Introduction
Antibody-drug conjugates (ADCs) combine the binding
specificity of a monoclonal antibody with the toxic effects of a
potent payload (1). The prototypal
ADC trastuzumab-emtansine (T-DM1) is now the standard of care in
advanced and adjuvant settings for breast cancer of the aggressive
ERBB2 subtype (2,3). T-DM1 overcomes pharmacological
resistance to trastuzumab (its unconjugated counterpart) and/or
pertuzumab (another ERBB2-specific antibody) and/or small molecules
(2). Moreover, it achieves better
efficacy at lower dosages than naked antibodies, and permits
reduction in the intensity of associated chemotherapy, drastically
improving toxicity profiles altogether (1).
However, like any other ADC and anticancer drug,
T-DM1 almost invariably induces pharmacological resistance by
adaptive selection through: (a) tumor antigen/epitope loss, and (b)
payload refractoriness (4). In
principle, (a) and (b) could be effectively tackled by preparing as
many ADCs as the number of possible combinations and permutations
of all the actionable cancer antigens and known active drugs for a
given tumor type. Then, these ADCs could be used in combination
and/or in sequence. However, this combinatorial approach is neither
conceivable nor applicable in practice, and for many reasons. Each
ADC is a unique product resulting from a dedicated and industrially
demanding manufacturing process, whereby a specific immunoglobulin
(Ig) is covalently attached (through a carefully designed chemical
linker) to a selected cytotoxic agent (e.g. a microtubule inhibitor
such as a maytansinoid) in defined stoichiometries (1). Therefore, an exponential expansion of
the ADC arsenal would result in hyperbolic costs, issues in
clinical trial design and regulatory clearance, and a likely
biomanufacturing crunch.
To tackle some of these challenges, we took
advantage of the degree of freedom that is only possible when
designing objects on the nanoscale, and combined several useful
drugging tools into a modular ‘box’ that, accordingly, was named
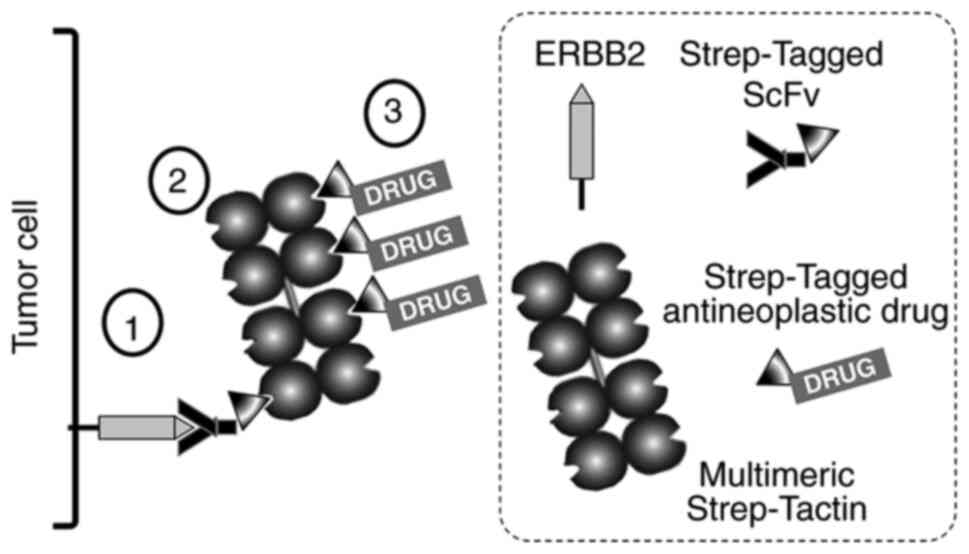
‘TOOLBOX’. TOOLBOX (Fig. 1) is a
multi-step platform comprising an affinity reagent of choice, that
in the selected example is a single chain fragment of variable
antibody region (ScFv) to the proto-oncogene product ERBB2, and a
small anticancer drug that per se has no specificity for
cancer cells. Both the ScFv and the drug are modified through the
addition (by genetic engineering and chemical synthesis,
respectively) of a so-called Strep-Tag, e.g. an 8-amino acid
WSHPQFEK moiety. The two Strep-Tagged reagents may then be bridged
by a universal multimeric adaptor protein (Strep-Tactin) that
mediates the selective deposition of a cytotoxic nano-complex onto
the surface of cancer cells. Since the affinity reagent and the
drug are non-covalently linked, the two can be variably combined to
generate a variety of tumor drugging systems.
This report describes the step-wise optimization of
one such TOOLBOX drugging systems, and its use to redirect two
different drugs onto ERBB2-high (susceptible to ERBB2 blockade) and
ERBB2-low (putatively insensitive/resistant) human breast cancer
cells, the latter grown as mouse tumor xenografts.
Materials and methods
For details and full descriptions of the Materials
and methods please refer to Data S1.
Cell lines
SK-BR-3 is a certified cell line obtained from the
American Type Culture Collection (ATCC). BRC230 cells (5) belong to a triple-negative breast cancer
(TNBC) subtype expressing low ERBB2 levels (6,7), and were
obtained from the originators. Cell lines were identity verified by
human leukocyte antigen (HLA) typing, as described (8), and were routinely assessed for
mycoplasma.
ScFv W6/800, antibodies and
reagents
The murine monoclonal antibody (mAb) W6/800 to ERBB2
and the corresponding ScFv are described (9,10).
Trastuzumab and oertuzumab (Herceptin® and
Perjeta® were generous gifts of Roche-Genentech. mAb 108
to the epidermal growth factor receptor (EGFR) was obtained from
the ATCC hybridoma HB-9764. Recombinant human epidermal growth
factor (EGF) was obtained from ImmunoTools GmbH. Human
NRG1-β1/HRG1-β1 EGF domain was obtained from R&D Systems, Inc.
PD168393 (Calbiochem/Merck) is an irreversible EGFR inhibitor.
Recombinant DNAs, Strep-Tag technology
and ScFv expression
All the tagging platforms and reagents are
commercially available from IBA Lifesciences, except those that
were developed for the purpose for this study, as noted.
Standard TOOLBOX flow cytometry
protocol
The standard 3-step TOOLBOX protocol involves
successive incubations with: i) tagged ScFv; ii) either
phycoerythrin-conjugated Strep-Tactin (Strept-Tactin-PE) or its
Strept-Tactin-Mult-PE variant; and iii) Strep-Tagged green
fluorescent protein (One-Strep-GFP). In the two-step protocol, the
concentrations of the reagents were identical, but Strep-Tactin-PE
and One-Strep-GFP were admixed. Two-color flow cytometry was
carried out in a FACScan (BD Biosciences). For further details see
Data S1.
Strep-Tagged drugs
The organic synthesis of DNA nanobinders (NAX
compounds) and their tagged derivatives (NAXT) is described
(11). For tagging, maleimide
derivatives of NAX compounds were mixed in equimolar amounts with
the WSHPQFEK peptide in the presence of dimethylformamide (DMF) at
room temperature for 4 h. DMF was vacuum-evaporated and the tagged
compound (typically about 50% of the total mix) was purified by
HPLC. Molecular weight was confirmed by MALDI-MS. Emtansine (DM1;
Abcam) was Strep-Tagged (DM1T) by adding an
N-(ε-maleimidocaproyloxy)succinimide ester (EMCS; MW 308.29) linker
(Thermo Fisher Scientific, Inc.) that introduces a 9.4 Å spacer
arm. Design and synthesis of NAXT and DM1T were carried out by Dr
Anette Jacob at Peps 4 Life Sciences (Peps4LS), Heidelberg,
Germany.
TOOLBOX treatments in vitro
The antiproliferative activity of NAX compounds was
tested by incubating SK-BR-3 cells for 45 min at 4°C in 96-well
plates with the indicated concentrations of the TOOLBOX components
in 100 µl of complete medium. The plates were then moved to a
CO2 incubator and grown for 72 h.
3[H]-Thymidine incorporation was measured (triplicates)
during the last 4 h of growth. The activity of DM1T was assessed by
assessing light emission of an ERBB2 pathway-dependent c-fos
promoter (12)/NanoLuc®
(Promega Corp.) luciferase reporter assay. The construct (details
in Data S1) was stably transfected into SK-BR-3 and BRC230 breast
carcinoma cells. Cells were pre-treated with the TOOLBOX components
for 45 min at 4°C, then grown for 72 h, lysed in the presence of
the furimazine substrate, and assessed for bio-luminescence in a
luminometer (13).
Animal experiments
nu/CD1 mice bearing BRC230-cfos xenotransplants were
treated by tail vein (i.v.) injection of either ScFv or ScFV-mut4
(10 mg/kg), followed 1 h later by a pre-mix of Strep-Tactin-Mult
(7.5 mg) and DM1T (2 mg/kg), as approved (prot. 665/2017-PR) by the
Animal Welfare Section of the Italian Ministry of Health. This
study was compliant with the 2010/63/EU directive. At selected time
points, the substrate furimazine was i.v. injected under mild
anaesthesia, as previously described (14), and light emitted by the NanoLuc
reporter was imaged in a IVIS Lumina (Perkin Elmer). Additional
details on anaesthesia and sacrifice may be found in Data S1.
Statistical analysis
GraphPAD Prism v.8.3 (GraphPad Software, Inc.) was
used for statistical elaboration.
Results
Prior to attempting full nano-assembly, the TOOLBOX
building blocks and protocols were optimized.
TOOLBOX: Optimizing step 1 (tagged
ScFv)
ScFv W6/800 to ERBB2 was selected for TOOLBOX since
its antigen binding site is remarkably resilient to reduction and
denaturation (10). Following removal
of the leftovers of previous DNA manipulations, a clean 284 amino
acid ScFv was obtained exclusively comprising murine Variable Heavy
and Variable Light chain Ig sequences connected by a glycine-serine
(G4S)3 linker. Following in silico
codon optimization for mammalian expression, this DNA was
whole-gene synthesized, cloned into the pESG142 and pESG144
double-tagging vectors (IBA Lifesciences), and expressed in a
secretable form in CHO transfectants. The two resulting variants
(ScFv142 and ScFv144) carry a Strep-Tag and a 6X His-tag arranged
in the two possible orientations at the N- and C-termini of an
otherwise identical ScFv backbone (Fig.
2A, left). ScFvs carrying mutated complementarity determining
regions (CDR) (Fig. 2A, right) were
also cloned in the pESG vectors.
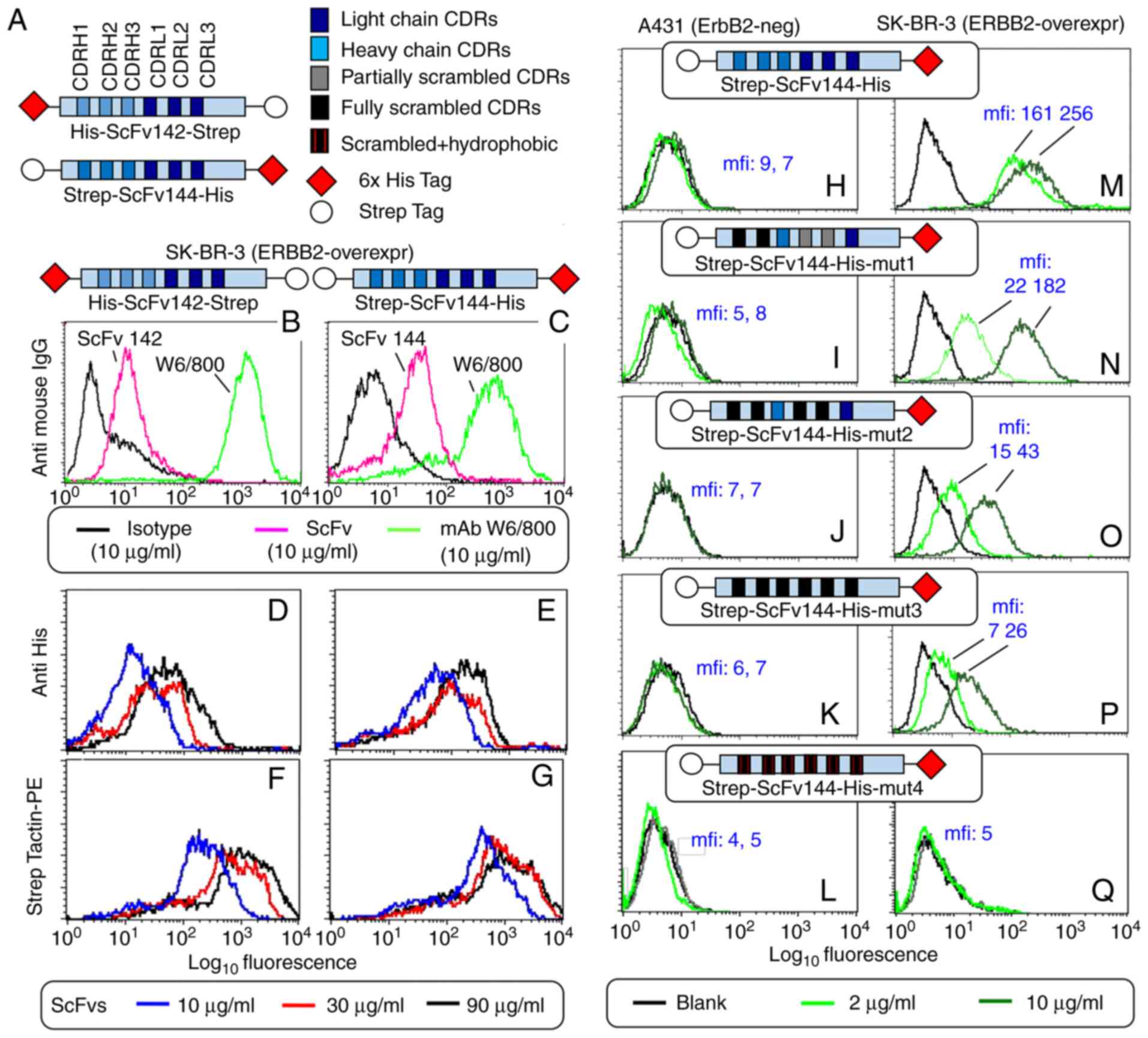
Wild-type (WT) ScFv142 and ScFv144 were found to
bind to ERBB2-overexpressing SK-BR-3 breast carcinoma cells
similarly (Fig. 2B and C, magenta),
but more weakly than the parental whole mAb W6/800 (green). This
was expected, since previous Scatchard plot experiments
demonstrated that monovalent binding causes a 4.5-fold drop in the
ScFv equilibrium binding constant as compared to the whole murine
antibody (10). Moreover, Fc epitopes
(recognized by the FITC-labelled secondary antibody) are available
on whole Igs but not the ScFvs. However, and remarkably, low ScFv
binding could be compensated using either anti-tag reagents such as
an FITC-labelled anti His-tag antibody (D and E), or a
Phycoerythrin (PE) conjugate of a Streptavidin derivative named
Strep-Tactin-PE (F and G). Both secondary reagents were able to
drive onto breast cancer cells a substantial amount of
fluorescence. Interestingly, although tags were accessible at both
ends of ScFv142 and ScFv144, binding was stronger and less affected
by ScFv dilution when the His-tag was mounted at the C terminus
(compare E to D), and the Strep-Tag was mounted at the N-terminus
(compare G to F), as in ScFv144. ScFv 144 performed better than
ScFv142 (F/G vs. D/E) and was therefore selected for further
studies.
Five ScFv144 variants (WT and 4 CDR mutants) were
generated and tested in parallel by flow cytometry on ERBB2-high
SK-BR-3 and ERBB2-negative A431 cells, using Strep-Tactin-PE as the
secondary reagent (Fig. 2H-Q). As
expected, A431 cells were unreactive (H-L), whereas parental
ScFv144 displayed the strongest SK-BR-3 binding (M). Binding was
gradually lost upon introduction of progressively more disabling
CDR mutations (N, O and P, and see graphical ScFv representation on
top of each panel and CDRs in panel A). However, consistent with a
resilient antigen binding site, both CDR scrambling (exchange in
amino acid order) and non-conservative substitutions at many
positions were necessary for complete inactivation (ScFv144-mut4;
Q; also see Table SI). In summary,
Strep-tagged ScFv144 (hitherto ScFv) was identified as the affinity
reagent of choice, and ScFv144-mut4 was set aside as a stringent
negative control.
TOOLBOX: Optimizing steps 3 and 4
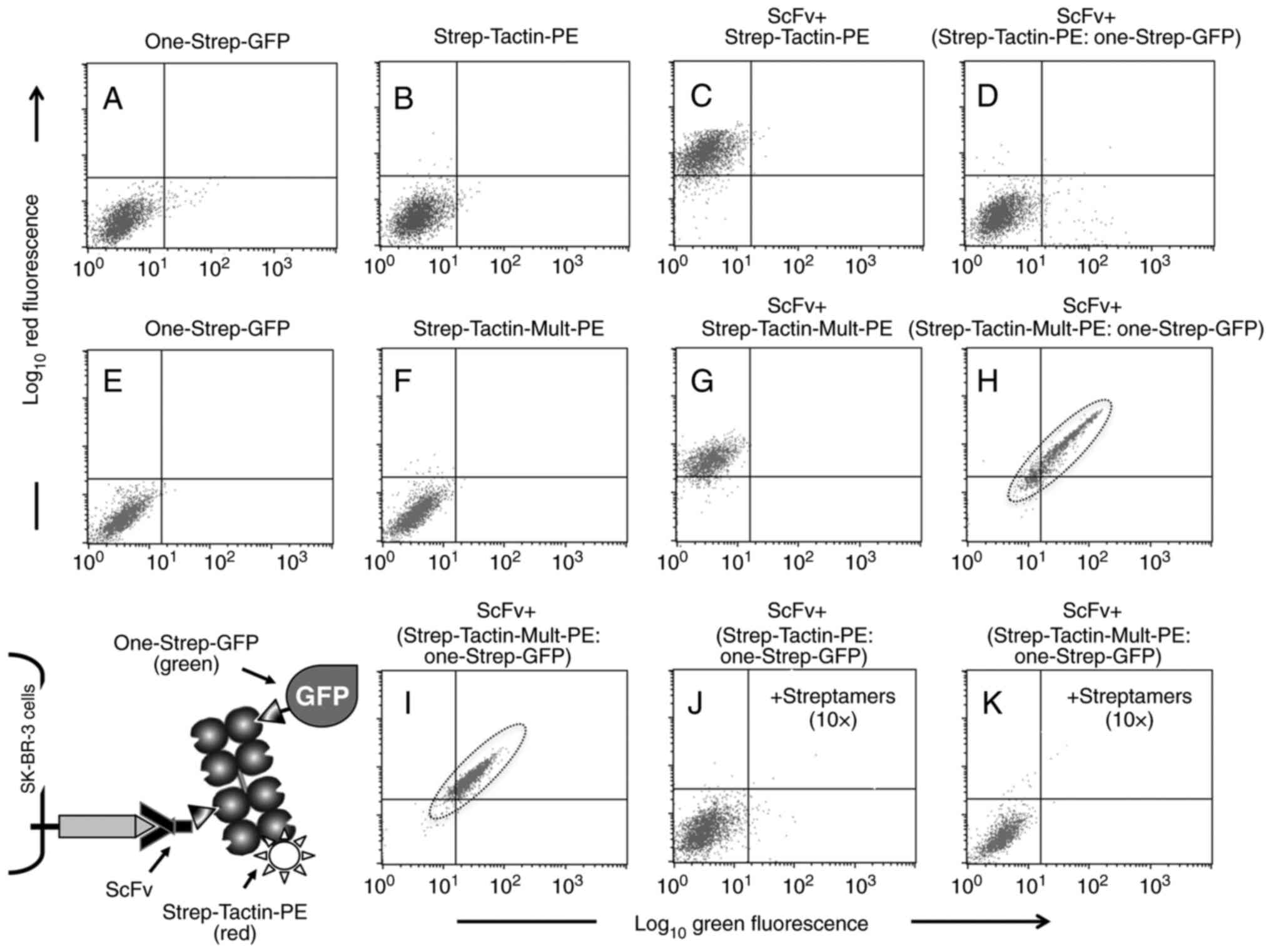
Two-color (red/green) flow cytometry was used to
individually detect cell surface binding of the TOOLBOX components,
and optimize their stoichiometry. As shown by the diagram in the
lower left of Fig. 3, Strep-Tactin-PE
was used to monitor TOOLBOX step 2 (adapter engagement) and,
indirectly, step 1 (ScFv binding). A Strep-Tagged variant of the
green fluorescent protein (GFP) called One-Strep-GFP was instead
used to monitor step 3, e.g. it was used as a convenient technical
surrogate of Strep-Tagged drugs. Two stepwise incubation formats
were tested, namely: a) ScFv + an optimal
Strep-Tactin:One-Strep-GFP pre-mix (2-step protocol); b) successive
addition of ScFv + Strep-Tactin + One-Strep-GFP (3-step protocol).
These protocols are described in detail in Data S1.
As shown by a representative 2-step experiment, we
consistently detected ScFv:Strep-Tactin-PE binding in the red
channel (Fig. 3C compared to A and
B). However, and paradoxically, One-Strep-GFP not only failed to
bind, but it did abrogate red fluorescence (Fig. 3D compared to C, and see A and B),
suggesting blocking of free Strep-Tactin valences. In the attempt
to identify a critical stoichiometric window compatible with double
Strep-Tactin engagement (with ScFv on the one hand and
One-Strep-GFP on the other), first we increased Strep-Tactin-PE
concentrations up to 8-fold, with no success (Fig. S1). Then, we diluted One-Strep-GFP,
but also in this case no green signal could be rescued in either
the 2-step protocol (Fig. S2A-D) or
the 3-step protocol (Fig. S2E-H).
However, and interestingly, a gradual rescue could be seen
exclusively in the red (Strep-Tactin-PE) channel, and was more
evident in the 3-step than the 2-step protocol (compare Fig. S2E-H to A-D), e.g. when One-Strep-GFP
was added after washing away unbound Strep-Tactin-PE. This strongly
indicated competition for a limiting number of binding sites of the
adapter, e.g. an intrinsic shortage in Strep-Tactin-PE valence
preventing the simultaneous engagement of ScFv and
One-Strep-GFP.
Since Strep-Tactin is routinely polymerized at a
predicted multiplicity of 4 (i.e. with 4 subunits carrying 4
nominal Strep-Tag binding sites), a novel Strep-Tactin was
oligomerized at a predicted multiplicity of 10 (Strep-Tact-Mult
hitherto). Strep-Tact-Mult displayed a slightly reduced ability to
detect ScFv binding (Fig. S3,
compare B and C), but as expected it enabled double staining,
particularly in the two-step protocol (Fig. 3H compared to E, F and G; and also
compared with D). Binding was specific because addition of a
non-fluorescent Strep-Tagged molecule (recombinant Streptamers, see
Materials and methods) in 10× excess totally abrogated fluorescence
in either or both the green and red channels (Fig. 3, compare J to C and K to I). Finally,
unconjugated Strep-Tactin-Mult, like Strep-Tactin-Mult-PE, was
found to also sustain multi-step TOOLBOX nano-complex formation
(Fig. S4, compare D to C). This
conclusively demonstrated specific interactions among the TOOLBOX
building blocks, and a minimum nominal Strep-Tactin valence
enabling TOOLBOX multimers. Then, we were ready to replace the
optimized amounts of One-Strep-GFP with equimolar amounts of tagged
drugs.
TOOLBOX assay development: Designing
and optimizing Strep-Tagged anticancer drugs
Toxic payload delivery onto breast cancer cells was
tested using the preferred 2-step protocol. Initially, DNA
nanobinders (berberine derivatives) were selected as the preferred
toxic payload since: a) they find application in many different
neoplasms including breast cancer (11); and b) in-house expertise was available
to design these chemical structures. Covalent modification with a
bulky Strep-Tag (WSHPQFEK) was expected to irreversibly inactivate
small drugs, including berberines. Then, we set out for a
systematic synthesis and screening study aimed at selecting the
rare berberine variants expected to retain their activity following
Strep-Tagging.
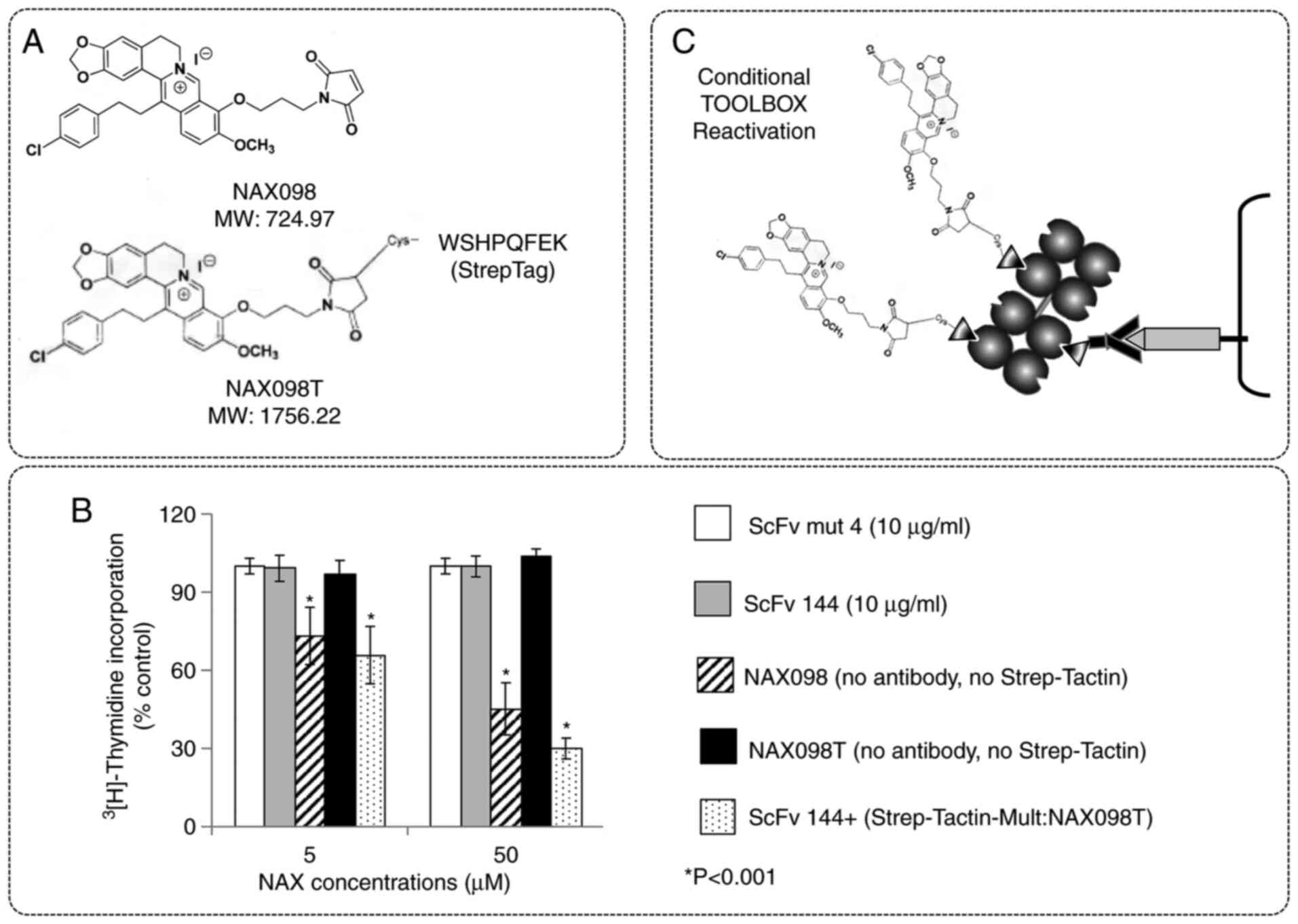
Ten pairs of NAX compounds and Strep-Tagged NAXT
counterparts were produced starting from the available (11) parent compounds. The organic synthesis
of 4 of these compounds (NAX063, NAX073, NAX098, and NAX110) is
outlined in Fig. S5. Their
Strep-Tagged counterparts (NAX063T, NAX073T, NAX098T, and NAX110T)
are displayed in Fig. S6. SK-BR-3
cells were treated for 72 h with two different concentrations of
the NAX/NAXT pairs at micromolar concentrations, in line with the
reported effective doses of these drugs (11), and then assessed for
3[H]-Thymidine incorporation. Fig. 4A displays the selected NAX098/NAX098T
pair, whereas results with all the 4 NAX compounds are shown and
described in Fig. S7. As shown in
Fig. 4B, NAX098 displayed the
anti-proliferative activity typical (11) of this class of nanobinders (compare
striped to white bars), but Strep-Tag addition resulted in complete
or nearly complete inactivation (compare black and white bars).
However, and unexpectedly, anti-proliferative activity was
surprisingly reinstated when NAX098T was administered by the
TOOLBOX protocol (stippled).
Thus, despite tagging turns berberins into inactive
(pro)drugs, the process is reversible, e.g. NAXT compounds may be
conditionally reactivated when they are incorporated into
Strep-Tactin nano-complexes, possibly because TOOLBOX funnels them
into an unknown cellular reactivation process. This makes the
original TOOLBOX concept (Fig. 4C)
feasible and (unexpectedly) even more appealing than
anticipated.
TOOLBOX refinement: The ERBB2-cfos
reporter, ERBB2-low cells and tagged emtansine
Next, to specifically assess the effects of TOOLBOX
treatment on ERBB2 signaling, we developed an ERBB2-dependent
genetic reporter system. In this construct, an improved luciferase
(NanoLuc®) was cloned under the control of the c-fos
promoter, a well-established downstream ERBB2 target (15). ERBB2 signaling was estimated by
measuring the light emitted following conversion of the substrate
furimazine. The construct was stably transfected not only in breast
cancer cells of the ERBB2-subtype (SK-BR-3-cfos), but also in TNBC
cells (BRC230-cfos) that do not host amplified ERBB2 (5). Low ERBB2 is a recognized feature of a
fraction of TNBCs and is held responsible, among other ill-defined
factors, for poor responsiveness of this molecular subtype to
therapeutic anti-ERBB2 antibodies (16). The two cell lines were found to differ
>10-fold in ERBB2 expression, as expected, but expressed similar
EGFR levels (Fig. S8). Accordingly,
ERBB2-low BRC230-cfos transfectants exhibited more limited (but
still detectable) response to the EGF, that mainly engages
EGFR:ERBB2 dimers, and signaling was counteracted by treatment with
the irreversible EGFR chemical inhibitor PD168393 (Fig. S9, compare A to B).
Having available an ERBB2-dependent promoter
reporter system in an ERBB2-low cell line, we were particularly
interested in assessing TOOLBOX performance under such demanding
conditions. Then, we switched to a more potent Strep-Tagged drug.
Emtansine (DM1) was selected, since this potent microtubule
inhibitor is a preferred ADC payload, as in the prototypic ADC
trastuzumab-emtansine (T-DM1). DM1 was attached to the Strep-Tag
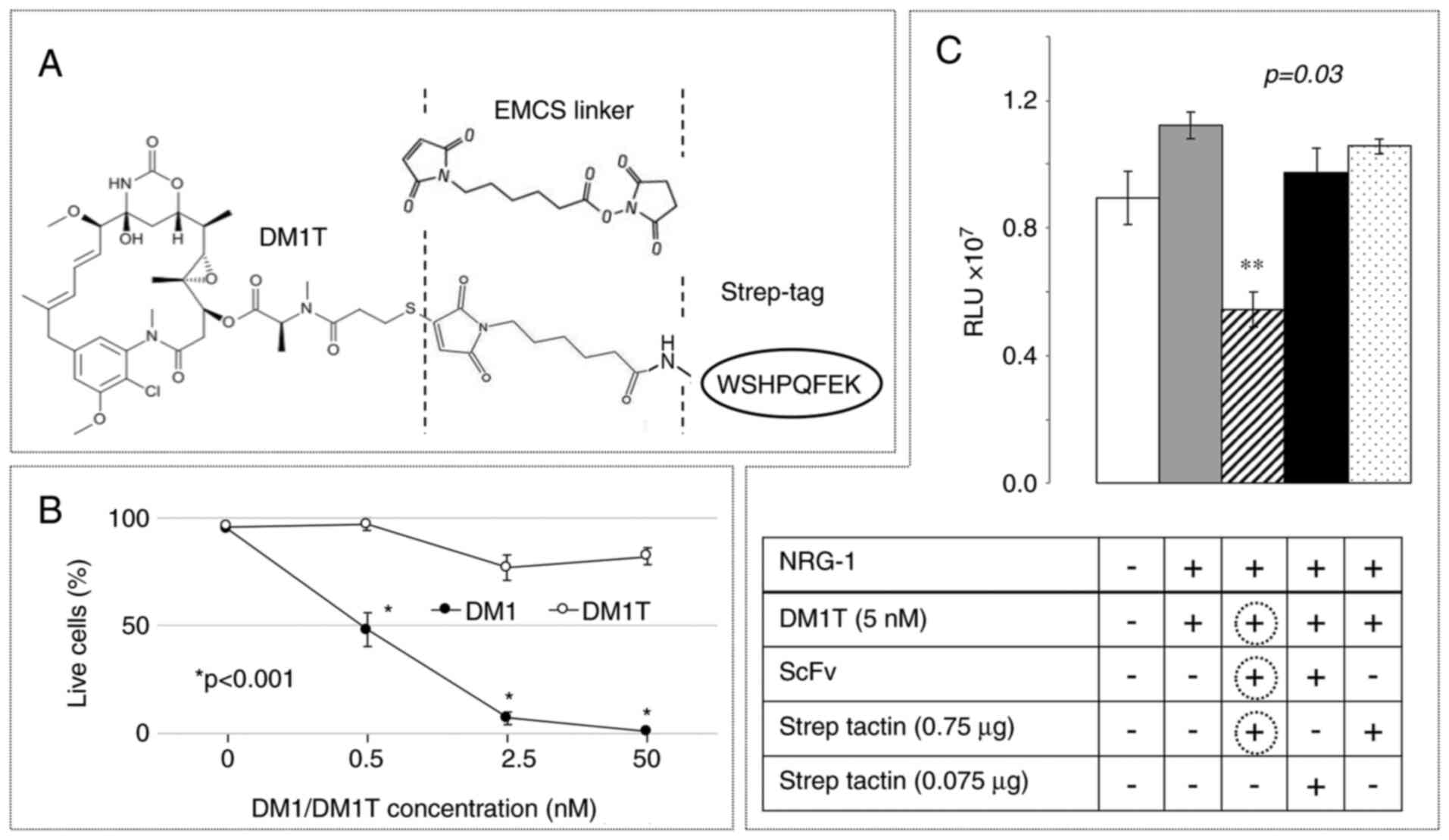
through a spacer (DM1T; Fig. 5A) and
the resulting DM1T derivative was used to treat BRC230-cfos
transfectants in a dose-response experiment (Fig. 5B). Cell viability was assessed by
propidium iodide (PI) exclusion. With this method, the half-maximal
inhibitory concentration (IC50) of DM1 was in the
sub-nanomolar range, as expected, and DM1T displayed limited
residual activity. Because essentially inactive, DM1T could be
incorporated in a 2-step TOOLBOX protocol at a concentration (2.5
nM) 5 times above the IC50 of DM1. Despite limited ERBB2
signaling, TOOLBOX treatment of BRC-230-cfos transfectants
(Fig. 5C) counteracted not only
NRG-1-mediated stimulation, but reduced light emission well below
baseline (striped bar). Remarkably, DM1T was active (in the TOOLBOX
protocol) at a concentration 103 times lower than
NAX098T (compare with Fig. 4), and
yet the pattern of conditional prodrug reactivation was very
similar. We conclude that also a preferred ADC payload can be
modified to fit the TOOLBOX approach.
TOOLBOX in vivo
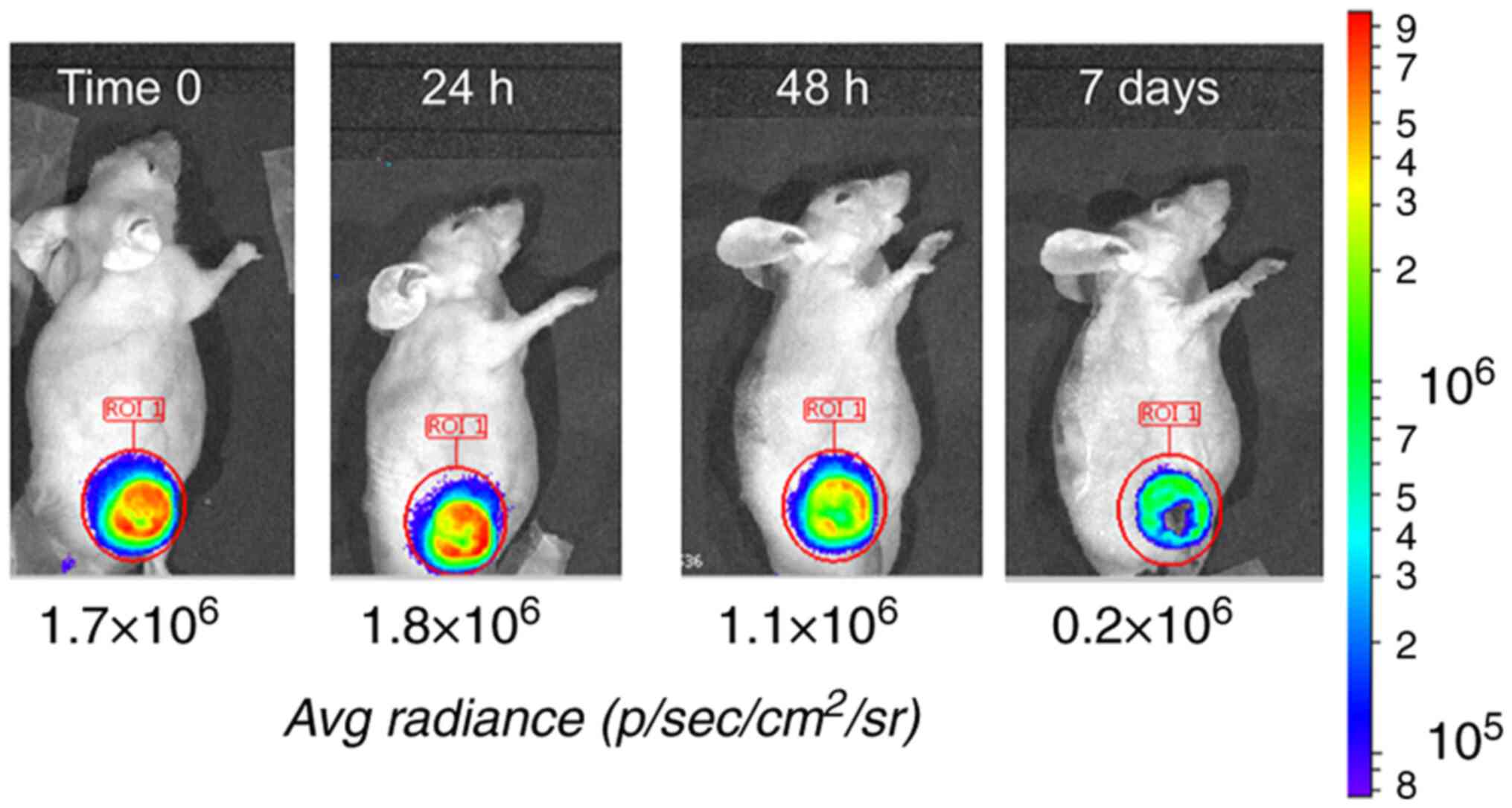
Finally, a TOOLBOX experiment with DM1T was carried
out in vivo on 6 nude mice (3 in the experimental group and
3 controls) bearing subcutaneous BRC230-cfos tumor xenografts.
Treatment was carried out by a single tail vein injection of the
TOOLBOX cocktail. The experimental group received ScFv +
Strep-Tactin-Mult followed, 60 min later, by DM1T (see Materials
and methods and Data S1 for details). Control littermates were
treated as above but wild-type ScFv was replaced by equal amounts
of inactive ScFv-mut4. ERBB2 activation was assessed by measuring
light emission after in vivo furimazine injection at time 0
(before treatment) and 7 days later. In a single mouse, two
additional intermediate determinations (24 and 48 h) were carried
out. The effect on promoter activity was detected (2-fold to 8-fold
reduction) in all mice (data not shown). In the mouse with multiple
time points it was gradual and became fully evident on day 7
(Fig. 6). ERBB2 signaling either
remained stable or slightly increased in the control mice (data not
shown). Remarkably, TOOLBOX-treated and control-treated mice did
not show appreciable signs of general toxicity upon injection of
DM1T. Body weight remained fairly stable, e.g. from 26.1 (±0.4 SD)
g at day 0, to 26.3 (±0.5 g) at day 7 in the active ScFv group, and
from 26.7 (±1.2 g) at time 0 to 27.3 (±0. 9 g) at day 7 in the
inactive ScFv-mut4 group.
Discussion
Altogether, our findings provide proof-of-principle
that TOOLBOX is a novel platform for antibody/drug
exchange/targeting and delivery. TOOLBOX has 5 favorable features.
It is a) specific, b) potent, c) modular, d) flexible, and e)
conditional.
Five favorable TOOLBOX features
Specificity (a) was demonstrated by an ERBB2
pathway-dependent promoter-reporter system. Potency (b) was
supported by i) the cytotoxic effects of two classes of anticancer
drugs (DNA nanobinders and DM1) at their respective optimal dosages
(micromolar and nanomolar), and ii) activity on
ERBB2-low/non-amplified TNBC cells, e.g. a molecular breast cancer
subtype that does not benefit from trastuzumab application
(7), but was very recently shown to
benefit from other ADCs (17).
Modularity (c) was supported by the accommodation of different drug
classes in TOOLBOX. Flexibility (d) in nanostructure assembly was
supported by the 2-step and 3-step options, although the 2-step
protocol (ScFv + pre-formed Strep-Tactin:Strep-Tagged drug
nano-assemblies) performed best in both flow cytometry and drugging
experiments (Fig. S2). This is not
surprising, since a second-order reaction is much more likely to
occur than a third-order reaction. Additionally, the 2-step
protocol requires a single 1 h interval between the two i.v.
administrations of ScFv and the preformed TOOLBOX nano-complex
(Fig. 6), which makes it more
convenient than the 3-step protocol, particularly in
vivo.
However, the most favorable built-in feature of
TOOLBOX is conditional reactivation (e) of NAXT and DM1T compounds,
e.g. the restoration of cytotoxic properties upon funneling into
TOOLBOX-mediated cellular dispatch. The mechanism is being
investigated. We favor the possibility that ERBB2:TOOLBOX complexes
are engulfed through the default endocytic receptor internalization
pathway, and then intracellular release of the active drugs,
possibly through cleavage/degradation of the Strep-Tag. This route
may be precluded to tagged prodrugs alone.
TOOLBOX technical limitations
Challenges were encountered of five types: Tagging,
stoichiometry, nano-assembly formation, Strep-Tactin valence, and
payload selection/adaption. Although there were tag position
preferences, Strep-Tagging was possible at both ScFv ends with
little if any interference with the small antigen-binding ScFv
domain. Production is underway of Strep-Tagged, humanized, whole
IgGs. Stoichiometry of the TOOLBOX building blocks, their order of
addition, and the identification of the right valence of the
Strep-Tactin adapter required careful troubleshooting. The critical
step was the development of a Strep-Tactin polymer with a high
nominal valence, to support simultaneous binding to tagged ScFv and
drugs. Improved Strep-Tactin scaffolds will have to be created for
controlled, optimized oligomerization.
The last and most crucial TOOLBOX step was
Strep-Tagging of small drugs. The process was rather inefficient (1
good candidate out of 10 screened) with berberine derivatives.
Building on this, we switched to DM1, that is a preferred ADC
payload. An appropriate linker-spacer was incorporated with the aim
of spatially separating the cytotoxic DM1 moiety from the
polypeptide tag. The first compound generated by this strategy was
found to work as intended.
Strep-Tactin nano-assemblies, that are the core
adapters of all our TOOLBOX drugs, are estimated to be ~9 nm in
size, e.g. smaller than most drug-entrapping systems such as
liposomes, synthetic membranes, and protein nanocages (18). This may favor tissue penetration, but
drug delivery is expected to be lesser than in systems based on
massive drug entrapment.
Altogether, the extensive trouble-shooting reported
herein clearly illustrates that TOOLBOX is innovative but, at the
same time, it is still at the proof-of-concept stage. As compared
to a conventional ADC, the use of multiple building
blocks/components potentially introduces additional issues such as
scaffold immunogenicity and unfavorable pharmacokinetics. These may
slow down or hamper its industrial exploitation, and will have to
be addressed in future studies.
TOOLBOX precedents
TOOLBOX bears some analogies with an avidin-biotin T
lymphocyte redirection system independently proposed long time ago
by two groups (19,20). These authors took advantage of
biotin-labelled antitumor antibodies, HLA class I tetramers
refolded around strong viral peptide antigens, and avidin, to
re-direct virus-specific T cells toward tumor cells. TOOLBOX can
similarly redirect HLA-I tetramers (e.g. HLA-A2 Streptamers, see
Fig. 3) and antigen-specific
cytotoxic T lymphocytes (data not shown). However, only TOOLBOX was
shown to redirect anticancer drugs.
A distinct modular method, possibly more relevant to
TOOLBOX, was developed by Metz et al using drug haptenized
by digoxigenin, and engineered bispecific antibodies that bind
tumor antigens with one arm, and digoxigenin-labelled doxorubicin
with the other (21). However, the
digoxigenin system requires extensive optimization for each
bispecific antibody, by juxtaposition and grafting of two
antigen-binding ‘halves’ onto selected Ig backbones to form
combinatorial sets of dedicated bispecifics (22). This is far more complicated than the
non-intrusive terminal addition of a very small tag to an affinity
reagent of choice, like in TOOLBOX. Most importantly, unlike our
NAXT and DM1T compounds, digoxigenin-labelled drugs are not known
to undergo, to our knowledge, any conditional
inactivation-reactivation process. Whereas a circulating active
drug may have minor toxic effects when activity is in the
micromolar range, like for doxorubicin (used in the digoxigenin
system) and NAX compounds (used herein), it may entirely preclude
the use of highly cytotoxic maytansinoids (DM1), since these work
in the nanomolar range, and failed a number of clinical trials as
free drugs in the 90s due to high-grade toxicity (1). Unlike all precedents, TOOLBOX
nano-assemblies incorporate a unique, non-dispensable safe lock
that protects host normal tissues even in the event of ‘leakage’ of
the cytotoxic substance from TOOLBOX nano-assemblies.
Clinical translational significance
Precision oncology is a fast moving field. Massive
parallel sequencing provides extensive catalogues of tumor
vulnerabilities and expanded therapeutic opportunities. Multiple
lines of targeted therapies are tolerated with fair to excellent
quality of life. Standard regimens of targeted and non-targeted
treatments (in sequence and in combination) are almost invariably
adopted to counter primary as well as secondary drug resistance.
Regulatory bodies authorize drugs for use at progressively earlier
cancer stages, e.g. in the neoadjuvant and adjuvant settings
(23). And yet, despite flexibility
in cancer targeting is clearly perceived as a priority, the issue
has not been really addressed at the biotech level due to a lack of
technical solutions. To our knowledge, TOOLBOX is one of the few
attempts to create a platform for the development of families of
ADC-like objects with no need to manufacture many distinct ADCs.
Switching and/or combining antibodies and/or anticancer agents (by
TOOLBOX, its improvements, or other unrelated technologies) will
make it possible to provide effective ADC-like target therapies to
different patients and/or the same patient through disease stages
and progression. This may effectively address some unmet needs of
precision drugging.
Supplementary Material
Supporting Data
Acknowledgements
Drs Rocco Fraioli and Adele Petricca are gratefully
acknowledged for skillful technical assistance and administrative
support, respectively. Special thanks to Dr Anette Jacob, Peps 4
Life Sciences for DM1T manufacturing.
Funding
This research was supported by the Eurostars E!5995
project (JB, LC, KF and PG), Associazione Italiana per la Ricerca
sul Cancro (AIRC) through I.G. 14204 and 19052 (PG) and Nuvenia
Fellowship 19503 (MA), the Thüringer Aufbaubank (European Regional
Development Fund grants FE 9034 and FE 9053) and the
Interdisziplinäres Zentrum für klinische Forschung (IZKF), Jena (KF
and NK).
Availability of data and materials
W6/800 and TOOLBOX are patent pending (WO2017167967,
WO2018138676) and their exploitation rights are owned by IBI
Lorenzini and the TOOLBOX Consortium, respectively. Restrictions
may apply to their distribution. Data sharing is not applicable to
this article as no datasets were generated or analyzed during the
current study.
Authors' contributions
ET and MA tested the TOOLBOX concept and performed
flow cytometry. LS, FC and LC designed and prepared the tagged
ScFv. NK and KF designed and prepared the NanoLuc promoter-reporter
construct. GH and JB designed and manufactured Strep-Tactins. ET
and IM performed the animal studies. PL designed and manufactured
NAX and NAXT compounds. PG conceptualized TOOLBOX and wrote the
manuscript with the contribution of all authors. All authors have
approved the final version of the manuscript.
Ethics approval and consent to
participate
In vivo experiments were approved (prot.
665/2017-PR) by the Veterinary Section of the Italian Ministry of
Health.
Patient consent for publication
Not applicable.
Competing interests
LC and JB are full-time employees of IBI Lorenzini
and IBA Lifesciences, that hold exploitation rights for W6/800,
Strep-Tag technologies and TOOLBOX. PG is directly supported by Ibi
Lorenzini funds. All the other coauthors declare no conflict of
interest.
Glossary
Abbreviations
Abbreviations:
|
ADCs
|
antibody-drug conjugates
|
|
T-DM1
|
trastuzumab emtansine
|
|
Ig
|
immunoglobulin
|
|
TNBC
|
triple-negative breast cancer
|
|
ATCC
|
American Type Culture Collection
|
|
HLA
|
human leukocyte antigens
|
|
mAb
|
monoclonal antibody
|
|
ScFv
|
single chain fragment of variable
antibody regions
|
|
EGFR
|
epidermal growth factor receptor
|
|
EGF
|
epidermal growth factor
|
|
PE
|
phycoerythrin
|
|
EMCS
|
N-(ε-maleimidocaproyloxy)succinimide
ester
|
|
GFP
|
green fluorescent protein
|
|
DMF
|
dimethylformamide
|
|
CDR
|
complementarity determining
regions
|
|
RLU
|
relative luminesce units
|
References
|
1
|
de Goeij BE and Lambert JM: New
developments for antibody-drug conjugate-based therapeutic
approaches. Curr Opin Immunol. 40:14–23. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Verma S, Miles D, Gianni L, Krop IE,
Welslau M, Baselga J, Pegram M, Oh DY, Diéras V, Guardino E, et al:
Trastuzumab emtansine for HER2-positive advanced breast cancer. N
Engl J Med. 367:1783–1791. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
von Minckwitz G, Huang CS, Mano MS, Loibl
S, Mamounas EP, Untch M, Wolmark N, Rastogi P, Schneeweiss A,
Redondo A, et al: Trastuzumab emtansine for residual invasive
HER2-positive breast cancer. N Engl J Med. 380:617–628. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Baselga J, Lewis Phillips GD, Verma S, Ro
J, Huober J, Guardino AE, Samant MK, Olsen S, de Haas SL and Pegram
MD: Relationship between tumor biomarkers and efficacy in EMILIA, a
phase iii study of trastuzumab emtansine in HER2-positive
metastatic breast cancer. Clin Cancer Res. 22:3755–3763. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Amadori D, Bertoni L, Flamigni A, Savini
S, De Giovanni C, Casanova S, De Paola F, Amadori A, Giulotto E and
Zoli W: Establishment and characterization of a new cell line from
primary human breast carcinoma. Breast Cancer Res Treat.
28:251–260. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chavez KJ, Garimella SV and Lipkowitz S:
Triple negative breast cancer cell lines: One tool in the search
for better treatment of triple negative breast cancer. Breast Dis.
32:35–48. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mayer IA, Abramson VG, Lehmann BD and
Pietenpol JA: New strategies for triple-negative breast
cancer-deciphering the heterogeneity. Clin Cancer Res. 20:782–790.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Giacomini P, Giorda E, Pera C and Ferrara
GB: An ID card for tumour cell lines: HLA typing can help. Lancet
Oncol. 2:6582001. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Digiesi G, Giacomini P, Fraioli R, Mariani
M, Nicotra MR, Segatto O and Natali PG: Production and
characterization of murine mAbs to the extracellular domain of
human neu oncogene product GP185HER2. Hybridoma. 11:519–527. 1992.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Galeffi P, Lombardi A, Pietraforte I,
Novelli F, Di Donato M, Sperandei M, Tornambé A, Fraioli R,
Martayan A, Natali PG, et al: Functional expression of a
single-chain antibody to ErbB-2 in plants and cell-free systems. J
Transl Med. 4:392006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Pierpaoli E, Fiorillo G, Lombardi P,
Salvatore C, Geroni C, Piacenza F and Provinciali M: Antitumor
activity of NAX060: A novel semisynthetic berberine derivative in
breast cancer cells. Biofactors. 44:443–452. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Schonthal A, Herrlich P, Rahmsdorf HJ and
Ponta H: Requirement for fos gene expression in the transcriptional
activation of collagenase by other oncogenes and phorbol esters.
Cell. 54:325–334. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Knutti N, Kuepper M and Friedrich K:
Soluble extracellular matrix metalloproteinase inducer (EMMPRIN,
EMN) regulates cancer-related cellular functions by homotypic
interactions with surface CD147. FEBS J. 282:4187–4200. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Stacer AC, Nyati S, Moudgil P, Iyengar R,
Luker KE, Rehemtulla A and Luker GD: NanoLuc reporter for dual
luciferase imaging in living animals. Mol Imaging. 12:1–13. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bravo R, Burckhardt J, Curran T and Muller
R: Stimulation and inhibition of growth by EGF in different A431
cell clones is accompanied by the rapid induction of c-fos and
c-myc proto-oncogenes. EMBO J. 4:1193–1197. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Daemen A and Manning G: HER2 is not a
cancer subtype but rather a pan-cancer event and is highly enriched
in AR-driven breast tumors. Breast Cancer Res. 20:82018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Modi S, Saura C, Yamashita T, Park YH, Kim
SB, Tamura K, Andre F, Iwata H, Ito Y, Tsurutani J, et al:
Trastuzumab deruxtecan in previously treated HER2-positive breast
cancer. N Engl J Med. 382:610–621. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Falvo E, Tremante E, Arcovito A, Papi M,
Elad N, Boffi A, Morea V, Conti G, Toffoli G, Fracasso G, et al:
Improved doxorubicin encapsulation and pharmacokinetics of
ferritin-fusion protein nanocarriers bearing proline, serine, and
alanine elements. Biomacromolecules. 17:514–522. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Robert B, Guillaume P, Luescher I, Romero
P and Mach JP: Antibody-conjugated MHC class I tetramers can target
tumor cells for specific lysis by T lymphocytes. Eur J Immunol.
30:3165–3170. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ogg GS, Dunbar PR, Cerundolo V, McMichael
AJ, Lemoine NR and Savage P: Sensitization of tumour cells to lysis
by virus-specific CTL using antibody-targeted MHC class I/peptide
complexes. Br J Cancer. 82:1058–1062. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Metz S, Haas AK, Daub K, Croasdale R,
Stracke J, Lau W, Georges G, Josel HP, Dziadek S, Hopfner KP, et
al: Bispecific digoxigenin-binding antibodies for targeted payload
delivery. Proc Natl Acad Sci USA. 108:8194–8199. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Stefan D, Claudio S and Ulrich B:
Engineered hapten-binding antibody derivatives for modulation of
pharmacokinetic properties of small molecules and targeted payload
delivery. Immunol Rev. 270:165–177. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Allegretti M, Fabi A, Buglioni S, Martayan
A, Conti L, Pescarmona E, Ciliberto G and Giacomini P: Tearing down
the walls: FDA approves next generation sequencing (NGS) assays for
actionable cancer genomic aberrations. J Exp Clin Cancer Res.
37:472018. View Article : Google Scholar : PubMed/NCBI
|