DNA repair gene products function together to
protect the destabilization of genetic material by errors that
occur during DNA replication. In this manner, they participate in
preventing the multistep process of the neoplastic transformation
of normal cells to the tumorigenic phenotype (1-7).
Genetic damage to the DNA mismatch repair (MMR) mechanism may lead
to microsatellite instability (MSI), a common finding in hereditary
forms of cancer, such as hereditary nonpolyposis colorectal cancer
(HNPCC) and hereditary endometrial cancer (8-16).
Defects in the DNA repair mechanism have been observed in
hereditary forms of cancer, and have been linked to specific
syndromes. Previous studies have demonstrated that MSI and/or the
loss of expression of MMR proteins or low levels of mRNA, are
common findings in a number of sporadic cancers, such as lung,
endometrial, ovarian and gastric cancers, where a loss of
expression of MMR proteins or low levels of mRNA are common results
of MMR gene promoter methylation (16-24).
Similarly, there have been reported cases of the increased
expression of MMR genes in sporadic colon, prostate or urinary
bladder cancers, supporting the theory of their complex role in
carcinogenesis (25-30).
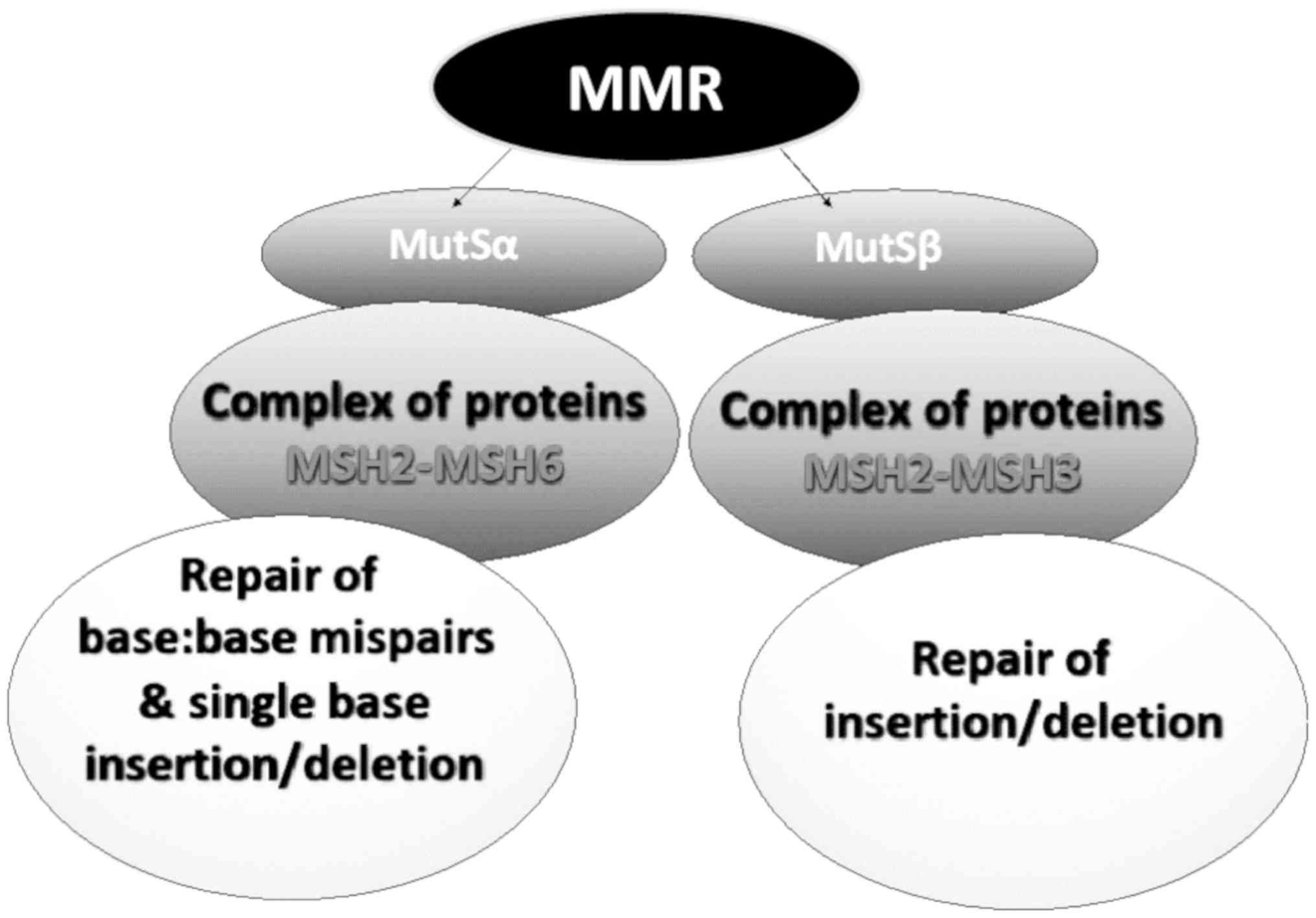
The main role of the MMR mechanism is to recognize
and repair single-base mismatch errors, such as insertion, deletion
and mis-incorporation that can occur during DNA replication. The
recognition of the error and subsequent activation of the mechanism
depends on the enzymatic complex of the proteins, MutS, MuH and
MutL (31). The MutS complex has
the ability to recognize mismatched nucleotides and bind to the
damaged DNA. The MuH complex attaches to the hemimethylated sites
along the impaired fragment, while the MutL complex activates the
MutH peptide, which acts as a mediator between MutS and MutH
(4-6).
Mismatch errors promote ATP hydrolysis, resulting in
a change in the configuration of the MSH2-MSH6 complex to slide
from its DNA binding site and to perform the repair. The complex
acts as ATPase by hydrolyzing ATP. The release of the complex from
the DNA does not depend on its activity as ATPase (1,30).
In addition to the DNA damage recognition complexes,
the mismatch DNA repair mechanism in humans includes MutL complexes
that relate to MLH1-MLH3 and MLH1-PMS2 heterodimers (possibly also
the MLH1-PMS1 complex) (32,33). MLH1-MLH3 binds to MutSa
(MSH2-MSH6) by converting it to a large complex. The MutL complexes
interact with the damage recognition complexes and with other
proteins that function in the action of the MMR mechanism, such as
exonucleases (for example EXO1), DNA polymerases (δ and ε),
replicating agents, helicases and PCNA to repair the damage
(32).
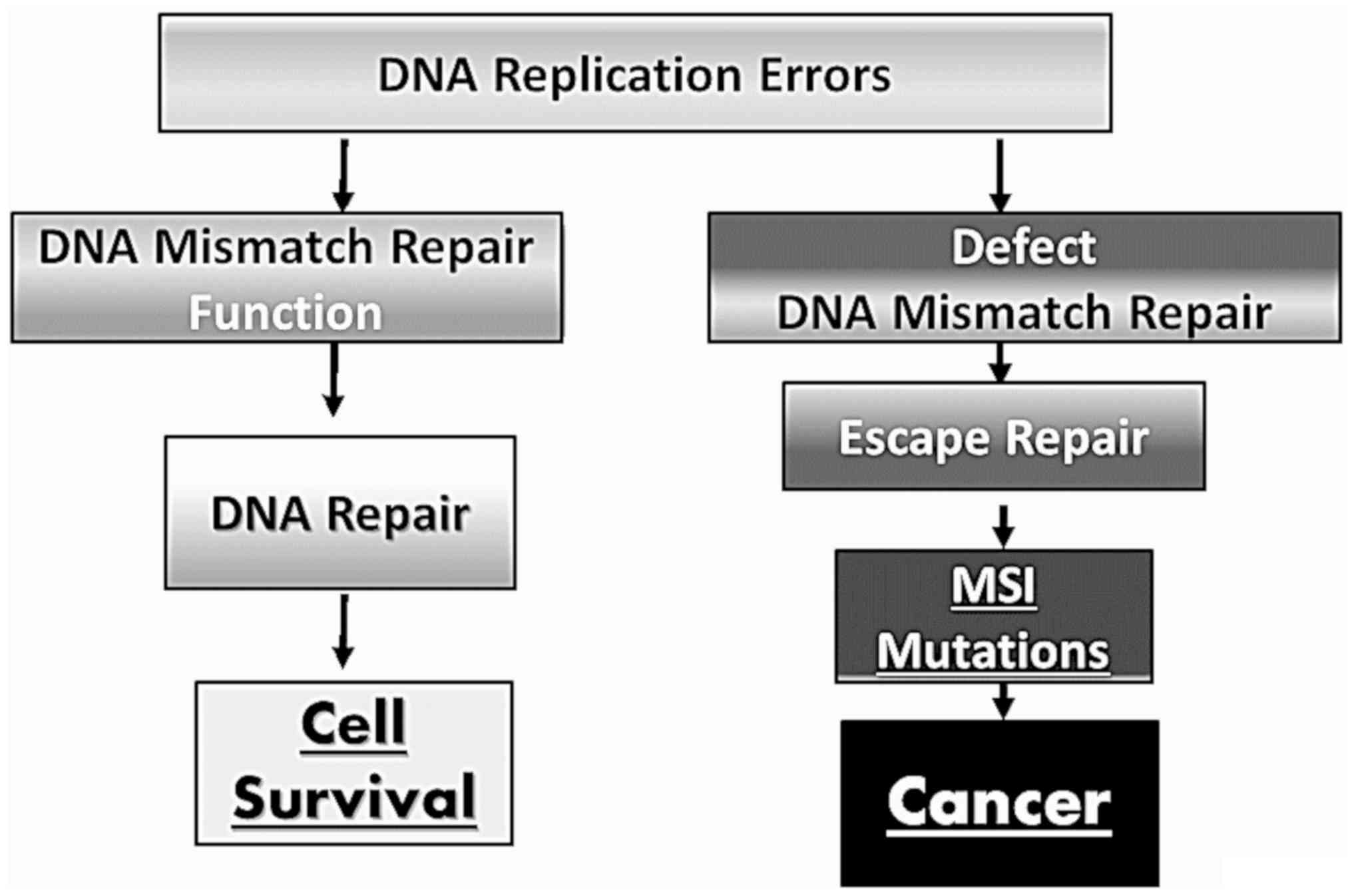
Defects in DNA recognition complexes of the MMR
system (MutSα and MutSβ) have been observed in humans.
Specifically, a deficiency in the expression of DNA MMR genes is
almost always followed by the alteration of the number of short
tandem repeats, known as MSI, and this leads to the development of
a number of known types of carcinomas (34-36)
(Fig. 2).
Recent studies have demonstrated that numerous
sporadic tumors exhibit MSI without harboring any mutations in the
repair genes (43), probably due
to epigenetic alterations in the transcription and translation of
MMR genes (30,44). Specifically, in sporadic cancers,
the loss or low levels of MMR protein expression or mRNA depicted
as increased MSI, due to the hypermethylation of hMLH1, or in some
cases the hMSH2 promoter, causes the suppression of their
expression and compromises the function of the MMR mechanism
(22,30).
In general, the loss or low expression of MMR is
evidence of a faulty repair mechanism constituting either a genetic
background of cancer transformation in hereditary cancers, allowing
for an increased MSI in cells, or promoting the tumorigenic pathway
in sporadic cancers, allowing for the survival of cells carrying a
significant amount of genetic alterations, some in proto-oncogenes
or genes regulating the cell cycle.
Defects that lead to the suppression of MSH6
expression, may result in an increase in MSH3 mRNA and protein
expression. It is clear that balanced expression levels of
components of the MMR system are essential for effective repair
functions (2,32,45). Vageli et al previously
demonstrated an association between an imbalanced mRNA phenotype of
MMR genes and cancer progression in human lung, colorectal and
urinary bladder cancer (18,46-49).
MSH2 protein, which is a major factor of base pair
recognition error in DNA, under physiological conditions, is
equated between the nucleus and cytoplasm. MSH2 has a high affinity
for binding to the damaged site, which can be repaired, while
having low affinity binding for non-severe damage that does not
allow for cell viability, leading to apoptosis. The process of the
initiation of apoptosis by MMR protein components appears to be
related to their concentration in the nucleus. The concentration of
MSH2 in the nucleus is a criterion for triggering apoptosis.
Therefore, the loss or decreased expression levels of MSH2, will
lead to an insufficient nuclear concentration of MMR and therefore,
in the inability of cells carrying extensive DNA damage to undergo
apoptosis (31,50).
A number of studies have demonstrated that there is
an association between the transcriptional activity of MMR
mechanisms and the development of resistance to chemotherapy.
Specifically, there is evidence that the inactivation of hMLH1 by
promoter hypermethylation promotes chemotherapeutic resistance
(36,51,52). On the other hand, an imbalance in
MMR mRNA phenotypes has been suggested to be of possible prognostic
value in adjuvant chemotherapy treatment in non-small cell lung
carcinomas (NSCLCs). Specifically, specific increased MMR mRNA
phenotypes exhibit a trend for improved survival following
chemotherapy, compared to other decreased mRNA phenotypes, which
appear to be more effective in combination with post-operative
chemotherapy (46).
The efficacy of the MMR mechanism can be strongly
influenced by various environmental factors and epigenetic
alterations that can significantly affect the ability of cells to
repair genetic damage. These risk factors are able to determine
profound epigenetic alterations, including the modulation of DNA
methylation levels and microRNA (miRNA or miR) expression levels
(53). The epigenetic regulation
of MMR may be also associated with the composition of gut
microbiota. This may strongly influence the development of several
pathologies, including cancer (54,55).
In particular, miRNAs have been shown to be involved
in the regulation of numerous genes associated with various
physiological processes, including MMR genes (67-72).
Mao et al demonstrated that MutLa could function as a
stimulatory factor for miRNA processing (73), while Valeri et al
demonstrated the capability of miR-155 to alter both the expression
and stability of the MMR pathway, supporting a regulatory role of
miR-155 in the MMR mechanism (74). Zhong et al suggested that
miRNAs play an important role in modulating the cell cycle by
targeting hMSH2 in lung cancer (75).
Overall, the unbalance of the MMR mechanism and the
acquisition of new oncogenic mutations are the result of different
genetic and epigenetic alterations. These alterations in expression
can be identified using innovative and high-sensitive techniques
(76), providing information with
which to predict the risk of cancer onset and to identify novel
biomarkers and therapeutic targets.
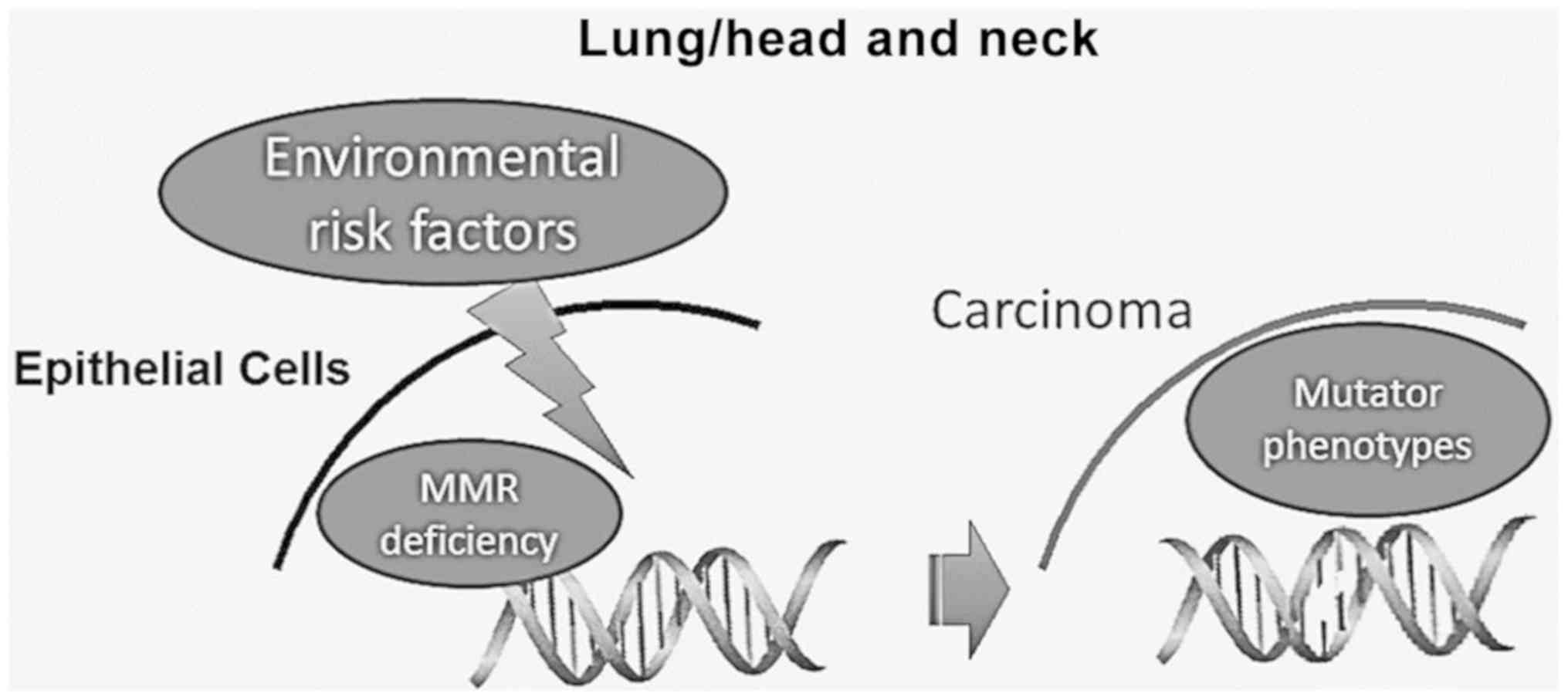
Lung and head and neck carcinogenesis is strongly
associated with known risk factors, such as alcohol, tobacco
smoking and oncoviruses, causing DNA damage. It is considered that
the extensive accumulation of genetic alterations in DNA by these
environmental risk factors may lead to an abnormal DNA damage
response (DDR), which can result in cell death, chromosomal
instability and unregulated proliferation (77-80).
Therefore, the proper function of the various DNA repair mechanisms
is essential for the elimination of these harmful effects,
maintaining the DNA integrity (Fig.
3).
Thus far, a number of studies have suggested that
polymorphisms of MMR are associated with an increased risk of
developing head and neck cancer (81-83).
A number of studies have shown that head and neck squamous cell
carcinomas (HNSCCs) very often exhibit MSI. Notably, it has been
observed that HNSCCs exhibit MSI at higher rates than other solid
tumors, such as esophageal, breast and gastric carcinomas (21-24,40,84,85).
Demokan et al, also showed that high levels of MSI in HNSCC
are strongly associated with hypermethylation of hMLH1 and
hMSH2 (81).
As has been discussed above, the dysfunction of the
MMR mechanism can lead to MSI and to the accumulation of mutations
in proto-oncogenes or tumor-suppressor genes, increasing the risk
of malignant development and progression. It has already been
demonstrated that the decreased expression of the MSH2 gene
causally increases the frequency of MSI (86,87). Furthermore, the investigation of
the MSH2 protein level in surgical specimens of head and neck
carcinoma have revealed an association between low MSH2 levels and
locoregional metastasis, as well as a worse survival (88).
Previous studies have demonstrated a reduced
expression of MSH2 or MLH1 genes at the protein or mRNA level in
>50% of lung adenocarcinomas, associated with a poor survival
and an increase in MSI (95-96). Kanellis
et al evaluated the protein expression levels of MMR genes
in fine-needle aspiration (FNA) specimens derived from various
types of lung cancer. Their study demonstrated that that NSCLCs,
and particularly squamous cell carcinomas, exhibited reduced MSH2
protein levels at relatively high rates compared to small cell
carcinomas (97).
Although in the majority of cases, the decreased
expression of MMR genes is attributed to epigenetic silencing,
other studies have indicated that MMR deficiency may act as a
‘second hit’, accelerating the development of lung tumors in mice
that carry the K-rasLA1/+ mutation (98). Specifically, Downey and Jirik,
using a murine animal model, recently demonstrated that a
deficiency in MMR genes can act in concert with the extremely
common K-ras mutation, enhancing tumor development (98).
More recently, some studies have focused on the
prognostic significance of a defective MMR mechanism and MSI in
several types of cancer, including NSCLC (104,105), and the responsiveness of
patients towards immune check-point inhibitors (106-108).
Overall, it is well-recognized that patients with MSI and/or
defective MMR have generally a durable complete response (106). Furthermore, MSI has been shown
to be a good predictive biomarker for immunotherapy efficacy in
several types of cancer treated with pembrolizumab or nivolumab,
including NSCLC, advanced melanoma, renal cell carcinoma, bladder
cancer, etc (107,108). As regards NSCLC, studies have
demonstrated a high response rate following treatment with the
immune check-point inhibitor, pembrolizumab, in the KEYNOTE-001 and
KEYNOTE-024 trials (109,110).
Further clinical trials consisting of a sufficient number of NSCLCs
patients and adequate follow-up are necessary to verify the
efficacy of pembrolizumab in patients with microsatellite
instability high/deficiency MMR (MSI-H/dMMR).
DNA repair deficiency is a hallmark in cancer
development and may affect the therapeutic outcomes. Sporadic head
and neck and lung tumors often exhibit genetic alterations due to
an inadequate mismatch DNA repair mechanism. The mechanisms through
which defects in the DNA MMR mechanism promote lung, and head and
neck cancer are not yet clear. To the best of our knowledge, the
present review article is the first attempt to summarize what is
known in the literature about the dysregulation of this mechanism
and its role in these types of cancer. This review supports the
further investigation of alterations in the expression of mismatch
DNA repair genes, at both the transcriptional and translational
level, in head and neck, and lung sporadic tumors, clarifying their
prognostic and diagnostic value, as well as their therapeutic
potential as novel targets.
Not applicable.
No funding was received.
Not applicable.
SGD and DPV were involved in the conceptualization
and in the design of this review article. TKN and GL, were involved
in searching the literature for the paragraphs ‘DNA mismatch repair
mechanism’ and ‘head and neck cancer and MMR’. LF was involved in
searching the literature for paragraphs ‘Epigenetic regulation of
MMR mechanism’ and ‘MMR deficiency affects the chemotherapy
treatment of NSCLC’. KK was involved in searching the literature
for the paragraph ‘the importance of MMR function as an indicator
of chemotherapeutic resistance’. AOD and AT were involved in
searching the relevant literature and databases for the paragraph
‘DNA MMR deficiency and carcinogenesis’. SGD and DPV were involved
in the preparation of the original draft and in the preparation of
the figures. SGD and DPV were involved in the writing of the
original draft. DPV, SGD, TKN, LF, AT and GL reviewed and edited
the article. All authors have read and approved the final
manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Peltomäki P: DNA mismatch repair and
cancer. Mutat Res. 488:77–85. 2001.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Peltomäki P: Role of DNA mismatch repair
defects in the pathogenesis of human cancer. J Clin Oncol.
21:1174–1179. 2003.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Li GM: DNA mismatch repair and cancer.
Front Biosci. 8:d997–d1017. 2003.PubMed/NCBI View
Article : Google Scholar
|
|
4
|
Yang W: Structure and mechanism for DNA
lesion recognition. Cell Res. 18:184–197. 2008.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Kolodner RD: Mismatch repair: Mechanisms
and relationship to cancer susceptibility. Trends Biochem Sci.
20:397–401. 1995.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Jiricny J and Nyström-Lahti M: Mismatch
repair defects in cancer. Curr Opin Genet Dev. 10:157–161.
2000.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Aarnio M, Sankila R, Pukkala E, Salovaara
R, Aaltonen LA, de la Chapelle A, Peltomäki P, Mecklin JP and
Järvinen HJ: Cancer risk in mutation carriers of
DNA-mismatch-repair genes. Int J Cancer. 81:214–218.
1999.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Bohr VA: DNA repair fine structure and its
relations to genomic instability. Carcinogenesis. 16:2885–2892.
1995.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Kruhøffer M, Jensen JL, Laiho P, Dyrskjøt
L, Salovaara R, Arango D, Birkenkamp-Demtroder K, Sørensen FB,
Christensen LL, Buhl L, et al: Gene expression signatures for
colorectal cancer microsatellite status and HNPCC. Br J Cancer.
92:2240–2248. 2005.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Leach FS, Nicolaides NC, Papadopoulos N,
Liu B, Jen J, Parsons R, Peltomäki P, Sistonen P, Aaltonen LA,
Nyström-Lahti M, et al: Mutations of a mutS homolog in hereditary
nonpolyposis colorectal cancer. Cell. 75:1215–1225. 1993.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Fishel R, Lescoe MK, Rao MRS, Copeland NG,
Jenkins NA, Garber J, Kane M and Kolodner R: The human mutator gene
homolog MSH2 and its association with hereditary nonpolyposis colon
cancer. Cell. 75:1027–1038. 1993.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Liu B, Nicolaides NC, Markowitz S, Willson
JKV, Parsons RE, Jen J, Papadopolous N, Peltomäki P, de la Chapelle
A, Hamilton SR, et al: Mismatch repair gene defects in sporadic
colorectal cancers with microsatellite instability. Nat Genet.
9:48–55. 1995.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Liu B, Parsons R, Papadopoulos N,
Nicolaides NC, Lynch HT, Watson P, Jass J, Dunlop M, Wyllie A,
Jessup JM, Peltomäki PT, et al: Mismatch repair gene analysis in
HNPCC patients. Nat Med. 2:169–174. 1996.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Kowalski LD, Mutch DG, Herzog TJ, Rader JS
and Goodfellow PJ: Mutational analysis of MLH1 and MSH2 in 25
prospectively-acquired RER+ endometrial cancers. Genes Chromosomes
Cancer. 18:219–227. 1997.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Chadwick RB, Pyatt RE, Niemann TH,
Richards SK, Johnson CK, Stevens MW, Meek JE, Hampel H, Prior TW
and de la Chapelle A: Hereditary and somatic DNA mismatch repair
gene mutations in sporadic endometrial carcinoma. J Med Genet.
38:461–466. 2001.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Soliman PT and Lu K: Endometrial cancer
associated with defective DNA mismatch repair. Obstet Gynecol Clin
North Am. 34:701–715, viii. 2007.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Rosen DG, Cai KQ, Luthra R and Liu J:
Immunohistochemical staining of hMLH1 and hMSH2 reflects
microsatellite instability status in ovarian carcinoma. Mod Pathol.
19:1414–1420. 2006.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Vageli D, Daniil Z, Dahabreh J, Karagianni
E, Vamvakopoulou DN, Ioannou MG, Scarpinato K, Vamvakopoulos NC,
Gourgoulianis KI and Koukoulis GK: Phenotypic mismatch repair hMSH2
and hMLH1 gene expression profiles in primary non-small cell lung
carcinomas. Lung Cancer. 64:282–288. 2009.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Wang YC, Lu YP, Tseng RC, Lin RK, Chang
JW, Chen JT, Shih CM and Chen CY: Inactivation of hMLH1 and hMSH2
by promoter methylation in primary non-small cell lung tumors and
matched sputum samples. J Clin Invest. 111:887–895. 2003.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Xinarianos G, Liloglou T, Prime W,
Sourvinos G, Karachristos A, Gosney JR, Spandidos DA and Field JK:
p53 status correlates with the differential expression of the DNA
mismatch repair protein MSH2 in non-small cell lung carcinoma. Int
J Cancer. 101:248–252. 2002.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Kitajima Y, Miyazaki K, Matsukura S,
Tanaka M and Sekiguchi M: Loss of expression of DNA repair enzymes
MGMT, hMLH1, and hMSH2 during tumor progression in gastric cancer.
Gastric Cancer. 6:86–95. 2003.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Tahara E: Genetic pathways of two types of
gastric cancer. IARC Sci Publ. 157:327–349. 2004.PubMed/NCBI
|
|
23
|
Nardone G, Rocco A and Budillon G:
Molecular alteration of gastric carcinoma. Minerva Gastroenterol
Dietol. 48:189–193. 2002.PubMed/NCBI
|
|
24
|
El-Rifai W, Powell SM and EI-Rifai W:
Molecular biology of gastric cancer. Semin Radiat Oncol.
12:128–140. 2002.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Sasaki S and Nakamura Y: Mutation of the
mismatch repair genes for carcinogenesis of sporadic colorectal
cancers with RER-positive phenotype. Nihon Rinsho. 54:1008–1013.
1996.(In Japanese). PubMed/NCBI
|
|
26
|
Wallis Y and Macdonald F: The genetics of
inherited colon cancer. Clin Mol Pathol. 49:M65–M73.
1996.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Burger M, Denzinger S, Hammerschmied CG,
Tannapfel A, Obermann EC, Wieland WF, Hartmann A and Stoehr R:
Elevated microsatellite alterations at selected tetranucleotides
(EMAST) and mismatch repair gene expression in prostate cancer. J
Mol Med (Berl). 84:833–841. 2006.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Kassem HS, Varley JM, Hamam SM and
Margison GP: Immunohistochemical analysis of expression and
allelotype of mismatch repair genes (hMLH1 and hMSH2) in bladder
cancer. Br J Cancer. 84:321–328. 2001.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Leach FS, Hsieh JT, Molberg K, Saboorian
MH, McConnell JD and Sagalowsky AI: Expression of the human
mismatch repair gene hMSH2: A potential marker for urothelial
malignancy. Cancer. 88:2333–2341. 2000.PubMed/NCBI
|
|
30
|
Kuismanen SA, Holmberg MT, Salovaara R, de
la Chapelle A and Peltomäki P: Genetic and epigenetic modification
of MLH1 accounts for a major share of microsatellite-unstable
colorectal cancers. Am J Pathol. 156:1773–1779. 2000.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Viale G, Trapani D and Curigliano G:
Mismatch repair deficiency as a predictive biomarker for
immunotherapy efficacy. BioMed Res Int.
2017(4719194)2017.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Hsieh P and Yamane K: DNA mismatch repair:
Molecular mechanism, cancer, and ageing. Mech Ageing Dev.
129:391–407. 2008.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Kijas AW, Studamire B and Alani E:
Msh2 separation of function mutations confer defects in the
initiation steps of mismatch repair. J Mol Biol. 331:123–138.
2003.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Gurin CC, Federici MG, Kang L and Boyd J:
Causes and consequences of microsatellite instability in
endometrial carcinoma. Cancer Res. 59:462–466. 1999.PubMed/NCBI
|
|
35
|
Hayashi M, Tamura G, Jin Z, Kato I, Sato
M, Shibuya Y, Yang S and Motoyama T: Microsatellite instability in
esophageal squamous cell carcinoma is not associated with
hMLH1 promoter hypermethylation. Pathol Int. 53:270–276.
2003.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Nikolouzakis TK, Vassilopoulou L,
Fragkiadaki P, Mariolis Sapsakos T, Papadakis GZ, Spandidos DA,
Tsatsakis AM and Tsiaoussis J: Improving diagnosis, prognosis and
prediction by using biomarkers in CRC patients (Review). Oncol Rep.
39:2455–2472. 2018.(Review). PubMed/NCBI View Article : Google Scholar
|
|
37
|
Lynch HT, Snyder CL, Shaw TG, Heinen CD
and Hitchins MP: Milestones of Lynch syndrome 1895-2015. Nat Rev
Canc. 15:181–194. 2015.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Antelo M, Golubicki M, Roca E, Mendez G,
Carballido M, Iseas S, Cuatrecasas M, Moreira L, Sanchez A,
Carballal S, et al: Lynch-like syndrome is as frequent as Lynch
syndrome in early-onset nonfamilial nonpolyposis colorectal cancer.
Int J Cancer. 145:705–713. 2019.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Carethers JM and Stoffel EM: Lynch
syndrome and Lynch syndrome mimics: The growing complex landscape
of hereditary colon cancer. World J Gastroenterol. 21:9253–9261.
2015.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Adar T, Friedman M, Rodgers LH, Shannon
KM, Zukerberg LR and Chung DC: Gastric cancer in Lynch syndrome is
associated with underlying immune gastritis. J Med Genet. doi:
10.1136/jmedgenet-2018-105757 (Epub ahead of print). PubMed/NCBI View Article : Google Scholar
|
|
41
|
Wang J, Greenberg S and Yates J: Lynch
syndrome-associated upper tract urothelial carcinoma. Urology.
121:19–21. 2018.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Cloyd JM, Chun YS, Ikoma N, Vauthey JN,
Aloia TA, Cuddy A, Rodriguez-Bigas MA and Nancy You Y: Clinical and
genetic implications of DNA mismatch repair deficiency in biliary
tract cancers associated with lynch syndrome. J Gastrointest
Cancer. 49:93–96. 2018.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Anacleto C, Leopoldino AM, Rossi B, Soares
FA, Lopes A, Rocha JC, Caballero O, Camargo AA, Simpson AJ and Pena
SD: Colorectal cancer ‘methylator phenotype’: Fact or artifact?
Neoplasia. 7:331–335. 2005.PubMed/NCBI
|
|
44
|
Imai K and Yamamoto H: Carcinogenesis and
microsatellite instability: The interrelationship between genetics
and epigenetics. Carcinogenesis. 29:673–680. 2008.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Chang DK, Ricciardiello L, Goel A, Chang
CL and Boland CR: Steady-state regulation of the human DNA mismatch
repair system. J Biol Chem. 275:18424–18431. 2000.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Vageli DP, Zaravinos A, Daniil Z, Dahabreh
J, Doukas SG, Spandidos DA, Gourgoulianis KI and Koukoulis GK:
hMSH2 and hMLH1 gene expression patterns differ between lung
adenocarcinoma and squamous cell carcinoma: Correlation with
patient survival and response to adjuvant chemotherapy treatment.
Int J Biol Markers. 27:e400–e404. 2013.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Vageli DP, Giannopoulos S, Doukas SG,
Kalaitzis C, Giannakopoulos S, Giatromanolaki A, Koukoulis GK and
Touloupidis S: Mismatch repair hMSH2, hMLH1, hMSH6 and hPMS2 mRNA
expression profiles in precancerous and cancerous urothelium. Oncol
Lett. 5:283–294. 2013.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Vageli DP, Doukas SG and Markou A:
Mismatch DNA repair mRNA expression profiles in oral melanin
pigmentation lesion and hamartomatous polyp of a child with
Peutz-Jeghers syndrome. Pediatr Blood Cancer. 60:E116–E117.
2013.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Vageli DP, Papamichali R, Kambosioras K,
Papandreou CN and Koukoulis GK: Mismatch DNA repair hMSH2,
hMLH1, hMSH6 and hPMS2 mRNA expression
profiles in colorectal carcinomas. J Genet Syndr Gene Ther.
View Article : Google Scholar
|
|
50
|
Christmann M and Kaina B: Nuclear
translocation of mismatch repair proteins MSH2 and MSH6 as a
response of cells to alkylating agents. J Biol Chem.
275:36256–36262. 2000.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Strathdee G, MacKean MJ, Illand M and
Brown R: A role for methylation of the hMLH1 promoter in loss of
hMLH1 expression and drug resistance in ovarian cancer. Oncogene.
18:2335–2341. 1999.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Carethers JM, Chauhan DP, Fink D, Nebel S,
Bresalier RS, Howell SB and Boland CR: Mismatch repair proficiency
and in vitro response to 5-fluorouracil. Gastroenterology.
117:123–131. 1999.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Bhattacharjee P, Sanyal T, Bhattacharjee S
and Bhattacharjee P: Epigenetic alteration of mismatch repair genes
in the population chronically exposed to arsenic in West Bengal,
India. Environ Res. 163:289–296. 2018.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Vivarelli S, Salemi R, Candido S, Falzone
L, Santagati M, Stefani S, Torino F, Banna GL, Tonini G and Libra
M: Gut Microbiota and Cancer: From Pathogenesis to Therapy. Cancers
(Basel). 11(E38)2019.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Banna GL, Torino F, Marletta F, Santagati
M, Salemi R, Cannarozzo E, Falzone L, Ferraù F and Libra M:
Lactobacillus rhamnosus GG: An Overview to explore the
rationale of its use in cancer. Front Pharmacol. doi:
10.3389/fphar.2017.00603. PubMed/NCBI View Article : Google Scholar
|
|
56
|
Westwood A, Glover A, Hutchins G, Young C,
Brockmoeller S, Robinson R, Worrilow L, Wallace D, Rankeillor K,
Adlard J, et al: Additional loss of MSH2 and MSH6 expression in
sporadic deficient mismatch repair colorectal cancer due to MLH1
promoter hypermethylation. J Clin Pathol. 72:443–447.
2019.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Kang Z, Zhu Y, Zhang QA, Dong L, Xu F,
Zhang X and Guan M: Methylation and expression analysis of mismatch
repair genes in extramammary Paget's disease. J Eur Acad Dermatol
Venereol. 33:874–879. 2019.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Mäki-Nevala S, Valo S, Ristimäki A,
Sarhadi V, Knuutila S, Nyström M, Renkonen-Sinisalo L, Lepistö A,
Mecklin JP and Peltomäki P: DNA methylation changes and somatic
mutations as tumorigenic events in Lynch syndrome-associated
adenomas retaining mismatch repair protein expression.
EBioMedicine. 39:280–291. 2019.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297.
2004.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Zimmerman AL and Wu S: MicroRNAs, cancer
and cancer stem cells. Cancer Lett. 300:10–19. 2011.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Falzone L, Lupo G, La Rosa GRM, Crimi S,
Anfuso CD, Salemi R, Rapisarda E, Libra M and Candido S:
Identification of novel microRNAs and their diagnostic and
prognostic significance in oral cancer. Cancers (Basel).
11(E610)2019.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Hafsi S, Candido S, Maestro R, Falzone L,
Soua Z, Bonavida B, Spandidos DA and Libra M: Correlation between
the overexpression of Yin Yang 1 and the expression levels of
miRNAs in Burkitt's lymphoma: A computational study. Oncol Lett.
11:1021–1025. 2016.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Falzone L, Romano GL, Salemi R, Bucolo C,
Tomasello B, Lupo G, Anfuso CD, Spandidos DA, Libra M and Candido
S: Prognostic significance of deregulated microRNAs in uveal
melanomas. Mol Med Rep. 19:2599–2610. 2019.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Falzone L, Candido S, Salemi R, Basile MS,
Scalisi A, McCubrey JA, Torino F, Signorelli SS, Montella M and
Libra M: Computational identification of microRNAs associated to
both epithelial to mesenchymal transition and NGAL/MMP-9 pathways
in bladder cancer. Oncotarget. 7:72758–72766. 2016.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Costa PM and Pedroso de Lima MC: MicroRNAs
as molecular targets for cancer therapy: On the modulation of
microRNA expression. Pharmaceuticals (Basel). 6:1195–1220.
2013.PubMed/NCBI View Article : Google Scholar
|
|
66
|
McCubrey JA, Fitzgerald TL, Yang LV,
Lertpiriyapong K, Steelman LS, Abrams SL, Montalto G, Cervello M,
Neri LM, Cocco L, et al: Roles of GSK-3 and microRNAs on epithelial
mesenchymal transition and cancer stem cells. Oncotarget.
8:14221–14250. 2017.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Reinhart BJ, Slack FJ, Basson M,
Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR and Ruvkun G:
The 21-nucleotide let-7 RNA regulates developmental timing in
Caenorhabditis elegans. Nature. 403:901–906. 2000.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Cheng AM, Byrom MW, Shelton J and Ford LP:
Antisense inhibition of human miRNAs and indications for an
involvement of miRNA in cell growth and apoptosis. Nucleic Acids
Res. 33:1290–1297. 2005.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Sasaki CT, Doukas SG and Vageli DP: In
vivo short-term topical application of BAY 11-7082 prevents the
acidic bile-induced mRNA and miRNA oncogenic phenotypes in exposed
murine hypopharyngeal mucosa. Neoplasia. 20:374–386.
2018.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Doukas SG, Vageli DP and Sasaki CT: NF-κB
inhibition reverses acidic bile-induced miR-21, miR-155, miR-192,
miR-34a, miR-375 and miR-451a deregulations in human hypopharyngeal
cells. J Cell Mol Med. 22:2922–2934. 2018.PubMed/NCBI View Article : Google Scholar
|
|
71
|
Doukas PG, Vageli DP, Doukas SG and Sasaki
CT: Temporal characteristics of NF-κB inhibition in blocking
bile-induced oncogenic molecular events in hypopharyngeal cells.
Oncotarget. doi: https://doi.org/10.18632/oncotarget.26917.
PubMed/NCBI View Article : Google Scholar
|
|
72
|
Landau DA and Slack FJ: MicroRNAs in
mutagenesis, genomic instability, and DNA repair. Semin Oncol.
38:743–751. 2011.PubMed/NCBI View Article : Google Scholar
|
|
73
|
Mao G, Lee S, Ortega J, Gu L and Li GM:
Modulation of microRNA processing by mismatch repair protein MutLα.
Cell Res. 22:973–985. 2012.PubMed/NCBI View Article : Google Scholar
|
|
74
|
Valeri N, Gasparini P, Fabbri M, Braconi
C, Veronese A, Lovat F, Adair B, Vannini I, Fanini F, Bottoni A, et
al: Modulation of mismatch repair and genomic stability by miR-155.
Proc Natl Acad Sci USA. 107:6982–6987. 2010.PubMed/NCBI View Article : Google Scholar
|
|
75
|
Zhong Z, Dong Z, Yang L, Chen X and Gong
Z: MicroRNA-31-5p modulates cell cycle by targeting human mutL
homolog 1 in human cancer cells. Tumour Biol. 34:1959–1965.
2013.PubMed/NCBI View Article : Google Scholar
|
|
76
|
Salemi R, Falzone L, Madonna G, Polesel J,
Cinà D, Mallardo D, Ascierto PA, Libra M and Candido S: MMP-9 as a
candidate marker of response to BRAF inhibitors in melanoma
patients with BRAFV600E mutation detected in circulating-free DNA.
Front Pharmacol. Aug 14 2018 (Epub ahead of print) doi:
10.3389/fphar.2018.00856. PubMed/NCBI View Article : Google Scholar
|
|
77
|
Pfeifer GP, Denissenko MF, Olivier M,
Tretyakova N, Hecht SS and Hainaut P: Tobacco smoke carcinogens,
DNA damage and p53 mutations in smoking-associated cancers.
Oncogene. 21:7435–7451. 2002.PubMed/NCBI View Article : Google Scholar
|
|
78
|
Zhong Y, Carmella SG, Upadhyaya P,
Hochalter JB, Rauch D, Oliver A, Jensen J, Hatsukami D, Wang J,
Zimmerman C, et al: Immediate consequences of cigarette smoking:
Rapid formation of polycyclic aromatic hydrocarbon diol epoxides.
Chem Res Toxicol. 24:246–252. 2011.PubMed/NCBI View Article : Google Scholar
|
|
79
|
Boda D, Docea AO, Calina D, Ilie MA,
Caruntu C, Zurac S, Neagu M, Constantin C, Branisteanu DE,
Voiculescu V, et al: Human papilloma virus: Apprehending the link
with carcinogenesis and unveiling new research avenues (Review).
Int J Oncol. 52:637–655. 2018.PubMed/NCBI View Article : Google Scholar
|
|
80
|
Dylawerska A, Barczak W, Wegner A,
Golusinski W and Suchorska WM: Association of DNA repair genes
polymorphisms and mutations with increased risk of head and neck
cancer: A review. Med Oncol. 34(197)2017.PubMed/NCBI View Article : Google Scholar
|
|
81
|
Demokan S, Suoglu Y, Demir D, Gozeler M
and Dalay N: Microsatellite instability and methylation of the DNA
mismatch repair genes in head and neck cancer. Ann Oncol.
17:995–999. 2006.PubMed/NCBI View Article : Google Scholar
|
|
82
|
Wang Y, Irish J, MacMillan C, Brown D,
Xuan Y, Boyington C, Gullane P and Kamel-Reid S: High frequency of
microsatellite instability in young patients with head-and-neck
squamous-cell carcinoma: Lack of involvement of the mismatch repair
genes hMLH1 and hMSH2. Int J Cancer. 93:353–360.
2001.PubMed/NCBI View Article : Google Scholar
|
|
83
|
Liu K, Huang H, Mukunyadzi P, Suen JY,
Hanna E and Fan CY: Promoter hypermethylation: An important
epigenetic mechanism for hMLH1 gene inactivation in head and neck
squamous cell carcinoma. Otolaryngol Head Neck Surg. 126:548–553.
2002.PubMed/NCBI View Article : Google Scholar
|
|
84
|
Murata H, Khattar NH, Kang Y, Gu L and Li
GM: Genetic and epigenetic modification of mismatch repair genes
hMSH2 and hMLH1 in sporadic breast cancer with
microsatellite instability. Oncogene. 21:5696–5703. 2002.PubMed/NCBI View Article : Google Scholar
|
|
85
|
Demokan S, Muslumanoglu M, Yazici H, Igci
A and Dalay N: Investigation of microsatellite instability in
Turkish breast cancer patients. Pathol Oncol Res. 8:138–141.
2002.PubMed/NCBI
|
|
86
|
Ruszkiewicz A, Bennett G, Moore J, Manavis
J, Rudzki B, Shen L and Suthers G: Correlation of mismatch repair
genes immunohistochemistry and microsatellite instability status in
HNPCC-associated tumours. Pathology. 34:541–547. 2002.PubMed/NCBI View Article : Google Scholar
|
|
87
|
Shia J, Ellis NA and Klimstra DS: The
utility of immunohistochemical detection of DNA mismatch repair
gene proteins. Virchows Arch. 445:431–441. 2004.PubMed/NCBI View Article : Google Scholar
|
|
88
|
Pereira CS, Oliveira MV, Barros LO,
Bandeira GA, Santos SH, Basile JR, Guimarães AL and De Paula AM:
Low expression of MSH2 DNA repair protein is associated with poor
prognosis in head and neck squamous cell carcinoma. J Appl Oral
Sci. 21:416–421. 2013.PubMed/NCBI View Article : Google Scholar
|
|
89
|
Nogueira GA, Lourenço GJ, Oliveira CB,
Marson FA, Lopes-Aguiar L, Costa EF, Lima TR, Liutti VT, Leal F,
Santos VC, et al: Association between genetic polymorphisms in DNA
mismatch repair-related genes with risk and prognosis of head and
neck squamous cell carcinoma. Int J Cancer. 137:810–818.
2015.PubMed/NCBI View Article : Google Scholar
|
|
90
|
Jha R, Gaur P, Sharma SC and Das SN:
Single nucleotide polymorphism in hMLH1 promoter and risk of
tobacco-related oral carcinoma in high-risk Asian Indians. Gene.
526:223–227. 2013.PubMed/NCBI View Article : Google Scholar
|
|
91
|
Liu K, Huang H, Mukunyadzi P, Suen JY,
Hanna E and Fan CY: Promoter hypermethylation: An important
epigenetic mechanism for hMLH1 gene inactivation in head and neck
squamous cell carcinoma. Otolaryngol Head Neck Surg. 126:548–553.
2002.PubMed/NCBI View Article : Google Scholar
|
|
92
|
Liu K, Zuo C, Luo QK, Suen JY, Hanna E and
Fan CY: Promoter hypermethylation and inactivation of hMLH1,
a DNA mismatch repair gene, in head and neck squamous cell
carcinoma. Diagn Mol Pathol. 12:50–56. 2003.PubMed/NCBI
|
|
93
|
Zuo C, Zhang H, Spencer HJ, Vural E, Suen
JY, Schichman SA, Smoller BR, Kokoska MS and Fan CY: Increased
microsatellite instability and epigenetic inactivation of the
hMLH1 gene in head and neck squamous cell carcinoma.
Otolaryngol Head Neck Surg. 141:484–490. 2009.PubMed/NCBI View Article : Google Scholar
|
|
94
|
Tawfik HM, El-Maqsoud NM, Hak BH and
El-Sherbiny YM: Head and neck squamous cell carcinoma: Mismatch
repair immunohistochemistry and promoter hypermethylation of
hMLH1 gene. Am J Otolaryngol. 32:528–536. 2011.PubMed/NCBI View Article : Google Scholar
|
|
95
|
Xinarianos G, Liloglou T, Prime W, Maloney
P, Callaghan J, Fielding P, Gosney JR and Field JK: hMLH1
and hMSH2 expression correlates with allelic imbalance on
chromosome 3p in non-small cell lung carcinomas. Cancer Res.
60:4216–4221. 2000.PubMed/NCBI
|
|
96
|
Hsu HS, Wen CK, Tang YA, Lin RK, Li WY,
Hsu WH and Wang YC: Promoter hypermethylation is the predominant
mechanism in hMLH1 and hMSH2 deregulation and is a
poor prognostic factor in nonsmoking lung cancer. Clin Cancer Res.
11:5410–5416. 2005.PubMed/NCBI View Article : Google Scholar
|
|
97
|
Kanellis G, Chatzistamou I, Koutselini H,
Politi E, Gouliamos A, Vlahos L and Koutselinis A: Expression of
DNA mismatch repair gene MSH2 in cytological material from lung
cancer patients. Diagn Cytopathol. 34:463–466. 2006.PubMed/NCBI View Article : Google Scholar
|
|
98
|
Downey CM and Jirik FR: DNA mismatch
repair deficiency accelerates lung neoplasm development in
K-ras(LA1/+) mice: A brief report. Cancer Med.
4:897–902. 2015.PubMed/NCBI View Article : Google Scholar
|
|
99
|
Scartozzi M, Franciosi V, Campanini N,
Benedetti G, Barbieri F, Rossi G, Berardi R, Camisa R, Silva RR,
Santinelli A, et al: Mismatch repair system (MMR) status correlates
with response and survival in non-small cell lung cancer (NSCLC)
patients. Lung Cancer. 53:103–109. 2006.PubMed/NCBI View Article : Google Scholar
|
|
100
|
Takahashi Y, Kondo K, Hirose T, Nakagawa
H, Tsuyuguchi M, Hashimoto M, Sano T, Ochiai A and Monden Y:
Microsatellite instability and protein expression of the DNA
mismatch repair gene, hMLH1, of lung cancer in chromate-exposed
workers. Mol Carcinog. 42:150–158. 2005.PubMed/NCBI View Article : Google Scholar
|
|
101
|
Falzone L, Salomone S and Libra M:
Evolution of cancer pharmacological treatments at the turn of the
third millennium. Front Pharmacol. doi:
doi.org/10.3389/fphar.2018.01300. PubMed/NCBI View Article : Google Scholar
|
|
102
|
Fink D, Nebel S, Norris PS, Aebi S, Kim
HK, Haas M and Howell SB: The effect of different chemotherapeutic
agents on the enrichment of DNA mismatch repair-deficient tumour
cells. Br J Cancer. 77:703–708. 1998.PubMed/NCBI View Article : Google Scholar
|
|
103
|
Vaisman A, Varchenko M, Umar A, Kunkel TA,
Risinger JI, Barrett JC, Hamilton TC and Chaney SG: The role of
hMLH1, hMSH3, and hMSH6 defects in cisplatin and oxaliplatin
resistance: Correlation with replicative bypass of platinum-DNA
adducts. Cancer Res. 58:3579–3585. 1998.PubMed/NCBI
|
|
104
|
Hause RJ, Pritchard CC, Shendure J and
Salipante SJ: Classification and characterization of microsatellite
instability across 18 cancer types. Nat Med. 22:1342–1350.
2016.PubMed/NCBI View Article : Google Scholar
|
|
105
|
Microsatellite Instability-Defective DNA
Mismatch Repair: ESMO Biomarker Factsheet. [https://oncologypro.esmo.org/Education-Library/Factsheets-on-Biomarkers/Microsatellite-Instability-Defective-DNA-Mismatch-Repair#eztoc1701983_0_0_8/.
|
|
106
|
Boyiadzis MM, Kirkwood JM, Marshall JL,
Pritchard CC, Azad NS and Gulley JL: Significance and implications
of FDA approval of pembrolizumab for biomarker-defined disease. J
Immunother Cancer. 6(35)2018.PubMed/NCBI View Article : Google Scholar
|
|
107
|
Chang L, Chang M, Chang HM and Chang F:
Microsatellite Instability: A Predictive Biomarker for Cancer
Immunotherapy. Appl Immunohistochem Mol Morphol. 26:e15–e21.
2018.PubMed/NCBI View Article : Google Scholar
|
|
108
|
Zhao P, Li L, Jiang X and Li Q: Mismatch
repair deficiency/microsatellite instability-high as a predictor
for anti-PD-1/PD-L1 immunotherapy efficacy. J Hematol Oncol.
12(54)2019.PubMed/NCBI View Article : Google Scholar
|
|
109
|
Garon EB, Rizvi NA, Hui R, Leighl N,
Balmanoukian AS, Eder JP, Patnaik A, Aggarwal C, Gubens M, Horn L,
et al: KEYNOTE-001 Investigators: Pembrolizumab for the treatment
of non-small-cell lung cancer. N Engl J Med. 372:2018–2028.
2015.PubMed/NCBI View Article : Google Scholar
|
|
110
|
Reck M, Rodríguez-Abreu D, Robinson AG,
Hui R, Csőszi T, Fülöp A, Gottfried M, Peled N, Tafreshi A, Cuffe
S, et al: KEYNOTE-024 Investigators: Pembrolizumab versus
Chemotherapy for PD-L1-Positive Non-Small-Cell Lung Cancer. N Engl
J Med. 375:1823–1833. 2016.PubMed/NCBI View Article : Google Scholar
|
|
111
|
Pylkkänen L, Karjalainen A, Anttila S,
Vainio H and Husgafvel-Pursiainen K: No evidence of microsatellite
instability but frequent loss of heterozygosity in primary resected
lung cancer. Environ Mol Mutagen. 30:217–223. 1997.PubMed/NCBI
|
|
112
|
Merlo A, Mabry M, Gabrielson E, Vollmer R,
Baylin SB and Sidransky D: Frequent microsatellite instability in
primary small cell lung cancer. Cancer Res. 54:2098–2101.
1994.PubMed/NCBI
|
|
113
|
Mao L, Lee DJ, Tockman MS, Erozan YS,
Askin F and Sidransky D: Microsatellite alterations as clonal
markers for the detection of human cancer. Proc Natl Acad Sci USA.
91:9871–9875. 1994.PubMed/NCBI View Article : Google Scholar
|
|
114
|
Chen XQ, Stroun M, Magnenat JL, Nicod LP,
Kurt AM, Lyautey J, Lederrey C and Anker P: Microsatellite
alterations in plasma DNA of small cell lung cancer patients. Nat
Med. 2:1033–1035. 1996.PubMed/NCBI View Article : Google Scholar
|
|
115
|
Hansen LT, Thykjaer T, Ørntoft TF,
Rasmussen LJ, Keller P, Spang-Thomsen M, Edmonston TB, Schmutte C,
Fishel R and Petersen LN: The role of mismatch repair in small-cell
lung cancer cells. Eur J Cancer. 39:1456–1467. 2003.PubMed/NCBI View Article : Google Scholar
|