Introduction
Calmodulin (CaM) is a ubiquitous, multifunctional
Ca2+-binding protein that is involved in the regulation
of numerous important physiological functions, including neural
activity, gene expression, enzyme regulation and muscle contraction
(1). A previous study demonstrated
that the elevation of CaM levels in transformed cells directly
affected the rate of cell proliferation (2). Numerous CaM-binding proteins have been
identified, a number of which have been shown to be critical for
the regulation of cell functions (3–5). CaM
regulates the activity of the majority of its binding partners in a
Ca2+-dependent manner (6). For example, CaM binds to L-type
Ca2+ channels, resulting in CaM-channel complexes that
are essential for Ca2+-dependent facilitation and
inactivation (7). Furthermore, CaM
plays a vital role in numerous diseases by participating in
signaling pathways that regulate multiple crucial physiological
processes. It has previously been reported that CaM may be an
important mediator for Ca2+ homeostasis in Alzheimer's
disease (8). In addition, defects in
CaM functions disrupt important calcium signaling events in the
heart, affecting membrane ion channels and inducing arrhythmias
(9).
CaM is a relatively small protein with only 148
amino acids. The protein is highly conserved across different
species, and comprises four EF hands that form two structurally
similar domains connected by a flexible central linker (10). A previous study demonstrated that CaM
is encoded by multiple genes in vertebrates and invertebrates, as
was first reported in chickens (11). Subsequently, CaM 1, 2 and 3 have been
cloned, sequenced and characterized in rats (12–14) and
humans (15–17). Furthermore, single-copy genes of CaM
have been identified and cloned in Dictyostelium discoideum
(18), Chlamydomonas
(19) and yeasts (20,21).
However, to the best of our knowledge, the genetic information of
CaM in guinea pigs has never been established.
Guinea pigs are one of the most widely used models
for various diseases, including pulmonary, gastrointestinal and
other life-threatening infections (22–24). The
electrophysiological features of cardiac Ca2+ channels
have been extensively studied in guinea pig cardiomyocytes
(25,26). Moreover, numerous findings have
highlighted the importance of CaM in the regulation of cardiac
Ca2+ channel-based activities (25,26).
Therefore, it is necessary to identify the molecular fundamentals
of CaM in guinea pig hearts.
In order to ascertain the CaM gene information in
the guinea pig genome, CaM genes were isolated from guinea pig
hearts and characterized. Therefore, the expression pattern of CaM
3 in guinea pigs was investigated with the aim to improve the
understanding of CaM 3 functions.
Materials and methods
Bacterial strains, vectors and
media
In order to clone the CaM gene from guinea pig
hearts, E. coli JM109 (Takara Bio Inc., Otsu, Japan) was
employed as the host cell, with the pGEM®-T Easy TA cloning vector
(Promega Corporation, Madison, WI, USA) used as the host-vector
system. The E. coli cells were grown at 37°C in lysogeny
broth agar plates containing ampicillin, with
5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside for the selection
of positive clones. The plasmid mini kit and gel extraction kit
were purchased from Axygen (Union City, CA, USA).
Molecular cloning of CaM cDNA from
guinea pig hearts
Experiments were carried out following approval from
the Committee of Animal Experimentation at China Medical University
(Shenyang, China). Six guinea pigs (either gender) were used in
this study. They were purchased from the Department of Laboratory
Animal, China Medical University (Shenyang, China). Following
anesthetization by ether (Tiangen Biotech Co., Ltd., Beijing,
China), adult guinea pigs (weight, 250–300 g) were sacrificed by
decapitation, and the left ventricular myocardium was quickly
removed, frozen in liquid nitrogen and stored at −80°C. Total RNA
from the tissue was isolated using TRIzol® reagent (Invitrogen Life
Technologies, Grand Island, NY, USA), and the RNA obtained was
reverse transcribed to cDNA using avian myeloblastosis virus
reverse transcriptase with an RNA polymerase chain reaction (PCR)
kit (version 3.0; Takara), oligo-(dT) and random primers, according
to the manufacturer's instructions. The cDNA was then subjected to
normal PCR amplification with Taq DNA polymerase (Takara), or rapid
amplification of the cDNA end (3′-RACE) with a 3′ full RACE kit
(Takara. Since the nucleotide sequences of CaM, including the
untranslated regions (UTRs), were known to be highly conserved
among mammals, nucleotide oligomers based on multiple alignments of
the highly conserved areas from humans and rats were employed as
primers for the PCR to amplify the coding region and the 5′-UTR.
With regard to the cloning of the 3′-UTR, 3′-RACE was carried out
with the gene-specific forward primer corresponding to the
N-terminal structure of the coding region, while GeneRacer Oligo dT
(Takara) was used as the reverse primer. The primers used are shown
in Table I. The amplification
conditions included an initial denaturation for 3 min at 94°C,
followed by 30 cycles of denaturation for 1 min at 94°C, annealing
for 1 min according to the melting temperature of the primers,
extension for 1 min at 72°C, and a final extension for 10 min at
72°C. PCR products of the expected size were purified from the
agarose gel using a gel extraction kit. The cDNA fragments obtained
were subcloned into the pGEM®-T Easy vector, and sequenced by
Sangon Biotech Co., Ltd. (Shanghai, China). The sequence of each
cDNA was determined from more than three independent clones, which
was subsequently used to deduce the full length cDNA sequence.
 | Table I.Nucleotide sequences of the primers
used in polymerase chain reaction amplification. |
Table I.
Nucleotide sequences of the primers
used in polymerase chain reaction amplification.
| Primers | Primer sequences |
|---|
| CaM 3 coding
region | Forward:
5′-ATGGCTGACCAGCTGAC-3′ |
|
| Reverse:
5′-CTTTGCAGTCATCATC-3′ |
| CaM 3 3′-UTR | Forward:
5′-ATGGCTGACCAACTGACTGAAGAG-3′ |
|
| Reverse:
5′-TACCGTCGTTCCACTAGTGATTT-3′ |
| CaM 3 5′-UTR | Forward:
5′-GCCGGAGGAACCTTG-3′ |
|
| Reverse:
5′-GCTCTGCTTCAGTGGG-3′ |
| β-actin | Forward:
5′-CCAACTGGGACGACATGGAG-3′ |
|
| Reverse:
5′-CGTAGCCCTCGTAGATGGGC-3′ |
Bioinformatics analysis
Analyses for nucleotide and protein sequence
similarities were conducted with the BLAST algorithm at the
National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/blast). Multiple
comparisons were conducted using DNASTAR software (DNASTAR,
Madison, WI, USA).
Reverse transcription PCR
(RT-PCR)
The mRNA expression levels of CaM 3 were analyzed
semi-quantitatively using RT-PCR. Total RNA was isolated from
50–100-mg tissue samples collected from the left ventricle,
cerebral cortex, cerebellum, small intestine, aorta, kidney, lung,
liver, skeletal muscle and spleen of the guinea pigs, as described
previously. Aliquots of the RNA solutions were added to the RT
mixture prepared from the RNA PCR kit, and following the RT
reaction, PCR was conducted for 30 cycles. The primer pairs were
specific for CaM 3, and the sequences were as follows: Forward,
5′-AAGGATGGAGATGGCACTATTACCA-3′; and reverse,
5′-AGGGGAGTGAAGGAGAGAAAAGAGC-3′; the gene product was 461 bp.
GAPDH, a constitutively expressed gene, was used as an internal
standard to verify the RT-PCR assay. The sequences of the GAPDH
primers were as follows: Forward, 5′-TTCCAGTATGATTCTACCCACG-3′; and
reverse, 5′-CCCTCCACAATGCCGAAG-3′. These primers were used to
amplify a 400-bp fragment of the guinea pig GAPDH cDNA.
Diethylpyrocarbonate-water for the replacement of the cDNA template
was used as a negative control. PCR products were analyzed on a
1.2% agarose gel.
Results
Cloning of the CaM 3 gene in guinea
pigs
In order to clone the CaM gene from guinea pigs, the
primers were designed based on the regions of the highest reported
homologous nucleotide sequences of CaM from humans, mice and rats,
as shown in Table I. The encoding
region of CaM cDNA from the guinea pig hearts was subsequently
isolated and amplified using RT-PCR, after which the genetic
information was inserted into the vectors. Nucleotide sequencing of
CaM was determined in three independent clones, which revealed
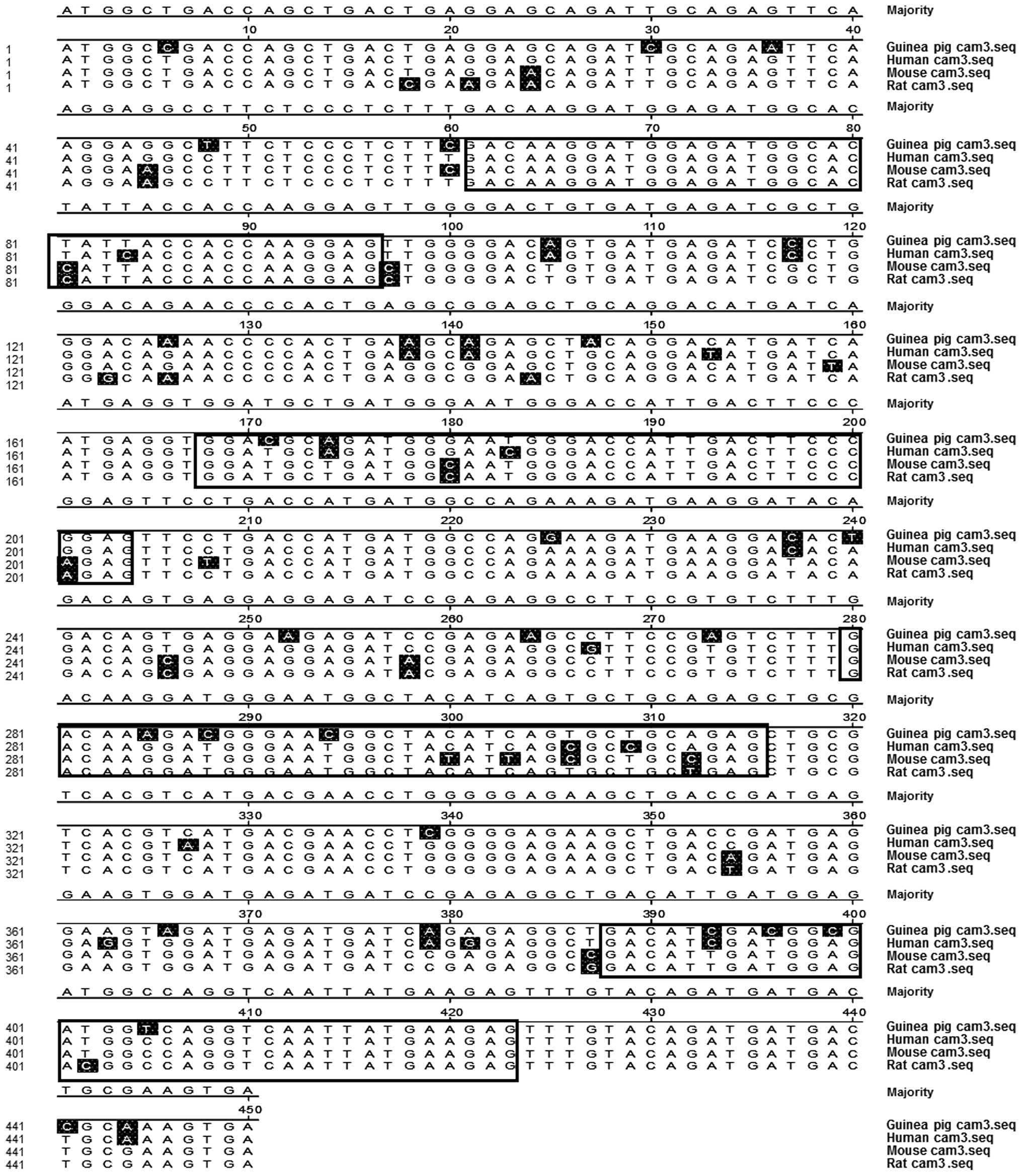
identical sequences (Fig. 1).
The comparison of CaM 3 sequences between different
species indicated that the coding regions of CaM were similar to
those from humans, mice and rats (Fig.
1). Bioinformatics analysis further indicated that the CaM gene
in guinea pigs shared high sequence homology with humans (93.1%
similarity), mice (89.8%) and rats (89.8%) (Fig. 2A). In addition, the phylogenetic tree
revealed close evolutionary associations between these groups of
CaM 3 genes (Fig. 2B). These results
indicated that the CaM gene isolated from the guinea pigs was
likely to be CaM 3.
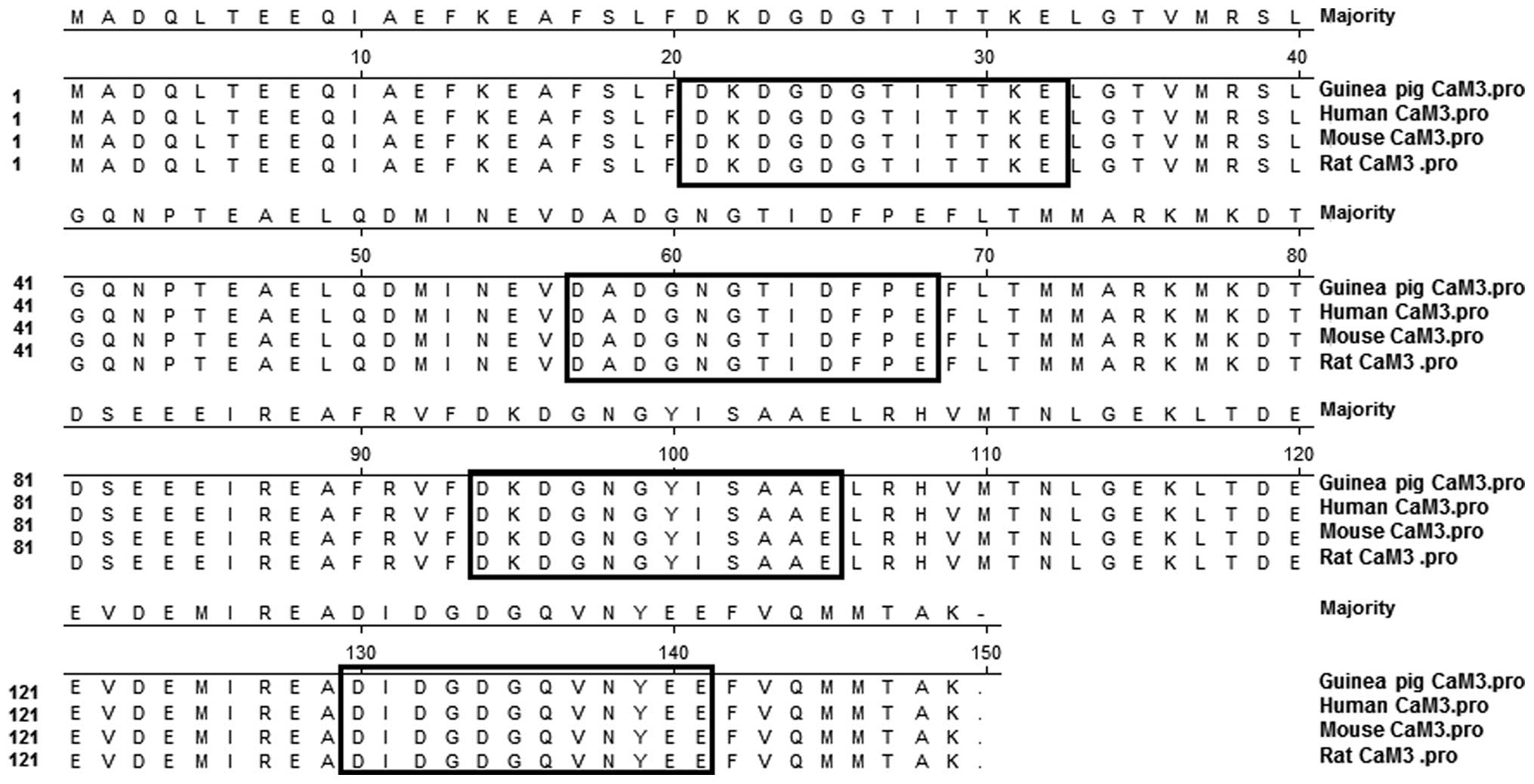
The deduced amino acid sequences of the CaM 3 coding
nucleotide sequences isolated from the guinea pigs revealed 100%
similarity to those products of the CaM 3 genes from humans, mice
and rats (Fig. 3). The sequences
contained four highly conserved Ca2+-binding domains
that were characteristic of CaM (Fig.
3). Based on these results, even though the base sequences were
not exactly the same, the predicted amino acid sequences of the
guinea pig CaM 3 showed 100% homology to the CaM proteins from
other species.
Cloning of the 5′- and 3′-UTR
sequences of CaM 3 in guinea pigs
To obtain more information on the CaM gene obtained
from guinea pig hearts, the 5′- and 3′-UTR sequences were
determined through the methods of RT-PCR and 3′-RACE PCR,
respectively. The cDNA fragments obtained were subsequently
inserted into vectors. The 5′-UTR of the cloned CaM cDNA was
relatively short (32 bp); however, the sequence showed high
homology with the CaM 3 genes in humans, mice and rats (data not
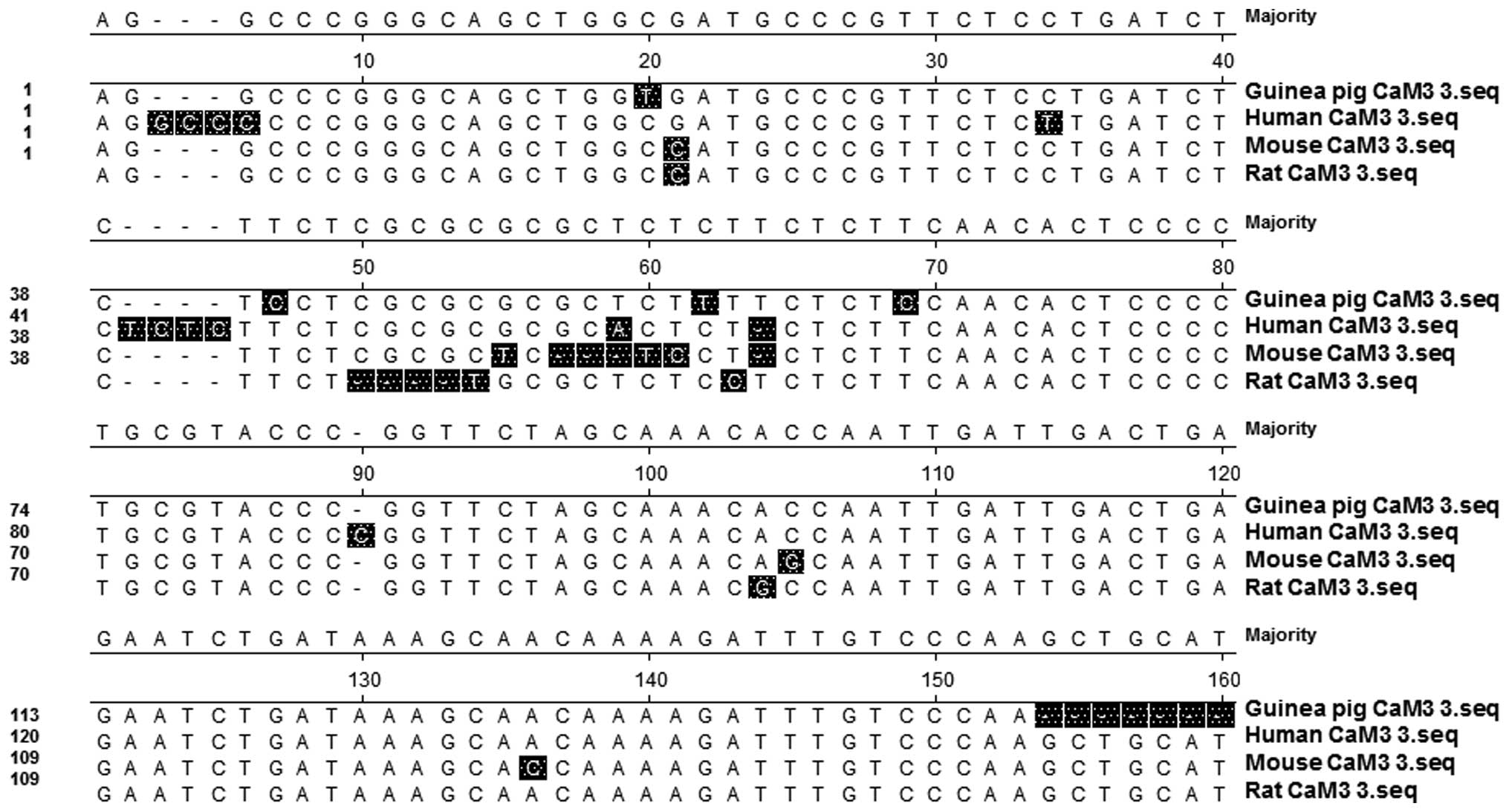
shown). In addition, the 3′-UTR sequence of the guinea pig CaM was
compared with those from the CaM 3 genes in humans, mice and rats,
and the nucleotide sequence homology similarities were determined
as 95.3, 93.2 and 94.7%, respectively (Fig. 4). These results demonstrated that the
guinea pig CaM cDNA clones (32, 450 and 154 bp for the 5′-UTR,
coding region and 3′-UTR, respectively) exhibited high homology
with the previously reported cDNA sequences of CaM 3 genes in
humans, mice and rats. Based on these results, sequence data of the
guinea pig CaM 3 gene, isolated in the current study, have been
registered in GenBank (accession no. FJ012165; 636 bp), and show
high homology with the counterparts from other animals.
Homology analysis of the guinea pig
CaM 3 gene with CaM 1 and 2 genes in other animals
Considering that there are three known CaM genes in
a number of species, the CaM 3 sequences obtained in the guinea pig
hearts were compared with the CaM 1 and 2 genes from humans, mice
and rats. As shown in Fig. 5, the
results demonstrated that the coding sequence similarities between
the guinea pig CaM 3 gene and the CaM 1 gene in humans, mice and
rats were 80.0, 82.9 and 82.5%, respectively. When compared with
the CaM 2 gene in humans, mice and rats, the nucleotide sequence
homologies were determined to be 81.4, 82.3 and 82.7%,
respectively. Therefore, the guinea pig CaM 3 gene was found to
share extensive homologies with the CaM 1 and 2 genes from other
animals, although the degree of homology was not as high as that
for the CaM 3 genes. However, the 5′- and 3′-UTRs of the CaM 3 mRNA
were highly diverged when compared with the respective CaM 1 and 2
sequences from other animals; the nucleotide sequence homologies
varied between 26 and 47%. These results indicated that the coding
regions of the guinea pig CaM 3 gene were highly conserved when
compared with the CaM 1, 2 and 3 genes in other animals; however,
the sequences of the UTRs were diverged among the CaM 1, 2, 3
genes. By contrast, the homologies of the CaM protein sequences
between CaMs 1, 2 and 3 were 100% (data not shown).
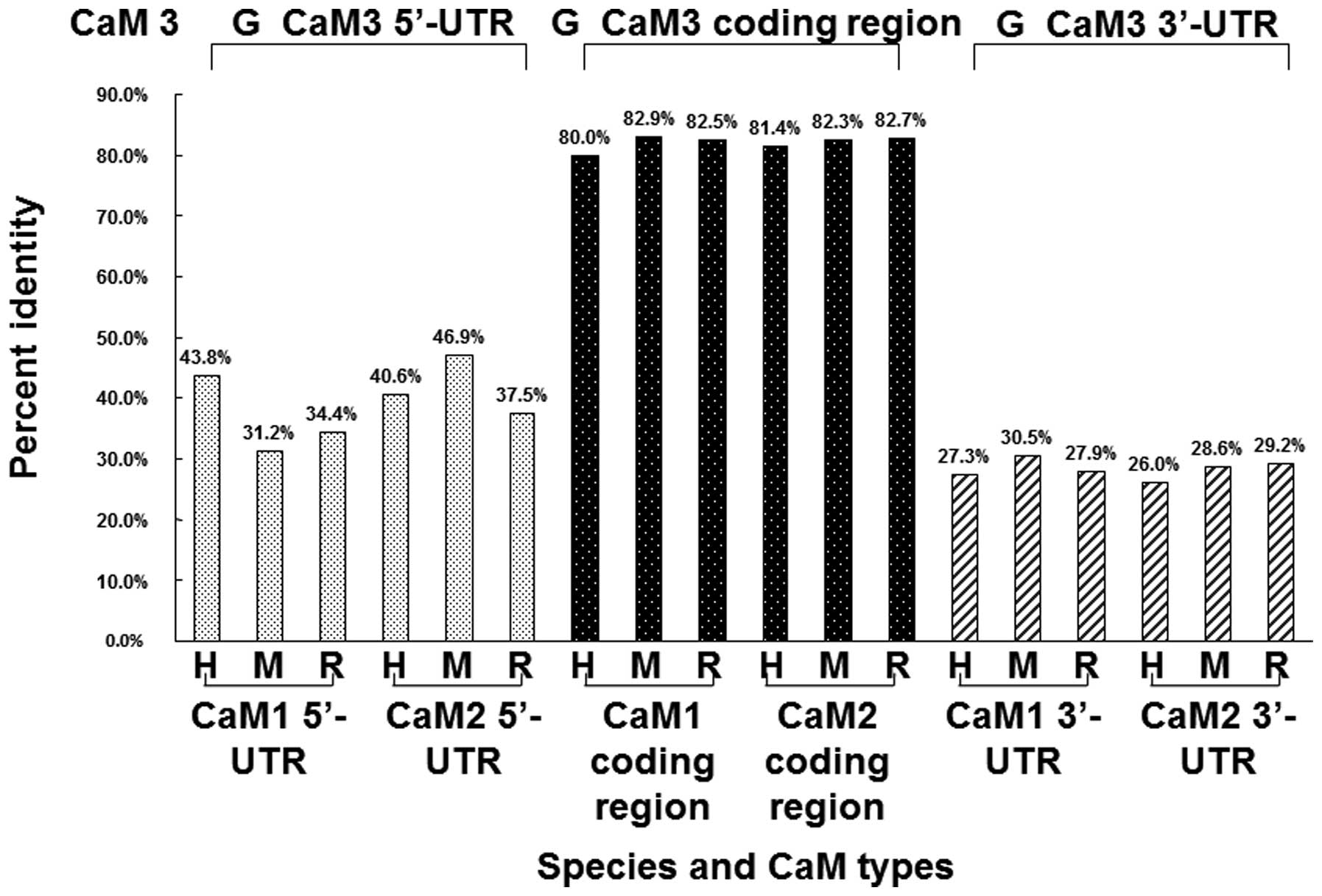 | Figure 5.Comparison of the CaM 3 sequence from
guinea pigs with the CaM 1 and CaM 2 sequences from humans, mice
and rats. The coding regions of CaM 1, CaM 2 and CaM 3 show high
homology, while the 5′- and 3′-UTRs of the CaM 3 gene from guinea
pigs show a low degree of homology with the CaM 1 and CaM 2 genes
from humans, mice and rats. H, humans; M, mice; R, rats; G, guinea
pigs; CaM, calmodulin; UTR, untranslated region. |
CaM 3 expression in different guinea
pig tissues
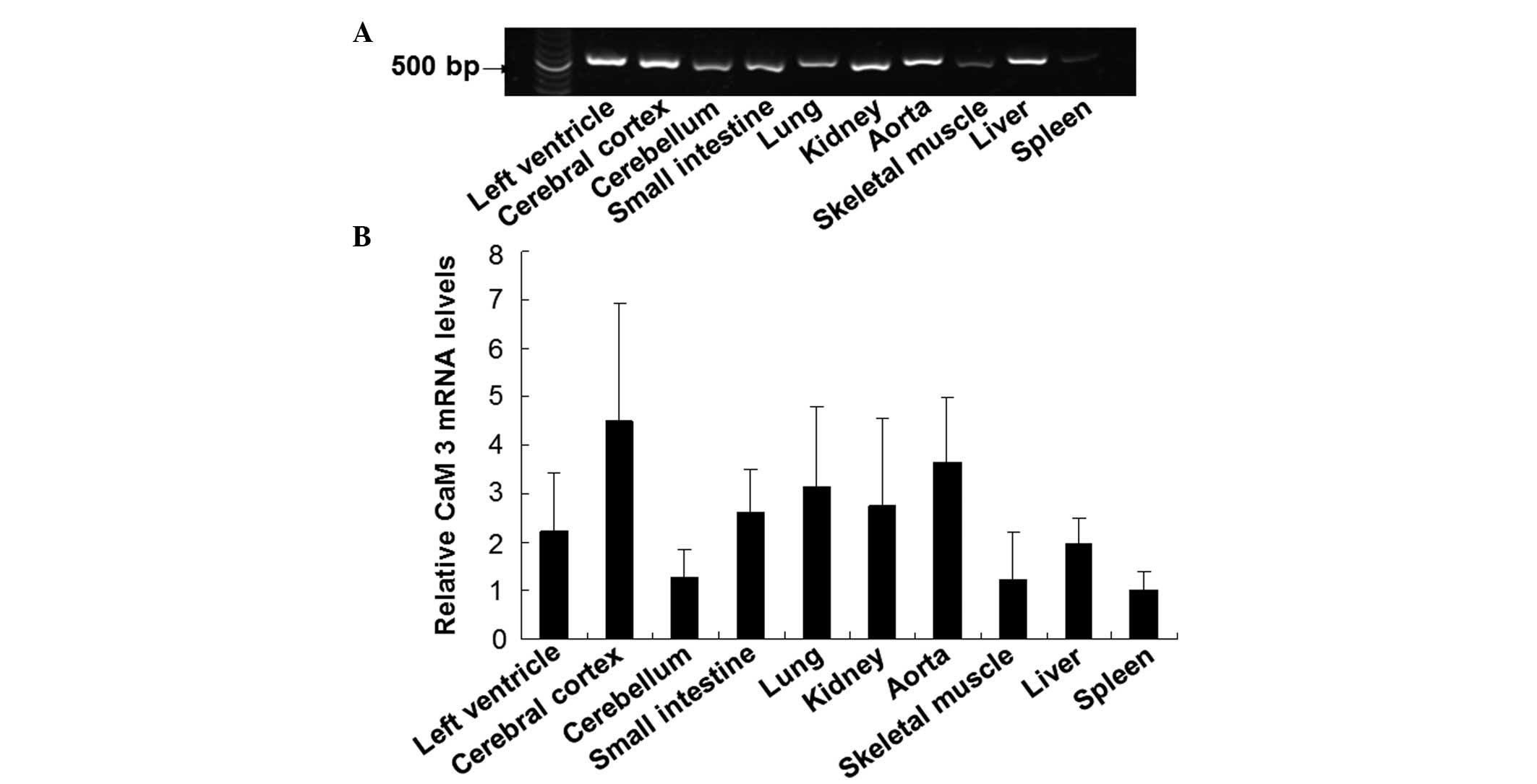
To investigate the expression pattern of CaM 3 in
guinea pig tissues, the mRNA expression levels of CaM 3 in various
tissues were detected using a RT-PCR method. As shown in Fig. 6, CaM 3 was widely distributed in the
guinea pig tissues, with expression at different levels. The
expression of CaM 3 was relatively abundant in the tissues of the
cerebral cortex, aorta and lung, while moderate levels of
expression were observed in the left ventricle, small intestine and
kidney. In addition, low mRNA expression levels of CaM 3 were
detected in the skeletal muscle, cerebellum and spleen. These
results demonstrated the wide, but differential distribution of CaM
3 in guinea pig tissues.
Discussion
CaM, a ubiquitous Ca2+-binding protein,
has a highly conserved amino acid sequence across a number of
species, indicating the pivotal role of the protein in the
regulation of basic cellular functions. In vertebrates and
invertebrates, CaM is always encoded by a multigene family,
exhibiting complex regulation. The same also holds true for plants
(27,28). One of the exceptions is Scoparia
dulcis, in which CaM protein is encoded by only one specific
gene (27). In mammals, CaM is
generally encoded by three different genes (29–31).
These CaM genes share a high degree of conservation with each
other, within a species, as well as across species. In the present
study, a CaM cDNA clone from guinea pig hearts was obtained and
characterized. The results demonstrated that the CaM cDNA clone
exhibited the highest degree of homology with the previously
reported cDNA of CaM 3 genes, indicating that the isolated gene was
CaM 3. Notably, the amino acid sequence of the CaM 3 cDNA clone was
identical to the previously reported sequences of the CaM 1, 2 and
3 proteins from other mammals.
It is well known that CaM functions as a key element
in the signaling mechanisms underlying the regulation of numerous
Ca2+-mediated cellular functions (32). The guinea pig is a widely used model
for diseases; however, little information is available with regard
to the genetic information of CaM in guinea pigs. To the best of
our knowledge, the present study was the first to clone the guinea
pig CaM 3 gene. When comparing the coding region of the CaM 3 gene
in guinea pigs with that from other animals (humans, mice and
rats), the homologies varied between 89 and 93%. In addition, the
sequences of the 5′- and 3′-UTRs of CaM 3 exhibited high homologies
across these species. These results indicated a high similarity in
CaM 3 genes among different species. Furthermore, the protein
product of the CaM 3 gene in guinea pigs was the same as that in
humans, mice and rats. In addition, the sequence of the CaM 3 gene
was compared with those of the CaM 1 and 2 genes. In the coding
regions, the nucleotide sequence homologies varied between 81 and
83%; however, the UTRs exhibited a lower degree of homology
(26–46.9%). Thus, the data indicated that the distinct types of CaM
were generally different from each other in the UTRs. The CaM gene
family has previously been reported to be comprised of three
non-allelic members in mammals, including humans and rats. By
contrast, in the non-coding regions, there were no marked sequence
similarities among these three CaM genes (33,34).
Therefore, the results of the present study were consistent with
the aforementioned studies. It can be hypothesized that there are
three CaM genes in guinea pig hearts, and the gene that was
isolated and characterized was the specific CaM 3 gene. Thus,
further studies are required to identify the genes corresponding to
CaM 1 and 2 in guinea pigs. In addition, the amino acid sequence of
CaM 3 in the guinea pigs was shown to be identical to those of the
CaM 1, 2 and 3 proteins in other mammals (humans, mice and rats).
Therefore, further investigation into whether multigene families
for the same CaM protein in guinea pigs, in the way that they do
for CaM in humans and rats, is required.
It is unknown whether the CaM 3 gene is
differentially expressed in various tissues of guinea pigs. In the
present study, the mRNA expression levels of CaM 3 in different
tissues from guinea pigs were detected by RT-PCR. Gene expression
occurred predominantly in the cerebral cortex, aorta and lung,
whilst lower expression was observed in the skeletal muscle,
cerebellum and spleen. In addition, CaM 3 was expressed at a
moderate level in the left ventricle, small intestine and kidney. A
previous study reported that distinct CaM genes are widely
expressed throughout the mid-brain stem region (35). Furthermore, Zhou et al
(36) studied the regional
distribution of CaM activity in the rat brain, while Solà et
al (37) investigated the
distribution pattern of CaM 1, 2 and 3 genes in the mouse brain.
The two studies found that CaM activity did not necessarily
correlate with the amount of CaM present. In the present study, the
results from the RT-PCR detection revealed the differences in the
relative abundance of CaM 3 mRNA expression levels among the
various tissues, which indicated that the CaM 3 gene may be
differentially regulated in these tissues. Therefore, CaM 3 may
have distinct functions according to the different tissues/regions
in guinea pigs.
In conclusion, the present study identified and
characterized a CaM 3 cDNA clone obtained from guinea pig hearts.
CaM is involved in the activation of CaM-dependent protein kinase
II, which is associated with the pathogenesis of
ischemia-reperfusion injury (12).
Future research should investigate the role of CaM 3 in
cardiovascular diseases. Therefore, the present study has provided
valuable information with regard to the cloning and expression of
CaM 3 in guinea pigs. CaM 3 was demonstrated to be expressed in
various tissues, indicating the extensive effects of the protein in
corresponding regions. The present results may help to improve the
understanding of CaM 3 function and the possible role of CaM 3 in
cardiovascular diseases.
Acknowledgements
The authors thank Dr Lewis Adler (Bioanalytical Mass
Spectrometry Facility, University of New South Wales, Sydney,
Australia) for the revision of the manuscript and Dr Fuyu Xu
(University of Maine, Orono, ME, USA). This study was supported by
grants from the National Natural Science Foundation of China (nos.
30870907, 31071004, 81100108 and 81001429).
References
|
1
|
Ben-Johny M and Yue DT: Calmodulin
regulation (calmodulation) of voltage-gated calcium channels. J Gen
Physiol. 143:679–692. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Rasmussen CD and Means AR: Calmodulin is
involved in regulation of cell proliferation. EMBO J. 6:3961–3968.
1987.PubMed/NCBI
|
|
3
|
Deb TB, Coticchia CM and Dickson RB:
Calmodulin-mediated activation of Akt regulates survival of
c-Myc-overexpressing mouse mammary carcinoma cells. J Biol Chem.
279:38903–38911. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
O'Day DH: CaMBOT: Profiling and
characterizing calmodulin-binding proteins. Cell Signal.
15:347–354. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yap KL, Kim J, Truong K, Sherman M, Yuan T
and Ikura M: Calmodulin target database. J Struct Funct Genomics.
1:8–14. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kim J, Ghosh S, Nunziato DA and Pitt GS:
Identification of the components controlling inactivation of
voltage-gated Ca2+ channels. Neuron. 41:745–754. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zühlke RD, Pitt GS, Deisseroth K, Tsien RW
and Reuter H: Calmodulin supports both inactivation and
facilitation of L-type calcium channels. Nature. 399:159–162. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Berrocal M, Sepulveda MR,
Vazquez-Hernandez M and Mata AM: Calmodulin antagonizes amyloid-β
peptides-mediated inhibition of brain plasma membrane
Ca2+-ATPase. Biochim Biophys Acta. 1822:961–969. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Crotti L, Johnson CN, Graf E, et al:
Calmodulin mutations associated with recurrent cardiac arrest in
infants. Circulation. 127:1009–1017. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Babu YS, Bugg CE and Cook WJ: Structure of
calmodulin refined at 2.2 A resolution. J Mol Biol. 204:191–204.
1988. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Stein JP, Munjaal RP, Lagace L, Lai EC,
O'Malley BW and Means AR: Tissue-specific expression of a chicken
calmodulin pseudogene lacking intervening sequences. Proc Natl Acad
Sci USA. 80:6485–6489. 1983. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Salas MA, Valverde CA, Sánchez G, et al:
The signalling pathway of CaMKII-mediated apoptosis and necrosis in
the ischemia/reperfusion injury. J Mol Cell Cardiol. 48:1298–1306.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Consolini AE and Bonazzola P: Energetics
of Ca2+ homeostasis during ischemia-reperfusion on
neonatal rat hearts under high-[K+] cardioplegia. Can J
Physiol Pharmacol. 86:866–879. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Farah C, Meyer G, André L, et al: Moderate
exercise prevents impaired Ca2+ handling in heart of
CO-exposed rat: implication for sensitivity to
ischemia-reperfusion. Am J Physiol Heart Circ Physiol.
299:H2076–H2081. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wawrzynczak EJ and Perham RN: Isolation
and nucleotide sequence of a cDNA encoding human calmodulin.
Biochem Int. 9:177–185. 1984.PubMed/NCBI
|
|
16
|
SenGupta B, Friedberg F and
Detera-Wadleigh SD: Molecular analysis of human and rat calmodulin
complementary DNA clones. Evidence for additional active genes in
these species. J Biol Chem. 262:16663–16670. 1987.PubMed/NCBI
|
|
17
|
Fischer R, Koller M, Flura M, et al:
Multiple divergent mRNAs code for a single human calmodulin. J Biol
Chem. 263:17055–17062. 1988.PubMed/NCBI
|
|
18
|
Goldhagen H and Clarke M: Identification
of the single gene for calmodulin in Dictyostelium discoideum. Mol
Cell Biol. 6:1851–1854. 1986.PubMed/NCBI
|
|
19
|
Zimmer WE, Schloss JA, Silflow CD,
Youngblom J and Watterson DM: Structural organization, DNA sequence
and expression of the calmodulin gene. J Biol Chem.
263:19370–19383. 1988.PubMed/NCBI
|
|
20
|
Davis TN, Urdea MS, Masiarz FR and Thorner
J: Isolation of the yeast calmodulin gene: calmodulin is an
essential protein. Cell. 47:423–431. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Takeda T and Yamamoto M: Analysis and in
vivo disruption of the gene coding for calmodulin in
Schizosaccharomyces pombe. Proc Natl Acad Sci USA. 84:3580–3584.
1987. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wei-xu H, Qin X, Zhu W, et al: Therapeutic
potential of anti-IL-1β IgY in guinea pigs with allergic asthma
induced by ovalbumin. Mol Immunol. 58:139–149. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Costa M, Dodds KN, Wiklendt L, Spencer NJ,
Brookes SJ and Dinning PG: Neurogenic and myogenic motor activity
in the colon of the guinea pig, mouse, rabbit, and rat. Am J
Physiol Gastrointest Liver Physiol. 305:G749–759. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Padilla-Carlin DJ, McMurray DN and Hickey
AJ: The guinea pig as a model of infectious diseases. Comp Med.
58:324–340. 2008.PubMed/NCBI
|
|
25
|
Shao D, Zhao M, Xu J, et al: The
individual N- and C-lobes of calmodulin tether to the Cav1.2
channel and rescue the channel activity from run-down in
ventricular myocytes of guinea-pig heart. FEBS Lett. 588:3855–3861.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Feng R, Xu J, Minobe E, et al: Adenosine
triphosphate regulates the activity of guinea pig Cav1.2 channel by
direct binding to the channel in a dose-dependent manner. Am J
Physiol Cell Physiol. 306:C856–863. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Saitoh D, Asakura Y, Nkembo MK, et al:
Cloning and expression of calmodulin gene in Scoparia dulcis. Biol
Pharm Bull. 30:1161–1163. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Al-Quraan NA, Locy RD and Singh NK:
Expression of calmodulin genes in wild type and calmodulin mutants
of Arabidopsis thaliana under heat stress. Plant Physiol Biochem.
48:697–702. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Palfi A, Kortvely E, Fekete E, Kovacs B,
Varszegi S and Gulya K: Differential calmodulin gene expression in
the rodent brain. Life Sci. 70:2829–2855. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Toutenhoofd SL, Foletti D, Wicki R, Rhyner
JA, Garcia F, Tolon R and Strehler EE: Characterization of the
human CALM2 calmodulin gene and comparison of the transcriptional
activity of CALM1, CALM2 and CALM3. Cell Calcium. 23:323–338. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Toutenhoofd SL and Strehler EE: The
calmodulin multigene family as a unique case of genetic redundancy:
multiple levels of regulation to provide spatial and temporal
control of calmodulin pools? Cell Calcium. 28:83–96. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Clapham DE: Calcium signaling. Cell.
131:1047–1058. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Nojima H: Structural organization of
multiple rat calmodulin genes. J Mol Biol. 208:269–282. 1989.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Berchtold MW, Egli R, Rhyner JA, Hameister
H and Strehler EE: Localization of the human bona fide calmodulin
genes CALM1, CALM2 and CALM3 to chromosomes 14q24-q31, 2p21.1-p21.3
and 19q13.2-q13.3. Genomics. 16:461–465. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Orojan I, Bakota L and Gulya K:
Differential calmodulin gene expression in the nuclei of the rat
midbrain-brain stem region. Acta Histochem. 108:455–462. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhou LW, Moyer JA, Muth EA, Clark B,
Palkovits M and Weiss B: Regional distribution of calmodulin
activity in rat brain. J Neurochem. 44:1657–1662. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Solà C, Tusell JM and Serratosa J:
Comparative study of the pattern of expression of calmodulin
messenger RNAs in the mouse brain. Neuroscience. 75:245–256. 1996.
View Article : Google Scholar : PubMed/NCBI
|