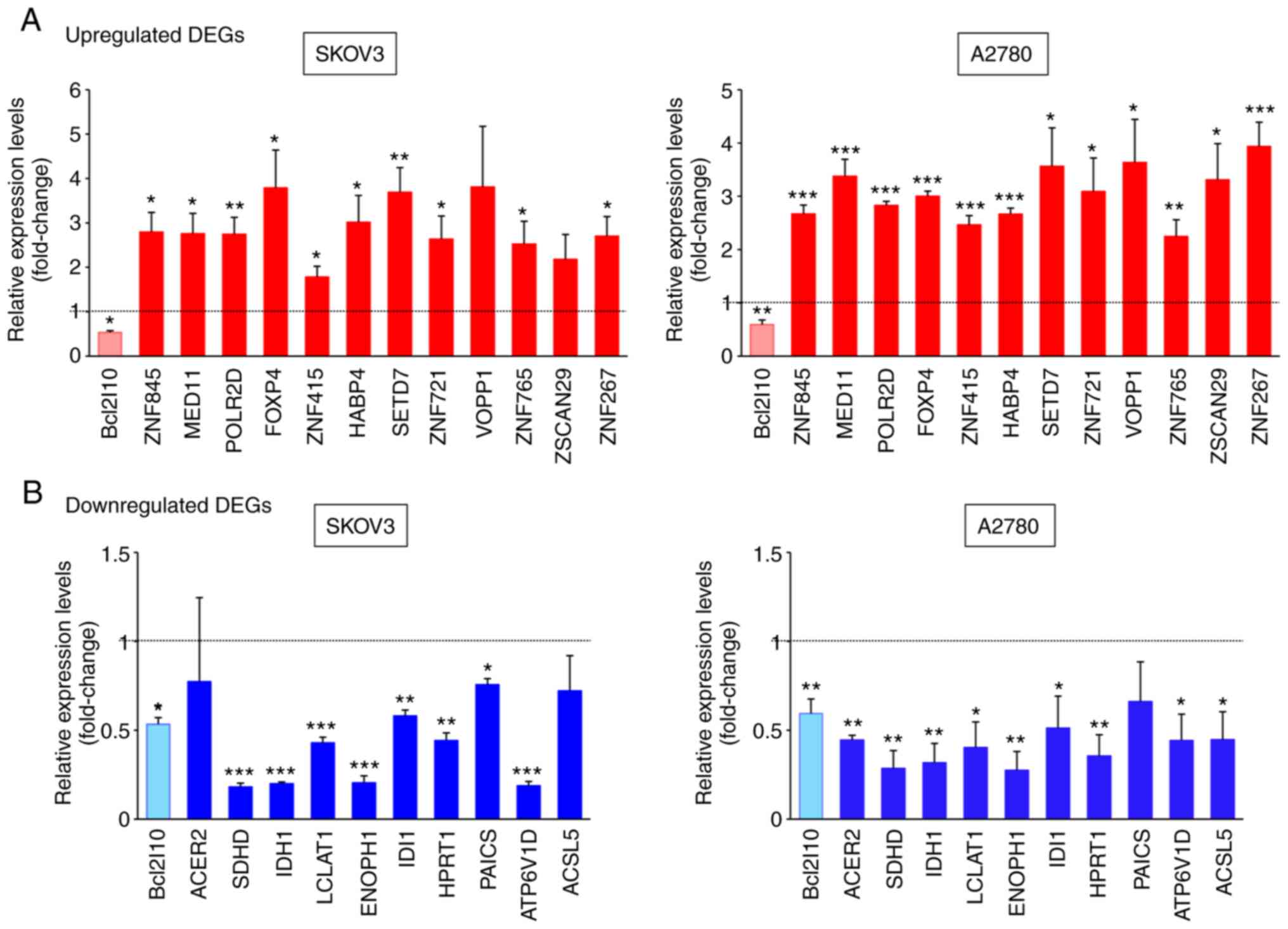
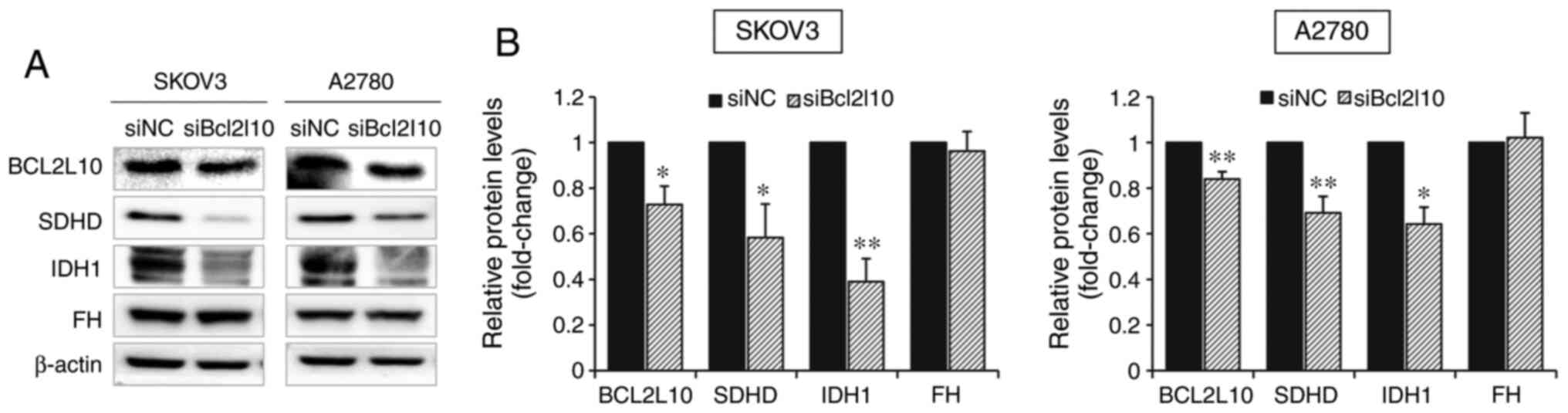
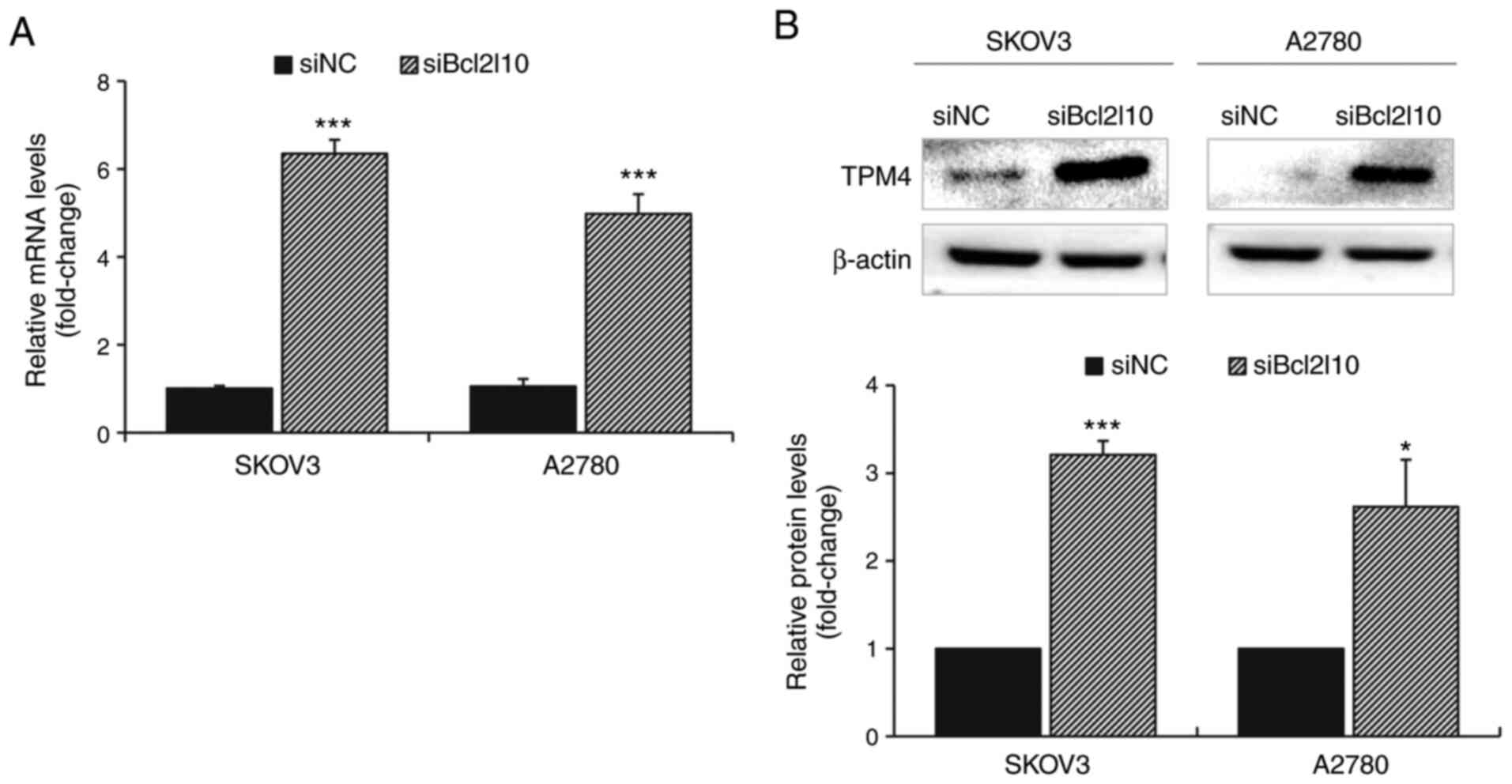
|
1
|
Siegel RL, Miller KD, Fuchs HE and Jemal
A: Cancer Statistics, 2021. CA Cancer J Clin. 71:7–33. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Markowska A, Sajdak S, Markowska J and
Huczynski A: Angiogenesis and cancer stem cells: New perspectives
on therapy of ovarian cancer. Eur J Med Chem. 142:87–94. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Boussios S, Abson C, Moschetta M, Rassy E,
Karathanasi A, Bhat T, Ghumman F, Sheriff M and Pavlidis N: Poly
(ADP-Ribose) polymerase inhibitors: Talazoparib in ovarian cancer
and beyond. Drugs R D. 20:55–73. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Levine B, Sinha SC and Kroemer G: Bcl-2
family members: Dual regulators of apoptosis and autophagy.
Autophagy. 4:600–606. 2008. View Article : Google Scholar
|
|
7
|
Inohara N, Gourley TS, Carrio R, Muñiz M,
Merino J, Garcia I, Koseki T, Hu Y, Chen S and Núñez G: Diva, a
Bcl-2 homologue that binds directly to Apaf-1 and induces
BH3-independent cell death. J Biol Chem. 273:32479–32486. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kang Y, Lee DC, Han J, Yoon S, Won M, Yeom
JH, Seong MJ, Ko JJ, Lee KA, Lee K and Bae J: NM23-H2 involves in
negative regulation of Diva and Bcl2L10 in apoptosis signaling.
Biochem Biophys Res Commun. 359:76–82. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Naumann U, Weit S, Wischhusen J and Weller
M: Diva/Boo is a negative regulator of cell death in human glioma
cells. FEBS Lett. 505:23–26. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang H, Holzgreve W and De Geyter C:
Bcl2-L-10, a novel anti-apoptotic member of the Bcl-2 family,
blocks apoptosis in the mitochondria death pathway but not in the
death receptor pathway. Hum Mol Genet. 10:2329–2339. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cluzeau T, Robert G, Mounier N, Karsenti
JM, Dufies M, Puissant A, Jacquel A, Renneville A, Preudhomme C,
Cassuto JP, et al: BCL2L10 is a predictive factor for resistance to
azacitidine in MDS and AML patients. Oncotarget. 3:490–501. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Nougarede A, Popgeorgiev N, Kassem L,
Omarjee S, Borel S, Mikaelian I, Lopez J, Gadet R, Marcillat O,
Treilleux I, et al: Breast cancer targeting through inhibition of
the endoplasmic reticulum-based apoptosis regulator Nrh/BCL2L10.
Cancer Res. 78:1404–1417. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bai Y, Wang J, Han J, Xie XL, Ji CG, Yin
J, Chen L, Wang CK, Jiang XY, Qi W and Jiang HQ: BCL2L10 inhibits
growth and metastasis of hepatocellular carcinoma both in vitro and
in vivo. Mol Carcinog. 56:1137–1149. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Xu JD, Furuya T, Cao XX, Liu XL, Li QQ,
Wang WJ, Xu JW, Xu ZD, Sasaki K and Liu XP: Loss of BCL2L10 protein
expression as prognostic predictor for poor clinical outcome in
gastric carcinoma. Histopathology. 57:814–824. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lee SY, Kim EY, Kim KH and Lee KA:
Bcl2l10, a new Tpx2 binding partner, is a master regulator of
Aurora kinase A in mouse oocytes. Cell Cycle. 15:3296–3305. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Damodaran AP, Vaufrey L, Gavard O and
Prigent C: Aurora a kinase is a priority pharmaceutical target for
the treatment of cancers. Trends Pharmacol Sci. 38:687–700. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yan M, Wang C, He B, Yang M, Tong M, Long
Z, Liu B, Peng F, Xu L, Zhang Y, et al: Aurora-a kinase: A potent
oncogene and target for cancer therapy. Med Res Rev. 36:1036–1079.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lee SY, Kwon J, Woo JH, Kim KH and Lee KA:
Bcl2l10 mediates the proliferation, invasion and migration of
ovarian cancer cells. Int J Oncol. 56:618–629. 2020.PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Trapnell C, Pachter L and Salzberg SL:
TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics.
25:1105–1111. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Quinlan AR and Hall IM: BEDTools: A
flexible suite of utilities for comparing genomic features.
Bioinformatics. 26:841–842. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Roberts A, Trapnell C, Donaghey J, Rinn JL
and Pachter L: Improving RNA-Seq expression estimates by correcting
for fragment bias. Genome Biol. 12:R222011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Laurenti G and Tennant DA: Isocitrate
dehydrogenase (IDH), succinate dehydrogenase (SDH), fumarate
hydratase (FH): Three players for one phenotype in cancer? Biochem
Soc Trans. 44:1111–1116. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bailey MJ, Shield-Artin KL, Oliva K, Ayhan
M, Reisman S and Rice GE: Stage-specific analysis of plasma protein
profiles in ovarian cancer: Difference in-gel electrophoresis
analysis of pooled clinical samples. J Carcinog. 12:102013.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Mikata R, Fukai K, Imazeki F, Arai M,
Fujiwara K, Yonemitsu Y, Zhang K, Nabeya Y, Ochiai T and Yokosuka
O: BCL2L10 is frequently silenced by promoter hypermethylation in
gastric cancer. Oncol Rep. 23:1701–1708. 2010.PubMed/NCBI
|
|
26
|
Xu JD, Cao XX, Long ZW, Liu XP, Furuya T,
Xu JW, Liu XL, De Xu Z, Sasaki K and Li QQ: BCL2L10 protein
regulates apoptosis/proliferation through differential pathways in
gastric cancer cells. J Pathol. 223:400–409. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Jen J and Wang YC: Zinc finger proteins in
cancer progression. J Biomed Sci. 23:532016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Cheng Y, Liang P, Geng H, Wang Z, Li L,
Cheng SH, Ying J, Su X, Ng KM, Ng MH, et al: A novel 19q13
nucleolar zinc finger protein suppresses tumor cell growth through
inhibiting ribosome biogenesis and inducing apoptosis but is
frequently silenced in multiple carcinomas. Mol Cancer Res.
10:925–936. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Harper J, Yan L, Loureiro RM, Wu I, Fang
J, D'Amore PA and Moses MA: Repression of vascular endothelial
growth factor expression by the zinc finger transcription factor
ZNF24. Cancer Res. 67:8736–8741. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hu R, Peng G, Dai H, Breuer EK,
Stemke-Hale K, Li K, Gonzalez-Angulo AM, Mills GB and Lin SY:
ZNF668 functions as a tumor suppressor by regulating p53 stability
and function in breast cancer. Cancer Res. 71:6524–6534. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Jen J, Lin LL, Chen HT, Liao SY, Lo FY,
Tang YA, Su WC, Salgia R, Hsu CL, Huang HC, et al: Oncoprotein
ZNF322A transcriptionally deregulates alpha-adducin, cyclin D1 and
p53 to promote tumor growth and metastasis in lung cancer.
Oncogene. 35:2357–2369. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Serra RW, Fang M, Park SM, Hutchinson L
and Green MR: A KRAS-directed transcriptional silencing pathway
that mediates the CpG island methylator phenotype. Elife.
3:e023132014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yang L, Zhang L, Wu Q and Boyd DD:
Unbiased screening for transcriptional targets of ZKSCAN3
identifies integrin beta 4 and vascular endothelial growth factor
as downstream targets. J Biol Chem. 283:35295–35304. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Bang HS, Choi MH, Kim CS and Choi SJ: Gene
expression profiling in undifferentiated thyroid carcinoma induced
by high-dose radiation. J Radiat Res. 57:238–249. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Omura N, Li CP, Li A, Hong SM, Walter K,
Jimeno A, Hidalgo M and Goggins M: Genome-wide profiling of
methylated promoters in pancreatic adenocarcinoma. Cancer Biol
Ther. 7:1146–1156. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Durinck S, Stawiski EW, Pavia-Jimenez A,
Modrusan Z, Kapur P, Jaiswal BS, Zhang N, Toffessi-Tcheuyap V,
Nguyen TT, Pahuja KB, et al: Spectrum of diverse genomic
alterations define non-clear cell renal carcinoma subtypes. Nat
Genet. 47:13–21. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Liu H, Li F, Zhu Y, Li T, Huang H, Lin T,
Hu Y, Qi X, Yu J and Li G: Whole-exome sequencing to identify
somatic mutations in peritoneal metastatic gastric adenocarcinoma:
A preliminary study. Oncotarget. 7:43894–43906. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Schnabl B, Hu K, Muhlbauer M, Hellerbrand
C, Stefanovic B, Brenner DA and Schölmerich J: Zinc finger protein
267 is up-regulated during the activation process of human hepatic
stellate cells and functions as a negative transcriptional
regulator of MMP-10. Biochem Biophys Res Commun. 335:87–96. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Schnabl B, Valletta D, Kirovski G and
Hellerbrand C: Zinc finger protein 267 is up-regulated in
hepatocellular carcinoma and promotes tumor cell proliferation and
migration. Exp Mol Pathol. 91:695–701. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Baras A and Moskaluk CA: Intracellular
localization of GASP/ECOP/VOPP1. J Mol Histol. 41:153–164. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Baras A, Yu Y, Filtz M, Kim B and Moskaluk
CA: Combined genomic and gene expression microarray profiling
identifies ECOP as an upregulated gene in squamous cell carcinomas
independent of DNA amplification. Oncogene. 28:2919–2924. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Gao C, Pang M, Zhou Z, Long S, Dong D,
Yang J, Cao M, Zhang C, Han S and Li L: Epidermal growth factor
receptor-coamplified and overexpressed protein (VOPP1) is a
putative oncogene in gastric cancer. Clin Exp Med. 15:469–475.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Baras AS, Solomon A, Davidson R and
Moskaluk CA: Loss of VOPP1 overexpression in squamous carcinoma
cells induces apoptosis through oxidative cellular injury. Lab
Invest. 91:1170–1180. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Wang H, Cao R, Xia L, Erdjument-Bromage H,
Borchers C, Tempst P and Zhang Y: Purification and functional
characterization of a histone H3-lysine 4-specific
methyltransferase. Mol Cell. 8:1207–1217. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Akiyama Y, Koda Y, Byeon SJ, Shimada S,
Nishikawaji T, Sakamoto A, Chen Y, Kojima K, Kawano T, Eishi Y, et
al: Reduced expression of SET7/9, a histone mono-methyltransferase,
is associated with gastric cancer progression. Oncotarget.
7:3966–3983. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Chen Y, Yang S, Hu J, Yu C, He M and Cai
Z: Increased expression of SETD7 promotes cell proliferation by
regulating cell cycle and indicates poor prognosis in
hepatocellular carcinoma. PLoS One. 11:e01549392016. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Manstein DJ and Mulvihill DP:
Tropomyosin-mediated regulation of cytoplasmic myosins. Traffic.
17:872–877. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Gunning PW, Hardeman EC, Lappalainen P and
Mulvihill DP: Tropomyosin-master regulator of actin filament
function in the cytoskeleton. J Cell Sci. 128:2965–2974. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Kabbage M, Trimeche M, Ben Nasr H, Hammann
P, Kuhn L, Hamrita B and Chahed K: Tropomyosin-4 correlates with
higher SBR grades and tubular differentiation in infiltrating
ductal breast carcinomas: An immunohistochemical and
proteomics-based study. Tumour Biol. 34:3593–3602. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Tang HY, Beer LA, Tanyi JL, Zhang R, Liu Q
and Speicher DW: Protein isoform-specific validation defines
multiple chloride intracellular channel and tropomyosin isoforms as
serological biomarkers of ovarian cancer. J Proteomics. 89:165–178.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Zhao X, Jiang M and Wang Z: TPM4 promotes
cell migration by modulating F-actin formation in lung cancer. Onco
Targets Ther. 12:4055–4063. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Lomnytska MI, Becker S, Bodin I, Olsson A,
Hellman K, Hellström AC, Mints M, Hellman U, Auer G and Andersson
S: Differential expression of ANXA6, HSP27, PRDX2, NCF2, and TPM4
during uterine cervix carcinogenesis: Diagnostic and prognostic
value. Br J Cancer. 104:110–119. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Yang R, Zheng G, Ren D, Chen C, Zeng C, Lu
W and Li H: The clinical significance and biological function of
tropomyosin 4 in colon cancer. Biomed Pharmacother. 101:1–7. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Li DQ, Wang L, Fei F, Hou YF, Luo JM,
Wei-Chen, Zeng R, Wu J, Lu JS, Di GH, et al: Identification of
breast cancer metastasis-associated proteins in an isogenic tumor
metastasis model using two-dimensional gel electrophoresis and
liquid chromatography-ion trap-mass spectrometry. Proteomics.
6:3352–3368. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Yoon SJ, Kim JW, Choi KH, Lee SH and Lee
KA: Identification of oocyte-specific diva-associated proteins
using mass spectrometry. Korean J Fertil Steril. 33:189–198.
2006.
|
|
56
|
Yamaguchi H and Condeelis J: Regulation of
the actin cytoskeleton in cancer cell migration and invasion.
Biochim Biophys Acta. 1773:642–652. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Gimenez-Cassina A and Danial NN:
Regulation of mitochondrial nutrient and energy metabolism by BCL-2
family proteins. Trends Endocrinol Metab. 26:165–175. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Gimenez-Cassina A, Martinez-Francois JR,
Fisher JK, Szlyk B, Polak K, Wiwczar J, Tanner GR, Lutas A, Yellen
G and Danial NN: BAD-dependent regulation of fuel metabolism and
K(ATP) channel activity confers resistance to epileptic seizures.
Neuron. 74:719–730. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Perciavalle RM, Stewart DP, Koss B, Lynch
J, Milasta S, Bathina M, Temirov J, Cleland MM, Pelletier S,
Schuetz JD, et al: Anti-apoptotic MCL-1 localizes to the
mitochondrial matrix and couples mitochondrial fusion to
respiration. Nat Cell Biol. 14:575–583. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Malia TJ and Wagner G: NMR structural
investigation of the mitochondrial outer membrane protein VDAC and
its interaction with antiapoptotic Bcl-xL. Biochemistry.
46:514–525. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Wu W and Zhao S: Metabolic changes in
cancer: Beyond the Warburg effect. Acta Biochim Biophys Sin
(Shanghai). 45:18–26. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Huang S and Millar AH: Succinate
dehydrogenase: The complex roles of a simple enzyme. Curr Opin
Plant Biol. 16:344–349. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Ashtekar A, Huk D, Magner A, La Perle K,
Zhang X, Piruat JI, López-Barneo J, Jhiang SM and Kirschner LS:
Sdhd ablation promotes thyroid tumorigenesis by inducing a
stem-like phenotype. Endocr Relat Cancer. 24:579–591. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Mu X, Zhao T, Xu C, Shi W, Geng B, Shen J,
Zhang C, Pan J, Yang J, Hu S, et al: Oncometabolite succinate
promotes angiogenesis by upregulating VEGF expression through
GPR91-mediated STAT3 and ERK activation. Oncotarget. 8:13174–13185.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Ricketts C, Woodward ER, Killick P, Morris
MR, Astuti D, Latif F and Maher ER: Germline SDHB mutations and
familial renal cell carcinoma. J Natl Cancer Inst. 100:1260–1262.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Aspuria PP, Lunt SY, Varemo L, Vergnes L,
Gozo M, Beach JA, Salumbides B, Reue K, Wiedemeyer WR, Nielsen J,
et al: Succinate dehydrogenase inhibition leads to
epithelial-mesenchymal transition and reprogrammed carbon
metabolism. Cancer Metab. 2:212014. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Zhang T, Wu X, Ke C, Yin M, Li Z, Fan L,
Zhang W, Zhang H, Zhao F, Zhou X, et al: Identification of
potential biomarkers for ovarian cancer by urinary metabolomic
profiling. J Proteome Res. 12:505–512. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Reitman ZJ and Yan H: Isocitrate
dehydrogenase 1 and 2 mutations in cancer: Alterations at a
crossroads of cellular metabolism. J Natl Cancer Inst. 102:932–941.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Turcan S, Rohle D, Goenka A, Walsh LA,
Fang F, Yilmaz E, Campos C, Fabius AW, Lu C, Ward PS, et al: IDH1
mutation is sufficient to establish the glioma hypermethylator
phenotype. Nature. 483:479–483. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Raynaud S, Carbuccia N, Colin C, Adélaïde
J, Mozziconacci MJ, Metellus P, Chinot O, Birnbaum D, Chaffanet M
and Figarella-Branger D: Absence of R140Q mutation of isocitrate
dehydrogenase 2 in gliomas and breast cancers. Oncol Lett.
1:883–884. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Rakheja D, Konoplev S, Medeiros LJ and
Chen W: IDH mutations in acute myeloid leukemia. Hum Pathol.
43:1541–1551. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Bleeker FE, Lamba S, Leenstra S, Troost D,
Hulsebos T, Vandertop WP, Frattini M, Molinari F, Knowles M,
Cerrato A, et al: IDH1 mutations at residue p.R132 (IDH1(R132))
occur frequently in high-grade gliomas but not in other solid
tumors. Hum Mutat. 30:7–11. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Dang L, White DW, Gross S, Bennett BD,
Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, et
al: Cancer-associated IDH1 mutations produce 2-hydroxyglutarate.
Nature. 462:739–744. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Prasad B, Tian Y and Li X: Large-Scale
analysis reveals gene signature for survival prediction in primary
glioblastoma. Mol Neurobiol. 57:5235–5246. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Calvert AE, Chalastanis A, Wu Y, Hurley
LA, Kouri FM, Bi Y, Kachman M, May JL, Bartom E, Hua Y, et al:
Cancer-associated IDH1 promotes growth and resistance to targeted
therapies in the absence of mutation. Cell Rep. 19:1858–1873. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Wahl DR, Dresser J, Wilder-Romans K,
Parsels JD, Zhao SG, Davis M, Zhao L, Kachman M, Wernisch S, Burant
CF, et al: Glioblastoma therapy can be augmented by targeting
IDH1-mediated NADPH biosynthesis. Cancer Res. 77:960–970. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Liu WS, Chan SH, Chang HT, Li GC, Tu YT,
Tseng HH, Fu TY, Chang HY, Liou HH, Ger LP and Tsai KW: Isocitrate
dehydrogenase 1-snail axis dysfunction significantly correlates
with breast cancer prognosis and regulates cell invasion ability.
Breast Cancer Res. 20:252018. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Selak MA, Armour SM, MacKenzie ED,
Boulahbel H, Watson DG, Mansfield KD, Pan Y, Simon MC, Thompson CB
and Gottlieb E: Succinate links TCA cycle dysfunction to
oncogenesis by inhibiting HIF-alpha prolyl hydroxylase. Cancer
Cell. 7:77–85. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Warburg O: On the origin of cancer cells.
Science. 123:309–314. 1956. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Warburg O: On respiratory impairment in
cancer cells. Science. 124:269–270. 1956.PubMed/NCBI
|
|
82
|
Barbosa IA, Machado NG, Skildum AJ, Scott
PM and Oliveira PJ: Mitochondrial remodeling in cancer metabolism
and survival: Potential for new therapies. Biochim Biophys Acta.
1826:238–254. 2012.PubMed/NCBI
|
|
83
|
Moreno-Sanchez R, Rodriguez-Enriquez S,
Marin-Hernandez A and Saavedra E: Energy metabolism in tumor cells.
FEBS J. 274:1393–1418. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Koppenol WH, Bounds PL and Dang CV: Otto
Warburg's contributions to current concepts of cancer metabolism.
Nat Rev Cancer. 11:325–337. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Phan LM, Yeung SC and Lee MH: Cancer
metabolic reprogramming: Importance, main features, and potentials
for precise targeted anti-cancer therapies. Cancer Biol Med.
11:1–19. 2014.PubMed/NCBI
|
|
86
|
Schwartzenberg-Bar-Yoseph F, Armoni M and
Karnieli E: The tumor suppressor p53 down-regulates glucose
transporters GLUT1 and GLUT4 gene expression. Cancer Res.
64:2627–2633. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Marbaniang C and Kma L: Dysregulation of
glucose metabolism by oncogenes and tumor suppressors in cancer
cells. Asian Pac J Cancer Prev. 19:2377–2390. 2018.PubMed/NCBI
|