Introduction
Breast cancer is the most common primary cancer in
woman, affecting approximately 12% of women worldwide (1). Breast cancer cases have markedly
increased since the 1970s, which has been attributed to the modern
lifestyle (2). There are various
subtypes of breast cancer, including triple-negative breast cancer
(TNBC), which lacks the expression of the genes encoding human
epidermal growth factor receptor-2, progesterone receptor, as well
as estrogen receptor. The majority of hormone and targeted
therapies target one of the three aforementioned receptors,
resulting in treatment resistance (2). In addition, TNBC is more common in young
women, demonstrating relapse and aggressive behaviors during the
invasive and metastatic states. Cancer relapse and distant
metastasis markedly increase morbidity and mortality, and are the
most formidable obstacles to successful treatment (3). Current medical treatments, such as
treatment with doxorubicin and toxoids, frequently lead to drug
resistance and cause severe side effects (4). As a result, researchers are exploring
novel treatment strategies for TNBC.
Natural products and their derivatives have recently
become an important source of drug and therapeutic candidates
(5). Between 1981 and 2010,
approximately 34% of US Food and Drug Administration-approved
novel-marketed drugs were derived from natural products (6). Curcumin, a golden color chemical derived
from Curcuma longa plants, has been widely used in
Traditional Chinese medicine in East Asia for centuries. Curcumin
has diverse pharmacological properties, including anti-bacterial
(7), anti-inflammatory (8), anti-oxidant (9), anti-depressant (9), anti-viral (10), anti-diabetes (11) and anticancer properties (12–14).
During the past 10 years, numerous studies have reported that
curcumin and its derivatives can effectively inhibit tumor cell
growth, and induce apoptosis, autophagy and cell cycle arrest
(4,12). Currently, numerous phase II and III
clinical trials have advocated for the application of curcumin in
patients with multiple myeloma, myelodysplastic syndromes,
pancreatic cancer, head and neck cancer and colon cancer (15). In a previous clinical study, curcumin
was proven to be safe even at doses of up to 8 g per day (16). Curcumin is, by all accounts, an ideal
medication for the inhibition of cancer growth through various
signaling pathways. However, certain animal and human
pharmacokinetic studies have reported a poor absorption of curcumin
in the gastrointestinal tract. The low systemic bioavailability of
curcumin prevents an adequate concentration from reaching the
target tissues to achieve pharmacological effects (17–20).
To overcome its poor bioavailability and increase
its absorption in vivo, a novel curcumin derivative
(1E,3Z,6E)-3-hydroxy-5-oxohepta-1,3,6-triene-1,7-diyl)bis(2-methoxy-4,1-phenylene)bis(3-hydroxy-2-hydroxymethyl)-2-methylpropanoate
(MTH-3), was designed and developed. Fig.
1A includes a schematic of MTH-3. In a previous study it was
demonstrated that, MTH-3 has superior hydrophilicity than curcumin.
The log P, calculated logarithmic partition coefficient, of MTH-3
is 1.73, and of curcumin it is 3.38 (21,22).
Furthermore, previous findings showed that MTH-3 inhibits tumor
proliferation and induces apoptosis in TNBC in vitro and
in vivo through cell cycle arrest and the autophagic pathway
(21). Chang et al (22) revealed that MTH-3 has a greater
inhibitory effect against TNBC cells compared with curcumin. That
study also reported a 10-fold higher potency of MTH-3 compared to
curcumin against doxorubicin-resistant MDA-MB-231 cell
proliferation.
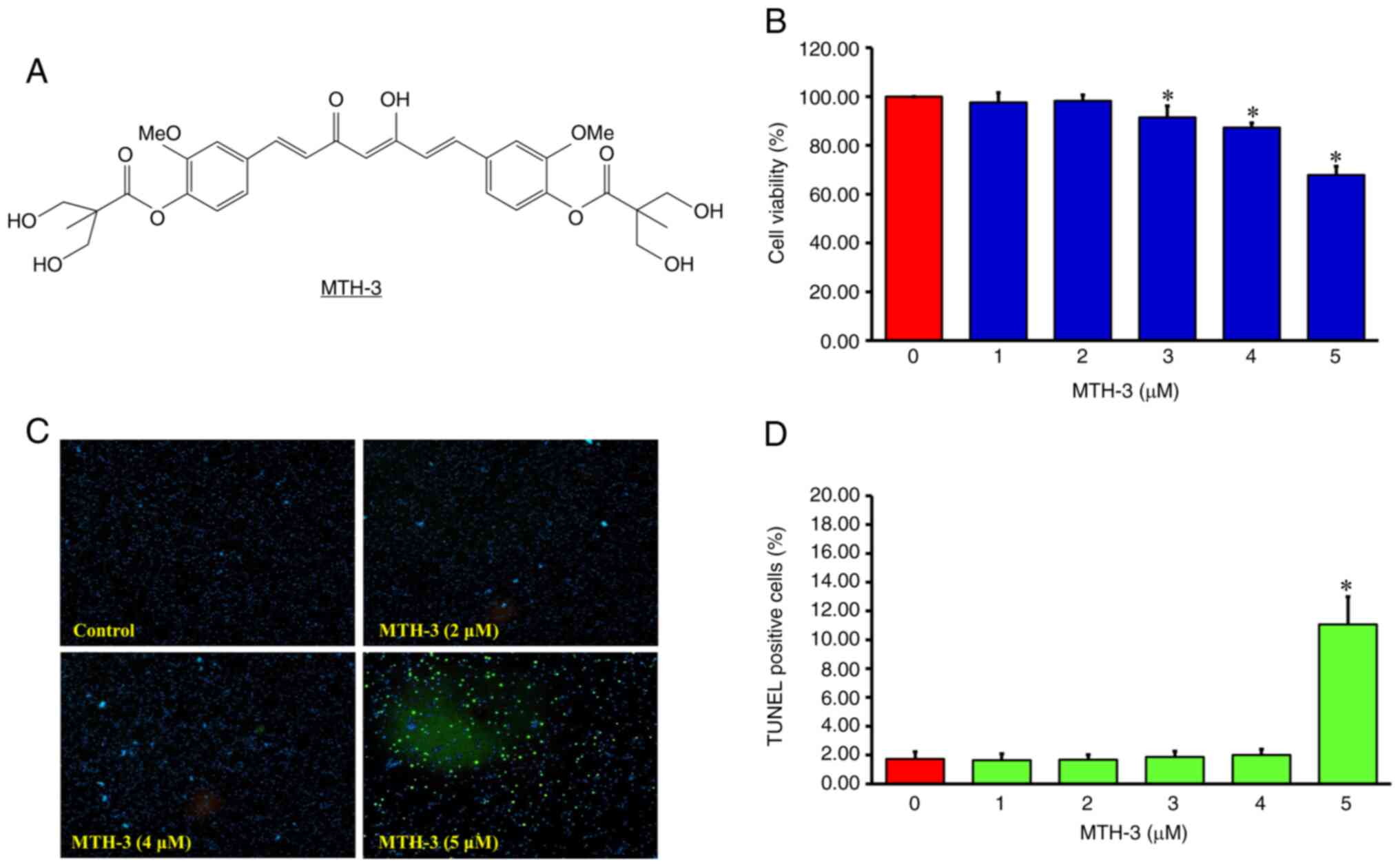 | Figure 1.(A) Chemical structure of MTH-3. (B)
Following treatment with 0, 1, 2, 3, 4 and 5 µM MTH-3 for 24 h, the
cell viability of MDA-MB-231 cells was evaluated by MTT assay. (C
and D) The proportions of apoptotic cell death was evaluated by
TUNEL assay. Data are presented as the mean ± standard deviation of
three experiments. *P<0.05. MTH-3,
(1E,3Z,6E)-3-hydroxy-5-oxohepta-1,3,6-triene-1,7-diyl)bis(2-methoxy-4,1-phenylene)bis(3-hydroxy-2-hydroxymethyl)-2-methyl
propanoate. |
In the present study, the ability of MTH-3 to
inhibit invasiveness in TNBC and the potential molecular signaling
pathways were investigated.
Materials and methods
Chemicals
MTH-3 was synthesized and designed as previously
described (21). Its chemical
structure is shown in Fig. 1A. Hsieh
et al (21) initially
nominated the novel curcuminoid derivative as compound 9a, and the
nomenclature was revised to MTH-3 in the study by Chang et
al (22). L-glutamine, fetal
bovine serum (FBS), streptomycin, Leibovitz's L-15 medium,
penicillin G, and trypsin-EDTA were purchased from Thermo Fisher
Scientific, Inc. Matrigel was obtained from Corning, Inc.
Antibodies were purchased from Cell Signaling Technology, Inc. All
other chemicals were purchased from Merck KGaA.
Cell culture
The MDA-MB-231 human breast adenocarcinoma cell line
was obtained from the Bioresource Collection and Research Center
(Hsinchu, Taiwan). MDA-MB-231 cells were cultured with 90%
Leibovitz's L-15 medium, 1% penicillin-streptomycin and 10% FBS in
75 cm2 culture flasks in an incubator with a humified 5%
CO2 atmosphere at 37°C (23).
Cell viability assay
MTT assay was conducted to evaluate the cytotoxicity
of MTH-3 in MDA-MB-231 cells. The initial concentration of tumor
cells was 1×105 cells/ml in a 96-well cell culture
plate. Tumor cells were treated with various concentrations of
MTH-3 (0, 1, 2, 3, 4 and 5 µM) at 37°C. After 24 h of cell culture,
MTT solution (0.5 mg/ml) was added, and the cells were incubated
for an additional 4 h at 37°C. Next, the formazan crystals were
dissolved in DMSO following the removal of the medium. The formazan
product was analyzed spectrophotometrically at a wavelength of 490
nm (24). This analysis was performed
in triplicate.
TUNEL assay
MDA-MB-231 cells were cultured in 12-well plates and
treated with different concentrations of MTH-3 (1, 2, 3, 4 and 5
µM) or with 0.1% DMSO in Leibovitz's L-15 medium at 37°C for 24 h.
Cells were collected and fixed with absolute ethanol. These cells
were subsequently stained with DAPI solution to detect DNA
breakdown using the In Situ Cell Death Detection Kit,
Fluorescein (Roche Diagnostics GmbH) as previously described
(25). This analysis was performed in
triplicate.
Wound healing assay
For the wound healing assay, MDA-MB-231 cells were
incubated untill they reached ~90% confluence in a tissue culture
plate. Next, to creat a 1 mm wound area, each well of the culture
plate was scratched using a micropipette tip. The tumor cells were
subsequently cultured in serum-free Leibovitz's L-15 medium with
MTH-3 at different concentrations (1, 2, 3 and 4 µM) or with 0.1%
DMSO at 37°C for 24 h. Tumor cells and the denuded zones were
photographed under a phase-contrast microscope (magnification,
×100) (26). This analysis was
performed in triplicate.
Transwell assay
To investigate tumor cell invasion, a Transwell
assay with a Matrigel®-coated invasion chamber was
performed. Firstly, to form a genuine reconstituted basement
membrane, the Transwell insert (polycarbonate filters with an 8-µm
porosity) was coated with 30 µg Engelbreth-Holm-Swarm sarcoma tumor
matrix (Matrigel®). Next, 1×106 MDA-MB-231
cells were seeded in serum-free Leibovitz's L-15 medium in a T-75
culture plate. After 24 h of incubation, these tumor cells were
suspended, and 5×104 cells/chamber were subsequently
added in the upper chamber of the Transwell insert with serum-free
medium. The lower chamber contained Leibovitz's L-15 medium with
10% FBS. The tumor cells were treated with different concentrations
of MTH-3 (1, 2, 3 and 4 µM) or with 0.1% DMSO at 37°C for 24 h.
After incubation for cancer cell invasion, the samples were fixed
with 4% formaldehyde for 15 min. Next, 2% crystal violet was used
for staining. Finally, the tumor cells in the upper chamber were
removed, and the invading cells in the lower chamber were
visualized under a light microscope (Leica Microsystems GmbH;
magnification, ×200) (26). This
analysis was performed in triplicate.
Gelatin zymography
Gelatin zymography assay was performed to
investigate the protein activity of matrix metalloproteinase
(MMP)-2 and MMP-9. Firstly, MDA-MB-231 cells were treated with
different concentrations of MTH-3 (1, 2, 3 and 4 µM) or with 0.1%
DMSO in serum-free Leibovitz's L-15 medium at 37°C for 24 h. These
tumor cells were then suspended in zymography sample buffer. Next,
the samples were added in loading buffer, and subsequently
electrophoresed on 10% sodium dodecyl sulfate-polyacrylamide gel
containing 0.1% gelatin. Subsequently, 2.5% Triton X-100 in
double-distilled H2O was used to wash the gels. The gels
were then stored in development buffer (0.02% Briji-35, 1 mM
ZnCl2, 5 mM CaCl2, 50 mM Tris, and 200 mM
NaCl at pH 7.5) at 37°C. After 18 h, the gels were stained with
0.5% Coomassie blue G-250. Then, the gels were de-stained. The
non-staining bands indicated proteolytic activities. These results
were analyzed using ImageJ software (National Institutes of Health)
(27). This analysis was performed in
triplicate.
3D spheroid invasion assay
Following the manufacturer's instructions,
MDA-MB-231 cells (5×103 cells/total) were seeded into
96-well round-bottom ultralow attachment plates for 3 days. After
the spheroid diameter reached 200-µm, MDA-MB-231 cells were treated
with different concentrations of MTH-3 (1, 2, 3 and 4 µM) or with
0.1% DMSO. Matrigel solution was added, and the plate was placed in
a 37°C incubator for 30 min to polymerize the Matrigel. The system
IncuCyte S3 ZOOM System instrument (Essen BioScience) was used to
monitor spheroid invasion (28,29). This
analysis was performed in triplicate.
Whole transcriptome sequencing
(next-generation sequencing analysis)
To evaluate the possible signaling pathways of MTH-3
in MDA-MB-231 cells, RNA sequencing analysis of MTH-3-exposed and
control groups was performed. Total RNA extraction, quantification,
as well as sequencing cluster generation and high throughput
sequencing were performed using Illumina (New England Biolabs),
according to the manufacturer's instructions. Every step was
performed under strict monitoring and quality control. After mixing
libraries based on the effective concentration and the required
sequencing data volume, high throughput sequencing was conducted
using a high throughput sequencing platform to capture and sequence
the entire mRNA pool. Bioinformatics analysis was performed after
obtaining the original sequence data, as previously described
(30). These individual libraries
were converted to the FASTQ format. The raw sequencing data
eliminated the adapter sequences. Short-read alignment was
performed using Hisat2 (v2.0.1) with the default parameters
(31). Using the pipeline on the
human reference genome from the UCSC Genome Browser, the paired-end
reads were mapped. Differential mRNA expression analysis was
conducted to identify over-represented functional terms presenting
in the background. Benjamini-Hochberg false discovery rate
correction and a Student's t-test were performed to determine
significantly differentially expressed genes (DEGs). The Kyoto
Encyclopedia of Genes and Genomes (KEGG) database with the
David/EASE tool was used for pathway analysis. Rich factor and
q-value were applied to evaluate the degree of KEGG enrichment. The
ratio of the number of DEGs to the number of total annotated genes
in a certain pathway is shown by Rich factor, and Q value (0–1) is
a multiple hypothesis-corrected P-value, which closer to 0 indicate
a greater enrichment. Differentially expressed genes (DEGs) between
MTH-3-exposed and control groups were selected using three
criteria: P<0.005, adj P<0.05 and absolute fold change of
≥1.2). The DEGs were selected by multiple systemic analysis
(30,32). This analysis was performed in
duplicate.
Antibodies microarray analysis
To further evaluate the correlation between mRNA and
protein expression levels, Kinex Antibody Microarray was used for
subsequent analysis. Briefly, MDA-MB-231 cells were seeded in
serum-free Leibovitz's L-15 medium at 37°C with 2 µM MTH-3 or with
0.1% DMSO for 24 h. Tumor cells were then collected. From each
sample, lysate proteins were labeled with proprietary fluorescent
dye combination. After blocking the non-specific binding sites, the
unbound proteins were washed away, and the chambers were displayed
on the microarray. Images were captured using a Perkin-Elmer
ScanArray Reader laser array scanner (Thermo Fisher Scientific,
Inc.). ImaGene 9.0 (BioDiscovery) was used for spot segmentation
and background correction. The overall average intensity of all
spots within the samples was subtracted from the raw intensity of
each spot to calculate the Z scores (33). Z ratios were calculated by dividing
the difference between the averages of the observed protein Z
scores by the standard deviations of all the differences for a
particular comparison. Z ratio values of ±1.2 were considered
statistically significant (33). This
analysis was performed in triplicate.
Western blot analysis
To further analyze the protein expression levels,
western blot analysis was performed. Briefly, MDA-MB-231 cells were
cultured in Leibovitz's L-15 medium at 37°C with 2 µM MTH-3 or with
0.1% DMSO for 24 h. Tumor cells were then collected to obtain the
protein lysate. Bio-Rad protein assay system (Bio-Rad Laboratories,
Inc.) was used for determination of protein concentrations. These
samples (35 µg per lane) were separated via 10–12% SDS-PAGE and
transferred to PVDF membranes. The membranes were blocked with 5%
skimmed dry milk at room temperature for 2 h. The membranes were
incubated overnight at 4°C with primary antibodies purchased from
Cell Signaling Technology, Inc., including AKT (cat. no. GTX121937;
dilution, 1:5,000), p-AKT (phospho Ser473, cat. no. GTX28932;
dilution, 1:400), ERK (cat. no. GTX59618; dilution, 1:10,000),
p-ERK (phospho Thr202/Tyr204, cat. no. GTX59568; dilution,
1:10,000), p38/MAPK (cat. no. GTX110720; dilution, 1:1,000),
p-p38/MAPK (cat. no. GTX48614; dilution, 1:500), JNK (cat. no.
GTX52360; dilution, 1:500), p-JNK (phospho Thr183/Tyr185, cat. no.
GTX52326; dilution, 1:500), and β-actin (cat. no. GTX109639;
dilution, 1:10,000). Subsequently, the membranes were incubated for
4 h at room temperature with horseradish peroxidase-conjugated
secondary antibodies, including anti-rabbit IgG (cat. no. 7074;
dilution, 1:10,000) and anti-mouse IgG (cat. no. 7076; dilution,
1:10,000). Finally, the protein bands were visualized using ECL
reagents (Cytiva) and scanned on a UMAX powerLook Scanner (UMAX
Technologies). The National Institutes of Health ImageJ 1.52v
program was used for quantitative analysis of the intensity of the
band signal. This analysis was performed in quadruplicate.
Statistical analysis
Data are presented as the mean ± standard deviation.
One-way analysis of variance followed by Dunnett's test and Tukey's
post hoc test was conducted to analyze the differences between two
groups and among multiple groups, respectively, using SPSS software
version 25.0 (IBM, Corp.). P<0.05 was considered to indicate a
statistically significant difference (34).
Results
MTH-3 inhibits the proliferation in
MDA-MB-231 human breast adenocarcinoma cells
The tumor cells were treated with different
concentrations of MTH-3 (1, 2, 3, 4 and 5 µM) or with 0.1% DMSO at
37°C for 24 h. The cells were suspended and analyzed by MTT method
to detect the cell viability. The results showed a significantly
reduced viability after 3, 4 and 5 µM MTH-3 at 91.45±4.69,
89.27±3.34 and 67.83±3.62%, respectively (Fig. 1B).
On the other hand, as shown in Fig. 1C and D, MTH-3 treatment at a
concentration of 1–4 µM did not significantly induce apoptotic cell
death, revealed by TUNEL assay. As a result, the concentrations of
1, 2, 3 and 4 µM of MTH-3 were subsequently selected to further
study the effects on MDA-MB-231 cell invasion and metastasis.
MTH-3 inhibits invasion and migration
in MDA-MB-231 human breast adenocarcinoma cells
The tumor cells were incubated with various
concentrations of MTH-3 (1, 2, 3 and 4 µM) or with 0.1% DMSO at
37°C for 24 h to determine their ability of cell migration and
invasion abilities by Transwell, wound healing and 3D spheroid
invasion assays. In Fig. 2A and B,
the tumor cells were found to be significantly decreased in the
lower chamber in the MTH-3 treatment groups. In Fig. 2C and D, the edge distance in MTH-3
treatment groups was significantly wider than that in the control
group, indicating that MTH-3 inhibited MDA-MB-231 cell motility in
a concentration-dependent manner. To develop a tumor cell invasion
and migration model, 3D spheroid invasion assay was applied to
recapitulate both biochemical and physical characteristics of the
tumor microenvironment. In Fig. 3A and
B, MTH-3 significantly inhibited tumor spheroid invasion and
migration in the Matrigel-coated area. These results suggested that
MTH-3 inhibited MDA-MB-231 cells invasion and migration in a
concentration-dependent manner. In combination, these data
suggested that a concentration of <5 µM MTH-3 predominantly
inhibited tumor cell migration and invasion. However,
MTH-3-exhibited toxicity in MDA-MB-231 cells may require higher
concentrations or longer incubation times.
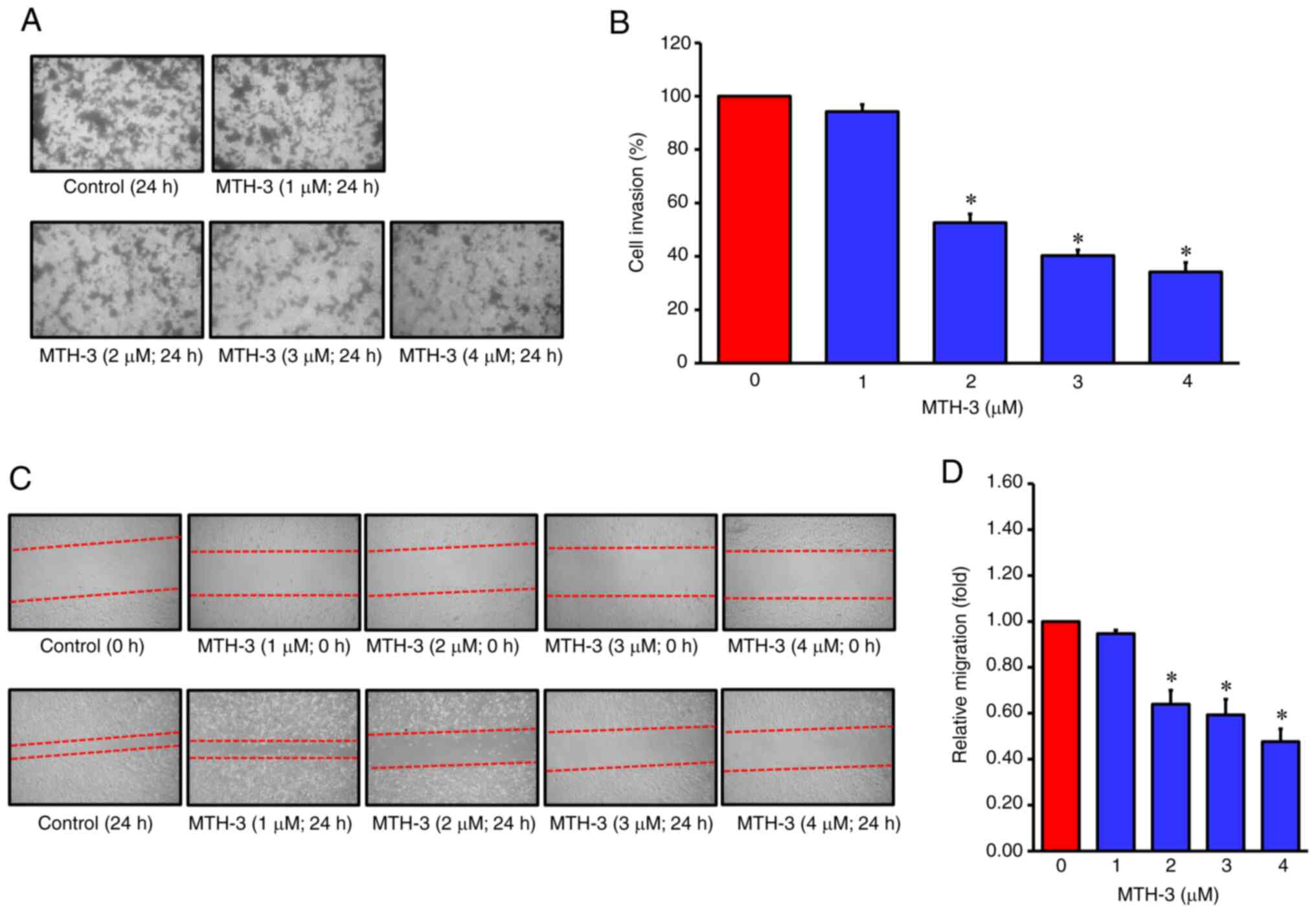 | Figure 2.MTH-3 suppressed invasion and
migration in human breast adenocarcinoma MDA-MB-231 cells. (A) The
invasion ability of MDA-MB-231 cells was evaluated using a
Matrigel®-coated invasion chamber. Following treatment
with various concentrations of MTH-3 for 24 h, the invading
MDA-MB-231 cells in the lower chamber were stained and subsequently
counted under a light microscope (magnification, ×200). (B) Tumor
cell invasion was semi-quantified. (C) The ability of migration of
MDA-MB-231 cells was evaluated by wound healing assay. Following
treatment with the various concentrations of MTH-3 for 24 h in
serum-free Leibovitz's L-15 medium, MDA-MB-231 cells were
photographed. (D) The migrated tumor cells were quantified. Data
are presented as the mean ± standard deviation of three
experiments. *P<0.05. MTH-3,
(1E,3Z,6E)-3-hydroxy-5-oxohepta-1,3,6-triene-1,7-diyl)bis(2-methoxy-4,1-phenylene)bis(3-hydroxy-2-hydroxymethyl)-2-methyl
propanoate. |
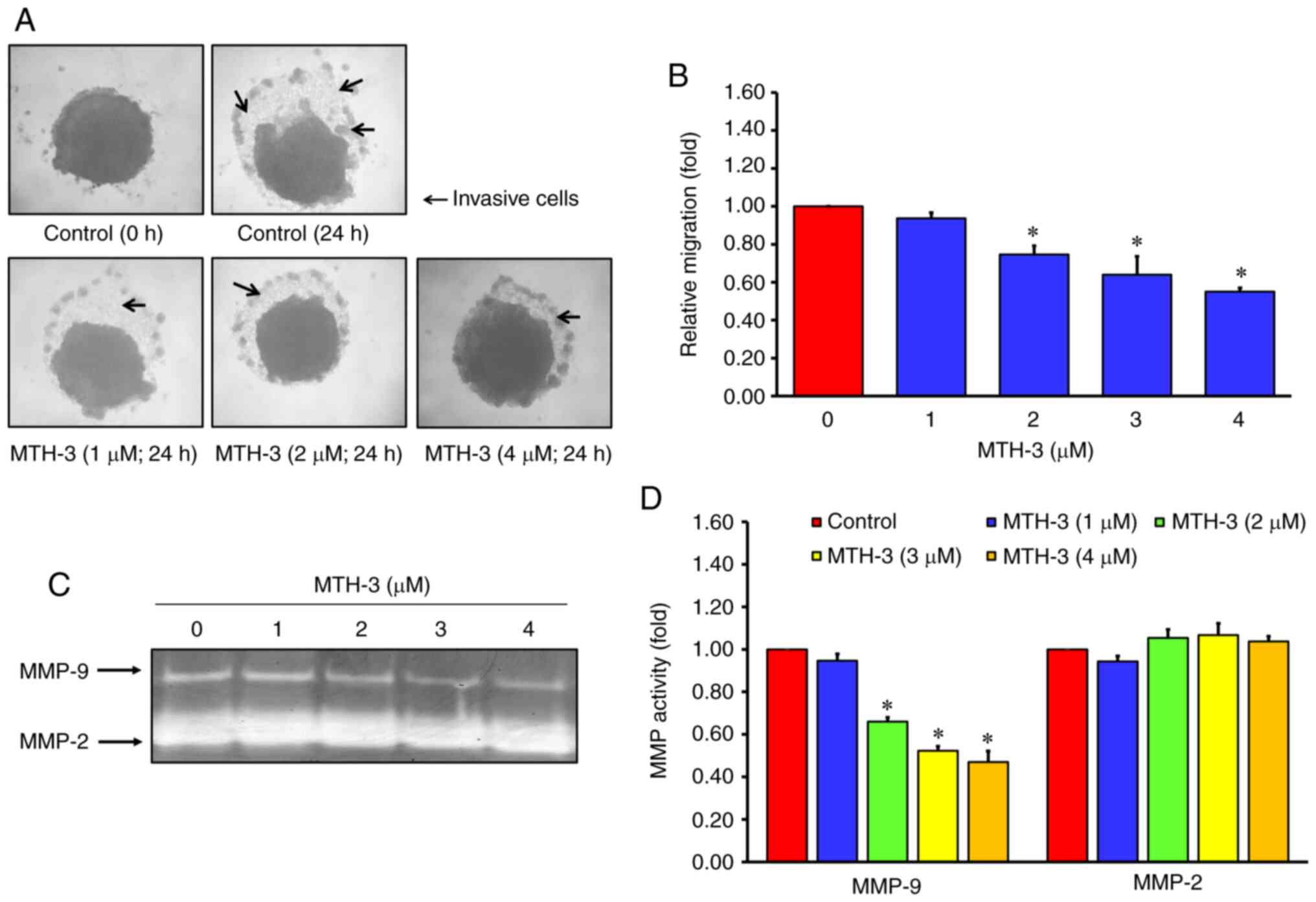 | Figure 3.MTH-3 inhibited MMP-9 activity, and
suppressed tumor spheroid invasion and migration in MDA-MB-231
human breast adenocarcinoma cells. (A) To mimic the complexity and
heterogeneity of clinical tumors, a 3D spheroid invasion assay was
performed. Following treatment with the indicated concentrations of
MTH-3 for 24 h, MTH-3 significantly inhibited MDA-MB-231 spheroid
invasion in the Matrigel-coated area, and (B) cell invasion was
semi-quantified. Arrows indicate invasive tumor cells. (C) The
activity of MMP-2 and −9 was evaluated by gelatin zymography, and
(D) the results were quantified. Data are presented as the mean ±
standard deviation of three experiments. *P<0.05. MTH-3,
(1E,3Z,6E)-3-hydroxy-5-oxohepta-1,3,6-triene-1,7-diyl)bis(2-methoxy-4,1-phenylene)bis(3-hydroxy-2-hydroxymethyl)-2-methyl
propanoate. |
MTH-3 suppresses the activity of MMP-9
in MDA-MB-231 human breast adenocarcinoma cells
Gelatin zymography was performed to further
determine the role of MMP-2/-9 activity in MTH-3-treated MDA-MB-231
cells. Following MTH-3 treatment for 24 h, the conditioned media of
tumor cells with different concentrations of MTH-3 (1, 2, 3 and 4
µM) or with 0.1% DMSO were collected to measure the gelatinase
activity of MMP-2 and −9. The results revealed that MTH-3 decreased
MMP-9 activity in a concentration-dependent manner in MDA-MB-231
cells in vitro (Fig. 3C and
D).
MTH-3 inhibits invasion through the
MAPK/ERK/AKT signaling pathway and causes cell cycle arrest in
MDA-MB-231 human breast adenocarcinoma cells
To gain insight into the biological activity of
MTH-3 in MDA-MB-231 cells, RNA sequencing transcriptional profile
analysis was performed. As shown in Fig.
4A, three replicates for normalized RNA-sequencing data from
MTH-3-treated samples and the control group were clustered
separately using unsupervised Principal Component Analysis,
indicating a significantly different Gene Expression Omnibus
analysis. Genes in blue were highly expressed, and those in red
were expressed at low levels. Fig. 4B
shows the differential expression of an MA plot. Red dots represent
significantly upregulated genes, and blue dots symbolized
significantly downregulated genes. Fig.
4C contains a bar graph of significantly up- or downregulated
genes between MTH-3-treated and control groups. A total of 315
genes were upregulated and 648 downregulated. To further determine
the physiological activities of the genes and associated functions,
the KEGG database were used (35).
KEGG pathway analysis and hypergeometric tests were performed to
identify the pathways of the DEGs and related pathways that were
significantly enriched compared to the transcriptome background. In
Fig. 4D, the scatter plot is used for
the graphical representation of the KEGG pathway enrichment
analysis, which is measured by the Rich factor, Q-value, and the
number of genes enriched in these pathways. The top 20 KEGG
pathways, most significantly enriched for the analysis, were
selected and shown. The enriched KEGG pathways are shown in
Fig. 5. Genes in red are upregulated
and those in blue are downregulated. Gene ontology (GO) enrichment
analysis revealed that the MTH-3-altered expression of genes was
largely associated with the MAPK/ERK/AKT signaling cascade and cell
cycle pathway. The sequencing raw data are shown in Table SI.
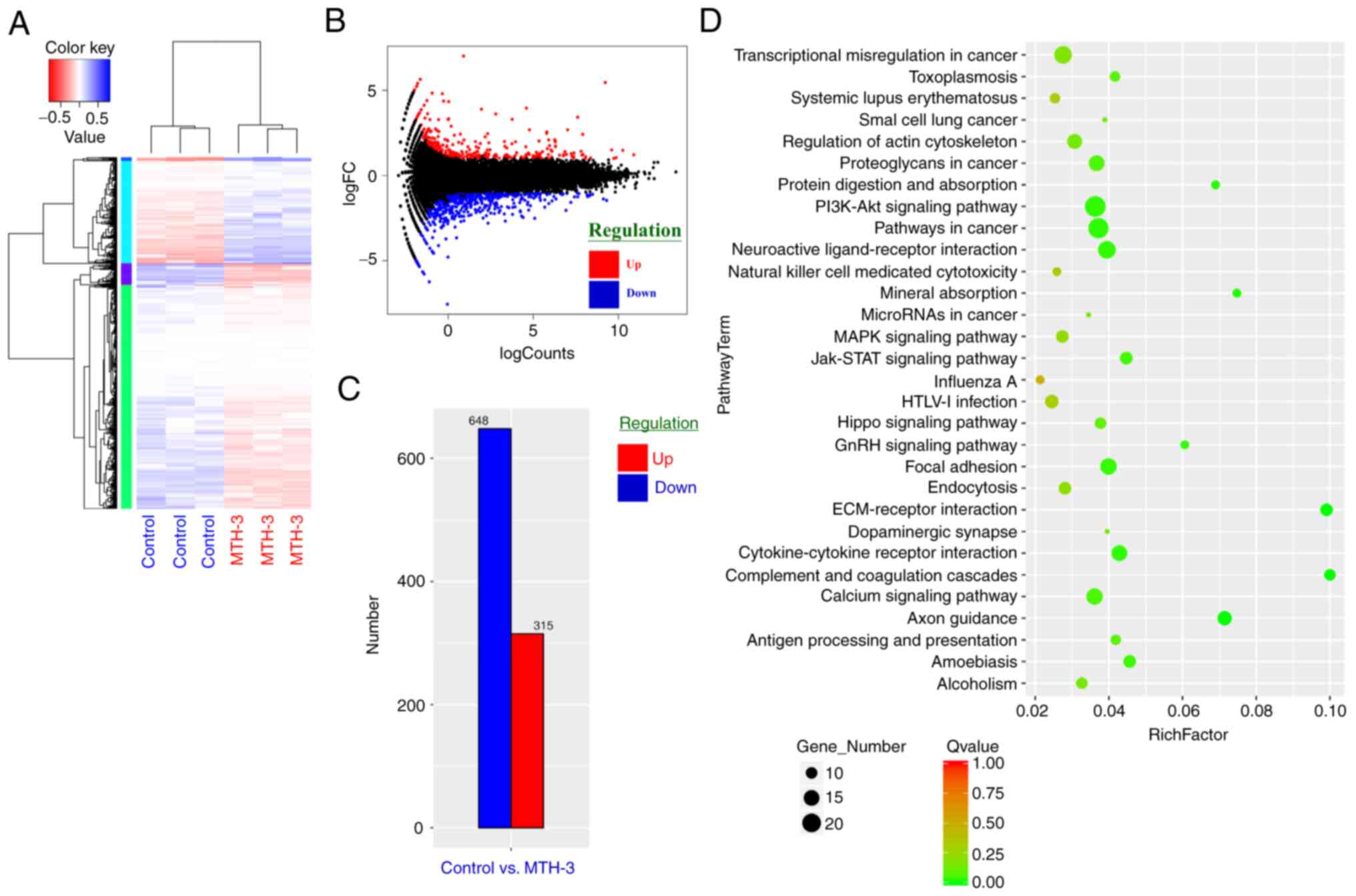 | Figure 4.RNA-sequencing transcriptional
profile analysis was performed to gain insight into the biologic
activity induced by MTH-3 in MDA-MB-231 cells. (A) The log10 (RPKM
+ 1) values of differentially expressed genes were used for cluster
analysis. Genes with a high expression are shown in blue and those
with a low expression in red. (B) Differential expression of MA
plot. Red dots represent significantly upregulated genes and blue
dots significantly downregulated genes. (C) Bar graph of genes that
were significantly up- or downregulated between the MTH-3-treated
and control groups. (D) Scatter plot representing the KEGG
enrichment of differentially expressed genes. x-axis, rich Factor;
y-axis, KEGG pathways. The number of differentially expressed genes
in the pathway is positively correlated to the size of these dots.
Color coding indicates different value ranges. KEGG, Kyoto
Encyclopedia of Genes and Genomes. MTH-3,
(1E,3Z,6E)-3-hydroxy-5-oxohepta-1,3,6-triene-1,7-diyl)bis(2-methoxy-4,1-phenylene)bis(3-hydroxy-2-hydroxymethyl)-2-methyl
propanoate. |
Subsequently, the protein expression levels were
examined using antibody microarray analysis and western blot
analysis to investigate the correlation between mRNA and protein
expression levels, since mRNA expression analysis can be inaccurate
and potentially misleading (33). The
results presented in Table I revealed
that MTH-3 treatment significantly downregulated phosphorylated
MAPK p38α, MAPK/ERK protein-serine kinase (MEK2), ERK1/2 and
proline-rich AKT substrate 40 kDa (PRAS40). Furthermore, MTH-3
significantly downregulated phosphorylated Cyclin A, Cyclin D1,
cell division control protein 42 (CDC42) and cyclin-dependent
protein-serine kinase 2 (CDK2). As shown in Fig. 6, the protein expression of p-AKT,
p-ERK, p-p38 and p-JNK was decreased. In combination, the present
results demonstrated that MTH-3 suppressed the MAPK/ERK/AKT
signaling pathway and cell cycle-related protein phosphorylation in
MDA-MB-231 human breast adenocarcinoma cells. The original images
of the integral western blot gels are shown in Fig. S1.
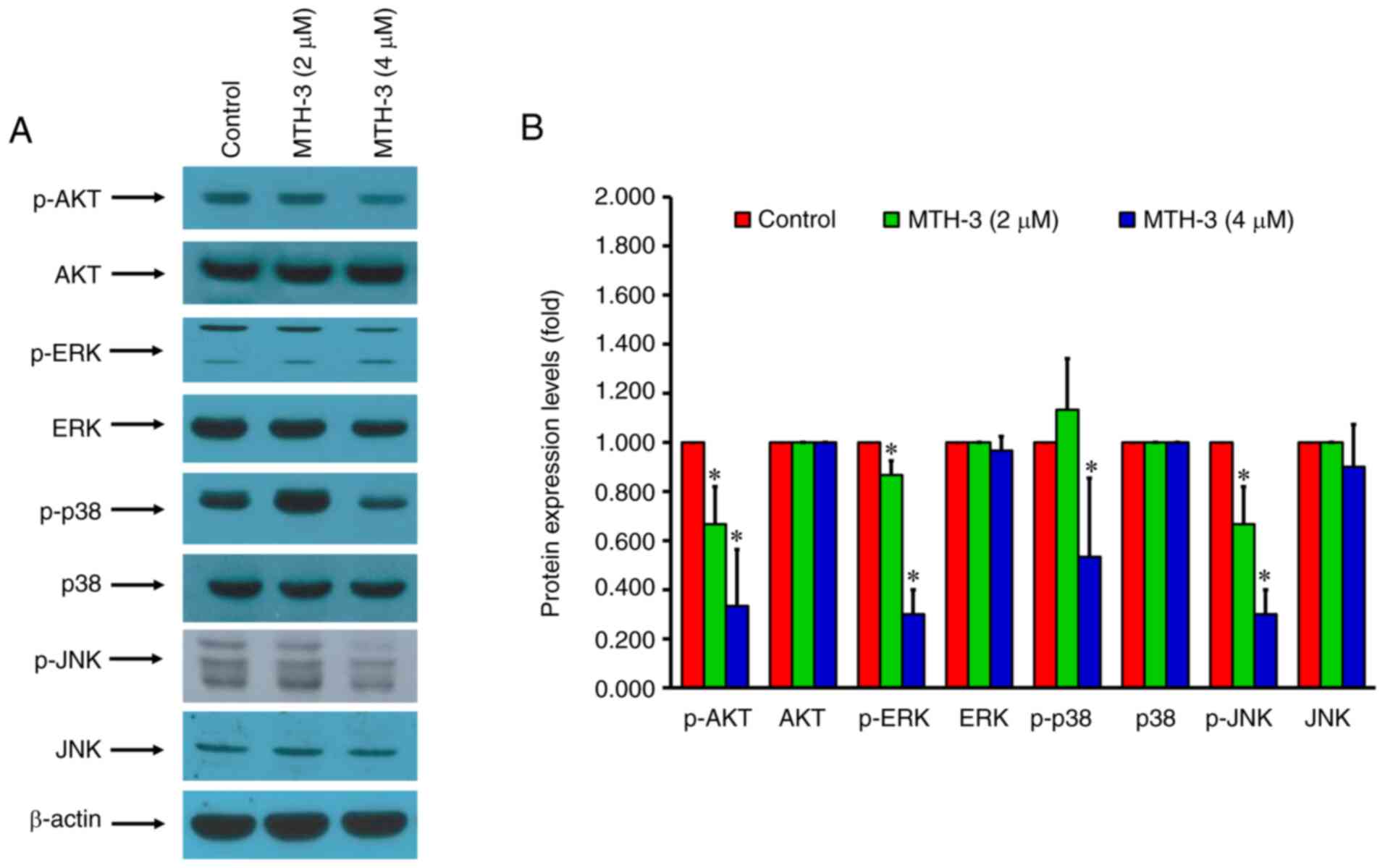 | Figure 6.MTH-3 modulates MAPK/ERK/AKT
signaling-related protein levels in MDA-MB-231 human breast
adenocarcinoma cells. (A) Western blot analysis shows a decreased
expression of p-AKT, p-ERK, p-p38/MAPK and p-JNK proteins. (B)
Quantitative analysis of the intensities of protein bands. MTH-3,
(1E,3Z,6E)-3-hydroxy-5-oxohepta-1,3,6-triene-1,7-diyl)bis(2-methoxy-4,1-phenylene)bis(3-hydroxy-2-hydroxymethyl)-2-methyl
propanoate; p-, phosphorylated. |
 | Table I.Summary of antibody microarray
analysesa. |
Table I.
Summary of antibody microarray
analysesa.
| Antibody codes | Target protein
name | Phospho site
(Human) | % CFC (MTH-3 from
CTL) | Z-ratio (MTH-3,
CTL) |
|---|
| NN024 | CDC42 | Pan-specific | −44 | −1.76 |
| NK026-3 | CDK2 | Pan-specific | −34 | −1.26 |
| NN028 | Cyclin A | Pan-specific | −51 | −2.03 |
| NN030-1 | Cyclin D1 | Pan-specific | −71 | −3.49 |
| PK170-PK171 | ERK1/2 | T202+T185 | −35 | −1.31 |
| NK120-2 | p38/MAPK | Pan-specific | −36 | −1.31 |
| NK120-4 | p38a/MAPK | Pan-specific | −43 | −1.68 |
| PK049-2 | MEK2 | T394 | −52 | −2.18 |
| PN062 | Proline-rich Akt
substrate 40 kDa (PRAS40) | T246 | −47 | −1.88 |
Discussion
MTH-3 is a novel bis(hydroxymethyl) alkanoate
curcuminoid derivative, designed by Hsieh et al (21). Findings of that study revealed that
MTH-3 was effective against numerous breast cancer cell lines and
induced limited toxicity to normal tissues in an established
xenograft nude mouse model of MDA-MB-231 cells. In addition, Chang
et al (22) identified a
synergistic activity of MTH-3 combined with doxorubicin in the
inhibition of MDA-MB-231 cell growth. MTH-3 treatment induced cell
cycle arrest, as well as the apoptotic and autophagic pathways in
MDA-MB-231 cells. To the best of our knowledge, the present study
was the first to demonstrate that MTH-3 inhibited the invasion of
MDA-MB-231 cells and elucidate the potential signaling pathways. In
anti-metastasis drug discovery, the compounds not only exhibit the
cytotoxic effect at high concentrations but also possess
anti-metastasis activity at low concentrations (36). Our previous findings demonstrated that
MTH-3 inhibited cell proliferation and induced G2/M
arrest, cell autophagy and apoptosis at a high concentration (>5
µM) in MDA-MB-231 cells. Results of the present study revealed that
a low concentration (2, 3 and 4 µM) of MTH-3 predominantly
inhibited MDA-MB-231 tumor cell migration and invasion (Figs. 2 and 3),
but did not induce cell apoptosis by TUNEL assay (Fig. 1C and D).
Curcumin has been proven to be safe and effective in
inhibiting cancer cell growth (16).
However, poor gastric absorption and low systemic bioavailability
prevent the pharmacological properties of curcumin from reaching
the target tissues. Researchers have developed several methods to
overcome the poor hydrophilicity of curcumin, including
nanoparticles, liposomes, phospholipid complexes, micelles, and
cyclodextrin encapsulation (37).
Furthermore, the half-life of curcumin is short, resulting in low
bioavailability. Several novel derivatives of curcumin have been
developed to delay the metabolism into glucuronides and sulfates
through a phase II transformation, replacing its phenolic OH groups
with ester (38,39). MTH-3, a novel curcumin derivative
designed by Hsieh et al (21),
was proven to have a superior solubility in water and alcohol than
curcumin in the previous study, with a 10-fold higher potency.
MTH-3 significantly inhibited MDA-MB-231 cell
invasion and migration in the present study. Cancer metastasis is a
complex process. The basement membrane and extracellular matrix are
major physical barriers to inhibiting cancer cell invasion and
migration (26). MMPs play a key role
in degrading the basement membrane and extracellular matrix.
Previous findings have shown that tumor cells can specifically
produce MMP-2 and −9 to destroy these natural barriers, resulting
in the invasion and migration of tumor cells into adjacent tissue
and blood vessels (40). MMP-2, which
can degrade type V, VI and X collagens, gelatins and IV collagen in
the basement membrane, was found to be constitutively expressed in
various tissues, including cancer cells, rather than as part of the
initial response to invasion (41).
MMP-9 can be secreted extracellularly to degrade type IV collagens
and fibronectins, which can be stimulated by various inflammatory
cytokines and growth factors during pathological processes,
including affecting the adhesion ability of tumor cells (42). The results of the present study showed
that MTH-3 inhibits MDA-MB-231 cell invasion and migration by
decreasing the activity of MMP-9, instead of that of MMP-2. These
results were consistent with those reported by Fan et al
(43), who demonstrated that casticin
inhibits MMP-9 protein expression and activity in breast cancer
cells.
Numerous studies have demonstrated the
anti-metastasis effects of curcumin in breast cancer cells.
Coker-Gurkan et al reported that curcumin inhibits invasion
and metastasis by targeting NF-κB signaling in breast cancer cell
lines, including MCF-7, MDA-MB-453 and MDA-MB-231 (45). Gallardo and Calaf (46) and Hu et al (47) reported curcumin induces
anti-metastasis activity through epithelial-mesenchymal transition.
Guan et al (48) reported that
curcumin suppresses migration in MDA-MB-231 cells through PI3K/AKT
signaling pathway. To the best of our knowledge, the present study
was the first to report that MTH-3, a novel curcuminoid derivative,
suppresses tumor invasion in breast cancer. The possible signal
transduction was further investigated.
Next-generation sequencing (NGS)-based molecular
diagnosis and analysis is becoming one of the major tools of
personalized treatment and drug development (49). RNA expression profile analysis
provides a more accurate analysis of the tumor phenotype compared
with genome analysis, which makes it the most powerful tool of high
throughput quantitative transcriptomics (50). The expression levels of targets of
molecular medicines were analyzed and profiling of the activation
of the relevant molecular pathways was used to enable the
personalized prescription of a wide range of molecular-targeted
therapies (51). To evaluate the
possible signaling pathways of MTH-3 in MDA-MB-231 cells, whole
transcriptome sequencing analysis of MTH-3-exposed and control
groups was performed in the present study. The results showed that
MTH-3-altered gene expressions were markedly associated with cell
invasion and MAPK/ERK/AKT signaling pathway. The MAPK/ERK/AKT
signaling pathway was suppressed in MDA-MB-231 human breast
adenocarcinoma cells after MTH-3 treatment, as shown in Fig. 5. Inhibition of the signaling
transduction was confirmed by western blot analysis, as shown in
Fig. 6. Furthermore, cell
cycle-related gene expression was also decreased. Since mRNA
expression analysis can be inaccurate and potentially misleading,
antibody microarray analysis was subsequently performed to
investigate the correlation between mRNA and protein expression
levels (52,53). The results in Table I revealed that treatment of MTH-3
significantly downregulated phosphorylated ERK1/2, p38/MAPK, MEK2
and proline-rich Akt substrate 40 kDa (PRAS40). MTH-3 also
significantly downregulated phosphorylated cyclins A and D1, CDC42
and CDK2.
The MAPK/ERK and PI3K/AKT signaling pathways play an
important role in cancer cell survival, proliferation, apoptosis,
invasion and metastasis (54,55). The aberrant activation of this
signaling pathway induces the survival, proliferation and
metastasis of breast cancer cells (54). Chen et al (56) reported that curcumin inhibits
doxorubicin-induced epithelial-mesenchymal transition through the
suppression of PI3K/AKT and TGF-β signaling transduction in TNBC.
Berrak et al (57) reported
that curcumin induced cell cycle arrest and inhibited PI3K
signaling transduction in MCF-7 breast cancer cells. Guan et
al (48) demonstrated that
curcumin suppresses proliferation and migration through
autophagy-dependent AKT degradation in MDA-MB-231 cells.
Furthermore, several studies have demonstrated that
cyclin proteins regulate tumor cell invasion and metastasis. Cell
cycle arrest inhibits tumor invasion and metastasis (58–61). Fusté
et al (58), Body et al
(62), and Chen et al
(63) reported that cyclin D1 fosters
tumor cell invasion and metastasis through cytoplasmic mechanisms.
CDC42 knockdown was shown to inhibit tumor cell migration and
invasion, and be associated with the downregulation of cyclins A,
D1 and E/CDK2 (14,59,60,64). In
previous studies, MTH-3 was found to inhibit TNBC cell
proliferation and induce apoptosis through the autophagic pathway
and cause cell cycle arrest with a higher potency than curcumin
(21,22). The present study demonstrated the
ability of MTH-3 to inhibit the invasion of TNBC cells through the
MAPK/ERK/AKT signaling pathways and cell cycle regulatory
cascade.
In conclusion, the present study revealed that MTH-3
inhibits tumor invasiveness via the MAPK/ERK/AKT signaling pathway
and cell cycle regulatory cascade in human adenocarcinoma
MDA-MB-231 cells. Significant information with respect to the
possible signal transduction of MTH-3 in TNBC was provided in the
current study, and the results suggested that MTH-3 may be used in
the treatment of breast cancer medication in the future.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
This study was supported in part through the Medical
Research Core Facilities, Office of Research and Development at
China Medical University (Taichung, Taiwan), and Yen Tjing Ling
Medical Foundation (Taipei, Taiwan). We also thank Mr. Kai-Hsiang
Chang and Mr. Chin-Chen Lin (Tekon Scientific Corp.) and Mr.
Chang-Wei Li (AllBio Science Incorporated, Taiwan) for their
assistance and support with the equipment in this study.
Funding
Funding for this study was provided in part by China
Medical University Hospital, Taichung, Taiwan (DMR-109-147),
Ministry of Science and Technology, Taiwan (MOST
109-2320-B-039-041-), Taipei Veterans General hospital, Taipei,
Taiwan (V110B-038), and Yen Tjing Ling Medical Foundation, Taipei,
Taiwan (CI-110-6). Chinese Medicine Research Center, China Medical
University from The Featured Areas Research Center Program within
the framework of the Higher Education Sprout Project by the
Ministry of Education (MOE) in Taiwan also provided partial
support.
Availability of data and materials
Data of transcriptome sequencing in this published
article have been uploaded to the European Nucleotide Archive.
Accession no.: ERP128028.
Authors' contributions
YJC, FJT, SCK and JSY contributed to the study
design. YJC, DTB, LCC, MTH and JSY conducted the experiments. YJC,
FJT, CCL and JSY analyzed the data. YJC, CCL and JSY confirmed the
authenticity of all the raw data. YJC, CCL, JSY, and SCK wrote and
revised the paper. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
TNBC
|
triple-negative breast cancer
|
|
ER
|
estrogen receptor
|
|
GO
|
gene ontology
|
|
(HER2/neu)
|
human epidermal growth factor
receptor-2
|
|
PR
|
progesterone receptor
|
|
FBS
|
fetal bovine serum
|
|
DEGs
|
differentially expressed genes
|
|
KEGG
|
Kyoto Encyclopedia of Genes and
Genomes
|
|
PI3K
|
phosphoinositide 3-kinase
|
|
RPKM
|
reads per kilobase per million mapped
reads
|
|
p38/MAPK
|
mitogen-activated protein-serin kinase
p38 α
|
|
MEK2
|
MAPK/ERK protein-serine kinase
|
|
PRAS40
|
proline-rich Akt substrate 40 kDa
|
|
CDC42
|
cell division control protein 42
|
|
CDK2
|
cyclin-dependent protein-serine kinase
2
|
|
ERK1/2
|
extracellular regulated protein-serine
kinase 1 and 2
|
|
MMP
|
matrix metalloproteinase
|
References
|
1
|
McGuire A, Brown JA, Malone C, McLaughlin
R and Kerin MJ: Effects of age on the detection and management of
breast cancer. Cancers (Basel). 7:908–929. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Feng Y, Spezia M, Huang S, Yuan C, Zeng Z,
Zhang L, Ji X, Liu W, Huang B, Luo W, et al: Breast cancer
development and progression: Risk factors, cancer stem cells,
signaling pathways, genomics, and molecular pathogenesis. Genes
Dis. 5:77–106. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Key TJ, Bradbury KE, Perez-Cornago A,
Sinha R, Tsilidis KK and Tsugane S: Diet, nutrition, and cancer
risk: What do we know and what is the way forward? BMJ.
368:m5112020. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wang X, Zhang H and Chen X: Drug
resistance and combating drug resistance in cancer. Cancer Drug
Resist. 2:141–160. 2019.
|
|
5
|
Chen T, Xiong H, Yang JF, Zhu XL, Qu RY
and Yang GF: Diaryl ether: A privileged scaffold for drug and
agrochemical discovery. J Agric Food Chem. 68:9839–9877. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Newman DJ and Cragg GM: Natural products
as sources of new drugs over the 30 years from 1981 to 2010. J Nat
Prod. 75:311–335. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Niwa T, Yokoyama Si, Mochizuki M and Osawa
T: Curcumin metabolism by human intestinal bacteria in vitro. J
Func Foods. 61:1034632019. View Article : Google Scholar
|
|
8
|
Yang H, Du Z, Wang W, Song M, Sanidad K,
Sukamtoh E, Zheng J, Tian L, Xiao H, Liu Z and Zhang G:
Structure-activity relationship of curcumin: Role of the methoxy
group in anti-inflammatory and anticolitis effects of curcumin. J
Agric Food Chem. 65:4509–4515. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jing S, Zou H, Wu Z, Ren L, Zhang T, Zhang
J and Wei Z: Cucurbitacins: Bioactivities and synergistic effect
with small-molecule drugs. J Func Foods. 72:1040422020. View Article : Google Scholar
|
|
10
|
Mathew D and Hsu WL: Antiviral potential
of curcumin. J func foods. 40:692–699. 2018. View Article : Google Scholar
|
|
11
|
Zaheri Z, Fahremand F, Rezvani ME,
Karimollah A and Moradi A: Curcumin exerts beneficial role on
insulin resistance through modulation of SOCS3 and Rac-1 pathways
in type 2 diabetic rats. J Func Foods. 60:1034302019. View Article : Google Scholar
|
|
12
|
Tomeh MA, Hadianamrei R and Zhao X: A
review of curcumin and its derivatives as anticancer agents. Int J
Mol Sci. 20:10332019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bondì ML, Emma MR, Botto C, Augello G,
Azzolina A, Di Gaudio F, Craparo EF, Cavallaro G, Bachvarov D and
Cervello M: Biocompatible lipid nanoparticles as carriers to
improve curcumin efficacy in ovarian cancer treatment. J Agric Food
Chem. 65:1342–1352. 2017. View Article : Google Scholar
|
|
14
|
DiMarco-Crook C, Rakariyatham K, Li Z, Du
Z, Zheng J, Wu X and Xiao H: Synergistic anticancer effects of
curcumin and 3′,4′-didemethylnobiletin in combination on colon
cancer cells. J Food Sci. 85:1292–1301. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Shehzad A, Wahid F and Lee YS: Curcumin in
cancer chemoprevention: Molecular targets, pharmacokinetics,
bioavailability, and clinical trials. Arch Pharm (Weinheim).
343:489–499. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Shanmugam MK, Rane G, Kanchi MM, Arfuso F,
Chinnathambi A, Zayed ME, Alharbi SA, Tan BK, Kumar AP and Sethi G:
The multifaceted role of curcumin in cancer prevention and
treatment. Molecules. 20:2728–2769. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yang KY, Lin LC, Tseng TY, Wang SC and
Tsai TH: Oral bioavailability of curcumin in rat and the herbal
analysis from Curcuma longa by LC-MS/MS. J Chromatogr B
Analyt Technol Biomed Life Sci. 853:183–189. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Marczylo TH, Verschoyle RD, Cooke DN,
Morazzoni P, Steward WP and Gescher AJ: Comparison of systemic
availability of curcumin with that of curcumin formulated with
phosphatidylcholine. Cancer Chemother Pharmacol. 60:171–177. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ma Y, Chen S, Liao W, Zhang L, Liu J and
Gao Y: Formation, physicochemical stability, and redispersibility
of curcumin-loaded rhamnolipid nanoparticles using the pH-Driven
method. J Agric Food Chem. 68:7103–7111. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cuomo F, Perugini L, Marconi E, Messia MC
and Lopez F: Enhanced curcumin bioavailability through nonionic
surfactant/caseinate mixed nanoemulsions. J Food Sci. 84:2584–2591.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hsieh MT, Chang LC, Hung HY, Lin HY, Shih
MH, Tsai CH, Kuo SC and Lee KH: New bis (hydroxymethyl) alkanoate
curcuminoid derivatives exhibit activity against triple-negative
breast cancer in vitro and in vivo. Eur J Med Chem. 131:141–151.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chang LC, Hsieh MT, Yang JS, Lu CC, Tsai
FJ, Tsao JW, Chiu YJ, Kuo SC and Lee KH: Effect of bis
(hydroxymethyl) alkanoate curcuminoid derivative MTH-3 on cell
cycle arrest, apoptotic and autophagic pathway in triple-negative
breast adenocarcinoma MDA-MB-231 cells: An in vitro study.
Int J Oncol. 52:67–76. 2018.PubMed/NCBI
|
|
23
|
Wu KM, Hsu YM, Ying MC, Tsai FJ, Tsai CH,
Chung JG, Yang JS, Tang CH, Cheng LY, Su PH, et al: High-density
lipoprotein ameliorates palmitic acid-induced lipotoxicity and
oxidative dysfunction in H9c2 cardiomyoblast cells via ROS
suppression. Nutr Metab (Lond). 16:362019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lin KH, Li CY, Hsu YM, Tsai CH, Tsai FJ,
Tang CH, Yang JS, Wang ZH and Yin MC: Oridonin, A natural
diterpenoid, protected NGF-differentiated PC12 cells against
MPP+-and kainic acid-induced injury. Food Chem Toxicol.
133:1107652019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ha HA, Chiang JH, Tsai FJ, Bau DT, Juan
YN, Lo YH, Hour MJ and Yang JS: Novel quinazolinone MJ-33 induces
AKT/mTOR-mediated autophagy-associated apoptosis in 5FU-resistant
colorectal cancer cells. Oncol Rep. 45:680–692. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chiu YJ, Hour MJ, Jin YA, Lu CC, Tsai FJ,
Chen TL, Ma H, Juan YN and Yang JS: Disruption of IGF-1R signaling
by a novel quinazoline derivative, HMJ-30, inhibits invasiveness
and reverses epithelial-mesenchymal transition in osteosarcoma U-2
OS cells. Int J Oncol. 52:1465–1478. 2018.PubMed/NCBI
|
|
27
|
Liu SC, Tsai CH, Wu TY, Tsai CH, Tsai FJ,
Chung JG, Huang CY, Yang JS, Hsu YM, Yin MC, et al:
Soya-cerebroside reduces IL-1β-induced MMP-1 production in
chondrocytes and inhibits cartilage degradation: Implications for
the treatment of osteoarthritis. Food Agric Immunol. 30:620–632.
2019. View Article : Google Scholar
|
|
28
|
Vinci M, Box C and Eccles SA:
Three-dimensional (3D) tumor spheroid invasion assay. J Vis Exp.
e526862015.PubMed/NCBI
|
|
29
|
Howes AL, Richardson RD, Finlay D and
Vuori K: 3-Dimensional culture systems for anti-cancer compound
profiling and high-throughput screening reveal increases in EGFR
inhibitor-mediated cytotoxicity compared to monolayer culture
systems. PLoS One. 9:e1082832014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Stacchiotti S, Pantaleo MA, Negri T,
Astolfi A, Tazzari M, Dagrada GP, Urbini M, Indio V, Maestro R,
Gronchi A, et al: Efficacy and biological activity of imatinib in
metastatic dermatofibrosarcoma protuberans (DFSP). Clin Cancer Res.
22:837–846. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kim D, Langmead B and Salzberg SL: HISAT:
A fast spliced aligner with low memory requirements. Nat Methods.
12:357–360. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kanehisa M: Toward understanding the
origin and evolution of cellular organisms. Protein Sci.
28:1947–1951. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Cheadle C, Vawter MP, Freed WJ and Becker
KG: Analysis of microarray data using Z score transformation. J Mol
Diagn. 5:73–81. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Chiu YJ, Yang JS, Hsu HS, Tsai CH and Ma
H: Adipose-derived stem cell conditioned medium attenuates
cisplatin-triggered apoptosis in tongue squamous cell carcinoma.
Oncol Rep. 39:651–658. 2018.PubMed/NCBI
|
|
35
|
Plaimas K, Mallm JP, Oswald M, Svara F,
Sourjik V, Eils R and König R: Machine learning based analyses on
metabolic networks supports high-throughput knockout screens. BMC
Syst Biol. 2:672008. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhang Z, Rui W, Wang ZC, Liu DX and Du L:
Anti-proliferation and anti-metastasis effect of barbaloin in
non-small cell lung cancer via inactivating p38MAPK/Cdc25B/Hsp27
pathway. Oncol Rep. 38:1172–1180. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Mirzaei H, Shakeri A, Rashidi B, Jalili A,
Banikazemi Z and Sahebkar A: Phytosomal curcumin: A review of
pharmacokinetic, experimental and clinical studies. Biomed
Pharmacother. 85:102–112. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Hoehle SI, Pfeiffer E, Sólyom AM and
Metzler M: Metabolism of curcuminoids in tissue slices and
subcellular fractions from rat liver. J Agric Food Chem.
54:756–764. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Vareed SK, Kakarala M, Ruffin MT, Crowell
JA, Normolle DP, Djuric Z and Brenner DE: Pharmacokinetics of
curcumin conjugate metabolites in healthy human subjects. Cancer
Epidemiol Biomarkers Prev. 17:1411–1417. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Lu CC, Chen HP, Chiang JH, Jin YA, Kuo SC,
Wu TS, Hour MJ, Yang JS and Chiu YJ: Quinazoline analog HMJ-30
inhibits angiogenesis: Involvement of endothelial cell apoptosis
through ROS-JNK-mediated death receptor 5 signaling. Oncol Rep.
32:597–606. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Shimokawa Ki K, Katayama M, Matsuda Y,
Takahashi H, Hara I, Sato H and Kaneko S: Matrix metalloproteinase
(MMP)-2 and MMP-9 activities in human seminal plasma. Mol Hum
Reprod. 8:32–36. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Nishio K, Motozawa K, Omagari D, Gojoubori
T, Ikeda T, Asano M and Gionhaku N: Comparison of MMP2 and MMP9
expression levels between primary and metastatic regions of oral
squamous cell carcinoma. J Oral Sci. 58:59–65. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Fan L, Zhang Y, Zhou Q, Liu Y, Gong B, Lü
J, Zhu H, Zhu G, Xu Y and Huang G: Casticin inhibits breast cancer
cell migration and invasion by down-regulation of PI3K/Akt
signaling pathway. Biosci Rep. 38:BSR201807382018. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Palange AL, Di Mascolo D, Singh J, De
Franceschi MS, Carallo C, Gnasso A and Decuzzi P: Modulating the
vascular behavior of metastatic breast cancer cells by curcumin
treatment. Front Oncol. 2:1612012. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Coker-Gurkan A, Celik M, Ugur M, Arisan
ED, Obakan-Yerlikaya P, Durdu ZB and Palavan-Unsal N: Curcumin
inhibits autocrine growth hormone-mediated invasion and metastasis
by targeting NF-κB signaling and polyamine metabolism in breast
cancer cells. Amino Acids. 50:1045–1069. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Gallardo M and Calaf GM: Curcumin inhibits
invasive capabilities through epithelial mesenchymal transition in
breast cancer cell lines. Int J Oncol. 49:1019–1027. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Hu C, Li M, Guo T, Wang S, Huang W, Yang
K, Liao Z, Wang J, Zhang F and Wang H: Anti-metastasis activity of
curcumin against breast cancer via the inhibition of stem cell-like
properties and EMT. Phytomedicine. 58:1527402019. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Guan F, Ding Y, Zhang Y, Zhou Y, Li M and
Wang C: Curcumin suppresses proliferation and migration of
MDA-MB-231 breast cancer cells through autophagy-dependent Akt
degradation. PLoS One. 11:e01465532016. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Turro E, Astle WJ, Megy K, Gräf S, Greene
D, Shamardina O, Allen HL, Sanchis-Juan A, Frontini M, Thys C, et
al: Whole-genome sequencing of patients with rare diseases in a
national health system. Nature. 583:96–102. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Collado-Torres L, Nellore A, Kammers K,
Ellis SE, Taub MA, Hansen KD, Jaffe AE, Langmead B and Leek JT:
Reproducible RNA-seq analysis using recount2. Nat Biotechnol.
35:319–321. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Vamathevan J, Clark D, Czodrowski P,
Dunham I, Ferran E, Lee G, Li B, Madabhushi A, Shah P, Spitzer M
and Zhao S: Applications of machine learning in drug discovery and
development. Nat Rev Drug Discov. 18:463–477. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Zhou W, Yui MA, Williams BA, Yun J, Wold
BJ, Cai L and Rothenberg EV: Single-cell analysis reveals
regulatory gene expression dynamics leading to lineage commitment
in early T cell development. Cell Syst. 9:321–337. e9. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Byrne A, Cole C, Volden R and Vollmers C:
Realizing the potential of full-length transcriptome sequencing.
Philos Trans R Soc Lond B Biol Sci. 374:201900972019. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Zhang Y, Moerkens M, Ramaiahgari S, de
Bont H, Price L, Meerman J and van de Water B: Elevated
insulin-like growth factor 1 receptor signaling induces
antiestrogen resistance through the MAPK/ERK and PI3K/Akt signaling
routes. Breast Cancer Res. 13:R522011. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Lin CC, Chen KB, Tsai CH, Tsai FJ, Huang
CY, Tang CH, Yang JS, Hsu YM, Peng SF and Chung JG: Casticin
inhibits human prostate cancer DU 145 cell migration and invasion
via Ras/Akt/NF-κB signaling pathways. J Food Biochem.
43:e129022019. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Chen WC, Lai YA, Lin YC, Ma JW, Huang LF,
Yang NS, Ho CT, Kuo SC and Way TD: Curcumin suppresses
doxorubicin-induced epithelial-mesenchymal transition via the
inhibition of TGF-β and PI3K/AKT signaling pathways in
triple-negative breast cancer cells. J Agric Food Chem.
61:11817–11824. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Berrak Ö, Akkoç Y, Arısan ED, Çoker-Gürkan
A, Obakan-Yerlikaya P and Palavan-Ünsal N: The inhibition of PI3K
and NFκB promoted curcumin-induced cell cycle arrest at G2/M via
altering polyamine metabolism in Bcl-2 overexpressing MCF-7 breast
cancer cells. Biomed Pharmacother. 77:150–160. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Fusté NP, Fernández-Hernández R, Cemeli T,
Mirantes C, Pedraza N, Rafel M, Torres-Rosell J, Colomina N,
Ferrezuelo F, Dolcet X and Garí E: Cytoplasmic cyclin D1 regulates
cell invasion and metastasis through the phosphorylation of
paxillin. Nat Commun. 7:115812016. View Article : Google Scholar
|
|
59
|
Maldonado MDM and Dharmawardhane S:
Targeting rac and Cdc42 GTPases in cancer. Cancer Res.
78:3101–3111. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Stengel K and Zheng Y: Cdc42 in oncogenic
transformation, invasion, and tumorigenesis. Cell Signal.
23:1415–1423. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Huang TY, Peng SF, Huang YP, Tsai CH, Tsai
FJ, Huang CY, Tang CH, Yang JS, Hsu YM, Yin MC, et al:
Combinational treatment of all-trans retinoic acid (ATRA) and
bisdemethoxycurcumin (BDMC)-induced apoptosis in liver cancer Hep3B
cells. J Food Biochem. 44:e131222020. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Body S, Esteve-Arenys A, Miloudi H,
Recasens-Zorzo C, Tchakarska G, Moros A, Bustany S, Vidal-Crespo A,
Rodriguez V, Lavigne R, et al: Cytoplasmic cyclin D1 controls the
migration and invasiveness of mantle lymphoma cells. Sci Rep.
7:139462017. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Chen K, Jiao X, Ashton A, Di Rocco A,
Pestell TG, Sun Y, Zhao J, Casimiro MC, Li Z, Lisanti MP, et al:
The membrane-associated form of cyclin D1 enhances cellular
invasion. Oncogenesis. 9:832020. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Du DS, Yang XZ, Wang Q, Dai WJ, Kuai WX,
Liu YL, Chu D and Tang XJ: Effects of CDC42 on the proliferation
and invasion of gastric cancer cells. Mol Med Rep. 13:550–554.
2016. View Article : Google Scholar : PubMed/NCBI
|