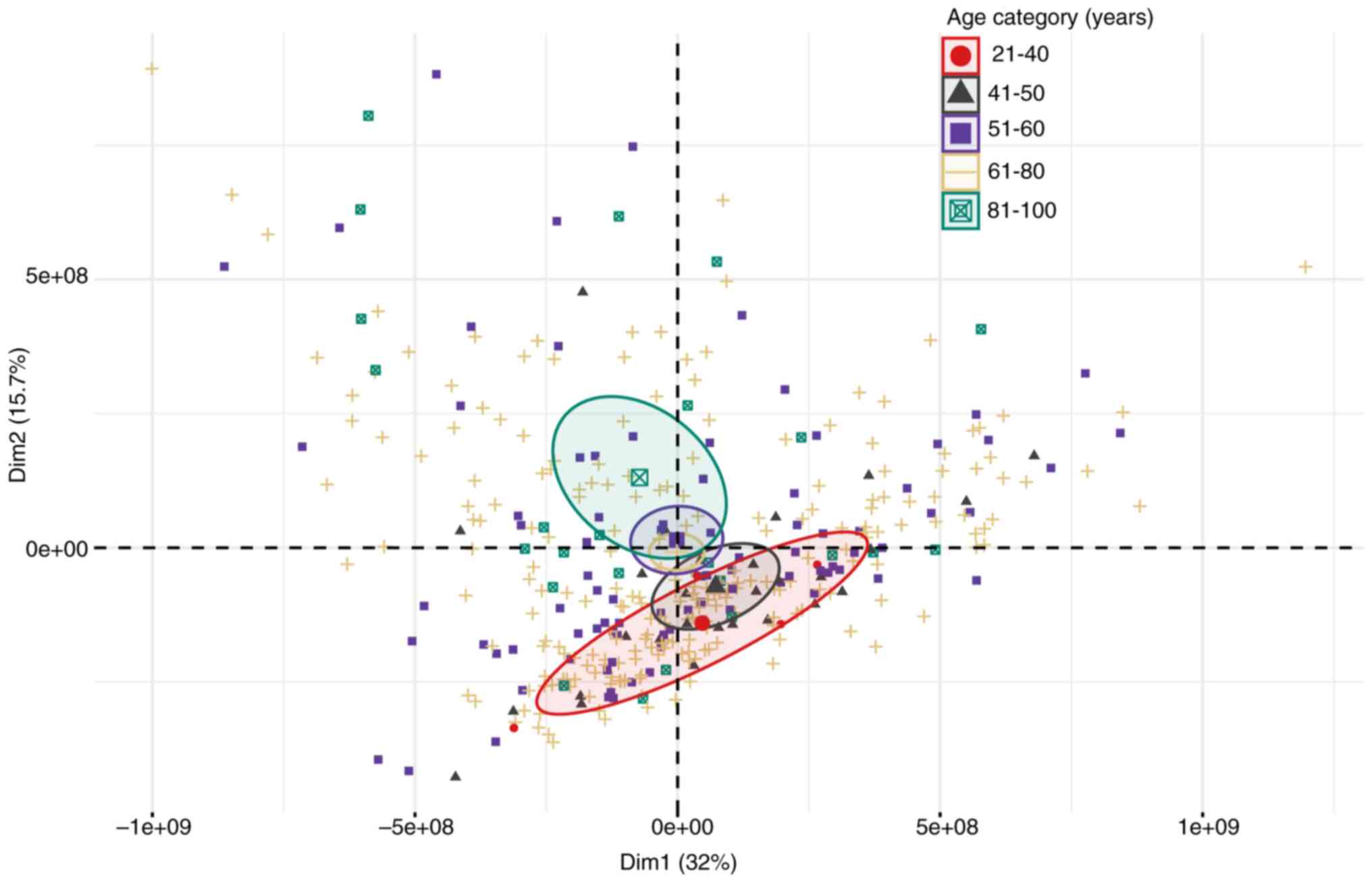
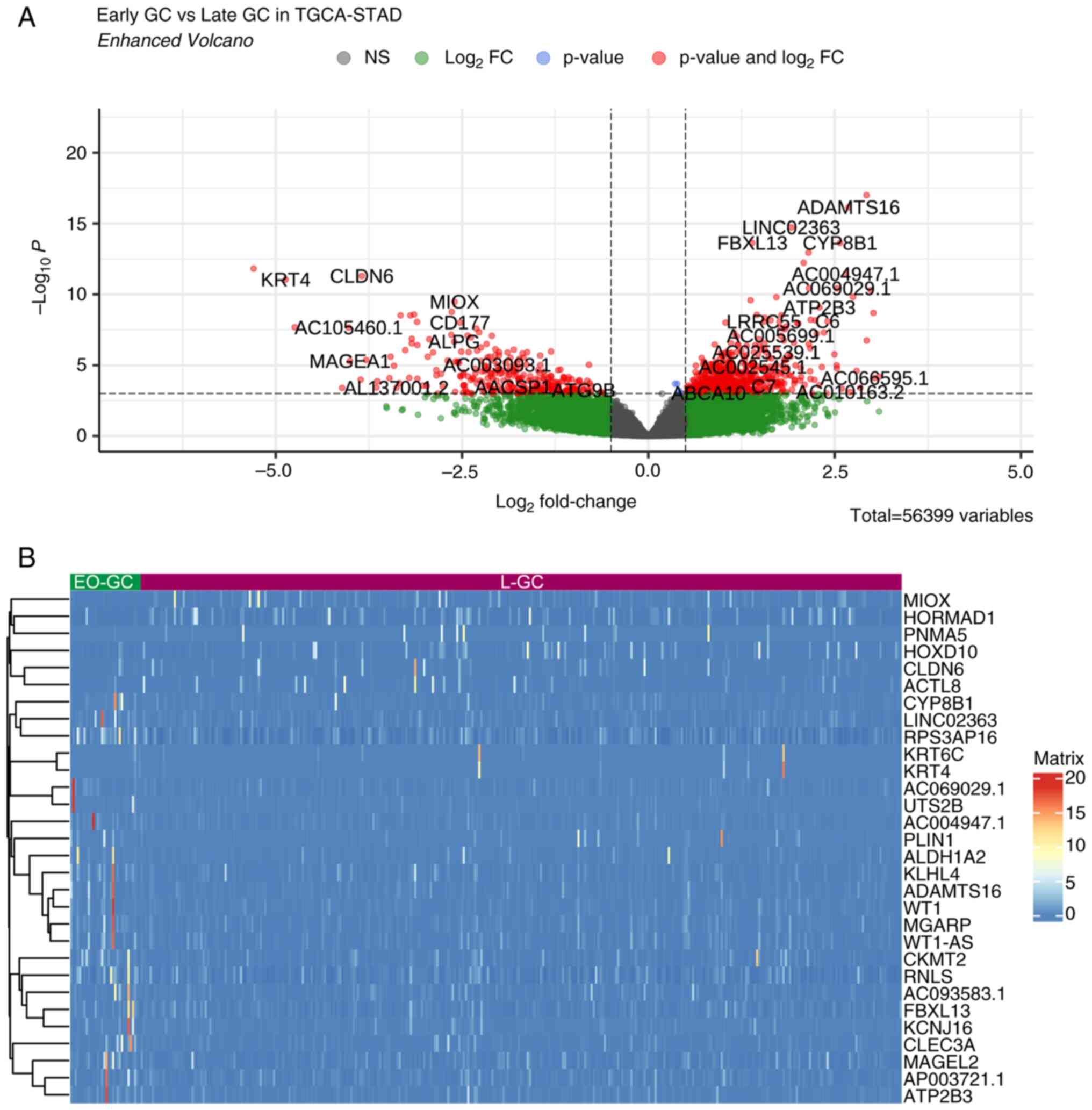
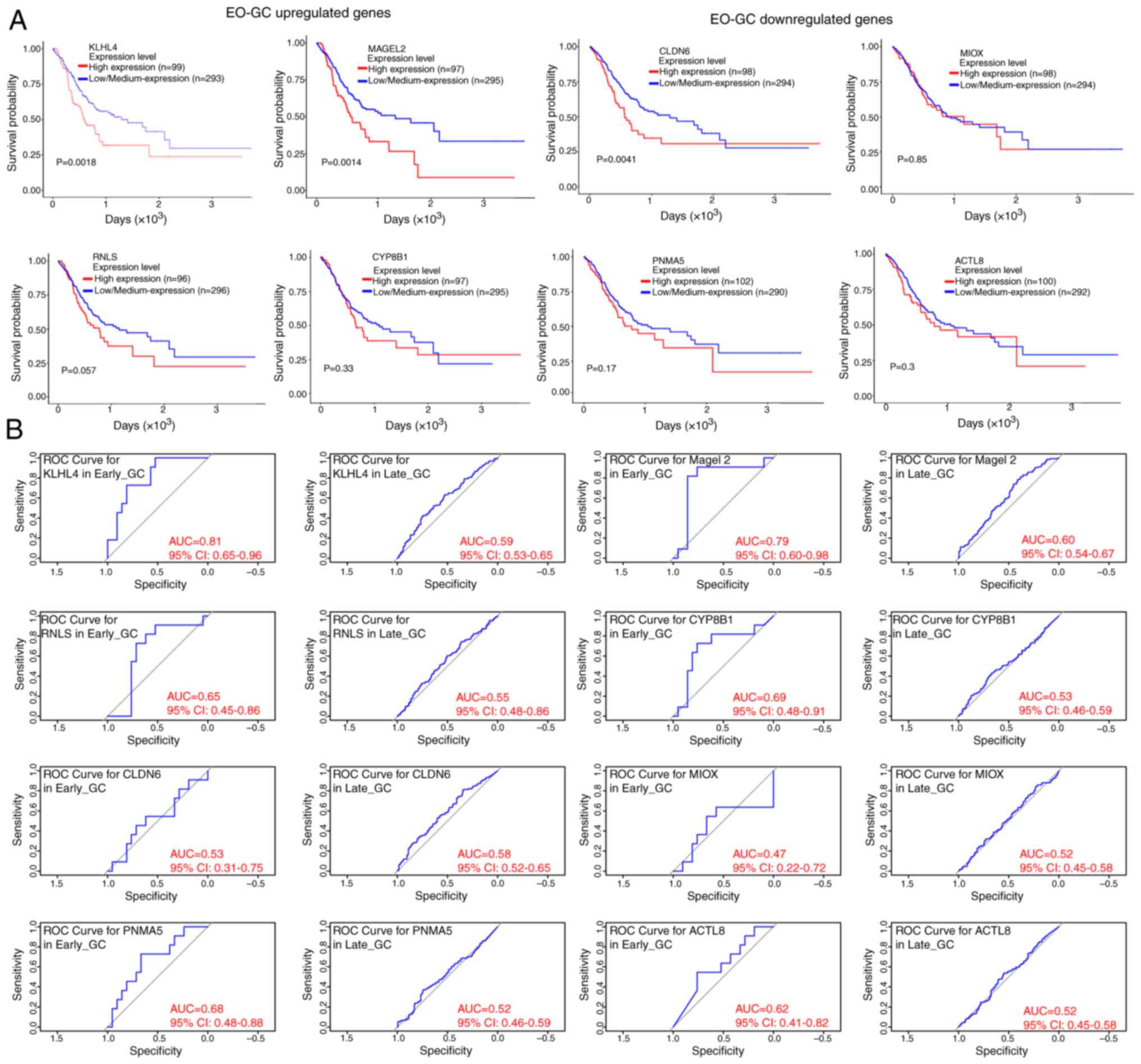
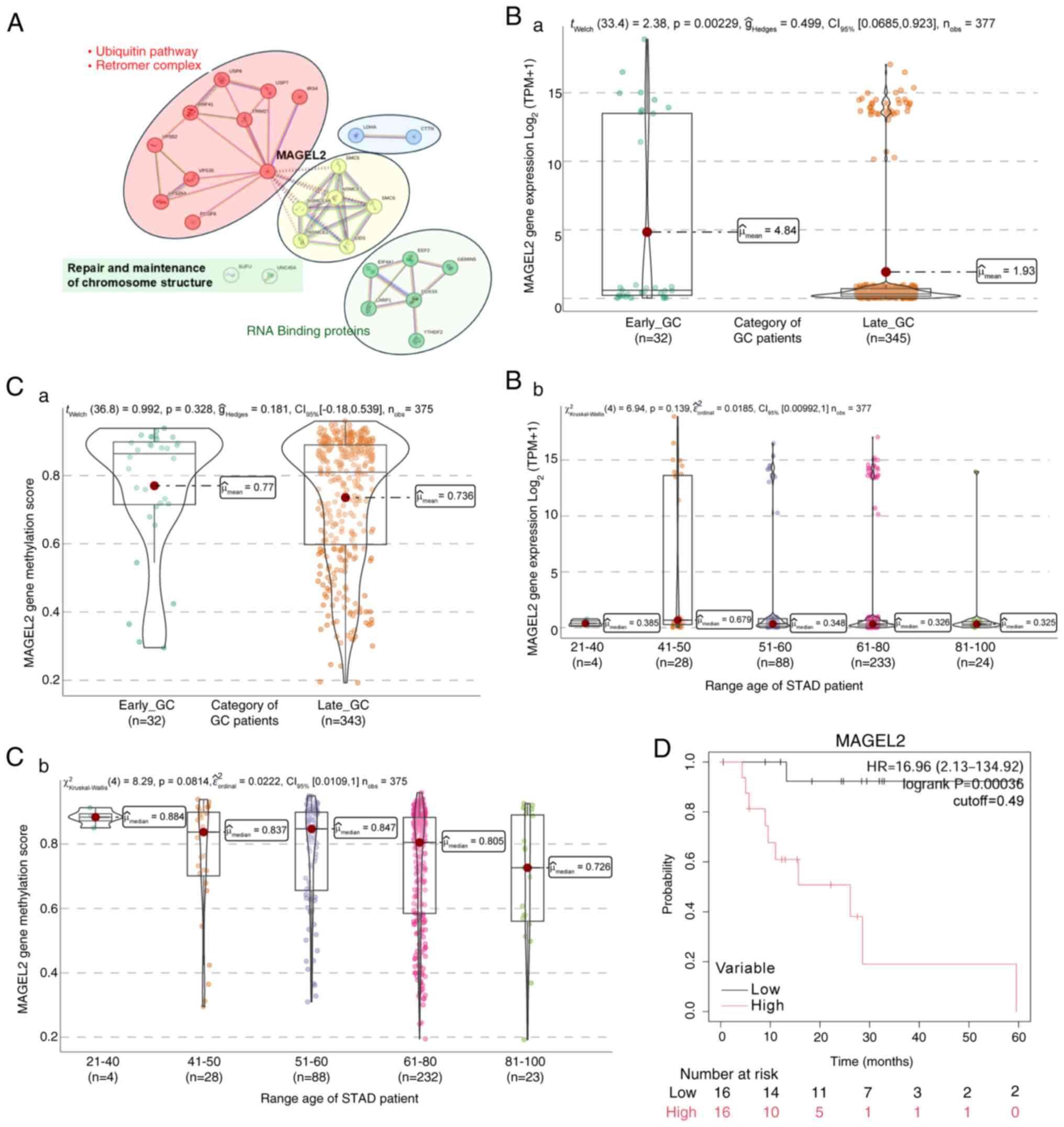
|
1
|
Zhang C, Tang R, Zhu H, Ge X, Wang Y, Wang
X and Miao L: Comparison of treatment strategies and survival of
early-onset gastric cancer: A population-based study. Sci Rep.
12:62882022. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bergquist JR, Leiting JL, Habermann EB,
Cleary SP, Kendrick ML, Smoot RL, Nagorney DM, Truty MJ and Grotz
TE: Early-onset gastric cancer is a distinct disease with worrisome
trends and oncogenic features. Surgery. 166:547–555. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Vishwanath A, Krishna S, Manudhane AP,
Hart PA and Krishna SG: Early-onset gastrointestinal malignancies:
An investigation into a rising concern. Cancers (Basel).
16:15532024. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Han X, Jia X, Sheng C, Li M, Han J, Duan F
and Wang K: A comparison analysis of the somatic mutations in
early-onset gastric cancer and traditional gastric cancer. Clin Res
Hepatol Gastroenterol. 48:1022872024. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Petrillo A, Federico P, Marte G, Liguori
C, Seeber A, Ottaviano M, Tufo A and Daniele B: Non-hereditary
early onset gastric cancer: An unmet medical need. Curr Opin
Pharmacol. 68:1023442023. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ben-Aharon I, van Laarhoven HWM, Fontana
E, Obermannova R, Nilsson M and Lordick F: Early-onset cancer in
the gastrointestinal tract is on the rise-evidence and
implications. Cancer Discov. 13:538–551. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Milne AN and Offerhaus GJ: Early-onset
gastric cancer: Learning lessons from the young. World J
Gastrointest Oncol. 2:59–64. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Triantafillidis JK, Georgiou K,
Konstadoulakis MM and Papalois AE: Early-onset gastrointestinal
cancer: An epidemiological reality with great significance and
implications. World J Gastrointest Oncol. 16:583–597. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhou Q, Tao F, Qiu L, Chen H, Bao H, Wu X,
Shao Y, Chi L and Song H: Somatic alteration characteristics of
early-onset gastric cancer. J Oncol. 2022:14980532022. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Machlowska J, Baj J, Sitarz M, Maciejewski
R and Sitarz R: Gastric cancer: Epidemiology, risk factors,
classification, genomic characteristics and treatment strategies.
Int J Mol Sci. 21:40122020. View Article : Google Scholar : PubMed/NCBI
|
|
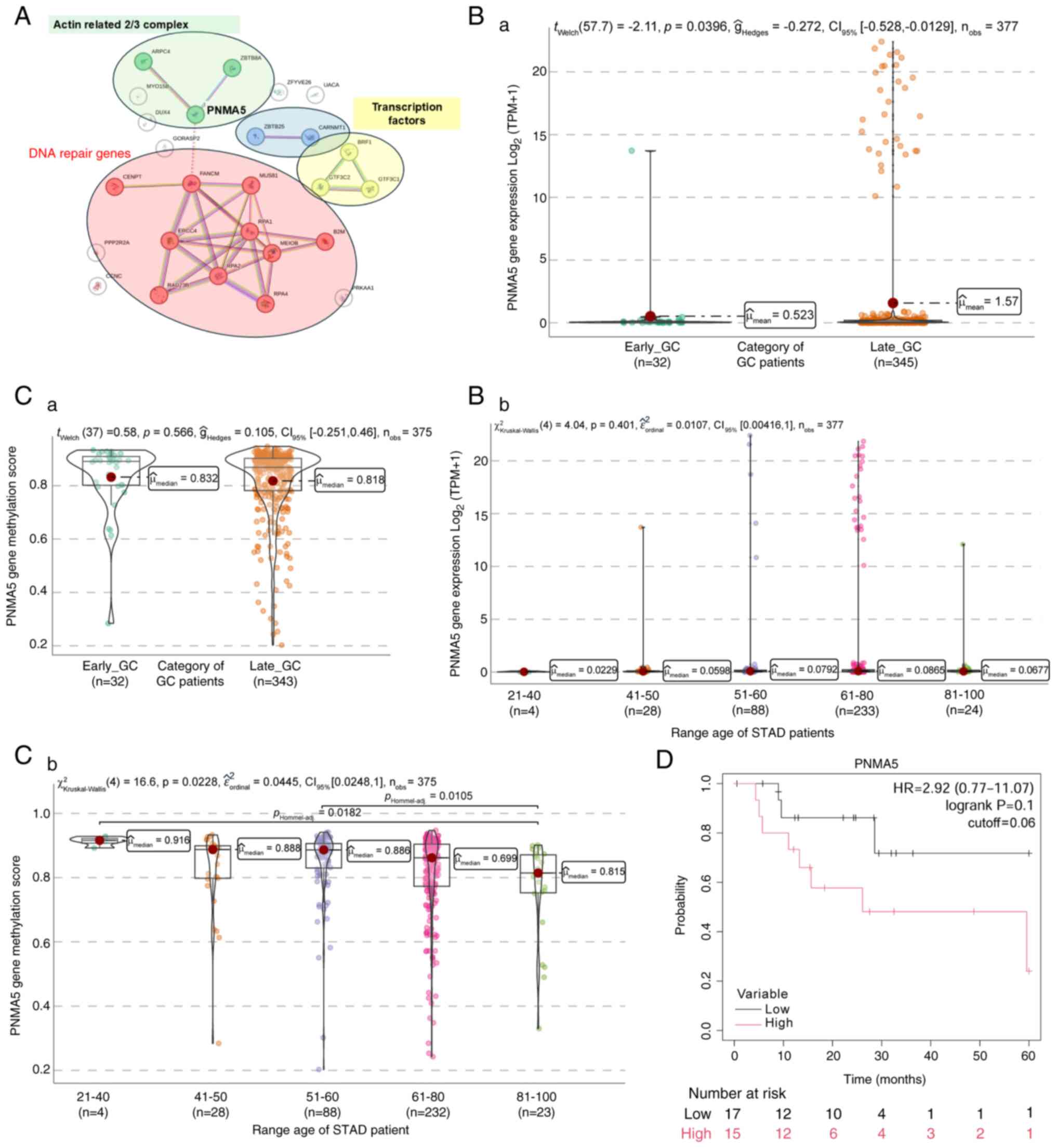
11
|
Ugai T, Sasamoto N, Lee HY, Ando M, Song
M, Tamimi RM, Kawachi I, Campbell PT, Giovannucci EL, Weiderpass E,
et al: Is early-onset cancer an emerging global epidemic? Current
evidence and future implications. Nat Rev Clin Oncol. 19:656–673.
2022. View Article : Google Scholar : PubMed/NCBI
|
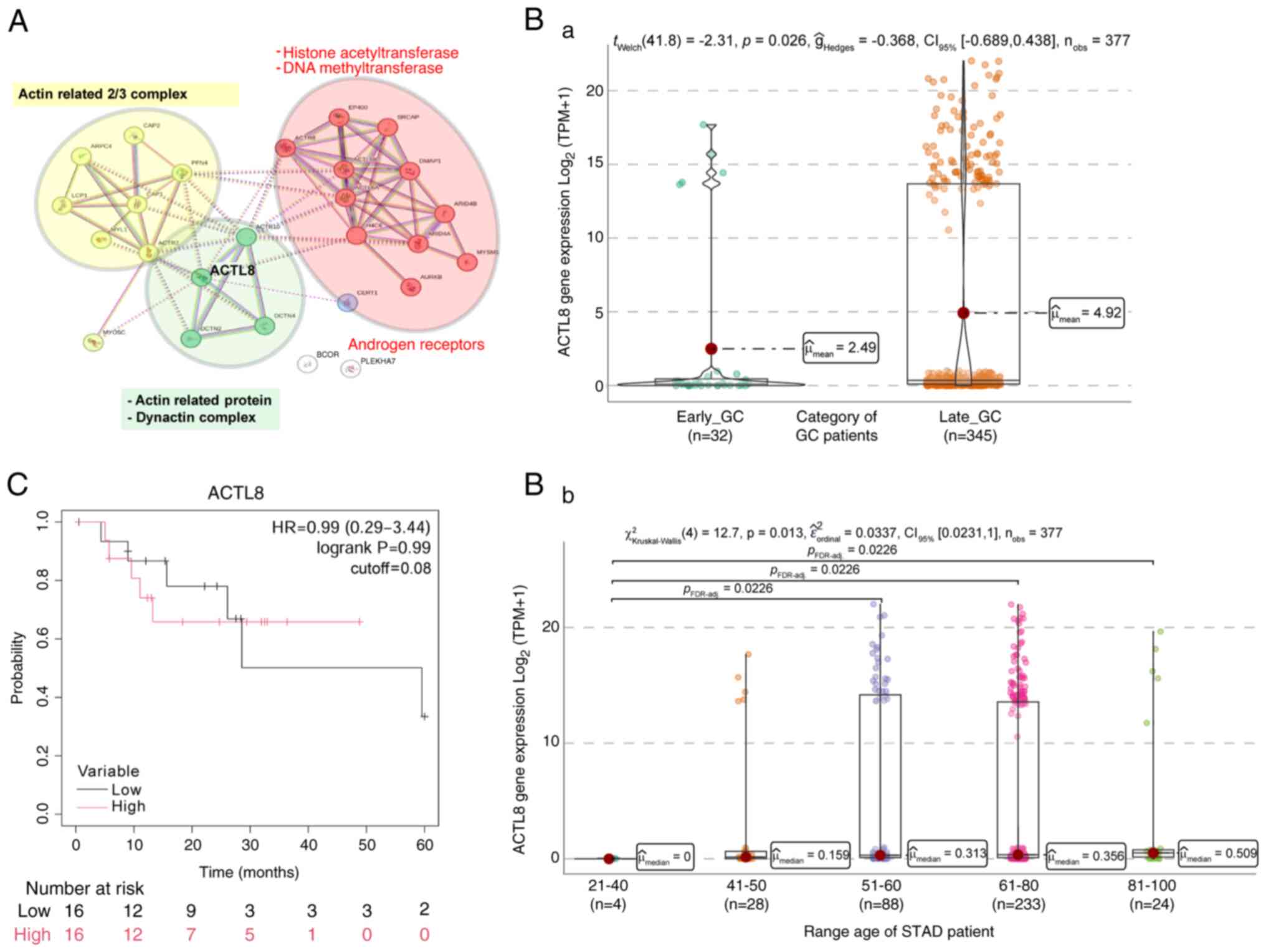
|
12
|
Colaprico A, Silva TC, Olsen C, Garofano
L, Cava C, Garolini D, Sabedot TS, Malta TM, Pagnotta SM,
Castiglioni I, et al: TCGAbiolinks: An R/bioconductor package for
integrative analysis of TCGA data. Nucleic Acids Res. 44:e712016.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lê S, Josse J and Husson F: FactoMineR: An
R package for multivariate analysis. J Stat Softw. 25:1–18. 2008.
View Article : Google Scholar
|
|
14
|
Love MI, Huber W and Anders S: Moderated
estimation of fold change and dispersion for RNA-seq data with
DESeq2. Genome Biol. 15:5502014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chandrashekar DS, Karthikeyan SK, Korla
PK, Patel H, Shovon AR, Athar M, Netto GJ, Qin ZS, Kumar S, Manne
U, et al: UALCAN: An update to the integrated cancer data analysis
platform. Neoplasia. 25:18–27. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Heagerty PJ and Saha P: SurvivalROC:
Time-dependent ROC curve estimation from censored survival data.
Biometrics. 2000.https://doi.org/10.32614/CRAN.package.survivalROC
View Article : Google Scholar
|
|
17
|
Wang X, Dong Y, Zhang H, Zhao Y, Miao T,
Mohseni G, Du L and Wang C: DNA methylation drives a new path in
gastric cancer early detection: Current impact and prospects. Genes
Dis. 11:847–860. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gao X, Liu H, Yu J and Nie Y: DNA
methylation biomarkers for early detection of gastric and
colorectal cancers. Cancer Biol Med. 20:955–962. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Necula L, Matei L, Dragu D, Neagu AI,
Mambet C, Nedeianu S, Bleotu C, Diaconu CC and Chivu-Economescu M:
Recent advances in gastric cancer early diagnosis. World J
Gastroenterol. 25:2029–2044. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Choi SH, Cho SY, Song J and Hur MW: KLHL4,
a novel p53 target gene, inhibits cell proliferation by activating
p21WAF/CDKN1A. Biochem Biophys Res Commun. 530:588–596.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gimeno RE, Ortegon AM, Patel S, Punreddy
S, Ge P, Sun Y, Lodish HF and Stahl A: Characterization of a
heart-specific fatty acid transport protein. J Biol Chem.
278:16039–16044. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hirokawa N, Noda Y, Tanaka Y and Niwa S:
Kinesin superfamily motor proteins and intracellular transport. Nat
Rev Mol Cell Biol. 10:682–696. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bellezza I, Giambanco I, Minelli A and
Donato R: Nrf2-Keap1 signaling in oxidative and reductive stress.
Biochim Biophys Acta Mol Cell Res. 1865:721–733. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Liu Y, Shi Y, Han R, Liu C, Qin X, Li P
and Gu R: Signaling pathways of oxidative stress response: The
potential therapeutic targets in gastric cancer. Front Immunol.
14:11395892023. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Baird L and Yamamoto M: The molecular
mechanisms regulating the KEAP1-NRF2 pathway. Mol Cell Biol.
40:e00099–20. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kobayashi A, Kang MI, Watai Y, Tong KI,
Shibata T, Uchida K and Yamamoto M: Oxidative and electrophilic
stresses activate Nrf2 through inhibition of ubiquitination
activity of Keap1. Mol Cell Biol. 26:221–229. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ulasov AV, Rosenkranz AA, Georgiev GP and
Sobolev AS: Nrf2/Keap1/ARE signaling: Towards specific regulation.
Life Sci. 291:1201112022. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Freigang S, Ampenberger F, Spohn G, Heer
S, Shamshiev AT, Kisielow J, Hersberger M, Yamamoto M, Bachmann MF
and Kopf M: Nrf2 is essential for cholesterol crystal-induced
inflammasome activation and exacerbation of atherosclerosis. Eur J
Immunol. 41:2040–1051. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kuhn AM, Tzieply N, Schmidt MV, von
Knethen A, Namgaladze D, Yamamoto M and Brüne B: Antioxidant
signaling via Nrf2 counteracts lipopolysaccharide-mediated
inflammatory responses in foam cell macrophages. Free Radic Biol
Med. 50:1382–1391. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang Y, Choksi S, Chen K, Pobezinskaya Y,
Linnoila I and Liu ZG: ROS play a critical role in the
differentiation of alternatively activated macrophages and the
occurrence of tumor-associated macrophages. Cell Res. 23:898–914.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Robledinos-Antón N, Fernández-Ginés R,
Manda G and Cuadrado A: Activators and inhibitors of NRF2: A review
of their potential for clinical development. Oxid Med Cell Longev.
2019:93721822019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tooze SA: Biogenesis of secretory granules
in the trans-Golgi network of neuroendocrine and endocrine cells.
Biochim Biophys Acta. 1404:231–244. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Štepihar D, Florke Gee RR, Hoyos Sanchez
MC and Fon Tacer K: Cell-specific secretory granule sorting
mechanisms: The role of MAGEL2 and retromer in hypothalamic
regulated secretion. Front Cell Dev Biol. 11:12430382023.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Chomez P, De Backer O, Bertrand M, De
Plaen E, Boon T and Lucas S: An overview of the MAGE gene family
with the identification of all human members of the family. Cancer
Res. 61:5544–5551. 2001.PubMed/NCBI
|
|
35
|
Hao YH, Doyle JM, Ramanathan S, Gomez TS,
Jia D, Xu M, Chen ZJ, Billadeau DD, Rosen MK and Potts PR:
Regulation of WASH-dependent actin polymerization and protein
trafficking by ubiquitination. Cell. 152:1051–1064. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Hoencamp C and Rowland BD: Genome control
by SMC complexes. Nat Rev Mol Cell Biol. 24:633–650. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Sanderson MR, Fahlman RP and Wevrick R:
The N-terminal domain of the Schaaf-Yang syndrome protein MAGEL2
likely has a role in RNA metabolism. J Biol Chem. 297:1009592021.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Beaupre BA, Hoag MR, Roman J, Försterling
FH and Moran GR: Metabolic function for human renalase: Oxidation
of isomeric forms of β-NAD(P)H that are inhibitory to primary
metabolism. Biochemistry. 54:795–806. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Beaupre BA, Carmichael BR, Hoag MR, Shah
DD and Moran GR: Renalase is an α-NAD(P)H oxidase/anomerase. J Am
Chem Soc. 135:13980–13987. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Pointer TC, Gorelick FS and Desir GV:
Renalase: A multi-functional signaling molecule with roles in
gastrointestinal disease. Cells. 10:20062021. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Guo X, Jessel S, Qu R, Kluger Y, Chen TM,
Hollander L, Safirstein R, Nelson B, Cha C, Bosenberg M, et al:
Inhibition of renalase drives tumour rejection by promoting T cell
activation. Eur J Cancer. 165:81–96. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Guo X, Hollander L, MacPherson D, Wang L,
Velazquez H, Chang J, Safirstein R, Cha C, Gorelick F and Desir GV:
Inhibition of renalase expression and signaling has antitumor
activity in pancreatic cancer. Sci Rep. 6:229962016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Hollander L, Guo X, Velazquez H, Chang J,
Safirstein R, Kluger H, Cha C and Desir GV: Renalase expression by
melanoma and tumor-associated macrophages promotes tumor growth
through a STAT3-mediated mechanism. Cancer Res. 76:3884–3894. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Pan Y, Wang X, He Y, Lin S, Zhu M, Li Y,
Wang J, Wang J, Ma X, Xu J, et al: Tumor suppressor ATP4B serve as
a promising biomarker for worsening of gastric atrophy and poor
differentiation. Gastric Cancer. 24:314–326. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Borghaei RC, Gorski G, Seutter S, Chun J,
Khaselov N and Scianni S: Zinc-binding protein-89 (ZBP-89)
cooperates with NF-κB to regulate expression of matrix
metalloproteinases (MMPs) in response to inflammatory cytokines.
Biochem Biophys Res Commun. 471:503–509. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Borghaei RC, Gorski G and Javadi M; Mariah
Chambers, : NF-kappaB and ZBP-89 regulate MMP-3 expression via a
polymorphic site in the promoter. Biochem Biophys Res Commun.
382:269–273. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Borghaei RC, Rawlings PL Jr, Javadi M and
Woloshin J: NF-kappaB binds to a polymorphic repressor element in
the MMP-3 promoter. Biochem Biophys Res Commun. 316:182–188. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Morán A, Iniesta P, de Juan C,
García-Aranda C, Díaz-López A and Benito M: Impairment of
stromelysin-1 transcriptional activity by promoter mutations in
high microsatellite instability colorectal tumors. Cancer Res.
65:3811–3814. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Kim SJ, Hwang JA, Ro JY, Lee YS and Chun
KH: Galectin-7 is epigenetically-regulated tumor suppressor in
gastric cancer. Oncotarget. 4:1461–1471. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Hou W, Pan M, Xiao Y and Ge W: Serum
extracellular vesicle stratifin is a biomarker of perineural
invasion in patients with colorectal cancer and predicts worse
prognosis. Front Oncol. 12:9125842022. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Jung JY, Koh SA, Lee KH and Kim JR: 14-3-3
Sigma protein contributes to hepatocyte growth factor-mediated cell
proliferation and invasion via matrix metalloproteinase-1
regulation in human gastric cancer. Anticancer Res. 42:519–530.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Chang WC, Huang SF, Lee YM, Lai HC, Cheng
BH, Cheng WC, Ho JY, Jeng LB and Ma WL: Cholesterol import and
steroidogenesis are biosignatures for gastric cancer patient
survival. Oncotarget. 8:692–704. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Cho LY, Yang JJ, Ko KP, Ma SH, Shin A,
Choi BY, Han DS, Song KS, Kim YS, Chang SH, et al: Genetic
susceptibility factors on genes involved in the steroid hormone
biosynthesis pathway and progesterone receptor for gastric cancer
risk. PLoS One. 7:e476032012. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Xu CY, Guo JL, Jiang ZN, Xie SD, Shen JG,
Shen JY and Wang LB: Prognostic role of estrogen receptor alpha and
estrogen receptor beta in gastric cancer. Ann Surg Oncol.
17:2503–2509. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Chandanos E, Rubio CA, Lindblad M, Jia C,
Tsolakis AV, Warner M, Gustafsson JA and Lagergren J: Endogenous
estrogen exposure in relation to distribution of histological type
and estrogen receptors in gastric adenocarcinoma. Gastric Cancer.
11:168–174. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Frycz BA, Murawa D, Borejsza-Wysocki M,
Wichtowski M, Spychała A, Marciniak R, Murawa P, Drews M and
Jagodziński PP: mRNA expression of steroidogenic enzymes, steroid
hormone receptors and their coregulators in gastric cancer. Oncol
Lett. 13:3369–3378. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Kameda C, Nakamura M, Tanaka H, Yamasaki
A, Kubo M, Tanaka M, Onishi H and Katano M: Oestrogen
receptor-alpha contributes to the regulation of the hedgehog
signalling pathway in ERalpha-positive gastric cancer. Br J Cancer.
102:738–747. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Correa P and Piazuelo MB: The gastric
precancerous cascade. J Dig Dis. 13:2–9. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
He Q, Liu L, Wei J, Jiang J, Rong Z, Chen
X, Zhao J and Jiang K: Roles and action mechanisms of bile
acid-induced gastric intestinal metaplasia: A review. Cell Death
Discov. 8:1582022. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Tatsugami M, Ito M, Tanaka S, Yoshihara M,
Matsui H, Haruma K and Chayama K: Bile acid promotes intestinal
metaplasia and gastric carcinogenesis. Cancer Epidemiol Biomarkers
Prev. 21:2101–2107. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Inoue Y, Yu AM, Yim SH, Ma X, Krausz KW,
Inoue J, Xiang CC, Brownstein MJ, Eggertsen G, Björkhem I and
Gonzalez FJ: Regulation of bile acid biosynthesis by hepatocyte
nuclear factor 4alpha. J Lipid Res. 47:215–227. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Tsukita S, Tanaka H and Tamura A: The
claudins: From tight junctions to biological systems. Trends
Biochem Sci. 44:141–152. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Singh AB, Uppada SB and Dhawan P: Claudin
proteins, outside-in signaling, and carcinogenesis. Pflugers Arch.
469:69–75. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Gao M, Li W, Wang H and Wang G: The
distinct expression patterns of claudin-10, −14, −17 and E-cadherin
between adjacent non-neoplastic tissues and gastric cancer tissues.
Diagn Pathol. 8:2052013. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Wang H and Yang X: The expression patterns
of tight junction protein claudin-1, −3, and −4 in human gastric
neoplasms and adjacent non-neoplastic tissues. Int J Clin Exp
Pathol. 8:881–887. 2015.PubMed/NCBI
|
|
66
|
Zhu J and Wang R, Cao H, Zhang H, Xu S,
Wang A, Liu B, Wang Y and Wang R: Expression of claudin-5, −7, −8
and −9 in cervical carcinoma tissues and adjacent non-neoplastic
tissues. Int J Clin Exp Pathol. 8:9479–9486. 2015.PubMed/NCBI
|
|
67
|
Lu YZ, Li Y, Zhang T and Han ST: Claudin-6
is down-regulated in gastric cancer and its potential pathway.
Cancer Biomark. 28:329–340. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Kohmoto T, Masuda K, Shoda K, Takahashi R,
Ujiro S, Tange S, Ichikawa D, Otsuji E and Imoto I: Claudin-6 is a
single prognostic marker and functions as a tumor-promoting gene in
a subgroup of intestinal type gastric cancer. Gastric Cancer.
23:403–417. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Łukaszewicz-Zając M and Mroczko B:
Claudins-promising biomarkers for selected gastrointestinal (GI)
malignancies? Cancers (Basel). 16:1522023. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Simon AG, Lyu SI, Laible M, Wöll S, Türeci
Ö, Şahin U, Alakus H, Fahrig L, Zander T, Buettner R, et al: The
tight junction protein claudin 6 is a potential target for
patient-individualized treatment in esophageal and gastric
adenocarcinoma and is associated with poor prognosis. J Transl Med.
21:5522023. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Torres-Martínez AC, Gallardo-Vera JF,
Lara-Holguin AN, Montaño LF and Rendón-Huerta EP: Claudin-6
enhances cell invasiveness through claudin-1 in AGS human
adenocarcinoma gastric cancer cells. Exp Cell Res. 350:226–235.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Thaler R, Rumpler M, Spitzer S, Klaushofer
K and Varga F: Mospd1, a new player in mesenchymal versus epidermal
cell differentiation. J Cell Physiol. 226:2505–2515. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Imoto Y, Raychaudhuri S, Ma Y, Fenske P,
Sandoval E, Itoh K, Blumrich EM, Matsubayashi HT, Mamer L,
Zarebidaki F, et al: Dynamin is primed at endocytic sites for
ultrafast endocytosis. Neuron. 110:2815–2835.e13. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Meng J: Distinct functions of dynamin
isoforms in tumorigenesis and their potential as therapeutic
targets in cancer. Oncotarget. 8:41701–41716. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Thorsell AG, Persson C, Voevodskaya N,
Busam RD, Hammarström M, Gräslund S, Gräslund A and Hallberg BM:
Structural and biophysical characterization of human myo-inositol
oxygenase. J Biol Chem. 283:15209–15216. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Meng L, Gao J, Mo W, Wang B, Shen H, Cao
W, Ding M, Diao W, Chen W, Zhang Q, et al: MIOX inhibits autophagy
to regulate the ROS-driven inhibition of STAT3/c-Myc-mediated
epithelial-mesenchymal transition in clear cell renal cell
carcinoma. Redox Biol. 68:1029562023. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Yang L, Li C, Qin Y, Zhang G, Zhao B, Wang
Z, Huang Y and Yang Y: A Novel Prognostic model based on
ferroptosis-related gene signature for bladder cancer. Front Oncol.
11:6860442021. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Liu W, Xiang J, Wu X, Wei S, Huang H, Xiao
Y, Zhai B and Wang T: Transcriptome profiles reveal a 12-signature
metabolic prediction model and a novel role of myo-inositol
oxygenase in the progression of prostate cancer. Front Oncol.
12:8998612022. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Xu Z, Zhang S, Nian F and Xu S:
Identification of a glycolysis-related gene signature associated
with clinical outcome for patients with lung squamous cell
carcinoma. Cancer Med. 10:4017–4029. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Cengiz B, Yumrutas O, Bozgeyik E, Borazan
E, Igci YZ, Bozgeyik I and Oztuzcu S: Differential expression of
the UGT1A family of genes in stomach cancer tissues. Tumor Biol.
36:5831–5837. 2015. View Article : Google Scholar
|
|
81
|
Pang SW, Lahiri C, Poh CL and Tan KO: PNMA
family: Protein interaction network and cell signalling pathways
implicated in cancer and apoptosis. Cell Signal. 45:54–62. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Lee YH, Pang SW, Poh CL and Tan KO:
Distinct functional domains of PNMA5 mediate protein-protein
interaction, nuclear localization, and apoptosis signaling in human
cancer cells. J Cancer Res Clin Oncol. 142:1967–1977. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Lin J, Zhang X, Meng F, Zeng F, Liu W and
He X: PNMA5 accelerated cellular proliferation, invasion and
migration in colorectal cancer. Am J Transl Res. 4:2231–2243.
2022.PubMed/NCBI
|
|
84
|
Cabarcas S and Schramm L: RNA polymerase
III trans-cription in cancer: The BRF2 connection. Mol Cancer.
10:472011. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Kang M, Lu S, Chong PK, Yeoh KG and Lim
YP: Comparative proteomic profiling of extracellular proteins
between normal and gastric cancer cells. Curr Cancer Drug Targets.
16:442–454. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Zhang Y, Wu H, Yang F, Ning J, Li M, Zhao
C, Zhong S, Gu K and Wang H: Prognostic value of the expression of
DNA repair-related biomarkers mediated by alcohol in gastric cancer
patients. Am J Pathol. 188:367–377. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Welch MD, DePace AH, Verma S, Iwamatsu A
and Mitchison TJ: The human Arp2/3 complex is composed of
evolutionarily conserved subunits and is localized to cellular
regions of dynamic actin filament assembly. J Cell Biol.
138:375–384. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Yoo Y, Wu X and Guan JL: A novel role of
the actin-nucleating Arp2/3 complex in the regulation of RNA
polymerase II-dependent transcription. J Biol Chem. 282:7616–7623.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Lee GE, Kim JH, Taylor M and Muller MT:
DNA methyltransferase 1-associated protein (DMAP1) is a
co-repressor that stimulates DNA methylation globally and locally
at sites of double strand break repair. J Biol Chem.
285:37630–37640. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Li B, Zhu J and Meng L: High expression of
ACTL8 is poor prognosis and accelerates cell progression in head
and neck squamous cell carcinoma. Mol Med Rep. 19:877–884.
2019.PubMed/NCBI
|
|
91
|
Han Q, Sun ML, Liu WS, Zhao HS, Jiang LY,
Yu ZJ and Wei MJ: Upregulated expression of ACTL8 contributes to
invasion and metastasis and indicates poor prognosis in colorectal
cancer. Onco Targets Ther. 12:1749–1763. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Mantilla MJ, Chaves JJ, Ochoa-Vera M,
Africano F, Parra-Medina R and Tovar-Fierro G: Clinical
characteristics of early-onset gastric cancer. A study in a
Colombian population. Rev Gastroenterol Peru. 43:236–241. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Liu H, Li Z, Zhang Q, Li Q, Zhong H, Wang
Y, Yang H, Li H, Wang X, Li K, et al: Multi-institutional
development and validation of a nomogram to predict prognosis of
early-onset gastric cancer patients. Front Immunol. 13:10071762022.
View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Umeyama K, Sowa M, Kamino K, Kato Y and
Satake K: Gastric carcinoma in young adults in Japan. Anticancer
Res. 2:283–286. 1982.PubMed/NCBI
|
|
95
|
LaPelusa M, Shen C, Gillaspie EA, Cann C,
Lambright E, Chakravarthy AB, Gibson MK and Eng C: Variation in
treatment patterns of patients with early-onset gastric cancer.
Cancers (Basel). 14:36332022. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Setia N, Wang CX, Lager A, Maron S, Shroff
S, Arndt N, Peterson B, Kupfer SS, Ma C, Misdraji J, et al:
Morphologic and molecular analysis of early-onset gastric cancer.
Cancer. 127:103–114. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Mun DG, Bhin J, Kim S, Kim H, Jung JH,
Jung Y, Jang YE, Park JM, Kim H, Jung Y, et al: Proteogenomic
characterization of human early-onset gastric cancer. Cancer Cell.
35:111–124.e10. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Ma Z, Liu X, Paul ME, Chen M, Zheng P and
Chen H: Comparative investigation of early-onset gastric cancer.
Oncol Lett. 21:3742021. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Skierucha M, Milne AN, Offerhaus GJ,
Polkowski WP, Maciejewski R and Sitarz R: Molecular alterations in
gastric cancer with special reference to the early-onset subtype.
World J Gastroenterol. 22:2460–2474. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Gao F, Li M, Xiang R, Zhou X, Zhu L and
Zhai Y: Expression of CLDN6 in tissues of gastric cancer patients:
Association with clinical pathology and prognosis. Oncol Lett.
17:4621–4625. 2019.PubMed/NCBI
|
|
101
|
Wu LH, Wang XX, Wang Y, Wei J, Liang ZR,
Yan X and Wang J: Construction and validation of a prognosis
signature based on the immune microenvironment in gastric cancer.
Front Surg. 10:10882922023. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Ajani JA, D'Amico TA, Bentrem DJ, Chao J,
Cooke D, Corvera C, Das P, Enzinger PC, Enzler T, Fanta P, et al:
Gastric cancer, version 2.2022, NCCN clinical practice guidelines
in oncology. J Natl Compr Canc Netw. 20:167–192. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Lordick F, Carneiro F, Cascinu S, Fleitas
T, Haustermans K, Piessen G, Vogel A and Smyth EC; ESMO Guidelines
Committee. Electronic address, : simpleclinicalguidelines@esmo.org:
Gastric cancer: ESMO clinical practice guideline for diagnosis,
treatment and follow-up. Ann Oncol. 33:1005–1020. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Gao S, Li J, Wang W, Wang Y, Shan Y and
Tan H: Rabdosia rubescens (Hemsl.) H. Hara: A potent
anti-tumor herbal remedy-Botany, phytochemistry, and clinical
applications and insights. J Ethnopharmacol. 340:1192002025.
View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Gao S, Shan Y, Wang Y, Wang W, Li J and
Tan H: Polysaccharides from Lonicera japonica Thunb.: Extraction,
purification, structural features and biological activities-A
review. Int J Biol Macromol. 281:1364722024. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Gao S, Gang J, Yu M, Xin G and Tan H:
Computational analysis for identification of early diagnostic
biomarkers and prognostic biomarkers of liver cancer based on GEO
and TCGA databases and studies on pathways and biological functions
affecting the survival time of liver cancer. BMC Cancer.
21:7912021. View Article : Google Scholar : PubMed/NCBI
|