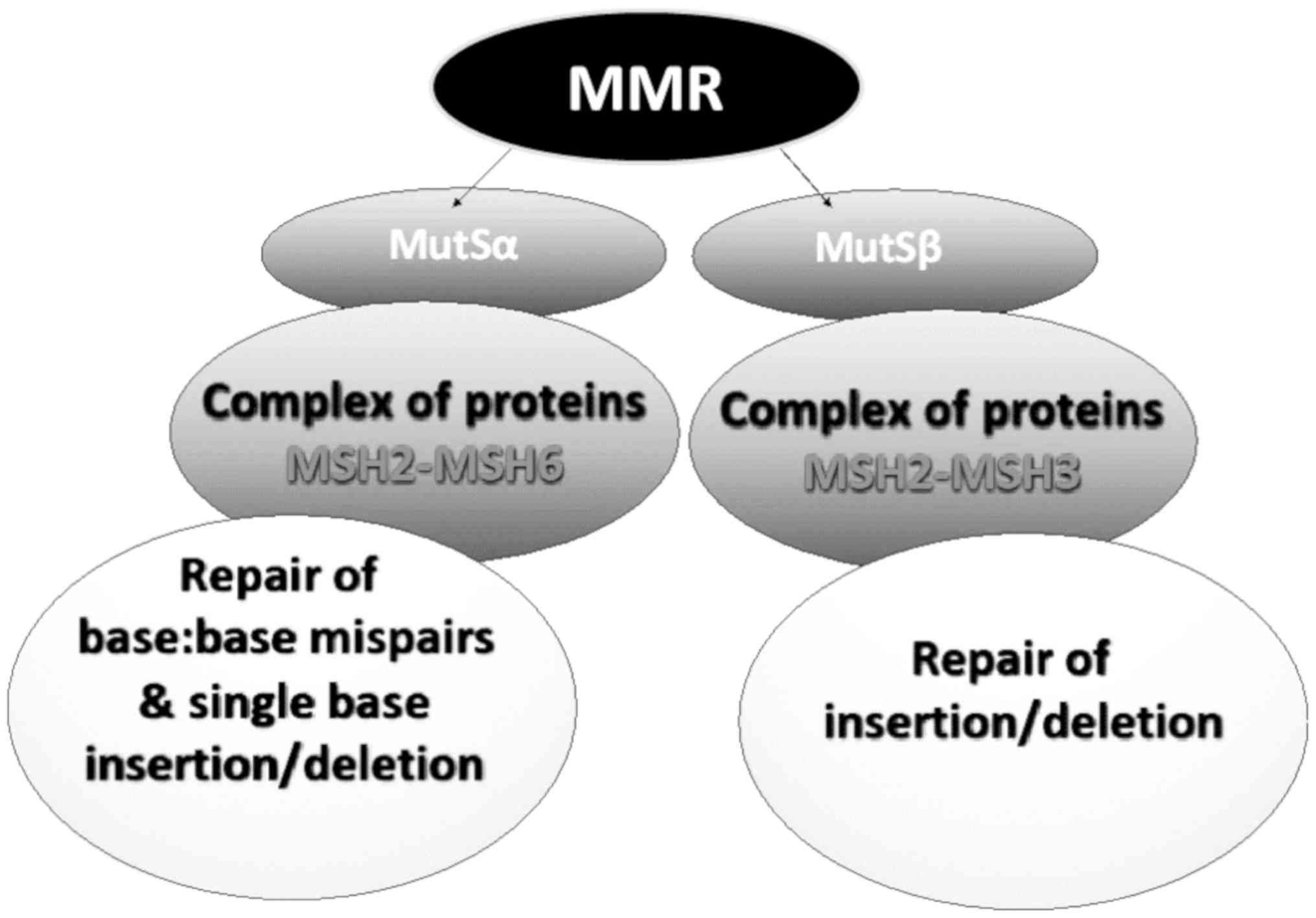
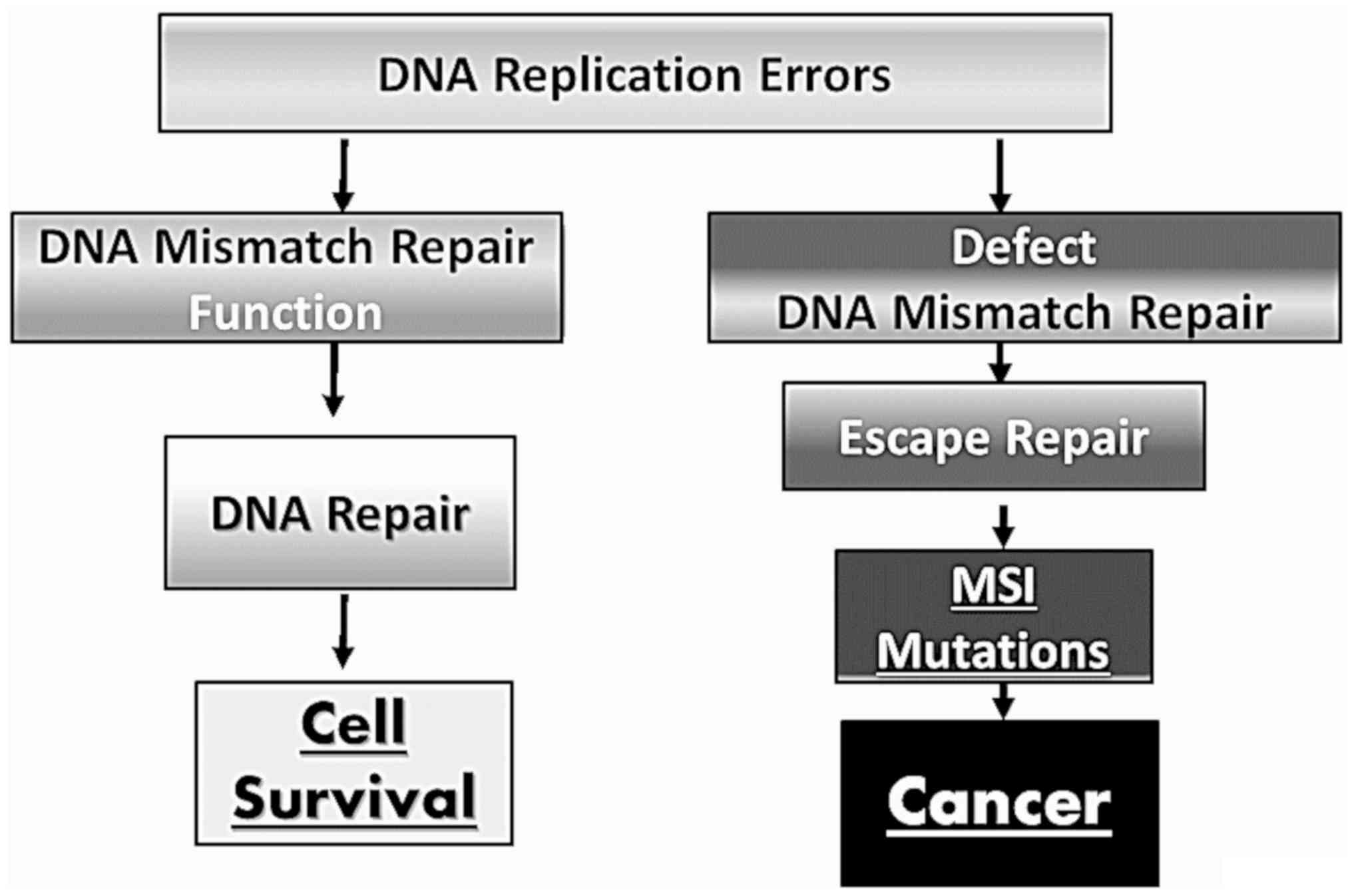
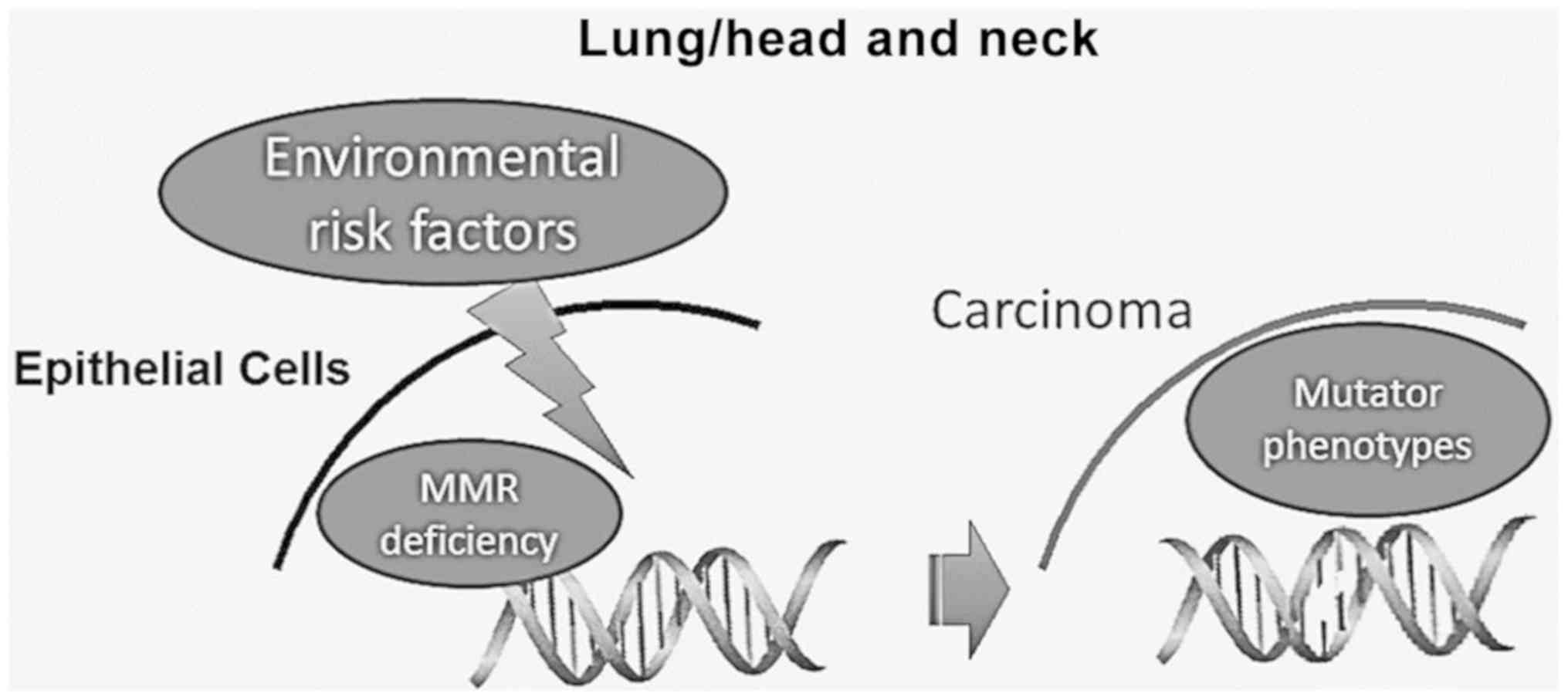
|
1
|
Peltomäki P: DNA mismatch repair and
cancer. Mutat Res. 488:77–85. 2001.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Peltomäki P: Role of DNA mismatch repair
defects in the pathogenesis of human cancer. J Clin Oncol.
21:1174–1179. 2003.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Li GM: DNA mismatch repair and cancer.
Front Biosci. 8:d997–d1017. 2003.PubMed/NCBI View
Article : Google Scholar
|
|
4
|
Yang W: Structure and mechanism for DNA
lesion recognition. Cell Res. 18:184–197. 2008.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Kolodner RD: Mismatch repair: Mechanisms
and relationship to cancer susceptibility. Trends Biochem Sci.
20:397–401. 1995.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Jiricny J and Nyström-Lahti M: Mismatch
repair defects in cancer. Curr Opin Genet Dev. 10:157–161.
2000.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Aarnio M, Sankila R, Pukkala E, Salovaara
R, Aaltonen LA, de la Chapelle A, Peltomäki P, Mecklin JP and
Järvinen HJ: Cancer risk in mutation carriers of
DNA-mismatch-repair genes. Int J Cancer. 81:214–218.
1999.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Bohr VA: DNA repair fine structure and its
relations to genomic instability. Carcinogenesis. 16:2885–2892.
1995.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Kruhøffer M, Jensen JL, Laiho P, Dyrskjøt
L, Salovaara R, Arango D, Birkenkamp-Demtroder K, Sørensen FB,
Christensen LL, Buhl L, et al: Gene expression signatures for
colorectal cancer microsatellite status and HNPCC. Br J Cancer.
92:2240–2248. 2005.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Leach FS, Nicolaides NC, Papadopoulos N,
Liu B, Jen J, Parsons R, Peltomäki P, Sistonen P, Aaltonen LA,
Nyström-Lahti M, et al: Mutations of a mutS homolog in hereditary
nonpolyposis colorectal cancer. Cell. 75:1215–1225. 1993.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Fishel R, Lescoe MK, Rao MRS, Copeland NG,
Jenkins NA, Garber J, Kane M and Kolodner R: The human mutator gene
homolog MSH2 and its association with hereditary nonpolyposis colon
cancer. Cell. 75:1027–1038. 1993.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Liu B, Nicolaides NC, Markowitz S, Willson
JKV, Parsons RE, Jen J, Papadopolous N, Peltomäki P, de la Chapelle
A, Hamilton SR, et al: Mismatch repair gene defects in sporadic
colorectal cancers with microsatellite instability. Nat Genet.
9:48–55. 1995.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Liu B, Parsons R, Papadopoulos N,
Nicolaides NC, Lynch HT, Watson P, Jass J, Dunlop M, Wyllie A,
Jessup JM, Peltomäki PT, et al: Mismatch repair gene analysis in
HNPCC patients. Nat Med. 2:169–174. 1996.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Kowalski LD, Mutch DG, Herzog TJ, Rader JS
and Goodfellow PJ: Mutational analysis of MLH1 and MSH2 in 25
prospectively-acquired RER+ endometrial cancers. Genes Chromosomes
Cancer. 18:219–227. 1997.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Chadwick RB, Pyatt RE, Niemann TH,
Richards SK, Johnson CK, Stevens MW, Meek JE, Hampel H, Prior TW
and de la Chapelle A: Hereditary and somatic DNA mismatch repair
gene mutations in sporadic endometrial carcinoma. J Med Genet.
38:461–466. 2001.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Soliman PT and Lu K: Endometrial cancer
associated with defective DNA mismatch repair. Obstet Gynecol Clin
North Am. 34:701–715, viii. 2007.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Rosen DG, Cai KQ, Luthra R and Liu J:
Immunohistochemical staining of hMLH1 and hMSH2 reflects
microsatellite instability status in ovarian carcinoma. Mod Pathol.
19:1414–1420. 2006.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Vageli D, Daniil Z, Dahabreh J, Karagianni
E, Vamvakopoulou DN, Ioannou MG, Scarpinato K, Vamvakopoulos NC,
Gourgoulianis KI and Koukoulis GK: Phenotypic mismatch repair hMSH2
and hMLH1 gene expression profiles in primary non-small cell lung
carcinomas. Lung Cancer. 64:282–288. 2009.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Wang YC, Lu YP, Tseng RC, Lin RK, Chang
JW, Chen JT, Shih CM and Chen CY: Inactivation of hMLH1 and hMSH2
by promoter methylation in primary non-small cell lung tumors and
matched sputum samples. J Clin Invest. 111:887–895. 2003.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Xinarianos G, Liloglou T, Prime W,
Sourvinos G, Karachristos A, Gosney JR, Spandidos DA and Field JK:
p53 status correlates with the differential expression of the DNA
mismatch repair protein MSH2 in non-small cell lung carcinoma. Int
J Cancer. 101:248–252. 2002.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Kitajima Y, Miyazaki K, Matsukura S,
Tanaka M and Sekiguchi M: Loss of expression of DNA repair enzymes
MGMT, hMLH1, and hMSH2 during tumor progression in gastric cancer.
Gastric Cancer. 6:86–95. 2003.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Tahara E: Genetic pathways of two types of
gastric cancer. IARC Sci Publ. 157:327–349. 2004.PubMed/NCBI
|
|
23
|
Nardone G, Rocco A and Budillon G:
Molecular alteration of gastric carcinoma. Minerva Gastroenterol
Dietol. 48:189–193. 2002.PubMed/NCBI
|
|
24
|
El-Rifai W, Powell SM and EI-Rifai W:
Molecular biology of gastric cancer. Semin Radiat Oncol.
12:128–140. 2002.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Sasaki S and Nakamura Y: Mutation of the
mismatch repair genes for carcinogenesis of sporadic colorectal
cancers with RER-positive phenotype. Nihon Rinsho. 54:1008–1013.
1996.(In Japanese). PubMed/NCBI
|
|
26
|
Wallis Y and Macdonald F: The genetics of
inherited colon cancer. Clin Mol Pathol. 49:M65–M73.
1996.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Burger M, Denzinger S, Hammerschmied CG,
Tannapfel A, Obermann EC, Wieland WF, Hartmann A and Stoehr R:
Elevated microsatellite alterations at selected tetranucleotides
(EMAST) and mismatch repair gene expression in prostate cancer. J
Mol Med (Berl). 84:833–841. 2006.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Kassem HS, Varley JM, Hamam SM and
Margison GP: Immunohistochemical analysis of expression and
allelotype of mismatch repair genes (hMLH1 and hMSH2) in bladder
cancer. Br J Cancer. 84:321–328. 2001.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Leach FS, Hsieh JT, Molberg K, Saboorian
MH, McConnell JD and Sagalowsky AI: Expression of the human
mismatch repair gene hMSH2: A potential marker for urothelial
malignancy. Cancer. 88:2333–2341. 2000.PubMed/NCBI
|
|
30
|
Kuismanen SA, Holmberg MT, Salovaara R, de
la Chapelle A and Peltomäki P: Genetic and epigenetic modification
of MLH1 accounts for a major share of microsatellite-unstable
colorectal cancers. Am J Pathol. 156:1773–1779. 2000.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Viale G, Trapani D and Curigliano G:
Mismatch repair deficiency as a predictive biomarker for
immunotherapy efficacy. BioMed Res Int.
2017(4719194)2017.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Hsieh P and Yamane K: DNA mismatch repair:
Molecular mechanism, cancer, and ageing. Mech Ageing Dev.
129:391–407. 2008.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Kijas AW, Studamire B and Alani E:
Msh2 separation of function mutations confer defects in the
initiation steps of mismatch repair. J Mol Biol. 331:123–138.
2003.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Gurin CC, Federici MG, Kang L and Boyd J:
Causes and consequences of microsatellite instability in
endometrial carcinoma. Cancer Res. 59:462–466. 1999.PubMed/NCBI
|
|
35
|
Hayashi M, Tamura G, Jin Z, Kato I, Sato
M, Shibuya Y, Yang S and Motoyama T: Microsatellite instability in
esophageal squamous cell carcinoma is not associated with
hMLH1 promoter hypermethylation. Pathol Int. 53:270–276.
2003.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Nikolouzakis TK, Vassilopoulou L,
Fragkiadaki P, Mariolis Sapsakos T, Papadakis GZ, Spandidos DA,
Tsatsakis AM and Tsiaoussis J: Improving diagnosis, prognosis and
prediction by using biomarkers in CRC patients (Review). Oncol Rep.
39:2455–2472. 2018.(Review). PubMed/NCBI View Article : Google Scholar
|
|
37
|
Lynch HT, Snyder CL, Shaw TG, Heinen CD
and Hitchins MP: Milestones of Lynch syndrome 1895-2015. Nat Rev
Canc. 15:181–194. 2015.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Antelo M, Golubicki M, Roca E, Mendez G,
Carballido M, Iseas S, Cuatrecasas M, Moreira L, Sanchez A,
Carballal S, et al: Lynch-like syndrome is as frequent as Lynch
syndrome in early-onset nonfamilial nonpolyposis colorectal cancer.
Int J Cancer. 145:705–713. 2019.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Carethers JM and Stoffel EM: Lynch
syndrome and Lynch syndrome mimics: The growing complex landscape
of hereditary colon cancer. World J Gastroenterol. 21:9253–9261.
2015.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Adar T, Friedman M, Rodgers LH, Shannon
KM, Zukerberg LR and Chung DC: Gastric cancer in Lynch syndrome is
associated with underlying immune gastritis. J Med Genet. doi:
10.1136/jmedgenet-2018-105757 (Epub ahead of print). PubMed/NCBI View Article : Google Scholar
|
|
41
|
Wang J, Greenberg S and Yates J: Lynch
syndrome-associated upper tract urothelial carcinoma. Urology.
121:19–21. 2018.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Cloyd JM, Chun YS, Ikoma N, Vauthey JN,
Aloia TA, Cuddy A, Rodriguez-Bigas MA and Nancy You Y: Clinical and
genetic implications of DNA mismatch repair deficiency in biliary
tract cancers associated with lynch syndrome. J Gastrointest
Cancer. 49:93–96. 2018.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Anacleto C, Leopoldino AM, Rossi B, Soares
FA, Lopes A, Rocha JC, Caballero O, Camargo AA, Simpson AJ and Pena
SD: Colorectal cancer ‘methylator phenotype’: Fact or artifact?
Neoplasia. 7:331–335. 2005.PubMed/NCBI
|
|
44
|
Imai K and Yamamoto H: Carcinogenesis and
microsatellite instability: The interrelationship between genetics
and epigenetics. Carcinogenesis. 29:673–680. 2008.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Chang DK, Ricciardiello L, Goel A, Chang
CL and Boland CR: Steady-state regulation of the human DNA mismatch
repair system. J Biol Chem. 275:18424–18431. 2000.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Vageli DP, Zaravinos A, Daniil Z, Dahabreh
J, Doukas SG, Spandidos DA, Gourgoulianis KI and Koukoulis GK:
hMSH2 and hMLH1 gene expression patterns differ between lung
adenocarcinoma and squamous cell carcinoma: Correlation with
patient survival and response to adjuvant chemotherapy treatment.
Int J Biol Markers. 27:e400–e404. 2013.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Vageli DP, Giannopoulos S, Doukas SG,
Kalaitzis C, Giannakopoulos S, Giatromanolaki A, Koukoulis GK and
Touloupidis S: Mismatch repair hMSH2, hMLH1, hMSH6 and hPMS2 mRNA
expression profiles in precancerous and cancerous urothelium. Oncol
Lett. 5:283–294. 2013.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Vageli DP, Doukas SG and Markou A:
Mismatch DNA repair mRNA expression profiles in oral melanin
pigmentation lesion and hamartomatous polyp of a child with
Peutz-Jeghers syndrome. Pediatr Blood Cancer. 60:E116–E117.
2013.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Vageli DP, Papamichali R, Kambosioras K,
Papandreou CN and Koukoulis GK: Mismatch DNA repair hMSH2,
hMLH1, hMSH6 and hPMS2 mRNA expression
profiles in colorectal carcinomas. J Genet Syndr Gene Ther.
View Article : Google Scholar
|
|
50
|
Christmann M and Kaina B: Nuclear
translocation of mismatch repair proteins MSH2 and MSH6 as a
response of cells to alkylating agents. J Biol Chem.
275:36256–36262. 2000.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Strathdee G, MacKean MJ, Illand M and
Brown R: A role for methylation of the hMLH1 promoter in loss of
hMLH1 expression and drug resistance in ovarian cancer. Oncogene.
18:2335–2341. 1999.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Carethers JM, Chauhan DP, Fink D, Nebel S,
Bresalier RS, Howell SB and Boland CR: Mismatch repair proficiency
and in vitro response to 5-fluorouracil. Gastroenterology.
117:123–131. 1999.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Bhattacharjee P, Sanyal T, Bhattacharjee S
and Bhattacharjee P: Epigenetic alteration of mismatch repair genes
in the population chronically exposed to arsenic in West Bengal,
India. Environ Res. 163:289–296. 2018.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Vivarelli S, Salemi R, Candido S, Falzone
L, Santagati M, Stefani S, Torino F, Banna GL, Tonini G and Libra
M: Gut Microbiota and Cancer: From Pathogenesis to Therapy. Cancers
(Basel). 11(E38)2019.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Banna GL, Torino F, Marletta F, Santagati
M, Salemi R, Cannarozzo E, Falzone L, Ferraù F and Libra M:
Lactobacillus rhamnosus GG: An Overview to explore the
rationale of its use in cancer. Front Pharmacol. doi:
10.3389/fphar.2017.00603. PubMed/NCBI View Article : Google Scholar
|
|
56
|
Westwood A, Glover A, Hutchins G, Young C,
Brockmoeller S, Robinson R, Worrilow L, Wallace D, Rankeillor K,
Adlard J, et al: Additional loss of MSH2 and MSH6 expression in
sporadic deficient mismatch repair colorectal cancer due to MLH1
promoter hypermethylation. J Clin Pathol. 72:443–447.
2019.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Kang Z, Zhu Y, Zhang QA, Dong L, Xu F,
Zhang X and Guan M: Methylation and expression analysis of mismatch
repair genes in extramammary Paget's disease. J Eur Acad Dermatol
Venereol. 33:874–879. 2019.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Mäki-Nevala S, Valo S, Ristimäki A,
Sarhadi V, Knuutila S, Nyström M, Renkonen-Sinisalo L, Lepistö A,
Mecklin JP and Peltomäki P: DNA methylation changes and somatic
mutations as tumorigenic events in Lynch syndrome-associated
adenomas retaining mismatch repair protein expression.
EBioMedicine. 39:280–291. 2019.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297.
2004.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Zimmerman AL and Wu S: MicroRNAs, cancer
and cancer stem cells. Cancer Lett. 300:10–19. 2011.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Falzone L, Lupo G, La Rosa GRM, Crimi S,
Anfuso CD, Salemi R, Rapisarda E, Libra M and Candido S:
Identification of novel microRNAs and their diagnostic and
prognostic significance in oral cancer. Cancers (Basel).
11(E610)2019.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Hafsi S, Candido S, Maestro R, Falzone L,
Soua Z, Bonavida B, Spandidos DA and Libra M: Correlation between
the overexpression of Yin Yang 1 and the expression levels of
miRNAs in Burkitt's lymphoma: A computational study. Oncol Lett.
11:1021–1025. 2016.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Falzone L, Romano GL, Salemi R, Bucolo C,
Tomasello B, Lupo G, Anfuso CD, Spandidos DA, Libra M and Candido
S: Prognostic significance of deregulated microRNAs in uveal
melanomas. Mol Med Rep. 19:2599–2610. 2019.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Falzone L, Candido S, Salemi R, Basile MS,
Scalisi A, McCubrey JA, Torino F, Signorelli SS, Montella M and
Libra M: Computational identification of microRNAs associated to
both epithelial to mesenchymal transition and NGAL/MMP-9 pathways
in bladder cancer. Oncotarget. 7:72758–72766. 2016.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Costa PM and Pedroso de Lima MC: MicroRNAs
as molecular targets for cancer therapy: On the modulation of
microRNA expression. Pharmaceuticals (Basel). 6:1195–1220.
2013.PubMed/NCBI View Article : Google Scholar
|
|
66
|
McCubrey JA, Fitzgerald TL, Yang LV,
Lertpiriyapong K, Steelman LS, Abrams SL, Montalto G, Cervello M,
Neri LM, Cocco L, et al: Roles of GSK-3 and microRNAs on epithelial
mesenchymal transition and cancer stem cells. Oncotarget.
8:14221–14250. 2017.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Reinhart BJ, Slack FJ, Basson M,
Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR and Ruvkun G:
The 21-nucleotide let-7 RNA regulates developmental timing in
Caenorhabditis elegans. Nature. 403:901–906. 2000.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Cheng AM, Byrom MW, Shelton J and Ford LP:
Antisense inhibition of human miRNAs and indications for an
involvement of miRNA in cell growth and apoptosis. Nucleic Acids
Res. 33:1290–1297. 2005.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Sasaki CT, Doukas SG and Vageli DP: In
vivo short-term topical application of BAY 11-7082 prevents the
acidic bile-induced mRNA and miRNA oncogenic phenotypes in exposed
murine hypopharyngeal mucosa. Neoplasia. 20:374–386.
2018.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Doukas SG, Vageli DP and Sasaki CT: NF-κB
inhibition reverses acidic bile-induced miR-21, miR-155, miR-192,
miR-34a, miR-375 and miR-451a deregulations in human hypopharyngeal
cells. J Cell Mol Med. 22:2922–2934. 2018.PubMed/NCBI View Article : Google Scholar
|
|
71
|
Doukas PG, Vageli DP, Doukas SG and Sasaki
CT: Temporal characteristics of NF-κB inhibition in blocking
bile-induced oncogenic molecular events in hypopharyngeal cells.
Oncotarget. doi: https://doi.org/10.18632/oncotarget.26917.
PubMed/NCBI View Article : Google Scholar
|
|
72
|
Landau DA and Slack FJ: MicroRNAs in
mutagenesis, genomic instability, and DNA repair. Semin Oncol.
38:743–751. 2011.PubMed/NCBI View Article : Google Scholar
|
|
73
|
Mao G, Lee S, Ortega J, Gu L and Li GM:
Modulation of microRNA processing by mismatch repair protein MutLα.
Cell Res. 22:973–985. 2012.PubMed/NCBI View Article : Google Scholar
|
|
74
|
Valeri N, Gasparini P, Fabbri M, Braconi
C, Veronese A, Lovat F, Adair B, Vannini I, Fanini F, Bottoni A, et
al: Modulation of mismatch repair and genomic stability by miR-155.
Proc Natl Acad Sci USA. 107:6982–6987. 2010.PubMed/NCBI View Article : Google Scholar
|
|
75
|
Zhong Z, Dong Z, Yang L, Chen X and Gong
Z: MicroRNA-31-5p modulates cell cycle by targeting human mutL
homolog 1 in human cancer cells. Tumour Biol. 34:1959–1965.
2013.PubMed/NCBI View Article : Google Scholar
|
|
76
|
Salemi R, Falzone L, Madonna G, Polesel J,
Cinà D, Mallardo D, Ascierto PA, Libra M and Candido S: MMP-9 as a
candidate marker of response to BRAF inhibitors in melanoma
patients with BRAFV600E mutation detected in circulating-free DNA.
Front Pharmacol. Aug 14 2018 (Epub ahead of print) doi:
10.3389/fphar.2018.00856. PubMed/NCBI View Article : Google Scholar
|
|
77
|
Pfeifer GP, Denissenko MF, Olivier M,
Tretyakova N, Hecht SS and Hainaut P: Tobacco smoke carcinogens,
DNA damage and p53 mutations in smoking-associated cancers.
Oncogene. 21:7435–7451. 2002.PubMed/NCBI View Article : Google Scholar
|
|
78
|
Zhong Y, Carmella SG, Upadhyaya P,
Hochalter JB, Rauch D, Oliver A, Jensen J, Hatsukami D, Wang J,
Zimmerman C, et al: Immediate consequences of cigarette smoking:
Rapid formation of polycyclic aromatic hydrocarbon diol epoxides.
Chem Res Toxicol. 24:246–252. 2011.PubMed/NCBI View Article : Google Scholar
|
|
79
|
Boda D, Docea AO, Calina D, Ilie MA,
Caruntu C, Zurac S, Neagu M, Constantin C, Branisteanu DE,
Voiculescu V, et al: Human papilloma virus: Apprehending the link
with carcinogenesis and unveiling new research avenues (Review).
Int J Oncol. 52:637–655. 2018.PubMed/NCBI View Article : Google Scholar
|
|
80
|
Dylawerska A, Barczak W, Wegner A,
Golusinski W and Suchorska WM: Association of DNA repair genes
polymorphisms and mutations with increased risk of head and neck
cancer: A review. Med Oncol. 34(197)2017.PubMed/NCBI View Article : Google Scholar
|
|
81
|
Demokan S, Suoglu Y, Demir D, Gozeler M
and Dalay N: Microsatellite instability and methylation of the DNA
mismatch repair genes in head and neck cancer. Ann Oncol.
17:995–999. 2006.PubMed/NCBI View Article : Google Scholar
|
|
82
|
Wang Y, Irish J, MacMillan C, Brown D,
Xuan Y, Boyington C, Gullane P and Kamel-Reid S: High frequency of
microsatellite instability in young patients with head-and-neck
squamous-cell carcinoma: Lack of involvement of the mismatch repair
genes hMLH1 and hMSH2. Int J Cancer. 93:353–360.
2001.PubMed/NCBI View Article : Google Scholar
|
|
83
|
Liu K, Huang H, Mukunyadzi P, Suen JY,
Hanna E and Fan CY: Promoter hypermethylation: An important
epigenetic mechanism for hMLH1 gene inactivation in head and neck
squamous cell carcinoma. Otolaryngol Head Neck Surg. 126:548–553.
2002.PubMed/NCBI View Article : Google Scholar
|
|
84
|
Murata H, Khattar NH, Kang Y, Gu L and Li
GM: Genetic and epigenetic modification of mismatch repair genes
hMSH2 and hMLH1 in sporadic breast cancer with
microsatellite instability. Oncogene. 21:5696–5703. 2002.PubMed/NCBI View Article : Google Scholar
|
|
85
|
Demokan S, Muslumanoglu M, Yazici H, Igci
A and Dalay N: Investigation of microsatellite instability in
Turkish breast cancer patients. Pathol Oncol Res. 8:138–141.
2002.PubMed/NCBI
|
|
86
|
Ruszkiewicz A, Bennett G, Moore J, Manavis
J, Rudzki B, Shen L and Suthers G: Correlation of mismatch repair
genes immunohistochemistry and microsatellite instability status in
HNPCC-associated tumours. Pathology. 34:541–547. 2002.PubMed/NCBI View Article : Google Scholar
|
|
87
|
Shia J, Ellis NA and Klimstra DS: The
utility of immunohistochemical detection of DNA mismatch repair
gene proteins. Virchows Arch. 445:431–441. 2004.PubMed/NCBI View Article : Google Scholar
|
|
88
|
Pereira CS, Oliveira MV, Barros LO,
Bandeira GA, Santos SH, Basile JR, Guimarães AL and De Paula AM:
Low expression of MSH2 DNA repair protein is associated with poor
prognosis in head and neck squamous cell carcinoma. J Appl Oral
Sci. 21:416–421. 2013.PubMed/NCBI View Article : Google Scholar
|
|
89
|
Nogueira GA, Lourenço GJ, Oliveira CB,
Marson FA, Lopes-Aguiar L, Costa EF, Lima TR, Liutti VT, Leal F,
Santos VC, et al: Association between genetic polymorphisms in DNA
mismatch repair-related genes with risk and prognosis of head and
neck squamous cell carcinoma. Int J Cancer. 137:810–818.
2015.PubMed/NCBI View Article : Google Scholar
|
|
90
|
Jha R, Gaur P, Sharma SC and Das SN:
Single nucleotide polymorphism in hMLH1 promoter and risk of
tobacco-related oral carcinoma in high-risk Asian Indians. Gene.
526:223–227. 2013.PubMed/NCBI View Article : Google Scholar
|
|
91
|
Liu K, Huang H, Mukunyadzi P, Suen JY,
Hanna E and Fan CY: Promoter hypermethylation: An important
epigenetic mechanism for hMLH1 gene inactivation in head and neck
squamous cell carcinoma. Otolaryngol Head Neck Surg. 126:548–553.
2002.PubMed/NCBI View Article : Google Scholar
|
|
92
|
Liu K, Zuo C, Luo QK, Suen JY, Hanna E and
Fan CY: Promoter hypermethylation and inactivation of hMLH1,
a DNA mismatch repair gene, in head and neck squamous cell
carcinoma. Diagn Mol Pathol. 12:50–56. 2003.PubMed/NCBI
|
|
93
|
Zuo C, Zhang H, Spencer HJ, Vural E, Suen
JY, Schichman SA, Smoller BR, Kokoska MS and Fan CY: Increased
microsatellite instability and epigenetic inactivation of the
hMLH1 gene in head and neck squamous cell carcinoma.
Otolaryngol Head Neck Surg. 141:484–490. 2009.PubMed/NCBI View Article : Google Scholar
|
|
94
|
Tawfik HM, El-Maqsoud NM, Hak BH and
El-Sherbiny YM: Head and neck squamous cell carcinoma: Mismatch
repair immunohistochemistry and promoter hypermethylation of
hMLH1 gene. Am J Otolaryngol. 32:528–536. 2011.PubMed/NCBI View Article : Google Scholar
|
|
95
|
Xinarianos G, Liloglou T, Prime W, Maloney
P, Callaghan J, Fielding P, Gosney JR and Field JK: hMLH1
and hMSH2 expression correlates with allelic imbalance on
chromosome 3p in non-small cell lung carcinomas. Cancer Res.
60:4216–4221. 2000.PubMed/NCBI
|
|
96
|
Hsu HS, Wen CK, Tang YA, Lin RK, Li WY,
Hsu WH and Wang YC: Promoter hypermethylation is the predominant
mechanism in hMLH1 and hMSH2 deregulation and is a
poor prognostic factor in nonsmoking lung cancer. Clin Cancer Res.
11:5410–5416. 2005.PubMed/NCBI View Article : Google Scholar
|
|
97
|
Kanellis G, Chatzistamou I, Koutselini H,
Politi E, Gouliamos A, Vlahos L and Koutselinis A: Expression of
DNA mismatch repair gene MSH2 in cytological material from lung
cancer patients. Diagn Cytopathol. 34:463–466. 2006.PubMed/NCBI View Article : Google Scholar
|
|
98
|
Downey CM and Jirik FR: DNA mismatch
repair deficiency accelerates lung neoplasm development in
K-ras(LA1/+) mice: A brief report. Cancer Med.
4:897–902. 2015.PubMed/NCBI View Article : Google Scholar
|
|
99
|
Scartozzi M, Franciosi V, Campanini N,
Benedetti G, Barbieri F, Rossi G, Berardi R, Camisa R, Silva RR,
Santinelli A, et al: Mismatch repair system (MMR) status correlates
with response and survival in non-small cell lung cancer (NSCLC)
patients. Lung Cancer. 53:103–109. 2006.PubMed/NCBI View Article : Google Scholar
|
|
100
|
Takahashi Y, Kondo K, Hirose T, Nakagawa
H, Tsuyuguchi M, Hashimoto M, Sano T, Ochiai A and Monden Y:
Microsatellite instability and protein expression of the DNA
mismatch repair gene, hMLH1, of lung cancer in chromate-exposed
workers. Mol Carcinog. 42:150–158. 2005.PubMed/NCBI View Article : Google Scholar
|
|
101
|
Falzone L, Salomone S and Libra M:
Evolution of cancer pharmacological treatments at the turn of the
third millennium. Front Pharmacol. doi:
doi.org/10.3389/fphar.2018.01300. PubMed/NCBI View Article : Google Scholar
|
|
102
|
Fink D, Nebel S, Norris PS, Aebi S, Kim
HK, Haas M and Howell SB: The effect of different chemotherapeutic
agents on the enrichment of DNA mismatch repair-deficient tumour
cells. Br J Cancer. 77:703–708. 1998.PubMed/NCBI View Article : Google Scholar
|
|
103
|
Vaisman A, Varchenko M, Umar A, Kunkel TA,
Risinger JI, Barrett JC, Hamilton TC and Chaney SG: The role of
hMLH1, hMSH3, and hMSH6 defects in cisplatin and oxaliplatin
resistance: Correlation with replicative bypass of platinum-DNA
adducts. Cancer Res. 58:3579–3585. 1998.PubMed/NCBI
|
|
104
|
Hause RJ, Pritchard CC, Shendure J and
Salipante SJ: Classification and characterization of microsatellite
instability across 18 cancer types. Nat Med. 22:1342–1350.
2016.PubMed/NCBI View Article : Google Scholar
|
|
105
|
Microsatellite Instability-Defective DNA
Mismatch Repair: ESMO Biomarker Factsheet. [https://oncologypro.esmo.org/Education-Library/Factsheets-on-Biomarkers/Microsatellite-Instability-Defective-DNA-Mismatch-Repair#eztoc1701983_0_0_8/.
|
|
106
|
Boyiadzis MM, Kirkwood JM, Marshall JL,
Pritchard CC, Azad NS and Gulley JL: Significance and implications
of FDA approval of pembrolizumab for biomarker-defined disease. J
Immunother Cancer. 6(35)2018.PubMed/NCBI View Article : Google Scholar
|
|
107
|
Chang L, Chang M, Chang HM and Chang F:
Microsatellite Instability: A Predictive Biomarker for Cancer
Immunotherapy. Appl Immunohistochem Mol Morphol. 26:e15–e21.
2018.PubMed/NCBI View Article : Google Scholar
|
|
108
|
Zhao P, Li L, Jiang X and Li Q: Mismatch
repair deficiency/microsatellite instability-high as a predictor
for anti-PD-1/PD-L1 immunotherapy efficacy. J Hematol Oncol.
12(54)2019.PubMed/NCBI View Article : Google Scholar
|
|
109
|
Garon EB, Rizvi NA, Hui R, Leighl N,
Balmanoukian AS, Eder JP, Patnaik A, Aggarwal C, Gubens M, Horn L,
et al: KEYNOTE-001 Investigators: Pembrolizumab for the treatment
of non-small-cell lung cancer. N Engl J Med. 372:2018–2028.
2015.PubMed/NCBI View Article : Google Scholar
|
|
110
|
Reck M, Rodríguez-Abreu D, Robinson AG,
Hui R, Csőszi T, Fülöp A, Gottfried M, Peled N, Tafreshi A, Cuffe
S, et al: KEYNOTE-024 Investigators: Pembrolizumab versus
Chemotherapy for PD-L1-Positive Non-Small-Cell Lung Cancer. N Engl
J Med. 375:1823–1833. 2016.PubMed/NCBI View Article : Google Scholar
|
|
111
|
Pylkkänen L, Karjalainen A, Anttila S,
Vainio H and Husgafvel-Pursiainen K: No evidence of microsatellite
instability but frequent loss of heterozygosity in primary resected
lung cancer. Environ Mol Mutagen. 30:217–223. 1997.PubMed/NCBI
|
|
112
|
Merlo A, Mabry M, Gabrielson E, Vollmer R,
Baylin SB and Sidransky D: Frequent microsatellite instability in
primary small cell lung cancer. Cancer Res. 54:2098–2101.
1994.PubMed/NCBI
|
|
113
|
Mao L, Lee DJ, Tockman MS, Erozan YS,
Askin F and Sidransky D: Microsatellite alterations as clonal
markers for the detection of human cancer. Proc Natl Acad Sci USA.
91:9871–9875. 1994.PubMed/NCBI View Article : Google Scholar
|
|
114
|
Chen XQ, Stroun M, Magnenat JL, Nicod LP,
Kurt AM, Lyautey J, Lederrey C and Anker P: Microsatellite
alterations in plasma DNA of small cell lung cancer patients. Nat
Med. 2:1033–1035. 1996.PubMed/NCBI View Article : Google Scholar
|
|
115
|
Hansen LT, Thykjaer T, Ørntoft TF,
Rasmussen LJ, Keller P, Spang-Thomsen M, Edmonston TB, Schmutte C,
Fishel R and Petersen LN: The role of mismatch repair in small-cell
lung cancer cells. Eur J Cancer. 39:1456–1467. 2003.PubMed/NCBI View Article : Google Scholar
|