Introduction
Colorectal cancer (CRC) is one of the most common
types of cancer worldwide, which is comparable to lung, liver and
stomach cancers (1). Each year,
there are ~2 million newly diagnosed cases in China, thereby making
CRC the third most common cancer and the fourth leading cause of
cancer-associated death (2). In
recent decades, although improvements have been made in health care
and screening programs (3), the
number of new cases of CRC and associated deaths is increasing. The
high incidence of CRC suggests that it is urgent to investigate the
mechanism of CRC in order to improve the current status of CRC
therapy (4).
Patients with CRC are classified into four risk
groups based on their tumor metastases, progression and biomarkers
(5). Briefly, group 0 patients have
no metastasis or lack signs of a poor prognosis. Group 1 patients
exhibit borderline resectable metastasis. Group 2 patients are
detected with disseminated and unresectable CRC. Patients with
unresectable CRC and a lack of intensive or sequential treatment
are classified as into group 3. The metastasis level and TNM stage
guide the therapeutic decision and the treatment purpose along with
the modified treatment strategy (6).
Generally, the fist-line chemotherapy approach is a cytotoxic agent
alone or in combination with a biological targeted drug, as
cytotoxic agents have been shown to increase the response rate of
CRC and biological targeted drugs to reduce the toxicity of the
treatment (7). The most commonly
used agents are fluorouracil (5-FU) alone or combined with
leucovorin (LV) (8). Due to the
limitations in the understanding of CRC biology, chemotherapy has
been challenged with chemo-resistance.
Recently, a series of small non-coding RNAs, which
are called microRNAs, has come to the attention of researchers.
MicroRNAs are short RNAs with about 18–24 nucleotides, which
regulate translation and the stability of the target mRNA (9). Decades ago, microRNAs were first
discovered in chronic lymphocytic leukemia and showed the antitumor
activity (10). Since then,
microRNAs have been investigated and found to be involved in the
initiation, development and progression of numerous cancer types,
as a tumor suppressor or oncogene (11). Extensive research is now aimed at
determining if microRNAs can be used as diagnostic biomarkers and
therapeutic targets for cancer. Genome-wide profiling demonstrated
that miRNA expression in both CRC cell lines and CRC tumors was
regulated by methylation, which might lead to a reduced expression
of miRNAs in CRC, including let-7, miR-34, miR-342, miR-345, miR-9
and miR-129 (12,13). In addition, it is well known that
miR-21 is commonly upregulated in a variety of cancers, including
lung, gastric, breast, colorectal, esophageal, pancreas and
hepatocellular carcinoma (14–22). As
the biological function of miR-21 has been well studied, it has
been reported as a robust and reproducible prognostic marker of CRC
(17,18). It has been reported that the
potential mechanisms of miR-21 as oncogenic microRNA include
downregulation of phosphatase and tensin homolog (PTEN), a decrease
in Bax/Bcl-2/caspase-3 activity, repression of PDCD4 and
downregulation of TIMP3 (23–25).
Additionally, in combination therapy, miR-21 inhibitors enhanced
the chemo-response to 5-FU (26).
Gambogic acid (GA) is a naturally occurring
molecule, which is commonly extracted from Garcinia hanburyi
trees (27). It has been reported
that GA showed numerous activities involving cell cycle arrest,
programmed cell death, autophagy, anti-proliferation, antioxidant,
anti-metastatic and anti-information (28). Currently, the molecular targets of GA
have been well studied and numerous molecular pathways have been
reported to be involved in the actions of GA, including PI3K/Akt,
caspase-3 apoptosis, ATR-Chk1, TNF-α/NF-κB and MET pathways
(29–32). In addition, Huang et al
(33)has reported that GA induced
HT-29 cell apoptosis through the mitochondrial pathway. However,
there are few reports regarding miRNA and GA, regardless of the
mechanism of GA anti-CRC. The present study demonstrated that GA
downregulated miR-21 in CRC and thus increased PTEN expression
causing migration inhibition and resulted in CRC cell apoptosis.
Taken together, these findings might provide strong evidence of GA
antitumor activity and a new insight for future CRC investigation
and treatment.
Materials and methods
Cell culture
Human CRC HT-29, SW480 and HCT116 cell lines were
obtained from Stem Cell Bank of Chinese Academy of Sciences
(Shanghai, China) and were cultured in complete RPMI-1640 medium
(Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) with 10% fetal
bovine serum (Thermo Fisher Scientific, Inc., Waltham, MA, USA),
penicillin (100 U/ml) and streptomycin (100 µg/ml; both
Sigma-Aldrich; Merck KGaA). Cells were maintained at 37°C with 5%
CO2 in a humidified cell culture incubator (Sanyo,
Tokyo, Janpan). For all experiments, five biological groups were
designed according to the concentrations of GA (0, 0.33, 1, 3.3 and
10 µM), which was purchased from Sigma-Aldrich; Merck KGaA. For
each indicated time points (24, 48, and 72 h), cells were sampled
for the following assay.
MTT assay of cell viability
An MTT assay kit (Beyotime Institute of
Biotechnology, Shanghai, China) was used to measure cell growth for
indicated time points following the protocol of manufacturer.
Briefly, HT-29 cells at the logarithmic growth phase were seeded
into 96-well plate at 5×103/well. 200 µl GA with dose
escalation (0, 0.33, 1, 3.3 and 10 µM) was added into each well in
triplicate when cells were totally adhered at 24 h. Cells were
cultured at 37°C (5% CO2) and sampled at 24, 48 and 72
h. MTT (5 mg/ml, Sigma-Aldrich; Merck KGaA) solution was added (20
µl/well) at each sampling time point and cells were further
incubated for additional 4 h in a cell incubator. At the end of
incubation, the supernatants were removed with a pipette. Before
reading under the microplate reader, 150 µl DMSO (Sigma-Aldrich;
Merck KGaA) was added to each well. The proliferation rate was
calculated with the absorbance value (OD) at a wavelength of 480
nm. The cell viability percentage was measured with the following
formula: [(drug treated group/control group) ×100]. Each assay was
performed triplicate and the results are presented as the mean ±
SD.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis
After incubation with GA (0, 1 or 3.3 µM) for 24, 48
or 72 h, CRC cells were collected and total RNA was isolated from
these cells using a PicoPure™ RNA Isolation kit (Arcturus,
Sunnyvale, CA, USA) according to the manufacturer's instructions.
Subsequently, 1 µg isolated RNA was transcribed into cDNA using
SuperScript III RNase H Reverse Transcriptase (Thermo Fisher
Scientific, Inc.). Following the reverse transcription reactions,
amplification was performed using a Vii™ 7 system (Bio-Rad
Laboratories, Hercules, CA, USA) according to the manufacturer's
instructions. Transcript quantities were compared by relative Ct
numbers, and the miR-21 expression level was normalized to an
endogenous reference gene, GAPDH, and then the relative miR-21 mRNA
levels to control sample was calculated using the 2−ΔΔCt
method. Both primers for miR-21and GAPDH were purchased from Thermo
Fisher Scientific, Inc.
Flow cytometric analysis of cell
apoptosis
Apoptotic cells were analyzed according to the
previously described method (34).
Briefly, HT-29 cells were exposed to 3.3 µM GA, GA combined with
miR-21 mimics or vehicle for 72 h. Subsequently, these three groups
of HT-29 cells were harvested and resuspended in PBS. Apoptotic
cells were identified with dual-staining of Annexin V-FITC and
propidium iodide (PI; Thermo Fisher Scientific, Inc.). For each
group, the experiments were performed in triplicate.
Wound healing assay of cell
migration
For the wound healing assay, the procedure was
performed according to the previous reports by Ni et al
(35). HT-29 cells were seeded into
6-well plates (6×104 cells/well) and treated with GA, GA
combined with miR-21 mimics or a vehicle. When the cells grew to
90% confluence, vertical scratches were induced down through the
monolayer cell surfaces using 1,000 µl pipette tips. The media and
cell debris were carefully aspirated before further culture with
serum-free RPMI-1640 medium at 37°C. At 24 h, images of each sample
were captured under a microscope at magnification, ×100 (Leica
DM500; Leica Buffalo Grove, IL, USA). The images were used to
analyze the distance of one side of the wound to the other side
using a scale bar.
Luciferase activity assay
MiR-21 mimics, scramble, wild-type and mutant 3′UTR
PTEN vectors were provided by Ambion (Thermo Fisher Scientific,
Inc.). Vectors carried the PTEN sequence, which contained the
predicted miR-21 binding sites with wild-type or mutant 3′UTR.
5×103 HT-29 cells were plated into 24-well plates, which
were then transfected with PTEN wild-type or mutant vector for 4 h
using Lipofectamine® 2000 transfection reagent (Thermo
Fisher Scientific, Inc.). Subsequently, the cells were treated with
100 nM miR-21 mimics or scramble miRNA. 48 h later, the cells were
lysed using 0.2% trypsin at 37°C and a dual luciferase assay kit
was used to detect the luciferase activities (Promega Corporation,
Madison, WI, USA).
Western blot analysis
Following treatment with GA, GA combined with miR-21
mimics or vehicle for 72 h, HT-29 cells were collected and lysed in
RIPA buffer (Sigma-Aldrich; Merck KGaA). The total protein
concentration was quantified using a BCA Protein Assay kit
(Beyotime Institute of Biotechnology), and 20 µg protein from each
group was separated with 10% Tris-SDS gel. After the
electrophoresis, the gel was electro-transferred onto
polyvinylidene fluoride membranes (Thermo Fisher Scientific, Inc.).
For the subsequent blocking, washing and antibody incubation at
room temperature, an iBind kit (Thermo Fisher Scientific, Inc.) was
used according the manufacturers' instructions. The following
primary antibodies were used: anti-PTEN (1:250), anti-PI3K (1:300),
anti-p-Erk (1:300), anti-matrix metalloproteinase (MMP2; 1:300),
anti-MMP9 (1:300; all Cell Signaling Technology, Inc., Danvers, MA,
USA) and anti-GAPDH (1:1,000; Santa Cruz Biotechnology, Inc., CA,
USA). Horseradish peroxidase-conjugated secondary antibodies
(1:3,000; anti-rabbit or anti-goat IgG; Santa Cruz Biotechnology,
Inc.) were used. The luminescent signal was detected by adding
super sensitive regent (Thermo Fisher Scientific, Inc.) and
quantified using the Image Lab™ system from Bio-Rad
Laboratories.
Transwell assay of cell invasion
The online protocol published by Justus et al
(36) was used in order to validate
the effect of GA on HT-29 invasion. Briefly, 100 µl HT-29 cells
(1×104 cells/well) mixed with 0.33 µM GA, GA combined
with miR-21 mimics or vehicle were added to the upper chamber of
Transwell chambers (Corning Life Science, Tewksbury, MA, USA) and
100 µl RPMI-1640 medium with 30% FBS was added to the lower
chamber. Following incubation for 72 h at 37°C, cells on the upper
surface of the microporous membrane were carefully removed with
cotton swabs, whereas cells on the lower surface of the membrane
were fixed and stained with crystal violet. The cells in five
selected views were counted under a microscope (Leica DM500; Leica)
at magnification, ×100. The average sum of the cells was used to
calculate the invasion rate.
Statistical analysis
All experiments were performed in triplicate and all
data are presented as the mean ± SD. One-way ANOVA followed by
Dunnett's test was used to determine the difference between groups
with P<0.05 being considered to indicate a statistically
significant difference (*P<0.05, **P<0.01).
Results
GA inhibited CRC cell proliferation
and downregulated miR-21 expression
The viability of HT-29 cells was detected at
indicated time points with dose escalation GA (Fig. 1A) from 0.33 to 10 µM using the MTT
assay. As shown in Fig. 1B, GA
inhibited the growth of HT-29 cells in a time- and dose-dependent
manner. GA at all doses significantly inhibited HT-29 cell
proliferation compared with vehicle at 72 h (P<0.01). miR-21 was
mostly found to be overexpressed in CRC. In order to explore the
effect of GA on miR-21 expression, the cells treated with 1 or 3.3
µM GA for 24, 48 and 72 h were subject to qPCR. The results
indicated that GA downregulated miR-21 RNA expression at both
doses, even at 24 h (Fig. 1C;
P<0.05). Moreover, we used the other two CRC cell lines, SW480
and HCT116, to investigate the involvement of miR-21 on the effects
of GA. The results were consistent with the data obtained using
HT-29 cells (Fig. 1D-F). All these
results indicated that GA exerted anti-proliferation effects on
HT-29 cancer cells through downregulation of miR-21 expression.
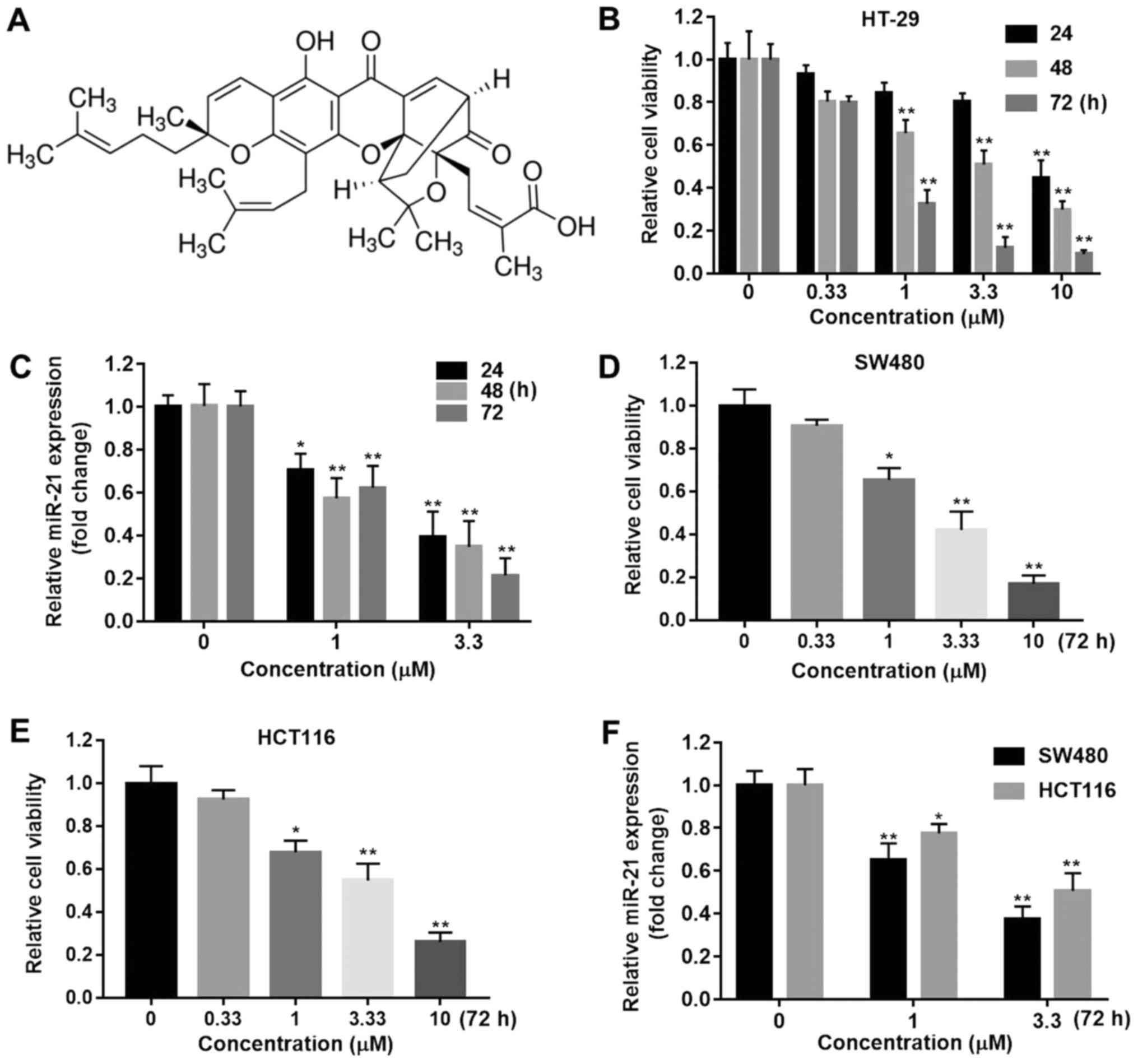 | Figure 1.Gambogic acid decreased CRC viability
and miR-21 expression. (A) The structure of GA. (B) Cell viability
was determined using an MTT assay in HT-29 cells treated with GA
(0, 0.33, 1, 3.3 or 10 µM) for 24, 48 or 72 h (**P<0.01 compared
with the 0 µM GA groups). (C) Expression of miR-21 in HT-29 cells
treated with GA (0, 1 or 3.3 µM) at the indicated time points was
determined by qPCR. The miR-21 expression level was normalizing to
the GAPDH level (*P<0.05, **P<0.01 compared with the 0 µM GA
groups. Cell viability was determined using an MTT assay in (D)
SW480 and (E) HCT116 cells treated with GA (0, 0.33, 1, 3.3 or 10
µM) for 72 h (*P<0.05, **P<0.01 compared with the 0 µM GA
group). (F) Expression of miR-21 in SW480 and HCT116 cells treated
with GA (0, 1 or 3.3 µM) at 72 h was determined by RT-qPCR
(*P<0.05 **P<0.01 compared with the 0 µM GA group). CRC,
colorectal cancer; GA, gambogic acid. |
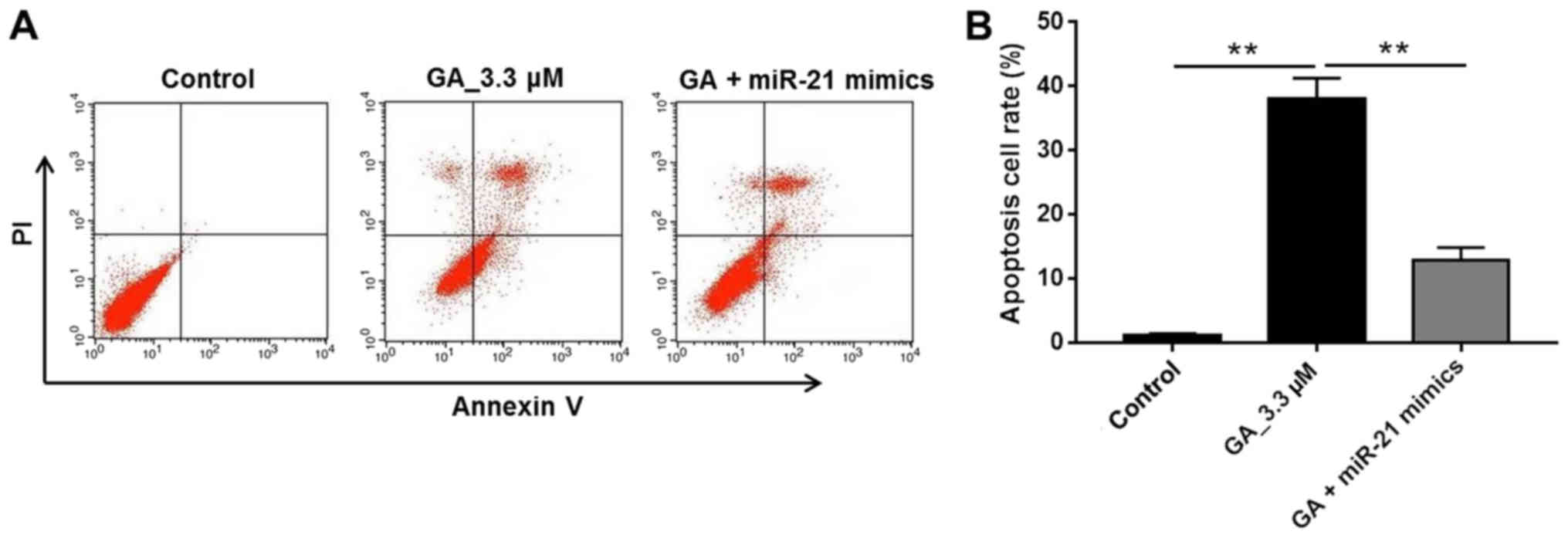
GA induced apoptosis of HT-29 cancer
cells
To further investigate how GA inhibited HT-29 cancer
cell growth and whether miR-21 was the effector of GA, apoptotic
cells were detected with FACS by double staining with Annexin V and
PI. In addition to the vehicle and GA (3.3 µM) treatment groups,
the GA with miR-21 mimics group was added to determine whether
miR-21mimics could interfere with the antitumor effect of GA. The
results in Fig. 2A and B
demonstrated that the majority of HT-29 cells were subject to
apoptosis following GA treatment compared with the control group
(P<0.01). Moreover, when miR-21 mimics were added to the GA 3.3
µM group, the apoptotic rate of the HT-29 cells was reduced to a
lower level, which was similar to that of the control group and was
significantly less than that of the GA 3.3 µM group (Fig. 2B; P<0.01). These results indicated
that GA inhibited HT-29 cancer cell growth by inducing cell
apoptosis via downregulation of miR-21 expression.
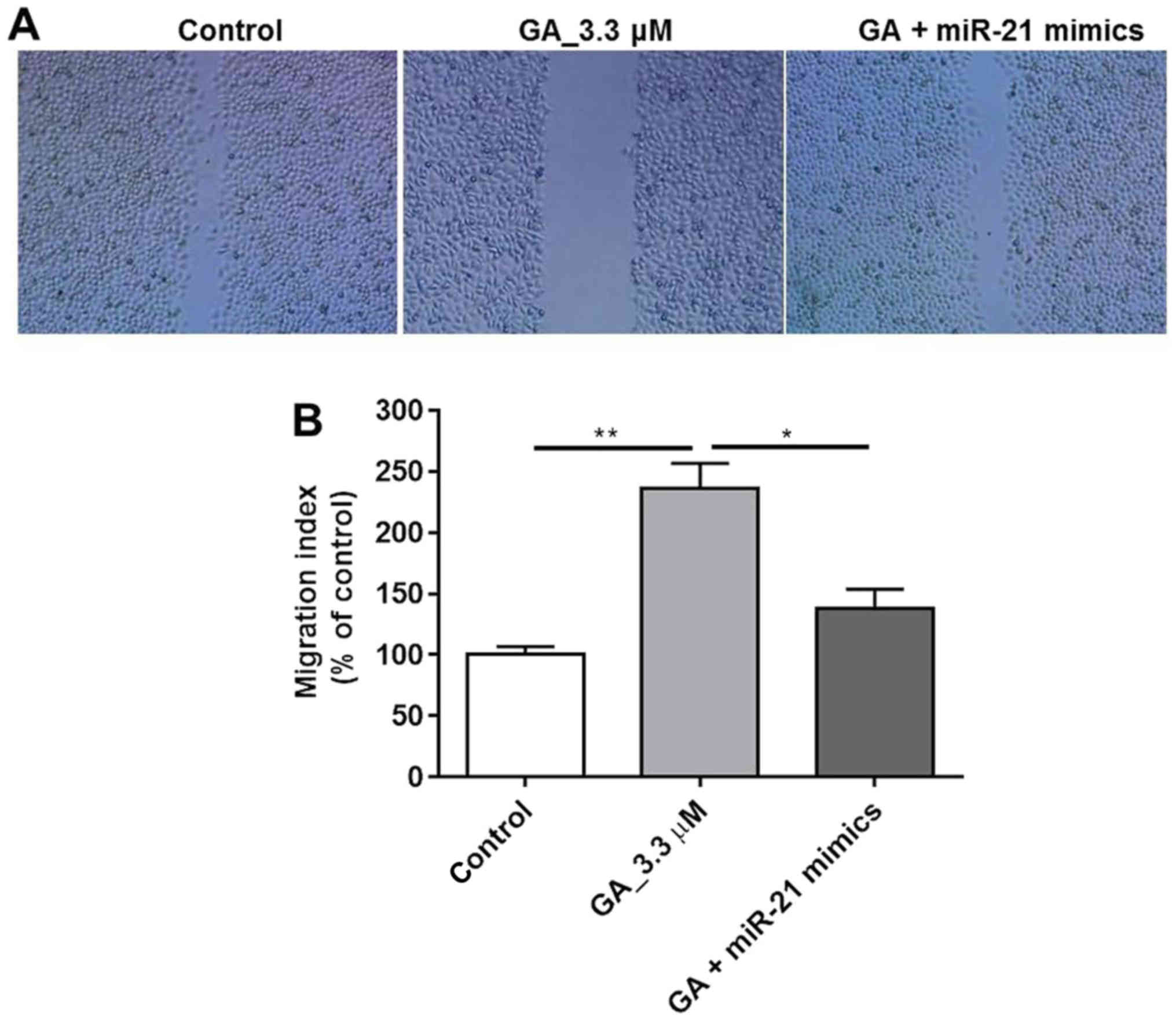
GA inhibited HT-29 cancer cell
migration
In order to validate the effect of GA on HT-29 cell
migration, a wound healing assay was performed. The results in
Fig. 3A indicated that cell
migration was significantly inhibited by 0.33 µM GA compared with
control group. Additionally, in the miR-21 mimics treated group,
the wound was similar to that in the control group, which further
confirmed that the anti-migration effect of GA was exerted through
miR-21. Quantification of the migration activity revealed
significant differences among the control, GA and GA with miR-21
groups (Fig. 3B, P<0.01,
P<0.05). Thus, this result suggested that GA inhibited HT-29
cell migration via blocking miR-21 activity.
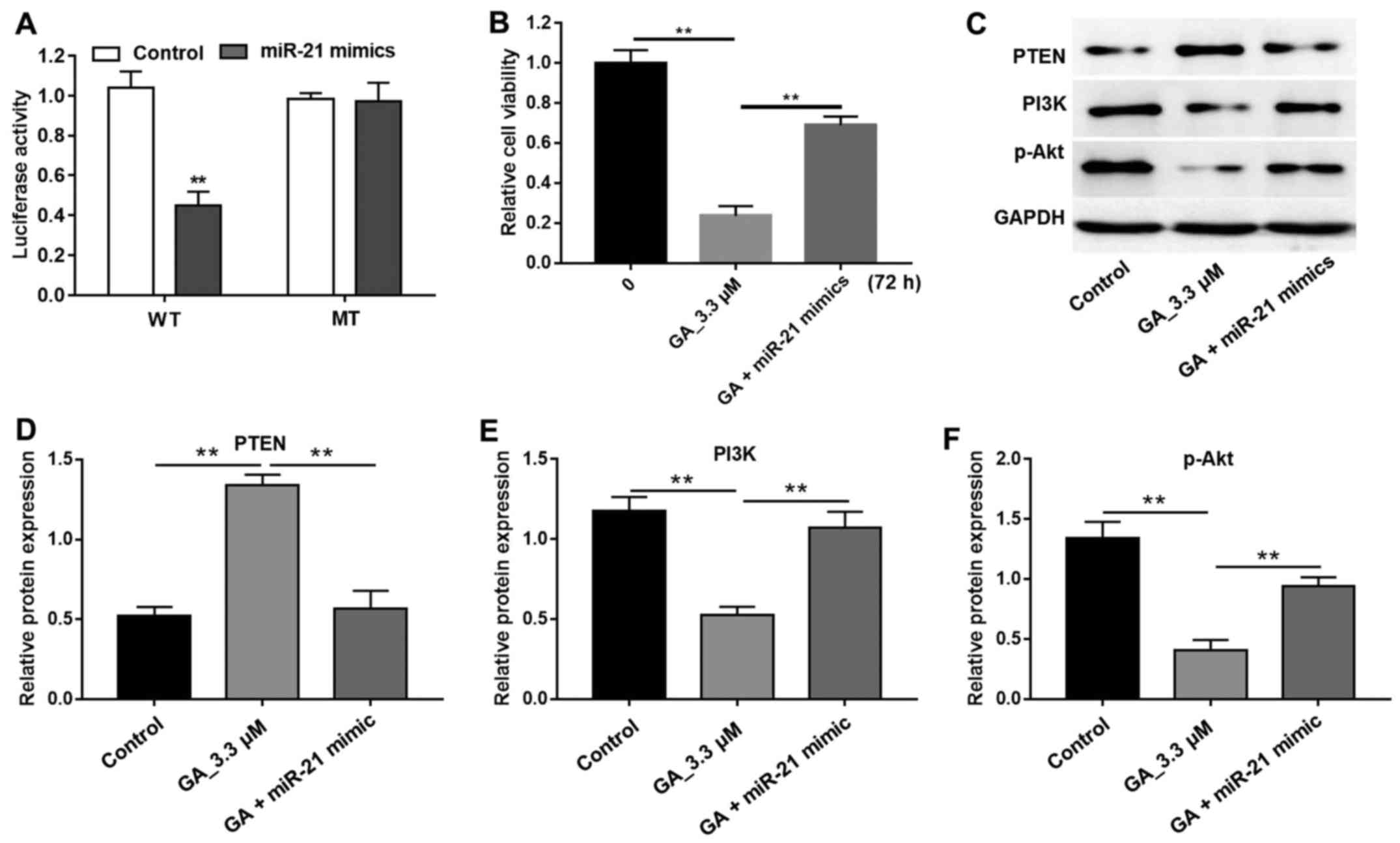
GA enhanced PTEN expression and
blocked the PI3K/Akt pathway
In the present study, miR-21 gene expression was
inhibited by GA (Fig. 1B) and, as
Zhang et al (15) had
previously reported that miR-21 was an onco-miRNA that
downregulated the tumor suppressor gene, PTEN in gastric cancer, we
needed to determine whether GA could affect PTEN expression in CRC.
From the luciferase assay, we found a statistically significant
decrease in firefly luciferase activity in the cells transfected
with miR-21 mimics and wild-type 3′UTR PTEN vectors (Fig. 4A). In addition, GA-induced cell
growth inhibition was reversed by miR-21 mimics (Fig. 4B). The western blot results further
confirmed that GA significantly increased the PTEN protein level
and miR-21 mimics suppressed the enhancement induced by GA
(Fig. 4C and D). Moreover, PTEN is a
suppressor of the PI3K/Akt pathway and therefore, the enhanced
expression of PTEN induced by GA would have definitely blocked the
PI3K/Akt signaling pathway. The western blot results shown in
Fig. 4C, E and F indicated that GA
significantly decreased PI3K and p-Akt protein expression
(P<0.01). All these findings demonstrated that GA showed
antitumor activity through targeting miR-21 and, in turn, blocking
of the PI3K/Akt signaling pathway.
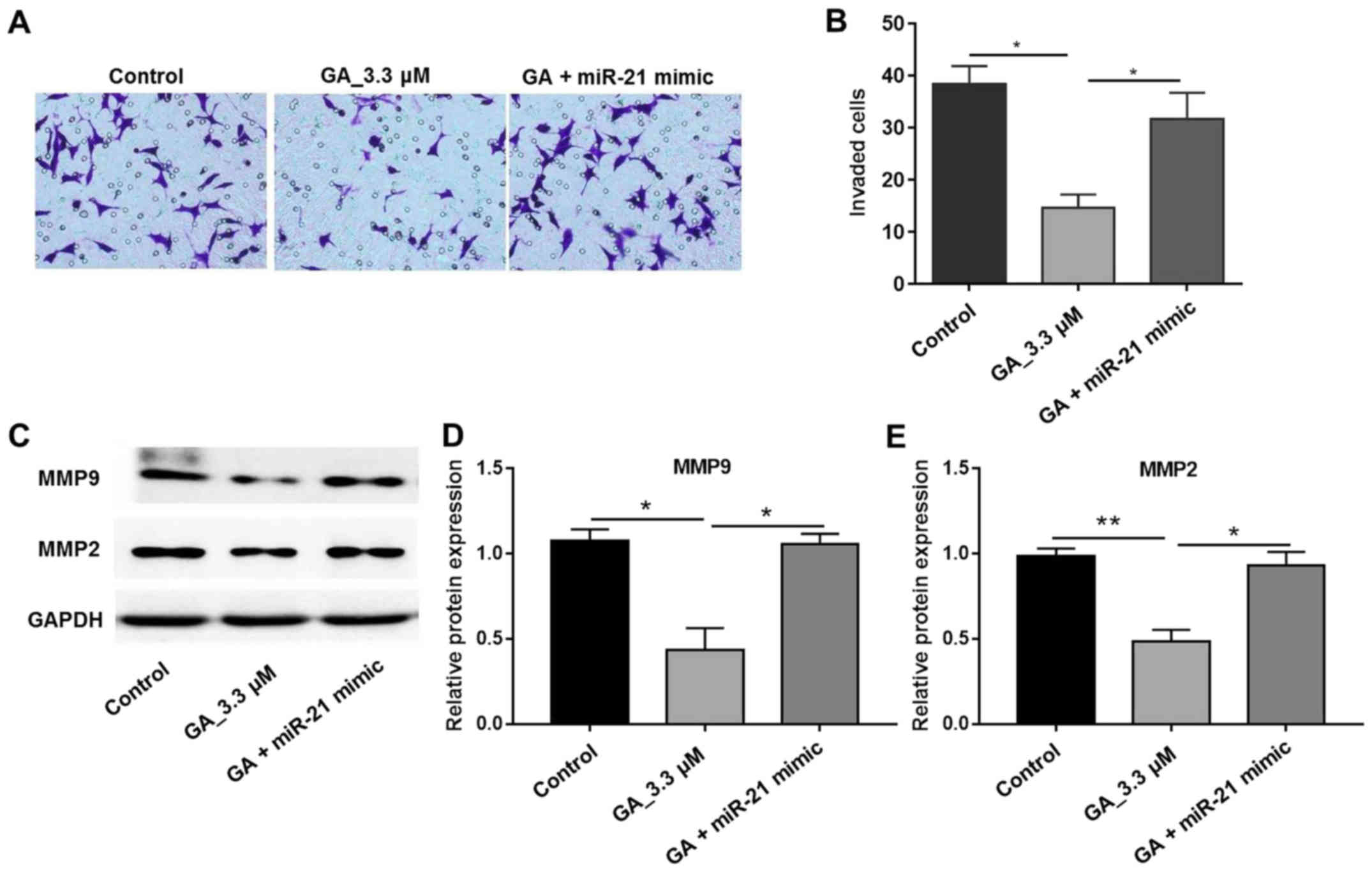
GA inhibited the invasion of HT-29
cells
A Transwell assay was used in order to further
investigate the biological function of GA on HT-29 cell invasion.
The invasion of HT-29 cells treated with 0.33 µM GA was markedly
less than that in the control group under the microscope view
(Fig. 5A). The invaded cell number
was quantified and is shown in Fig.
5B, which reveals a significantly smaller number of invading
cells compared with the vehicle group. Moreover, in the GA and
miR-21 mimics treated group, the cell number was comparable with
that in the control group (Fig. 5B).
As MMP2 and MMP-9 are closely associated with cell migration and
invasion, the expression of these factors was also detected. The
results revealed that MMP-2 and MMP-9 expression was significantly
decreased following treatment with GA (Fig. 5C-E). These results indicated that GA
inhibited the activity of MMP2 and MMP-9 and thus showed its
anti-invasion activity in HT-29 cancer cells.
Discussion
CRC has become a global public health problem with
its high incidence and mortality rate worldwide (4). One review published recently has
summarized discoveries and newest findings in CRC diagnosis and
treatment (5). Based on genetic
screening tests, most of the major cancer genes involved in CRC
have been well characterized, such as APC, KRAS, PI3K and TP53
(37–39). Epigenetic instability, mainly with
respect to CpG island methylation, was also a common feature in CRC
(38). Non-coding RNAs (ncRNA) are
RNA molecules without an open reading frame and are not translated
into proteins. This results in the deregulation of miRNAs
associated with CRC. Previously, researchers have reported that
miRNAs function as potential diagnostic and prognostic biomarkers
in CRC (9). For instance, 12 miRNAs
showed higher expression levels in CRC patient samples than in
those from healthy controls, such as miR-7, −17, −20a, −21, −92a,
−96, −106a, −134, −183, −196a, −199a-3p and −214. Meanwhile, 8
miRNAs were shown to be downregulated, including miR-9, −29b,
−127-5p, −138, −143, −146a, −222 and −938 (40). At present, an extensive amount of
research regarding CRC is focused on the development of new less
aggressive and more effective therapies than conventional ones.
It has been shown that GA inhibited the growth of a
wide variety of cancers, including hepatocarcinoma, gastric
carcinoma, lung cancer and breast cancer (41–45). The
mechanisms of GA antitumor activity have been well studied.
However, few studies about the effect of GA on miRNA expression
have been reported. In this article, we not only validated the
antitumor effect of GA via MTT assay for proliferation, wound
healing assay for migration, Transwell assay for invasion and FACS
for cell apoptosis, but we also first demonstrated that miR-21 was
down regulated by GA in CRC. In addition, miR-21 expression was
suppressed by GA in HT-29 cells, as determined by RT-qPCR. Based on
this knowledge, we proposed that miR-21 might be an effector of GA
in HT-29 cells. For the potential mechanisms of miR-21 as an
onco-miRNA, several reports have been published. Meng et al
(46) has found that PTEN was
regulated by miR-21 in hepatocellular carcinoma. Similarly, PTEN
was repressed by miR-21 in non-small cell lung cancer (47).
PTEN was revealed to be an essential tumor
suppressor and modulator of cell growth and proliferation (48). PTEN was reported to be under the
control of various transcription factors involving EGR-1, p53, ATF2
and PPARγ (48). miRNA-guided
degradation would control the process of PTEN mRNA splicing, which
is translocated to the cytosol (48). The prominent miRNAs of PTEN-targeting
included miR-19, miR-21 and miR-221 (46). Based on these, we investigated the
protein expression level of PTEN, PI3K and p-Akt in GA-treated
HT-29 cells. As expected, PTEN was increased by GA, while PI3K and
p-AKT proteins were decreased in HT-29 cells.
Taken together, the anti-proliferation effect of GA
on HT-29 cancer cells may be mediated via decreasing miR-21
expression and blocking the PI3K/Akt pathway. Therefore, our
article might open up a new pathway toward CRC pharmacological
intervention.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
GG, YB, HQ, MY, JH and LL collaborated in the
experiment design, sample collection and experiments execution. GG,
LY and BL analyzed and interpreted the patient data and were the
major contributors in developing the first draft of the manuscript.
BL reviewed and edited the manuscript. XQ analyzed and interpreted
the data and reviewed and approved the final draft of the
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Haggar FA and Boushey RP: Colorectal
cancer epidemiology: Incidence, mortality, survival, and risk
factors. Clin Colon Rectal Surg. 22:191–197. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Connell O JB, Maggard MA and Ko CY: Colon
cancer survival rates with the new American Joint Committee on
Cancer sixth edition staging. J Natl Cancer Inst. 96:1420–1425.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kraus S, Nabiochtchikov I, Shapira S and
Arber N: Recent advances in personalized colorectal cancer
research. Cancer Lett. 347:15–21. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ferlay J, Shin HR, Bray F, Forman D,
Mathers C and Parkin DM: Estimates of worldwide burden of cancer in
2008: GLOBOCAN, 2008. Int J Cancer. 127:2893–2917. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mármol I, Sánchez-de-Diego C, Dieste
Pradilla A, Cerrada E and Yoldi Rodriguez MJ: Colorectal carcinoma:
A general overview and future perspectives in colorectal cancer.
Int J Mol Sci. 18:pii: E197. 2017. View Article : Google Scholar
|
|
6
|
Venook A: Critical evaluation of current
treatments in metastatic colorectal cancer. Oncologist. 10:250–261.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Van Cutsem E, Cervantes A, Nordlinger B
and Arnold D: ESMO guidelines working group: Metastatic colorectal
cancer: ESMO clinical practice guidelines for diagnosis, treatment
and follow-up. Ann Oncol. 3 25 Suppl:iii1–iii9. 2014. View Article : Google Scholar
|
|
8
|
Lee JJ, Beumer JH and Chu E: Therapeutic
drug monitoring of 5-fluorouracil. Cancer Chemother Pharmacol.
78:447–464. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Liu M and Chen H: The role of microRNAs in
colorectal cancer. J Genet Genomics. 37:347–358. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kempin S: Update on chronic lymphocytic
leukemia: Overview of new agents and comparative analysis. Curr
Treat Options Oncol. 14:144–155. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Di Leva G, Cheung DG and Croce CM: miRNA
clusters as therapeutic targets for hormone-resistant breast
cancer. Expert Rev Endocrinol Metab. 10:607–617. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Grady WM and Carethers JM: Genomic and
epigenetic instability in colorectal cancer pathogenesis.
Gastroenterology. 135:1079–1099. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Karakatsanis A, Papaconstantinou I,
Gazouli M, Lyberopoulou A, Polymeneas G and Voros D: Expression of
microRNAs, miR-21, miR-31, miR-122, miR-145, miR-146a, miR-200c,
miR-221, miR-222, and miR-223 in patients with hepatocellular
carcinoma or intrahepatic cholangiocarcinoma and its prognostic
significance. Mol Carcinog. 52:297–303. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li Y, Li W, Ouyang Q, Hu S and Tang J:
Detection of lung cancer with blood microRNA-21 expression levels
in Chinese population. Oncol Lett. 2:991–994. 2011.PubMed/NCBI
|
|
15
|
Zhang BG, Li JF, Yu BQ, Zhu ZG, Liu BY and
Yan M: microRNA-21 promotes tumor proliferation and invasion in
gastric cancer by targeting PTEN. Oncol Rep. 27:1019–1026. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Papagiannakopoulos T, Shapiro A and Kosik
KS: MicroRNA-21 targets a network of key tumor-suppressive pathways
in glioblastoma cells. Cancer Res. 68:8164–8172. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu GH, Zhou ZG, Chen R, Wang MJ, Zhou B,
Li Y and Sun XF: Serum miR-21 and miR-92a as biomarkers in the
diagnosis and prognosis of colorectal cancer. Tumour Biol.
34:2175–2181. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Asangani IA, Rasheed SA, Nikolova DA,
Leupold JH, Colburn NH, Post S and Allgayer H: MicroRNA-21 (miR-21)
post-transcriptionally downregulates tumor suppressor Pdcd4 and
stimulates invasion, intravasation and metastasis in colorectal
cancer. Oncogene. 27:2128–2136. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang HL, Yang LF, Zhu Y, Yao XD, Zhang
SL, Dai B, Zhu YP, Shen YJ, Shi GH and Ye DW: Serum miRNA-21:
Elevated levels in patients with metastatic hormone-refractory
prostate cancer and potential predictive factor for the efficacy of
docetaxel-based chemotherapy. Prostate. 71:326–331. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Dillhoff M, Liu J, Frankel W, Croce C and
Bloomston M: MicroRNA-21 is overexpressed in pancreatic cancer and
a potential predictor of survival. J Gastrointest Surg.
12:2171–2176. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Faragalla H, Youssef YM, Scorilas A,
Khalil B, White NM, Mejia-Guerrero S, Khella H, Jewett MA, Evans A,
Lichner Z, et al: The clinical utility of miR-21 as a diagnostic
and prognostic marker for renal cell carcinoma. J Mol Diagn.
14:385–392. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kumar S, Keerthana R, Pazhanimuthu A and
Perumal P: Overexpression of circulating miRNA-21 and miRNA-146a in
plasma samples of breast cancer patients. Indian J Biochem Biophys.
50:210–214. 2013.PubMed/NCBI
|
|
23
|
Zhu Q, Wang Z, Hu Y, Li J, Li X, Zhou L
and Huang Y: miR-21 promotes migration and invasion by the
miR-21-PDCD4-AP-1 feedback loop in human hepatocellular carcinoma.
Oncol Rep. 27:1660–1668. 2012.PubMed/NCBI
|
|
24
|
Liu CZ, Liu W, Zheng Y, Su JM, Li JJ, Yu
L, He XD and Chen SS: PTEN and PDCD4 are bona fide targets of
microRNA-21 in human cholangiocarcinoma. Chin Med Sci J. 27:65–72.
2012.PubMed/NCBI
|
|
25
|
Wang N, Zhang CQ, He JH, Duan XF, Wang YY,
Ji X, Zang WQ, Li M, Ma YY, Wang T and Zhao GQ: MiR-21
down-regulation suppresses cell growth, invasion and induces cell
apoptosis by targeting FASL, TIMP3, and RECK genes in esophageal
carcinoma. Dig Dis Sci. 58:1863–1870. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Feng YH, Wu CL, Shiau AL, Lee JC, Chang
JG, Lu PJ, Tung CL, Feng LY, Huang WT and Tsao CJ:
MicroRNA-21-mediated regulation of Sprouty2 protein expression
enhances the cytotoxic effect of 5-fluorouracil and metformin in
colon cancer cells. Int J Mol Med. 29:920–926. 2012.PubMed/NCBI
|
|
27
|
Zhang HZ, Kasibhatla S, Wang Y, Herich J,
Guastella J, Tseng B, Drewe J and Cai SX: Discovery,
characterization and SAR of gambogic acid as a potent apoptosis
inducer by a HTS assay. Bioorg Med Chem. 12:309–317. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang X and Chen W: Gambogic acid is a
novel anti-cancer agent that inhibits cell proliferation,
angiogenesis and metastasis. Anticancer Agents Med Chem.
12:994–1000. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ishaq M, Khan MA, Sharma K, Sharma G,
Dutta RK and Majumdar S: Gambogic acid induced oxidative stress
dependent caspase activation regulates both apoptosis and autophagy
by targeting various key molecules (NF-κB, Beclin-1, p62 and NBR1)
in human bladder cancer cells. Biochim Biophys Acta.
1840:3374–3384. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lü L, Tang D, Wang L, Huang LQ, Jiang GS,
Xiao XY and Zeng FQ: Gambogic acid inhibits TNF-α-induced invasion
of human prostate cancer PC3 cells in vitro through PI3K/Akt and
NF-κB signaling pathways. Acta Pharmacol Sin. 33:531–541. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang LH, Li Y, Yang SN, Wang FY, Hou Y,
Cui W, Chen K, Cao Q, Wang S, Zhang TY, et al: Gambogic acid
synergistically potentiates cisplatin-induced apoptosis in
non-small-cell lung cancer through suppressing NF-κB and MAPK/HO-1
signalling. Br J Cancer. 110:341–352. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhang H, Lei Y, Yuan P, Li L, Luo C, Gao
R, Tian J, Feng Z, Nice EC and Sun J: ROS-mediated autophagy
induced by dysregulation of lipid metabolism plays a protective
role in colorectal cancer cells treated with gambogic acid. PLoS
One. 9:e964182014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Huang GM, Sun Y, Ge X, Wan X and Li CB:
Gambogic acid induces apoptosis and inhibits colorectal tumor
growth via mitochondrial pathways. World J Gastroenterol.
21:6194–6205. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hu Y, Yang Y, You QD, Liu W, Gu HY, Zhao
L, Zhang K, Wang W, Wang XT and Guo QL: Oroxylin A induced
apoptosis of human hepatocellular carcinoma cell line HepG2 was
involved in its antitumor activity. Biochem Biophys Res Commun.
351:521–527. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ni W, Fang Y, Tong L, Tong Z, Yi F, Qiu J,
Wang R and Tong X: Girdin regulates the migration and invasion of
glioma cells via the PI3K-Akt signaling pathway. Mol Med Rep.
12:5086–5092. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Justus CR, Leffler N, Ruiz-Echevarria M
and Yang LV: In vitro cell migration and invasion assay. J Vis Exp.
88:2014.
|
|
37
|
Li W, Qiu T, Zhi W, Shi S, Zou S, Ling Y,
Shan L, Ying J and Lu N: Colorectal carcinomas with KRAS codon 12
mutation are associated with more advanced tumor stages. BMC
Cancer. 15:3402015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ogino S, Nosho K, Kirkner GJ, Kawasaki T,
Meyerhardt JA, Loda M, Giovannucci EL and Fuchs CS: CpG island
methylator phenotype, microsatellite instability, BRAF mutation and
clinical outcome in colon cancer. Gut. 58:90–96. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Munro AJ, Lain S and Lane DP: P53
abnormalities and outcomes in colorectal cancer: A systematic
review. Br J Cancer. 92:434–444. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ahmed FE, Ahmed NC, Vos PW, Bonnerup C,
Atkins JN, Casey M, Nuovo GJ, Naziri W, Wiley JE, Mota H and
Allison RR: Diagnostic microRNA markers to screen for sporadic
human colon cancer in stool: I. Proof of principle. Cancer Genomics
Proteomics. 10:93–113. 2013.PubMed/NCBI
|
|
41
|
Kashyap D, Mondal R, Tuli HS, Kumar G and
Sharma AK: Molecular targets of gambogic acid in cancer: Recent
trends and advancements. Tumor Biol. 37:12915–12925. 2016.
View Article : Google Scholar
|
|
42
|
Yang Y, Sun X, Yang Y, Yang X, Zhu H, Dai
S, Chen X, Zhang H, Guo Q, Song Y, et al: Gambogic acid enhances
the radiosensitivity of human esophageal cancer cells by inducing
reactive oxygen species via targeting Akt/mTOR pathway. Tumor Biol.
37:1853–1862. 2016. View Article : Google Scholar
|
|
43
|
Yu J, Guo QL, You QD, Zhao L, Gu HY, Yang
Y, Zhang HW, Tan Z and Wang X: Gambogic acid-induced G2/M phase
cell-cycle arrest via disturbing CDK7-mediated phosphorylation of
CDC2/p34 in human gastric carcinoma BGC-823 cells. Carcinogenesis.
28:632–638. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Yu J, Guo QL, You QD, Lin SS, Li Z, Gu HY,
Zhang HW, Tan Z and Wang X: Repression of telomerase reverse
transcriptase mRNA and hTERT promoter by gambogic acid in human
gastric carcinoma cells. Cancer Chemother Pharmacol. 58:434–443.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Felth J, Lesiak-Mieczkowska K, D'Arcy P,
Haglund C, Gullbo J, Larsson R, Linder S, Bohlin L, Fryknäs M and
Rickardson L: Gambogic acid is cytotoxic to cancer cells through
inhibition of the ubiquitin-proteasome system. Invest New Drugs.
31:587–598. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Meng F, Henson R, Wehbe-Janek H, Ghoshal
K, Jacob ST and Patel T: MicroRNA-21 regulates expression of the
PTEN tumor suppressor gene in human hepatocellular cancer.
Gastroenterology. 133:647–658. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Cao L, Chen J, Ou B, Liu C, Zou Y and Chen
Q: GAS5 knockdown reduces the chemo-sensitivity of non-small cell
lung cancer (NSCLC) cell to cisplatin (DDP) through regulating
miR-21/PTEN axis. Biomed Pharmacother. 93:570–579. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Salmena L: PTEN: History of a tumor
suppressor. Methods Mol Biol. 1388:3–11. 2016. View Article : Google Scholar : PubMed/NCBI
|