Introduction
Colorectal cancer ranks as the fourth leading cause
of mortality worldwide, with ~900,000 deaths recorded annually
(1). It is the second most common
type of cancer in women and third most common in men (1,2).
Currently, combinatorial treatment based on surgery and adjuvant
chemotherapy or radiotherapy remains the main treatment approach
for colorectal cancer (3,4). Despite recent developments in targeted
therapy and immunotherapy, which have almost doubled the overall
survival time for non-metastatic cases to three years, clinicians
are still facing significant challenges when dealing with advanced
or treatment-resistant tumors (2,3).
Understanding the mechanism of tumor progression, metastasis and
treatment resistance in colorectal cancer is crucial to improving
prognosis.
The insulin-like growth factor 1 receptor (IGF1R) is
a membrane-based receptor tyrosine kinase (RTK) that plays various
roles in multiple biological events, including cell growth,
transformation, apoptosis, migration and invasion in both
physiological and pathological conditions (5–8). Similar
to other RTKs, the conventional activation for IGF1R requires
ligand-receptor binding with IGF1, which in turn activates the
PI3K-Akt and MAPK pathways (9,10). Our
previous studies reported that upon SUMOylation, membranous IGF1R
could undergo nuclear translocation and serve as a transcription
co-factor, thus regulating various cellular functions (11–14). Other
previous studies have revealed an association between nuclear IGF1R
(nIGF1R) and cell proliferation, tumorigenicity, resistance to EGFR
inhibition and DNA repair (6,15–17);
however, a global network study for protein function is still
required.
DNA double-strand breaks (DSBs) arise regularly in
cells and, when left unrepaired, cause senescence or cell death.
Homologous recombination (HR) and non-homologous end-joining (NHEJ)
are the two major DNA-repair pathways (18). While HR allows successful DSB repair
and healthy cell growth, NHEJ is more likely to contribute to
mutations and malignancy (18–20). When
DSBs are detected, the histone variant H2AX is phosphorylated by
ataxia-telangiectasia mutated kinase, which leads to the
recruitment of mediator of DNA damage checkpoint protein 1 (MDC1)
and activation of ring finger protein (RNF)8/RNF168-dependent
chromatin ubiquitination. p53-binding protein 1 (53BP1) then binds
to the ubiquitinated histone and recruits RAP1-interacting factor 1
(RIF1), thus preventing the activation of the HR pathway induced by
the association between breast cancer 1 and MRE11 homolog, double
strand break repair nuclease-RAD50 double strand break repair
protein-nibrin complex-bound CtBP-interacting protein (21,22). NHEJ
is initiated by the rapid binding of the X-ray repair cross
complementing 6-X-ray repair cross complementing 5 (Ku80)
heterodimer to the DNA ends, followed by the recruitment and
activation of DNA-dependent protein kinase catalytic subunit
(DNA-PKcs) (20). During DSB repair,
53BP1 serves not only as an important DSB-responsive factor, but a
key determinant of DSB repair pathway choice. It has been reported
that 53BP1 colocalizes and interacts with the structural protein
nuclear mitotic apparatus protein (NuMA) through the nucleoplasm.
In response to ionizing radiation (IR), the interaction is reduced
to allow 53BP1 to accumulate at DSB sites (23). Studies by Chitnis et al have
indicated that nIGF1R plays a pivotal role in regulating DSB repair
by both NHEJ and HR pathways (15,24);
however, the regulatory mechanism of this process remains
unclear.
In the present study, we investigated the
interactome of nIGF1R in colorectal cancer cell line SW480 using
immunoprecipitation-mass spectrometry (IP-MS) method. Validation of
protein-protein interaction between NuMA and nIGF1R was conducted
using co-immunoprecipitation and in situ proximity ligation
assay (PLA). The role of nIGF1R in modulating the NuMA-53BP1
complex and NHEJ repair pathway was further illustrated by in
situ PLA and immunofluorescence. The clinical significance of
NuMA-53BP1 and IGF1R-NuMA colocalization in colorectal cancer was
investigated using PLA in FFPE tissue samples.
Materials and methods
Cell culture and transfection
IGF1R-negative [(R−); mouse embryonic
fibroblast (MEF) igf1r−/−] and IGF1R-positive
(R+; R-overexpressing IGF1R) cells were obtained from Dr
R. Baserga (Thomas Jefferson University; Philadelphia, PA, USA).
The SW480 colorectal cancer cell line was purchased from the
American Type Culture Collection. All cell lines were cultured in
high-glucose DMEM (cat. no. 41965039; Thermo Fisher Scientific,
Inc.) supplemented with 10% FBS (cat. no. 16000044; Thermo Fisher
Scientific, Inc.). All cell lines were maintained at 37°C in a
humidified atmosphere containing 5% CO2 and controlled
for mycoplasma contamination using a Mycoalert™ kit (cat. no.
LT07-418; Lonza Group Ltd.). All human cell lines were short tandem
repeat-authenticated using an AmpFLSTR®
Identifiler® Plus kit (cat. no. A26182; Thermo Fisher
Scientific, Inc.).
The IGF1R cDNA sequence was subcloned into a
pBABE-puro vector (Cell Biolabs, Inc.) and transfected into a
Platinum-A cell line (Cell Biolabs, Inc.) for lentivirus packaging,
according to the manufacturer's instructions. To establish a stable
IGF1R-overexpressing cell line, SW480 cells were seeded into
24-well plates and infected with IGF1R-coding lentivirus particles
24 and 48 h following seeding. A total of 96 h after infection,
cells were sorted into single cells in 96-well plates and selected
with 2.5 µg/ml puromycin. After 2 weeks, the cell colonies were
selected and IGF1R overexpression was verified by western blot
analysis. The newly established IGF1R-overexpressing colorectal
cancer cell line was defined as SW480-OE.
Nuclear protein extraction and IP
Nuclear protein extraction was conducted in cells,
as previously described (25), and is
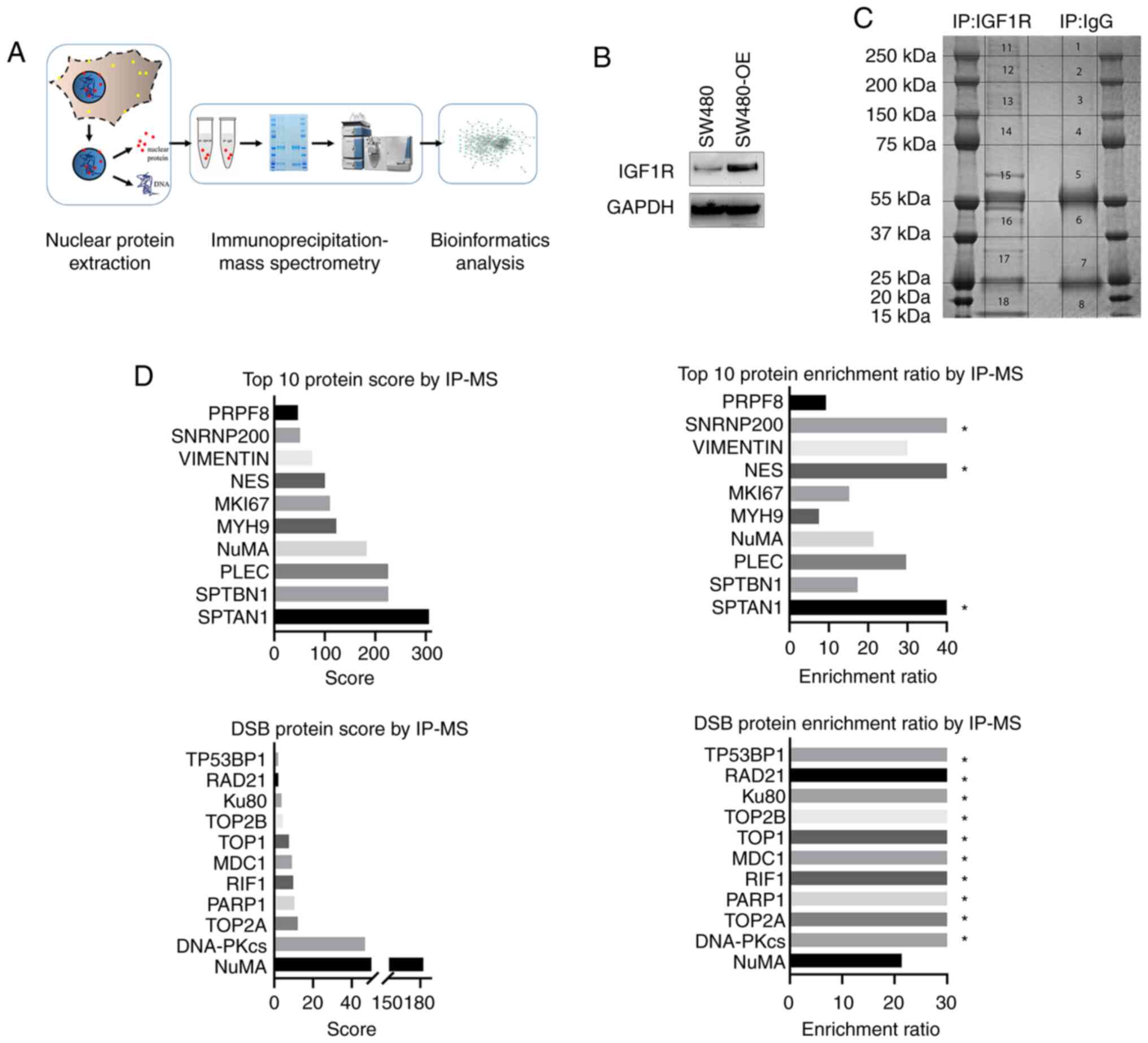
schematically shown in Fig. 1A. Cells
were removed and successively lysed in hypotonic lysis buffer (10
mM HEPES, pH 7.4, 10 mM KCl and 0.05% Nonidet P-40). Then, RIPA
lysis buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 1% Nonidet P-40, 1
mM EDTA and 0.25% sodium deoxycholate) containing Protease and
Phosphatase Inhibitor Cocktail (cat. no. 78447; Thermo Fisher
Scientific, Inc.) was used to extract the nuclear extract. Protein
concentration was measured using a Pierce™ BCA Protein Assay kit
(cat. no. 23227; Thermo Fisher Scientific, Inc.). Nuclear protein
(2–4 mg) was incubated with 4–7 µg mouse anti-IGF1R (cat. no.
556000; BD Biosciences) antibody coupled with Dynabeads™ Protein A
(cat. no. 10002D; Thermo Fisher Scientific, Inc.) overnight at 4°C.
The immune complexes were washed three times with lysis buffer and
eluted by boiling in SDS sample buffer (cat. no. NP0007; Thermo
Fisher Scientific, Inc.).
In-gel digestion and sample
preparation
The eluted proteins were separated by SDS-PAGE on a
NuPAGE 4–12% Bis-Tris protein Gel (Thermo Fisher Scientific, Inc.).
The proteins were visualized using the Colloidal Blue Staining kit
(Thermo Fisher Scientific, Inc.). Gels were cut into eight bands
according to the molecular mass. Each gel band was cut into
1-mm2 pieces and placed in a microcentrifuge tube. The
gels were destained [1:1 (v/v) mixture of 50 mM triethylammonium
bicarbonate buffer (TEAB) and 100% acetonitrile (ACN)] for 10 min,
which was repeated until the solution was clear. Subsequently, the
gels were incubated with 5 mM TCEP [0.5 M bond-breaker TCEP
solution (Thermo Fisher Scientific, Inc.)] (in 50 mM TEAB) for 30
min at 65°C, followed by a 30-min incubation at 37°C with 15 mM
chloroacetamide (in 50 mM TEAB). Lys-C [lysyl endopeptidase (Wako
Chemicals Ltd.)] and trypsin were prepared in 50 mM acetic acid and
added to the sample (50:1 protein:enzyme ratio). The incubation
time of Lys-C and trypsin was 4 h and overnight at 37°C,
respectively. The supernatants were transferred to a clean tube and
gel pieces were washed with 60% ACN and 5% trifluoroacetic acid
(TFA) in Milli-Q water. This step was repeated, and supernatants
were collected and dried in a vacuum centrifuge for 2–3 h. The
samples were resuspended in 0.1% (v/v) formic acid in Milli-Q for
liquid chromatography-mass spectrometry (LC-MS).
LC-MS/MS and database search
Online chromatography was performed using Dionex
UltiMate 3000 UPLC system coupled to a Q Exactive HF mass
spectrometer (Thermo Fisher Scientific, Inc.). Each sample was
separated on a 50 cm ×75 µm EASY-Spray analytical column (Thermo
Fisher Scientific, Inc.) using a 120-min gradient of a programmed
mixture of solvents A (0.1% formic acid in water) and B (95% ACN
and 5% water with 0.1% formic acid). MS data were acquired using a
Top 12 data-dependent acquisition method. Full Scan MS spectra were
acquired at 300-1,600 m/z at a resolution of 70,000 and AGC target
of 3e6; Top12 ddMS2 35,000 and 1e5 with an isolation window of 1.6
m/z.
Proteome Discoverer 2.1 software (Thermo Fisher
Scientific, Inc.) was used to analyze the Xcalibur® raw
files for subsequent protein identification and quantification.
Both Mascot 2.6.0 (Matrix Science, Inc.) and Sequest HT (Thermo
Fisher Scientific, Inc.) search engines were used to search against
the human-reviewed UniProt database. MS precursor mass tolerance
was set at 20 ppm, fragment mass tolerance at 0.05 Da and maximum
missed cleavage sites at 3. Only the spectrum peaks with a
signal-to-noise ratio of >4 were chosen for searches. The false
discovery rate was set to 1% at both the peptide-spectrum match
(PSM) and peptide levels. The Mascot score threshold for PSM was
set at 10.
Bioinformatics analysis of MS
data
Gene Ontology (GO) term and Reactome pathway
enrichment analyses were performed using the Search Tool for the
Retrieval of Interacting Genes/Proteins (STRING) database in
Cytoscape software (https://string-db.org/), and the enrichment was
calculated using the human genome as a reference (26,27).
Western blot analysis
Eluted proteins were separated by SDS-PAGE on NuPage
4–12% Bis-Tris Protein Gels (Thermo Fisher Scientific, Inc.) and
incubated with the following primary antibodies: Rabbit anti-IGF1R
(1:1,000, cat. no. 3024; Cell Signaling Technology, Inc.), rabbit
anti-NuMA (1:800, cat. no. 8967; Cell Signaling Technology, Inc.),
mouse anti-β-actin (1:5,000, cat. no. A5441; Merck KGaA) and rabbit
anti-53BP1 (1:1,000, cat. no. NB100-904; Novus Biologicals).
Following the primary antibody incubation, the membranes were
incubated with secondary anti-rabbit (1:2,000, cat. no. NA934; GE
Healthcare), -mouse (1:2,000, cat. no. NA931; GE Healthcare) or
-goat (1:2,000, cat. no. 31402; Thermo Fisher Scientific, Inc.) IgG
HRP-conjugated antibodies, followed by signal detection using an
iBright FL1500 imaging system (Thermo Fisher Scientific, Inc.). At
least three independent experiments were performed.
Fluorescent in situ proximity ligation
assay (PLA) in cell slides
Cells were seeded onto coverslips, fixed with 4%
PBS-buffered paraformaldehyde and permeabilized with 0.1% Triton
X-100. Following blocking for 30 min in blocking buffer (5% BSA, 5%
donkey serum and 0.3% Triton X-100 in PBS), cells were stained
according to the manufacturer's instructions of Duolink®
In Situ Detection reagents (Merck KGaA). Protein-protein
interactions were visualized as foci using a Zeiss LSM710 confocal
microscope (Carl Zeiss AG) and analyzed using ImageJ software
(V_1.8.0_172, National Institutes of Health). The antibodies used
in PLA were diluted in antibody diluent as follows: For IGF1R-NuMA
colocalization, mouse anti-IGF1R (cat. no. sc-390130; dilution,
1:50; Santa Cruz Biotechnology, Inc.) and rabbit anti-NuMA (cat.
no. 8967; dilution, 1:100; Cell Signaling Technology, Inc.); for
NuMA-53BP1 colocalization, rabbit anti-53BP1 (cat. no. NB100-904;
dilution, 1:150; Novus Biologicals) and mouse anti-NuMA (cat. no.
sc-56325; dilution, 1:50; Santa Cruz Biotechnology, Inc.). At least
three independent experiments were performed.
Patient selection and tissue
microarray (TMA) preparation
This study was approved by the Medical Ethics
Committee of Xiangya Hospital of Central South University
(Changsha, China; approval. no. 201403168). All patients provided
written informed consent for the use of their surgical specimens
for pathological examination. No personal information was disclosed
in this article. Between January 2014 and December 2016, 73
colorectal cancer and paired adjacent non-tumor tissues and related
clinical information were collected from patients who underwent
radical colorectal surgery at the Department of General Surgery,
Xiangya Hospital of Central South University. All tissues collected
were clinically and pathologically diagnosed as colorectal
cancer.
TMA preparation was conducted as previously
described (28). Tissues were excised
and fixed in 10% neutral-buffered formalin and then embedded in
paraffin blocks. Each paraffin-embedded section was cut into 4-µm
thick sections, deparaffinized and rehydrated. Hematoxylin and
eosin staining was performed to detect and mark typical tumor
sections in colorectal cancer tissues and the normal colorectal
mucosa in adjacent tissues, and was evaluated by a professional
pathologist. Paraffin-embedded sections measuring 2-mm in diameter
were separated from the original section, arranged and re-embedded
into the tissue microarray. A total of 73 colorectal cancer and
normal colorectal mucosa tissues were collected from different
patients in each tissue microarray slide.
PLA scoring for TMA
The slides were rehydrated by incubation in xylene
for 10 min, graded ethanol solutions (3×99, 2×95 and 1×70%) for 2
min each and washed with running water for 2 min. For antigen
retrieval, the sections were incubated in citrate antigen retrieval
solution (cat. no. S1699; Dako; Agilent Technologies, Inc.) and
microwaved at 750 W for 8 min and then at 350 W for 20 min
(sub-boiling). After cooling the tissue sections in a water bath
with running water for 10 min, the intrinsic peroxidase activity
was quenched by incubation in an H2O2
solution (dilution, 1:60; Merck KGaA) and incubated in the dark at
room temperature for 30 min. The following steps were performed
according to the Duolink® PLA BrightField Protocol
(Merck KGaA).
The TMAs were scored blindly by a clinical
pathologist. Total and nuclear PLA signals were evaluated for both
IGF1R-NuMA and NuMA-53BP1. Tumors were arbitrarily classified for
statistical comparisons: Tumors with no or very few signals were
scored as 0–1 (negative/weak); tumors with moderate signals (5–10
per cell/nuclei in the majority of cells) were scored as 2
(intermediate); and tumors with abundant signals (>10 signals
per cell/nuclei in the majority of cells) were scored as 3
(strong).
The clinical implication of IGF1R and NuMA was
further assessed using The Cancer Genome Atlas (TCGA, http://portal.gdc.cancer.gov/).
Statistical analysis
Statistical significance was assessed using an
unpaired Student's t-test using GraphPad Prism 8 (GraphPad
Software, Inc.). To assess the prognostic significance of PLA
staining in the TMAs, a χ2 test was used. Overall
survival was compared by Kaplan-Meier estimator and differences
were calculated using a log-rank test. P<0.05 was considered
statistically significant.
Results
Characterizing the nIGF1R interactome
in colorectal cancer cells
To further understand the function of IGF1R in the
colorectal cancer cell nuclei, immunoprecipitation-coupled mass
spectrometry (IP-MS) was conducted on the nuclear protein extract
of the IGF1R-overexpressing SW480-OE cell line (Fig. 1A and B). All IGF1R and IgG pulled-down
proteins were eluted and separated by SDS-PAGE electrophoresis, and
the gel was cut into pieces for enrichment in MS detection
(Fig. 1C). A total of 328
IGF1R-pulldown proteins were identified following the initial
database search; ≥1.5-fold higher abundance was required for the
proteins in the IGF1R pulled-down group compared with the IgG group
to qualify as potential nIGF1R interactors, to distinguish from
background binding proteins. In addition, common contaminants in
the mass spectrometer were eliminated as previously described
(29). Within these criteria, 197
potential nIGF1R interacting proteins were categorized and included
in the following network analysis (Tables SI and SII). The top 10 enriched interacting
proteins ranked by Score (defined as sum ion score of
identified protein peptide by IP-MS) out of all the protein targets
and those from the DSB repair pathway are listed in Fig. 1D.
Network analysis of nIGF1R
interactome
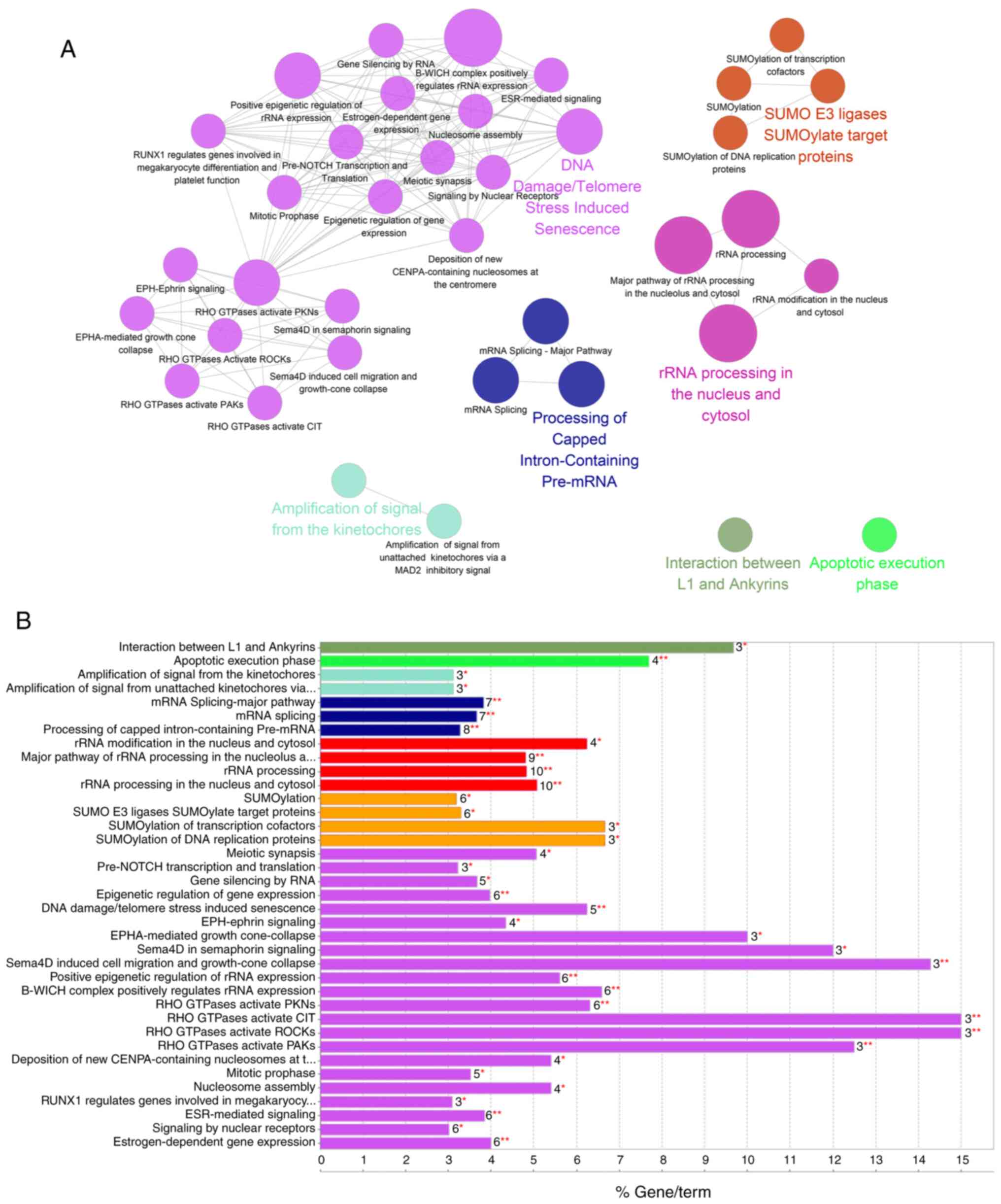
GO term and Reactome database enrichment analyses
were performed using the STRING database in Cytoscape software.
Highly enriched functional pathways revealed by Biological Process
analysis included ‘RNA process’, ‘Nucleic metabolic process’ and
‘Nucleobase-containing compound metabolic process’, along with
several less enriched functional pathways, such as ‘Cellular
component organization or biogenesis’, ‘RNA splicing’ and ‘DNA
metabolic process pathways’. The most enriched pathways as
identified by Reactome analysis were ‘RNA processing’, ‘Cell
cycle’, ‘SUMOylation’ and ‘DNA repair’ (Fig. 2A and B). This was in line with our
previous finding that IGF1R serves as a transcription cofactor
(11), and that IGF1R SUMOylation
leads to its nuclear translocation (12,14).
The DNA repair pathway was among the most enriched
pathways in the network analysis. Key components of DSB repair
pathways were detected (Table SII),
including the 53BP1-RIF1 complex, Ku80 and DNA-PKcs, which are
considered to be the key regulators in the NHEJ pathway (30). Other identified key regulators
included poly[ADP-ribose] polymerase 1 (PARP1), and DNA
topoisomerases (TOP) I and II (31–33).
IGF1R facilitates the binding between
NuMA and 53BP1
Although the role of nIGF1R in the DNA repair
pathway has been reported by previous studies to involve the
promotion of DSB repair by IGF1R through both NHEJ and HR (15,24), the
underlying mechanism remains unclear. Based on the present IP-MS
data, NuMA was identified as an nIGF1R co-localizing partner
(Fig. 3A) and stood out as one of the
most enriched targets in the list (Fig.
1D and Table SI). The
interaction between IGF1R and NuMA was further validated by co-IP
and in situ PLA (Fig. 3B and
C). The dynamics of the IGF1R-NuMA interaction in both
R+ and SW480 cells was also investigated, but no
significant change was reported in any of the cells within 16 h
from IR (Fig. S1).
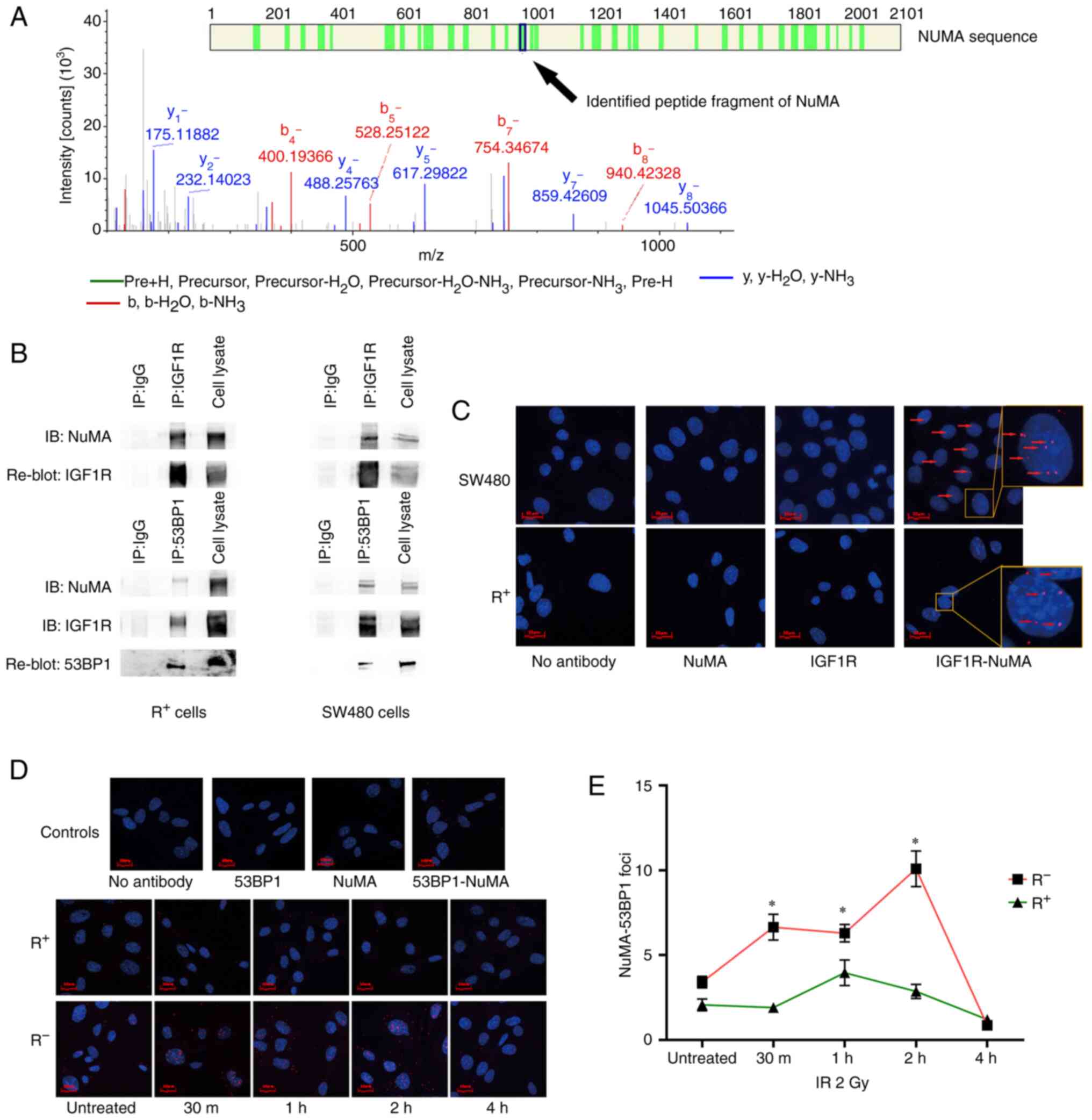 | Figure 3.nIGF1R interacts with NuMA and
facilitates NuMA-53BP1 colocalization in response to IR. (A)
Candidate peptides of NuMA, from which theoretical spectra were
sequentially generated and compared against experimental spectra
identified by IP-MS. (B) Co-IP with anti-IGF1R antibody validated
nIGF1R colocalization with NuMA in SW480 colorectal cancer and
R+ (R-overexpressing IGF1R) cell lines (upper panel).
Co-IP with anti-53BP1 antibody validated 53BP1 colocalization with
NuMA and IGF1R in SW480 and R+ cell lines (lower panel).
IP antibodies were replaced with normal mouse IgG as the negative
control. (C) In situ PLA validated nIGF1R colocalization
with NuMA using anti-IGF1R and anti-NuMA antibodies in SW480 (upper
panel) and R+ (lower panel) cell lines. Red fluorescence dots
(arrows) indicate the colocalizations. Either primary antibody or
both were removed from the experiment to generate negative controls
(first three images). Cell nuclei were stained with DAPI (blue).
(D) In situ PLA showed that NuMA-53BP1 colocalization (red
dots) was increased in response to IR (2 Gy) in R− (MEF
igf1r−/-) cell line but remained unchanged in the
R+ cell line. Cell nuclei were stained with DAPI (blue).
(E) Dynamic change of NuMA-53BP1 colocalization in response to IR
(2 Gy) in R+ and R− cell lines at 30 min, and
1, 2 and 4 h after treatment. Number of NuMA-53BP1 foci represents
the average in situ PLA signal per cell from at least 50
cells in each condition. *P<0.05. nIGF1R, nuclear insulin-like
growth factor 1 receptor; NuMA, nuclear mitotic apparatus protein;
53BP1, NuMA-p53-binding protein 1; IP-MS,
immunoprecipitation-coupled mass spectrometry; IR, ionizing
radiation; PLA, proximity ligation assay; R+,
IGF1R-positive; R−, IGF1R-negative; MEF, mouse embryonic
fibroblast. |
Despite its various roles in mitotic activities
(34–36), NuMA has been associated with multiple
other biological processes, including DSB repair. Most recently,
Salvador Moreno et al (23)
reported that NuMA retained 53BP1 mobility outside the repairing
foci to control 53BP1 distribution, which prevented 53BP1
accumulation at the DSBs. We hypothesized that the IGF1R-NuMA
interaction might regulate the NuMA-53BP1 complex to regulate DSB
repair. In order to validate this hypothesis, R− (MEF
igf1r−/−) and R+ (R-overexpressing IGF1R)
cell lines were used to examine the cellular response to IR in the
presence and absence of IGF1R. The baseline NuMA-53BP1
colocalization level was approximately the same between the
R− and R+ cell lines. In response to IR,
colocalizing signals in the R− cells were markedly
increased, while PLA signals in the R+ cells were barely
changed in the cell nuclei (Fig. 3D and
E). The NuMA-53BP1 colocalization dynamic was also examined in
SW480 and SW480-OE cells following IR. However, no significant
difference between SW480 and SW480-OE cells was observed (Fig. S2). This was interpreted as a sign
that the existence of endogenous nIGF1R in SW480 had already
reached the saturation point.
NuMA-53BP1 colocalization in the
nucleus predicts poor survival in patients with colorectal
cancer
Next, it was investigated whether the IGF1R-NuMA and
NuMA-53BP1 interactions carry clinical significance in tissue
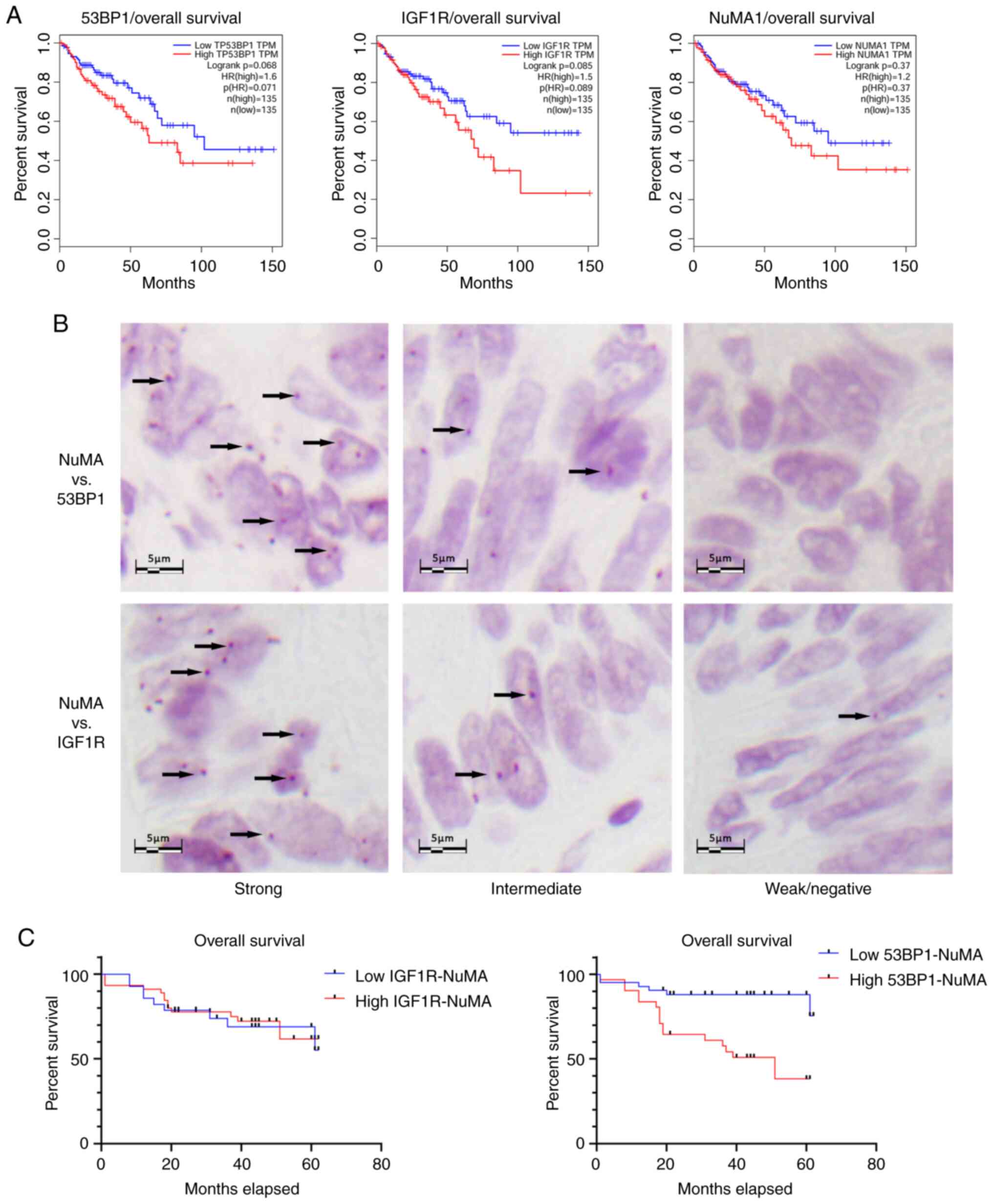
samples from patients with colorectal cancer. According to survival
analysis based on TCGA (http://gepia.cancer-pku.cn), none of the three
analyzed proteins (IGF1R, NuMA and 53BP1) exhibited a significant
association with overall or disease-free survival in patients with
colorectal cancer (Fig. 4A), despite
their well-known oncogenic functions in various molecular
biological studies. Therefore, PLA-based survival analysis was
conducted to examine whether protein-protein interactions in tumor
samples indicated a significant prognostic value. The clinical
characteristics of the cohort are presented in Table I. The present study cohort consisted
of 73 patients with colorectal cancer (43 males and 30 females aged
33–86 years). Complete clinical follow-up information was available
for 56 patients.
 | Table I.Correlation between the PLA staining
of IGF1R-NuMA and 53BP1-NuMA colocalization (in the whole cell and
in the nucleus) and clinicopathologic characteristics in the 73
cases of human colorectal cancer tissues. |
Table I.
Correlation between the PLA staining
of IGF1R-NuMA and 53BP1-NuMA colocalization (in the whole cell and
in the nucleus) and clinicopathologic characteristics in the 73
cases of human colorectal cancer tissues.
|
|
| IGF1R-NuMA in whole
cell | IGF1R-NuMA in cell
nucleus | 53BP1-NuMA in whole
cell | 53BP1-NuMA in cell
nucleus |
|---|
|
|
|
|
|
|
|
|---|
| Parameters | n | Low | High |
P-valuea | Low | High |
P-valuea | Low | High |
P-valuea | Low | High |
P-valuea |
|---|
| Total | 73 | 13 | 60 |
| 28 | 45 |
| 28 | 45 |
| 42 | 31 |
|
| Age (years) |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
≤60 | 34 | 5 | 29 | 0.518 | 15 | 19 | 0.345 | 15 | 19 | 0.345 | 19 | 15 | 0.790 |
|
>60 | 39 | 8 | 31 |
| 13 | 26 |
| 13 | 26 |
| 23 | 16 |
|
| Sex |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Male | 43 | 7 | 36 | 0.683 | 18 | 25 | 0.461 | 18 | 25 | 0.461 | 24 | 19 | 0.722 |
|
Female | 30 | 6 | 24 |
| 10 | 20 |
| 10 | 20 |
| 18 | 12 |
|
| Histologic
type |
|
|
|
|
|
|
|
|
|
|
|
|
|
| Poor
and undifferentiated | 16 | 3 | 13 | 0.911 | 6 | 10 | 0.936 | 9 | 7 | 0.096 | 12 | 4 | 0.110 |
| Well
and moderately differentiated | 57 | 10 | 47 |
| 22 | 35 |
| 19 | 38 |
| 30 | 27 |
|
| Depth of tumor
invasion |
|
|
|
|
|
|
|
|
|
|
|
|
| T1,
T2 | 12 | 3 | 9 | 0.039 | 5 | 7 | 0.150 | 5 | 7 | 0.500 | 7 | 5 | 0.288 |
| T3,
T4 | 61 | 5 | 56 |
| 18 | 43 |
| 21 | 40 |
| 31 | 30 |
|
| Regional lymph node
metastasis |
|
|
|
|
|
|
|
|
|
|
|
|
| No | 48 | 9 | 39 | 0.771 | 19 | 29 | 0.765 | 20 | 28 | 0.420 | 29 | 19 | 0.490 |
|
Yes | 25 | 4 | 21 |
| 9 | 16 |
| 8 | 17 |
| 13 | 12 |
|
| Regional nerve
metastasis |
|
|
|
|
|
|
|
|
|
|
|
|
| No | 70 | 12 | 58 | 0.473 | 26 | 44 | 0.303 | 25 | 45 | 0.025 | 39 | 31 | 0.129 |
|
Yes | 3 | 1 | 2 |
| 2 | 1 |
| 3 | 0 |
| 3 | 0 |
|
| Regional vascular
metastasis |
|
|
|
|
|
|
|
|
|
|
|
| No | 61 | 9 | 52 | 0.124 | 22 | 39 | 0.364 | 22 | 39 | 0.364 | 34 | 27 | 0.484 |
|
Yes | 12 | 4 | 8 |
| 6 | 6 |
| 6 | 6 |
| 8 | 4 |
|
| Cancer relapse (in
12 months) |
|
|
|
|
|
|
|
|
|
|
|
| No | 66 | 12 | 54 | 0.798 | 25 | 41 | 0.797 | 26 | 40 | 0.576 | 40 | 26 | 0.103 |
|
Yes | 7 | 1 | 6 |
| 3 | 4 |
| 2 | 5 |
| 2 | 5 |
|
| TNM staging |
|
|
|
|
|
|
|
|
|
|
|
|
|
| I,
II | 48 | 9 | 39 | 0.771 | 19 | 29 | 0.765 | 20 | 28 | 0.420 | 29 | 19 | 0.490 |
| III,
IV | 25 | 4 | 21 |
| 9 | 16 |
| 8 | 17 |
| 13 | 12 |
|
In general, the PLA signals in paraffin-embedded TMA
slides exhibited a different pattern than that of fixed cells on
cover slides. More cytoplasmic IGF1R-NuMA colocalizing signals were
observed in the TMAs (Fig. 4B). A
higher IGF1R-NuMA and NuMA-53BP1 colocalization was observed in
tumor vs. adjacent non-tumor tissues in the whole cell (P=0.0153
and P=0.0316, respectively) but not in the nucleus (P=0.1587 and
P=0.8707, respectively).
Semi-quantitative scoring revealed that 45/73
(61.6%) tumors had strong nuclear IGF1R-NuMA signals and 31/73
(42.5%) had strong nuclear NuMA-53BP1 signals. Total IGF1R-NuMA
colocalization was found to be significantly associated with tumor
invasiveness (T stage; P=0.039), while total NuMA-53BP1
colocalization was shown to be associated with regional nerve
metastasis (P=0.025; Table I).
Survival analysis showed that strong nuclear 53BP1-NuMA
colocalization was associated with poor survival (P<0.001;
low-rank test; Fig. 4C).
Discussion
Despite advances in the study of the various
oncogenic roles of nuclear insulin-like growth factor 1 receptor
(IGF1R), the targeting of the IGF axis in cancer treatment has
yielded disappointing results (37–40). One
plausible strategy includes the use of combination therapy, for
which an in-depth understanding of nIGF1R function is essential.
Although nIGF1R is known for its various functions in cell growth
and proliferation, as well as metastasis and DSB repair (13,17,25,41–43),
understanding of the regulatory mechanisms remains limited. In the
present study, proteomics and global network analyses were
performed to identify the functional partners of nIGF1R in
colorectal cancer cells. The identification of 197 potential nIGF1R
colocalizing proteins suggested that nIGF1R was functionally
associated with various biological pathways.
The global network analysis identified significant
enrichment in RNA processing, which is a large and complex pathway
that includes RNA transcription, pre-mRNA splicing, and RNA
editing, transport, translation and degradation (44). Our and other previous studies have
implicated nIGF1R in RNA regulation, including transcription
activation (13,14,16).
Aleksic et al (41) showed
that IGF1R may be recruited to chromatin, directly binding DNA and
interacting with RNA polymerase II. In this study, nIGF1R was found
to interact with key proteins of the spliceosomes, including
pre-mRNA processing factor (PRPF)8, PRPF6, spliceosome associated
factor 1, recruiter of U4/U6.U5 tri-snRNP and DEAD-box helicase 5.
However, additional studies are required to investigate the
potential role of nIGF1R in the function of spliceosomes.
Following proteomic screening, the large nuclear
mitotic apparatus protein 1 (NuMA) was identified and validated as
an nIGF1R colocalizing partner. NuMA is a coiled-coil nuclear
structural protein identified ~40 years ago, which plays essential
roles in mitotic spindle maintenance (45). A previous study reported that an
isoform of NuMA could be located in the cytoplasm (46). In the present study, a few cytosolic
IGF1R-NuMA colocalization signals and a higher number of cytosolic
NuMA1-53BP1 colocalization signals were identified. As the focus
was DNA repair, the present analyses were based on nuclear signals
only.
Previous studies have indicated that IGF1R
facilitates treatment resistance and enhances DSB repair through
both the HR and NHEJ pathways (24).
p53-binding protein 1 (53BP1) is a key determinant in DNA
double-strand break (DSB) repair pathway choice and was verified to
colocalize with nIGF1R. Based on the current analysis, MDC1, RIF1,
DNA-PKcs and Ku80 were identified by IP-MS as nIGF1R interactors.
Several key components of DSB repair, such as PARP1, DNA
topoisomerase I, DNA topoisomerase II (TOPII)α and TOPIIβ were also
identified, suggesting that nIGF1R plays an important role in DSB
repair pathways, which prompted the current study to investigate
the potential participation of nIGF1R in the regulation of the
NuMA-53BP1 complex. The present data confirmed the regulatory role
of IGF1R. The nuclear NuMA-53BP1 complex was increased in
R− compared with R+ cells [significant
changes observed from 30 min to 2 h post-ionizing radiation (IR)].
During DSB repair, 53BP1 serves as a molecular scaffold that
recruits additional DSB-responsive proteins to damaged chromatin
(47). 53BP1 needs to bind to NuMA in
a storage-like capacity to be readily available for DNA repair in
case of DNA damage (23). Therefore,
a decreased in NuMA-53BP1 colocalization was expected in
R+ cells following IR. However, the possibility that
other complementary mechanisms were activated, which recruited more
53BP1 to NuMA to form a dynamic 53BP1 turnover as a response, could
not be excluded.
The present results did not provide any information
about the location of IGF1R-NuMA interaction and the mechanism
behind the IGF1R-NuMA-53BP1 interaction. The NuMA-53BP1 interaction
was reported to be located in the nucleoplasm when there is no DSB
repair (23). The fact that
IGF1R-NuMA colocalization was not inducible by IR in the present
study suggested that the interaction most likely does not occur at
the chromatin surrounding the DSB site, despite the structural and
chromosome-binding roles of NuMA (48), as well as the transcription regulatory
function of nIGF1R through DNA binding (14). The mechanism behind IGF1R-NuMA-53BP1
interactions could be either that IGF1R-NuMA and NuMA-53BP1
interactions are mutually exclusive or that they form a tripartite
complex. Considering the non-dynamic feature of IGF1R-NuMA
interaction, the tripartite complex hypothesis is likely; the
dynamic turnover of 53BP1, which kept the NuMA-53BP1 colocalization
relatively stable in R+ cells, was interrupted,
resulting in an elevation of nuclear NuMA-53BP1 colocalization in
IGF1R-negative R− cells following IR. However, further
experimental evidence is required to confirm this hypothesis.
In addition to cellular evidence, a correlation
between IGF1R and the NuMA-53BP1 interaction in the present study
in a clinical cohort of patients with colorectal cancer would
further support the current findings. However, this information
could not be obtained due to technical issues. The PLA results
indicated that high levels of nuclear NuMA-53BP1 colocalization in
the tumor cells were significantly associated with poor overall
survival. Patients with a high level of NuMA-53BP1 colocalization
were prone to exhibiting a disruption of normal 53BP1-dependent
DSB-repair, which could lead to more rapid tumor progression.
Although it should be noted that the present cohort only involved
patients that had not received chemotherapy or radiotherapy prior
to surgical intervention, the significance of NuMA-53BP1 in
treatment resistance could not be sufficiently explained. No
significant association was identified between IGF1R-NuMA
colocalization and patient survival in the current cohort. Based on
our hypothesis from the cellular experiment, the IGF1R-NuMA and
IGF1R-53BP1 interactions may be consistent with the dynamic change
of NuMA-53BP1 colocalization. Future well-designed studies that
focus on the dynamic changes and interactive functions of the
IGF1R-NuMA-53BP1 complex in post-IR tissue samples are warranted to
obtain a deeper understanding of this mechanism.
In conclusion, the interactome of nIGF1R in
colorectal cancer was presented herein. nIGF1R interacted with NuMA
and appeared to regulate the NuMA-53BP1 interaction. In clinical
colorectal cancer tissues, the NuMA-53BP1 interaction was
associated with poor overall survival and could therefore serve as
a molecular treatment target for those patients. The present study
results might shed light on the DNA repair-related function of
nIGF1R and benefit the development of novel IGF1R-related cancer
treatments.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This study was supported by the Swedish Cancer
Foundation, Swedish Research, the Cancer Society in Stockholm,
Swedish Children Cancer Society, Stockholm County Council,
Karolinska Institute, China Scholarship Council (grant no.
CSC201706370014) and the National Natural Science Foundation of
China (grant nos. 81201904 and 81974386).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YL, ZC, OL and FH conceived the study. CY, YZ, NS
and CH were responsible for data analysis. OL, ZC, FH and CY were
responsible for funding acquisition. CY, YZ, NS, PZ, XT, LM and ZC
were responsible for performing the experiments. PZ, XT, LM and ZC
were responsible for obtaining the resources. YL and ZC supervised
the study. CY wrote the original draft. YL, OL and FH wrote,
reviewed and edited the manuscript. All authors read and approved
the final manuscript.
Ethics approval and consent to
participate
The study was conducted according to the guidelines
of the Declaration of Helsinki and approved by the Ethics Committee
of Xiangya Hospital of Central South University (Changsha, China;
approval no. 201403168). Informed consent was obtained from all
patients involved in the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
nIGF1R
|
nuclear insulin-like growth factor 1
receptor
|
|
NuMA
|
nuclear mitotic apparatus protein
1
|
|
53BP1
|
p53-binding protein 1
|
|
DSB
|
DNA double-strand break
|
|
SUMO
|
small ubiquitin-related modifier
|
|
PLA
|
proximity ligation assay
|
|
NHEJ
|
non-homologous end-joining
|
|
HR
|
homologous recombination
|
|
TEAB
|
triethylammonium bicarbonate
buffer
|
|
IP-MS
|
immunoprecipitation coupled-mass
spectrometry
|
|
LC/MS
|
liquid chromatography mass
spectrometry
|
|
CAA
|
chloroacetamide
|
|
CAN
|
acetonitrile
|
|
TCEP
|
0.5 M bond-breaker TCEP solution
|
|
Lys-C
|
lysyl endopeptidase
|
|
TFA
|
trifluoroacetic acid
|
References
|
1
|
Dekker E, Tanis PJ, Vleugels JLA, Kasi PM
and Wallace MB: Colorectal cancer. Lancet. 394:1467–1480. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mahar AL, Compton C, Halabi S, Hess KR,
Weiser MR and Groome PA: Personalizing prognosis in colorectal
cancer: A systematic review of the quality and nature of clinical
prognostic tools for survival outcomes. J Surg Oncol. 116:969–982.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Fakih MG: Metastatic colorectal cancer:
Current state and future directions. J Clin Oncol. 33:1809–1824.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Martini G, Troiani T, Cardone C, Vitiello
P, Sforza V, Ciardiello D, Napolitano S, Della Corte CM, Morgillo
F, Raucci A, et al: Present and future of metastatic colorectal
cancer treatment: A review of new candidate targets. World J
Gastroenterol. 23:4675–4688. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Dyer AH, Vahdatpour C, Sanfeliu A and
Tropea D: The role of insulin-like growth factor 1 (IGF-1) in brain
development, maturation and neuroplasticity. Neuroscience.
325:89–99. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lin Y, Liu H, Waraky A, Haglund F, Agarwal
P, Jernberg-Wiklund H, Warsito D and Larsson O: SUMO-modified
insulin-like growth factor 1 receptor (IGF-1R) increases cell cycle
progression and cell proliferation. J Cell Physiol. 232:2722–2730.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Heidegger I, Kern J, Ofer P, Klocker H and
Massoner P: Oncogenic functions of IGF1R and INSR in prostate
cancer include enhanced tumor growth, cell migration and
angiogenesis. Oncotarget. 5:2723–2735. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Riedemann J and Macaulay VM: IGF1R
signalling and its inhibition. Endocr Relat Cancer. 13 (Suppl
1):S33–S43. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Rodrigues Alves APN, Fernandes JC,
Fenerich BA, Coelho-Silva JL, Scheucher PS, Simões BP, Rego EM,
Ridley AJ, Machado-Neto JA and Traina F: IGF1R/IRS1 targeting has
cytotoxic activity and inhibits PI3K/AKT/mTOR and MAPK signaling in
acute lymphoblastic leukemia cells. Cancer Lett. 456:59–68. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zorea J, Prasad M, Cohen L, Li N, Schefzik
R, Ghosh S, Rotblat B, Brors B and Elkabets M: IGF1R upregulation
confers resistance to isoform-specific inhibitors of PI3K in
PIK3CA-driven ovarian cancer. Cell Death Dis. 9:9442018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Warsito D, Lin Y, Gnirck AC, Sehat B and
Larsson O: Nuclearly translocated insulin-like growth factor 1
receptor phosphorylates histone H3 at tyrosine 41 and induces SNAI2
expression via Brg1 chromatin remodeling protein. Oncotarget.
7:42288–42302. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Packham S, Warsito D, Lin Y, Sadi S,
Karlsson R, Sehat B and Larsson O: Nuclear translocation of IGF-1R
via p150(Glued) and an importin-β/RanBP2-dependent pathway in
cancer cells. Oncogene. 34:2227–2238. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Warsito D, Sjostrom S, Andersson S,
Larsson O and Sehat B: Nuclear IGF1R is a transcriptional
co-activator of LEF1/TCF. EMBO Rep. 13:244–250. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sehat B, Tofigh A, Lin Y, Trocmé E,
Liljedahl U, Lagergren J and Larsson O: SUMOylation mediates the
nuclear translocation and signaling of the IGF-1 receptor. Sci
Signal. 3:ra102010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Aleksic T, Verrill C, Bryant RJ, Han C,
Worrall AR, Brureau L, Larré S, Higgins GS, Fazal F, Sabbagh A, et
al: IGF-1R associates with adverse outcomes after radical
radiotherapy for prostate cancer. Br J Cancer. 117:1600–1606. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang Y, Yuan JL, Zhang YT, Ma JJ, Xu P,
Shi CH, Zhang W, Li YM, Fu Q, Zhu GF, et al: Inhibition of both
EGFR and IGF1R sensitized prostate cancer cells to radiation by
synergistic suppression of DNA homologous recombination repair.
PLoS One. 8:e687842013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Guerard M, Robin T, Perron P, Hatat AS,
David-Boudet L, Vanwonterghem L, Busser B, Coll JL, Lantuejoul S,
Eymin B, et al: Nuclear translocation of IGF1R by intracellular
amphiregulin contributes to the resistance of lung tumour cells to
EGFR-TKI. Cancer Lett. 420:146–155. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Her J and Bunting SF: How cells ensure
correct repair of DNA double-strand breaks. J Biol Chem.
293:10502–10511. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wright WD, Shah SS and Heyer WD:
Homologous recombination and the repair of DNA double-strand
breaks. J Biol Chem. 293:10524–10535. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chang HHY, Pannunzio NR, Adachi N and
Lieber MR: Non-homologous DNA end joining and alternative pathways
to double-strand break repair. Nat Rev Mol Cell Biol. 18:495–506.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Escribano-Diaz C, Orthwein A,
Fradet-Turcotte A, Xing M, Young JT, Tkáč J, Cook MA, Rosebrock AP,
Munro M, Canny MD, et al: A cell cycle-dependent regulatory circuit
composed of 53BP1-RIF1 and BRCA1-CtIP controls DNA repair pathway
choice. Mol Cell. 49:872–883. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chapman JR, Barral P, Vannier JB, Borel V,
Steger M, Tomas-Loba A, Sartori AA, Adams IR, Batista FD and
Boulton SJ: RIF1 is essential for 53BP1-dependent nonhomologous end
joining and suppression of DNA double-strand break resection. Mol
Cell. 49:858–871. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Salvador Moreno N, Liu J, Haas KM, Parker
LL, Chakraborty C, Kron SJ, Hodges K, Miller LD, Langefeld C,
Robinson PJ, et al: The nuclear structural protein NuMA is a
negative regulator of 53BP1 in DNA double-strand break repair.
Nucleic Acids Res. 47:2703–2715. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chitnis MM, Lodhia KA, Aleksic T, Gao S,
Protheroe AS and Macaulay VM: IGF-1R inhibition enhances
radiosensitivity and delays double-strand break repair by both
non-homologous end-joining and homologous recombination. Oncogene.
33:5262–5273. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Waraky A, Lin Y, Warsito D, Haglund F,
Aleem E and Larsson O: Nuclear insulin-like growth factor 1
receptor phosphorylates proliferating cell nuclear antigen and
rescues stalled replication forks after DNA damage. J Biol Chem.
292:18227–18239. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Doncheva NT, Morris JH, Gorodkin J and
Jensen LJ: Cytoscape StringApp: Network analysis and visualization
of proteomics data. J Proteome Res. 18:623–632. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bindea G, Mlecnik B, Hackl H, Charoentong
P, Tosolini M, Kirilovsky A, Fridman WH, Pagès F, Trajanoski Z and
Galon J: ClueGO: A Cytoscape plug-in to decipher functionally
grouped gene ontology and pathway annotation networks.
Bioinformatics. 25:1091–1093. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Huang C, Yuan W, Lai C, Zhong S, Yang C,
Wang R, Mao L and Chen Z and Chen Z: EphA2-to-YAP pathway drives
gastric cancer growth and therapy resistance. Int J Cancer.
146:1937–1949. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
ten Have S, Boulon S, Ahmad Y and Lamond
AI: Mass spectrometry-based immuno-precipitation proteomics-the
user's guide. Proteomics. 11:1153–1159. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kakarougkas A and Jeggo PA: DNA DSB repair
pathway choice: An orchestrated handover mechanism. Br J Radiol.
87:201306852014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Xu Y and Her C: Inhibition of
topoisomerase (DNA) I (TOP1): DNA damage repair and anticancer
therapy. Biomolecules. 5:1652–1670. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Rocha JC, Busatto FF, Guecheva TN and
Saffi J: Role of nucleotide excision repair proteins in response to
DNA damage induced by topoisomerase II inhibitors. Mutat Res Rev
Mutat Res. 768:68–77. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
de Lange T: Shelterin-mediated telomere
protection. Ann Rev Genet. 52:223–247. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Maiato H and Pereira AJ: Cell division:
NuMA bears the load in the spindle. Curr Biol. 27:R765–R767. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Gallini S, Carminati M, De Mattia F,
Pirovano L, Martini E, Oldani A, Asteriti IA, Guarguaglini G and
Mapelli M: NuMA phosphorylation by Aurora-A orchestrates spindle
orientation. Curr Biol. 26:458–469. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kotak S and Gonczy P: Mechanisms of
spindle positioning: Cortical force generators in the limelight.
Curr Opin Cell Biol. 25:741–748. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Werner H, Sarfstein R and Bruchim I:
Investigational IGF1R inhibitors in early stage clinical trials for
cancer therapy. Expert Opin Investig Drugs. 28:1101–1112. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Qu X, Wu Z, Dong W, Zhang T, Wang L, Pang
Z, Ma W and Du J: Update of IGF-1 receptor inhibitor (ganitumab,
dalotuzumab, cixutumumab, teprotumumab and figitumumab) effects on
cancer therapy. Oncotarget. 8:29501–29518. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Yee D: Anti-insulin-like growth factor
therapy in breast cancer. J Mol Endocrinol. 61:T61–T68. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Pollak M: The insulin and insulin-like
growth factor receptor family in neoplasia: An update. Nat Rev
Cancer. 12:159–169. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Aleksic T, Gray N, Wu X, Rieunier G, Osher
E, Mills J, Verrill C, Bryant RJ, Han C, Hutchinson K, et al:
Nuclear IGF1R interacts with regulatory regions of chromatin to
promote RNA Polymerase II recruitment and gene expression
associated with advanced tumor stage. Cancer Res. 78:3497–3509.
2018.PubMed/NCBI
|
|
42
|
Solomon-Zemler R, Pozniak Y, Geiger T and
Werner H: Identification of nucleolar protein NOM1 as a novel
nuclear IGF1R-interacting protein. Mol Genet Metab. 126:259–265.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Solomon-Zemler R, Sarfstein R and Werner
H: Nuclear insulin-like growth factor-1 receptor (IGF1R) displays
proliferative and regulatory activities in non-malignant cells.
PLoS One. 12:e01851642017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Wickramasinghe VO and Venkitaraman AR: RNA
processing and genome stability: Cause and consequence. Mol Cell.
61:496–505. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Radulescu AE and Cleveland DW: NuMA after
30 years: The matrix revisited. Trends Cell Biol. 20:214–222. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Wu J, Xu Z, He D and Lu G: Identification
and characterization of novel NuMA isoforms. Biochem Biophys Res
Commun. 454:387–392. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Panier S and Boulton SJ: Double-strand
break repair: 53BP1 comes into focus. Nat Rev Mol Cell Biol.
15:7–18. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Serra-Marques A, Houtekamer R, Hintzen D,
Canty JT, Yildiz A and Dumont S: The mitotic protein NuMA plays a
spindle-independent role in nuclear formation and mechanics. J Cell
Biol. 219:e2020042022020. View Article : Google Scholar : PubMed/NCBI
|